Abstract
Selenomethionine incorporation has proven useful in x-ray crystallography of proteins to obtain phase information. In nucleic acids, the introduction of selenium to different positions is beneficial for solving the phase problem as well, but its addition to the 2' position also significantly enhances crystal formation. The selenium modification in a single nucleotide shows a preference towards 2'-endo sugar puckering, which is in conflict with existing crystal structures where the duplex incorporated 2'-selenium modified nucleotide is exclusively found in a 3'-endo conformation. Our work provides a rationale why 2'-selenium modifications facilitate crystallization despite this contradictory behavior.
Introduction
X-ray crystallography has been used to determine the structures of many biological macromolecules, but the phase determination problem forces crystallographers to use heavy-atom soakings or covalent introduction of heavy atoms. For nucleic acids, an additional problem is that some modifications may render the nucleic acids unstable or can affect the structure (Jiang, Sheng, Carrasco, & Huang, 2007). The incorporation of selenium in proteins has been shown to facilitate the phase determination, and the use of selenium in nucleic acids crystallography has been successfully demonstrated by Huang et al (Du et al., 2002; Höbartner & Micura, 2004; Sheng & Huang, 2010; Sheng, Jiang, Salon, & Huang, 2007). Only minor structural differences in the A-form double helical structures of unmodified DNA oligomers and derivatives with 2'-methylseleno and 5-bromine uracil modifications were reported. Interestingly, the Se-modified nucleic acids exhibited a much more rapid rate of crystal formation than either the bromine derivative or the unmodified control. Notably, in x-ray crystal structures, the 2’-methylseleno modified nucleotide is always found in the 3’-endo sugar conformation. (Jiang et al., 2007; Sheng et al., 2007; Sheng & Huang, 2010)
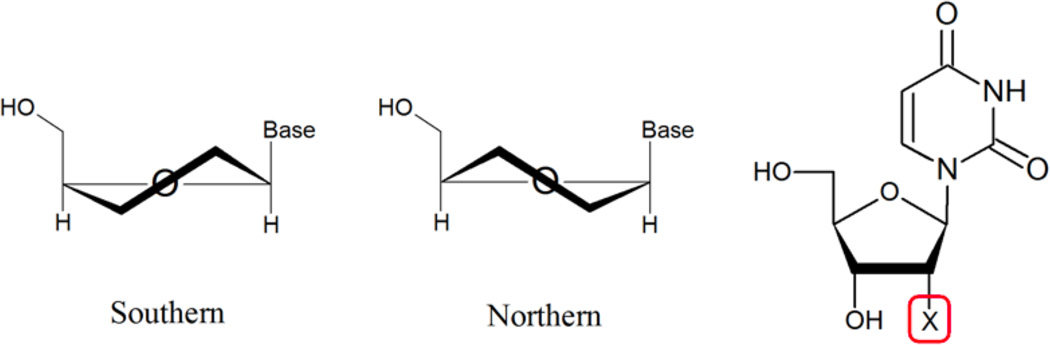
It has been well established that the nature of the 2’ substituent is a determinant of the sugar conformation and its dynamics. (Rozners, 2006) This study addresses the structural origin for the facilitated crystal formation by determining the conformational bias imparted by a 2'-methylseleno derivative of uridine and other 2’-modifications in nucleosides (Figure 1). The consequence of such a conformational bias is then examined in the context of A- or B-form helical structures. Table 1 contains the sequences used in duplex studies.
Figure 1.
Compounds in the nucleoside studies and sugar puckering nomenclature within the context of nucleic acids. Compounds investigated in this study: X= H (deoxyuridine, 1); OH (uridine, 2); OCH3 (3); F (4); SCH3 (5); SeCH3 (6).
Table 1.
List of Sequences.
| I | 5’-d(CATGCATG) |
| II | 5’-d(GCGAATTCGC) |
| III | 5’-d(GCGAAUSeTCGC) |
| IV | 5’-d(CGCGAATTCGCG) |
Materials and Methods
Nucleoside Studies
All experiments were recorded on a Bruker Avance 500 MHz spectrometer equipped with a triple resonance cryogenic probe. Samples (1.0 mM) were prepared in D2O, 10 mM sodium phosphate at pH* 6.0. DSS was used as an internal standard. Double quantum filter COSY experiments (32 scans) were recorded to estimate the individual couplings and confirm assignments. A low-flip angle COSY was recorded for 1 to clarify couplings caused by the 2' and 2" protons.
Sugar Pucker Computational Parameters
DAISYSIM, a component of Topspin 2.1 (Bruker) was used to simulate spectra from the acquired NMR data in order to determine the individual couplings and chemical shifts. The refined coupling constants were used as the input into PSEUROT 6.2 to calculate the pseudorotation parameters (Frank A A M De Leeuw & Altona, 1983; Houseknecht, Altona, Hadad, & Lowary, 2002; Rosemeyer et al., 1997; Watts, Sadalapure, Choubdar, Pinto, & Damha, 2006). In addition, a Matlab based pseudorotation GUI was used for substantiation (Hendrickx & Martins, 2008). The computation for each compound was initially set up with the following conditions: PN=18.0°, PS=153.6°, ΦM=35°, %S = 0.2 – 0.8. Each of these initial states was refined during the computation. The change in electronegativity of the 2' substitutions was accounted for in the input file; the values are derived from a Huggin's based electronegativity scale referenced to hydrogen specifically for use with generalized Haasnoot-Karplus equation (Altona et al., 1989; Donders, De Leeuw, & Altona, 1989; Haasnoot, de Leeuw, de Leeuw, & Altona, 1981).
TM Determination
The thermal denaturation of the control and modified duplexes (II and III, respectively) were monitored at 274 nm in 1 cm cuvettes on a Cary 100 Bio UV/VIS spectrometer. The temperature was ramped from 20°C to 90°C at 0.3°C / min. Samples were at 8 µM strand concentration in 400 mM sodium chloride, 10 mM sodium phosphate, and 0.1 mM EDTA at pH 6.5. A second selenium sample III was prepared in the same buffer with 32 µM DNA.
Ethidium Bromide Fluorescence
Samples were 15 µM in nucleotides or ~ 0.8 µM in duplex concentration; the individual samples were incubated in 1 µg/mL ethidium bromide concentrations in 100 mM sodium chloride, 10 mM sodium phosphate, and 0.1 mM EDTA at pH 6.5 in PCR tubes. The tubes were imaged on a Typhoon 9400 Variable Mode Imager from Amersham Biosciences. Excitation for imaging occurred at 532 nm and emission was measured at 610 nm.
Imino Proton Observation
NMR samples of sequences II and III were prepared at 50 mM sodium chloride, 10 mM sodium phosphate, 0.1 mM EDTA and pH 6.4 in 9:1 H2O:D2O. Imino proton spectra were recorded on an Avance 600 MHz NMR spectrometer (Bruker) using jump and return water suppression (Plateau & Gueron, 1982). Selenium samples (III) were prepared at strand concentrations of 100 and 20 µM (designated high and low, respectively). The control sequence was prepared at 250 µM.
Model Development
Standard A- and B-form DNA helical models of sequence III were built within Spartan06 (Wavefunction) to estimate the position of 2’-Se-modification inside each secondary structure. The A-form set the sugar puckering as N–type and with a rise and twist of 2.548 Å and 32.7° per base, respectively, as described in the Spartan manual. The second model was made to be B-form (S - type, 3.375 Å, 36°). Energy minimization was only applied to the –SeCH3 group to find an optimal orientation within an ideal helical structure.
Ab Initio Calculations
Spartan10 (Wavefunction) was used for all ab initio calculations. Model systems of S-sugar or N-sugar 2’-methoxy and 2’-methylseleno uracil nucleosides were built in Spartan and minimized to a DFT RB3LYP / HF6-31G(d) level as follows: MMFF → HF 3-21 → HF 6-31G(d) → DFT RB3LYP / HF 6-31G(d). Single point energy calculations at the same DFT level were then performed on the minimized structures. All minimizations and single point energy calculations were conducted using the SM8 solvation model for water and subsequently replicated in vacuum (Marenich, Olson, Kelly, Cramer, & Truhlar, 2007).
Molecular Dynamics
The AMBER parm99 force field was parameterized to include the 2’-methylseleno modification (Cheatham, Cieplak, & Kollman, 1999). The Se-carbon bond distance and C-Se-C bond angle of 1.94 Å and 96°, respectively, were taken from microwave spectrum studies of dimethyl selenide (Beecher, 1966). Bond length force constants for Se-C were approximated from comparisons to O-C and S-C force constants contained in the general amber force field (GAFF) (Wang, Wolf, Caldwell, Kollman, & Case, 2004). Force constants for 3-bond angles and torsion angles involving Se were taken from similar sulfur based values. RESP charges were determined using an iterative process as described previously (Johnson, Spring, Sergueev, Shaw, & Germann, 2011). Briefly, a model of the 2’-methylseleno sugar was built and minimized to the HF 6-31G(d) level using Gaussian03 (Frisch et al.). The RED script was then used in conjunction with Gaussian03 and AMBER 9 to calculate the RESP charges (Case et al., 2006; Dupradeau et al., 2010; Pigache, Cieplak, & Dupradeau, 2004). This process was repeated for a similar sugar containing a 2’-methoxy modification, which was compared to 2’-methoxy cytosine RESP charges determined by the Case group (Case & Meyer). The Case group 2’-methoxy charges were applied to the 2’-methylseleno uracil except for H2’, C2’, and 2-methylseleno group; for these atoms, the derived charges were used and the residue was balanced for an overall charge of −1.
Sequences for molecular dynamics (MD) were built using the AMBER 9 NUCGEN and XLEaP utilities (Case et al., 2006). For the 2’-methylseleno uracil modification, the sugar was biased to either S or N conformations within XLEaP and subsequently minimized using AMBER 9; initially minimized structures were visually inspected for accuracy and replicated using Spartan 10. Within XLEaP, Na+ counterions were added to neutralize the charge and the system was solvated using TIP3P water in an octahedral box. The water and counterions were then minimized in Amber 9 while the DNA was held rigid followed by an energy minimization of the entire system. MD annealing simulations were conducted on all systems as follows: the system was heated to 500 K over 100 ps, held at 500 K for 100 ps, cooled to 300 K over 50 ps, and held at 300 K for 250 ps giving a total MD simulation of 500 ps and subsequently minimized.
Results and Discussion
The conformational state of the 5-membered ribose is described by the phase angle (P) and puckering amplitude (Φm) (Altona & Sundaralingam, 1972). Two main conformations occur in nucleic acids: North (3'-endo, N) and South (2'-endo, S), which can be in dynamic equilibrium. (Figure 1) In A-form DNA and RNA, the sugar conformations are of the N type, while in B-form DNA the southern conformations are favored. The dominant sugar conformations and distribution can be derived from the 3JHH NMR coupling constants following well-established protocols (Altona & Sundaralingam, 1973; Karplus, 1963).
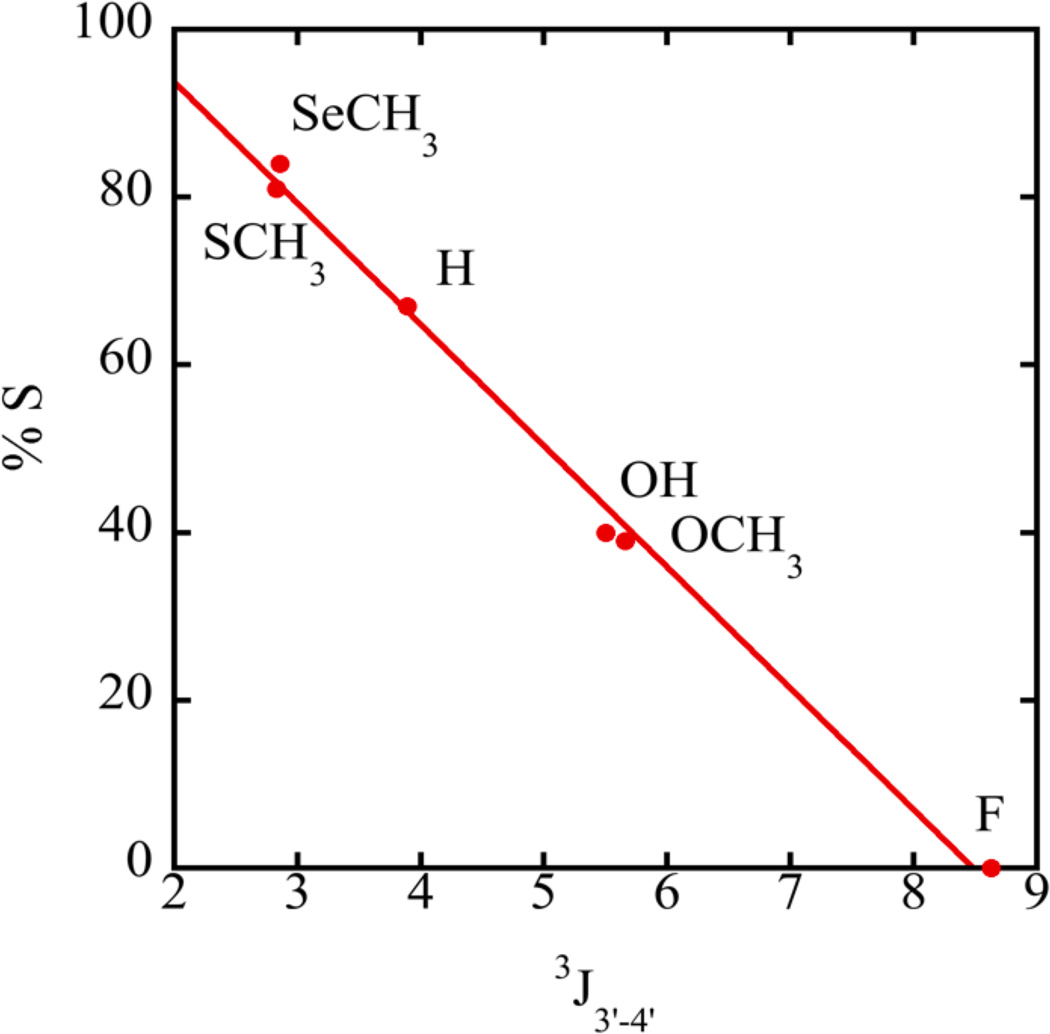
We first determined the sugar conformation of single nucleosides containing modifications at the 2’ ribose position. The experimentally measured ribose proton coupling constants of compounds 1 – 6 (Figure 1) obtained from DAISYSIM fitting were used as input for PSEUROT 6.2 and a similar Matlab program (Frank A A M De Leeuw & Altona, 1983; Houseknecht et al., 2002; Rosemeyer et al., 1997; Watts et al., 2006). The optimized conformations as well as the equilibrium between them, expressed as %S-conformer, were comparable for each program and agreed with published results (Table 2). An increase in the electronegativity of the substituents generally drives the system into a state with a higher %N conformation (Altona et al., 1989; Frank A A M De Leeuw & Altona, 1983; Donders et al., 1989; Haasnoot et al., 1981). For example, compound 1 (X=H) favors an S conformation while –OH, -OCH3 and –F 2’-modifications (compounds 2, 3, and 4) progressively shift the equilibrium to N. (Figure 1, Table 2) However, despite the somewhat higher electronegativity of the -SCH3 and -SeCH3 modifications (compounds 5, 6) compared to H, both were found to more strongly favor the S conformation in solution. Highly favored S-conformations for -SCH3 and -SeCH3 modified nucleosides are also evident directly from raw NMR data. The 3J3’-4’ couplings are affected by the ring dynamics, and would be only minimally impacted by the 2'-substituent identity. A linear relationship was observed between 3J3’-4’ and %S (Figure 2), which can be used as a qualitative indicator of sugar puckering. This expands on the graphical method of Rinkel and Altona, which uses the sum of 3’ couplings and is generally resorted to in oligomers (Rinkel & Altona, 1987). Spartan 10 was used to determine the single point energies of the minimized S and N sugars for the –SeCH3 modified nucleoside. Consistent with NMR observations, the S sugar was found to be favored over the N conformation with a difference of energy of 2.65 kJ/mol between the two forms. For an –OCH3 nucleoside, the N sugar was more stable by 0.63 kJ/mol in agreement with the NMR data. These results clearly establish that an isolated 2'-SeCH3 modified nucleoside prefers a 2'-endo conformation.
Table 2.
Compiled NMR and PSEUROT 6.2 Data. Exp. And Lit. refer to experimentally determined values and literature references, respectively.
| H | OH | OCH3 | F* | SCH3 | SeCH3 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exp. | Lit. a | Exp. | Lit. a | Exp. | Lit. b | Exp. | Lit. a,c | Exp. | Lit. d,e | Exp. | ||
| J 1’-2’ | 7.2 | 6.3 | 4.5 | 4.8 | 3.9 | 4.0 | 1.4 | 1.5 | 8.3 | 8.5 | 8.7 | |
| J 1’-2” | 6.1 | 6.4 | 19.7 | 19.7 | ||||||||
| J 2’-3’ | 6.9 | 6.3 | 5.3 | 5.2 | 5.2 | 5.4 | 5.0 | 5.1 | 5.8 | 5.5 | 5.7 | |
| J 2”-3’ | 3.9 | 4.3 | 21.5 | 21.6 | ||||||||
| J 3’-4’ | 3.9 | 4.0 | 5.5 | 5.4 | 5.7 | 5.9 | 8.6 | 8.7 | 2.8 | 2.0 | 2.9 | |
| Exp. | Lit.a | Exp. | Lit.a | Exp. | Lit.b | Exp. | Lit. a | Exp. | Lit.e | Exp. | ||
| I. | ||||||||||||
| North | ||||||||||||
| P | 18.0 | 18.0 | 32.4 | 16.0 | 12.5 | 11.0 | 28.6‡ | 21.0 | −22.5 | - | −13.3 | |
| ΦM | 38.0 | - | 32.0 | 35.0 | 32.0 | 35.0 | 32.0 | 41 | 32.0 | - | 32.0 | |
| II. | ||||||||||||
| South | ||||||||||||
| P | 141.5 | 162.0 | 156.6 | 167.0 | 144.7 | 171.0 | 38.6‡ | 159.0 | 138.7 | - | 137.6 | |
| ΦM | 32.3 | - | 35.0 | 38.0 | 35.0 | 37.0 | 35.0 | 41 | 35.0 | - | 35.0 | |
| %S | 67 | 60 | 40 | 47 | 38 | 40 | 32‡ | 13 | 81 | 76 | 84 | |
The J1’-2’’ and J2’’-3’ values in this column designate H1’-F2’ and H3’-F2’ coupling, respectively.
These values do not represent a %S conformer, rather a percent of a second starting northern conformer. Neither Pseurot nor the Matlab script were able to fit the data of 4 to both northern and southern conformations; despite varying input conditions, the programs consistently returned two northern conformers.
Table references:
Figure 2.
Relationship between coupling constants and %S determined by NMR data.
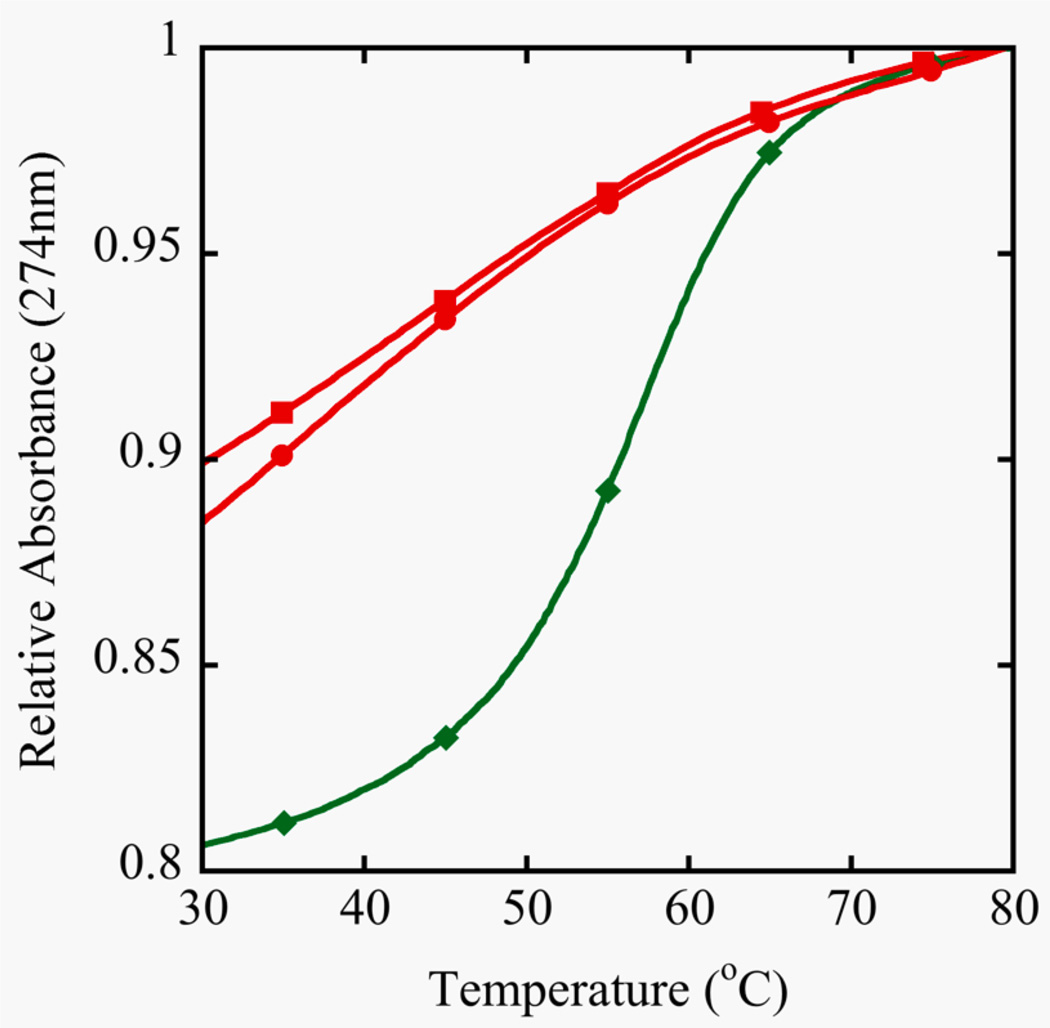
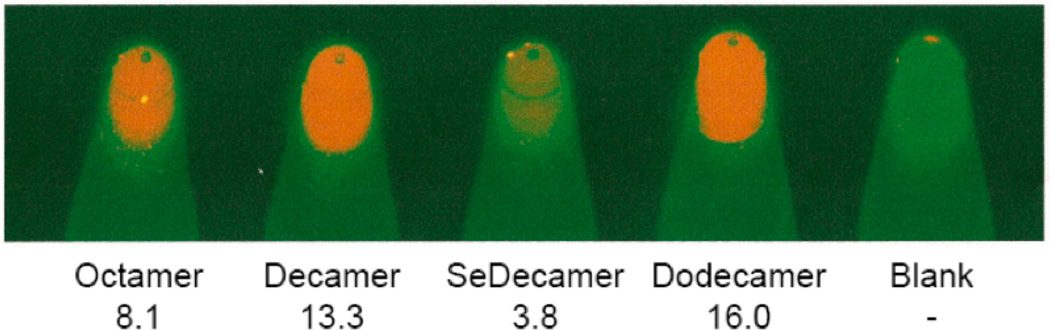
However, this is exactly the opposite of what is observed in the crystal structures. In order to gain perspective on the physical effects of the Se-modification to a double stranded DNA molecule in solution, TM, fluorescence and NMR data were acquired. The stability of the self-complementary sequence III containing one 2’-SeCH3-modification and its control, sequence II, was determined by UV melting (Figure 3). The control duplex forms a standard B-type helical structure and exhibits a regular melting profile with an expected stability (Aramini, Kalisch, Pon, van de Sande, & Germann, 1996; Aramini, Mujeeb, & Germann, 1998). On the other hand, the shifted and shallow melting curve for the DNA strand III containing a single 2’-Se-modification per strand demonstrates that duplex formation was seriously impaired. The observation that the denaturation was not concentration dependent in the range studied also suggests the involvement of intramolecularly formed hairpin structures that are stabilized by only a few G-C basepairs. A decreased amount of duplex formation for this oligomer was also indicated from the ethidium bromide staining assay. Upon intercalation in a duplex structure, the stain becomes fluorescent; a higher overall fluorescence is expected for segments containing more base pairs. This is evident in Figure 4, where the fluorescence signal increases upon increasing oligonucleotide size for the control oligomers (sequences I, II, IV). For sequence III, only marginal fluorescence is obtained. This is consistent with the low stability observed in the melting curves and signifies the presence of just a few base pairs / intercalation sites.
Figure 3.
Duplex stability from UV Melting curves. Samples were prepared in 400 mM NaCl, 10 mM NaP, 0.1 mM EDTA at pH 6.5. The unmodified control decamer (II, ●) at 8.5 µM showed a TM of 59°C and two different concentrations of the selenium decamer (III) were compared, 8.5 µM (■) and 32 µM (♦). Absorbance monitoring occurred at 274 nm because of the increase in hyperchromicity compared to 260 nm.
Figure 4.
Duplex stability from ethidium bromide fluorescence. DNA samples (Octamer: I, Decamer: II, SeDecamer: III, Dodecamer: VI) containing 1µg/mL ethidium bromide were placed in PCR tubes and imaged. Relative fluorescence data, corrected for the blank, is indicated for each sample.
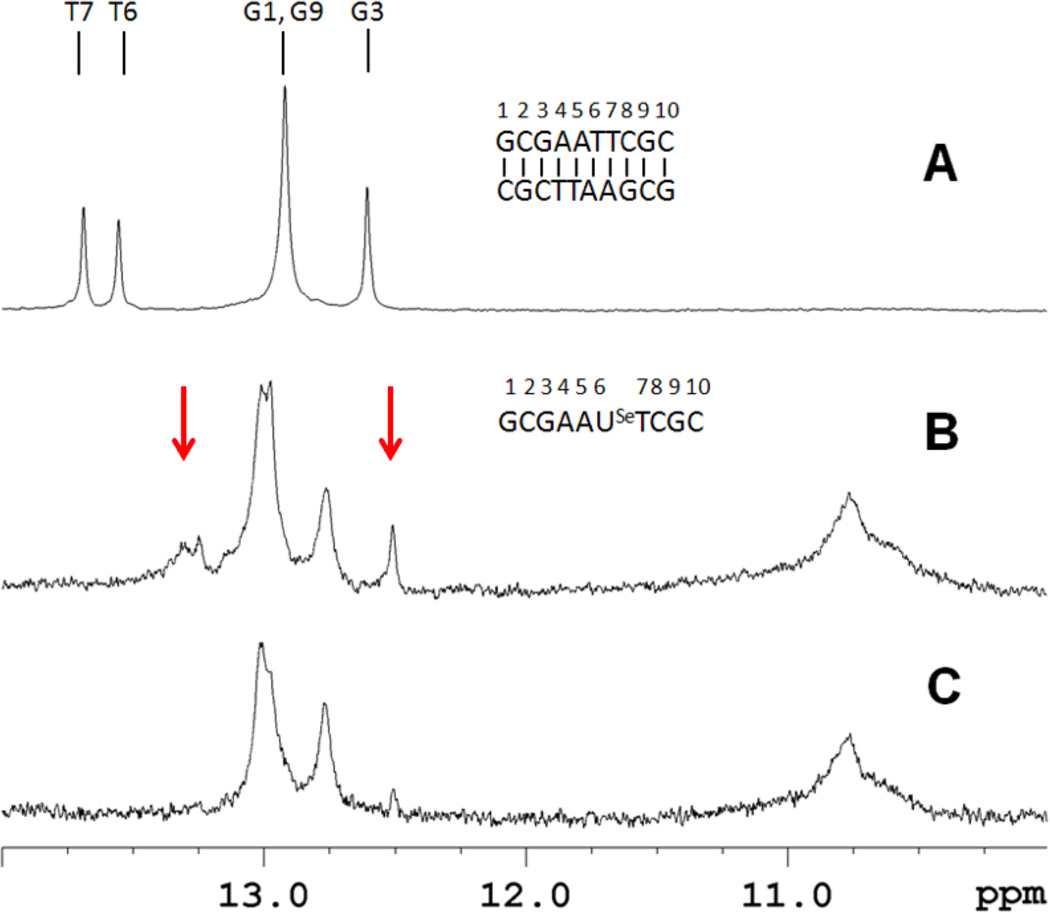
Base pair formation was directly probed from the imino proton spectra of the oligomers. For the self-complementary duplex control (sequence II), five imino proton signals are observed, corresponding to the expected 3 GC and 2 AT base pairs of the DNA duplex (Figure 5A). In contrast, sequence III at 100 µM strand concentration showed more imino proton resonances than would be expected for a self-complementary duplex, suggesting formation of multiple species (Figure 5B). Additionally, there are resonances near 10.8 ppm that are generally associated with unpaired hairpin loop resonances (Germann, Kalisch, Lundberg, Vogel, & van de Sande, 1990). At lower concentrations, intramolecular hairpin formation is thermodynamically favored. When sequence III is examined at 20 µM strand concentration, the spectrum simplifies and essentially only 3 GC base pairs are observed in addition to the hairpin loop resonances (Figure 5C). Under these conditions the predominant species is a hairpin structure with a 3 GC base pair stem; also, it is noted that this lower concentration is comparable to the UV melting studies. Taken together, these spectra indicate that the higher concentration sample contains a mixture of duplex and hairpin forms. We have previously demonstrated complete base pairing for a non-self-complementary duplex containing a single 2’-Se-modification (Salon, Sheng, Gan, & Huang, 2010). However, the stability was compromised in this construct as well, and homoduplex formation of the individual strands was observed at elevated temperatures (data not shown).
Figure 5.
Imino proton spectra of sequences II and III at 288 K referenced to DSS. (A) The control decamer (II, 250 µM strand concentration) spectrum shows the presence of five base pairs. (B) and (C) are the selenium-containing decamer (III) at high and low strand concentration (100 and 20 µM, respectively). Arrows highlight resonances that disappear upon dilution. These signals are also sensitive to increased temperatures.
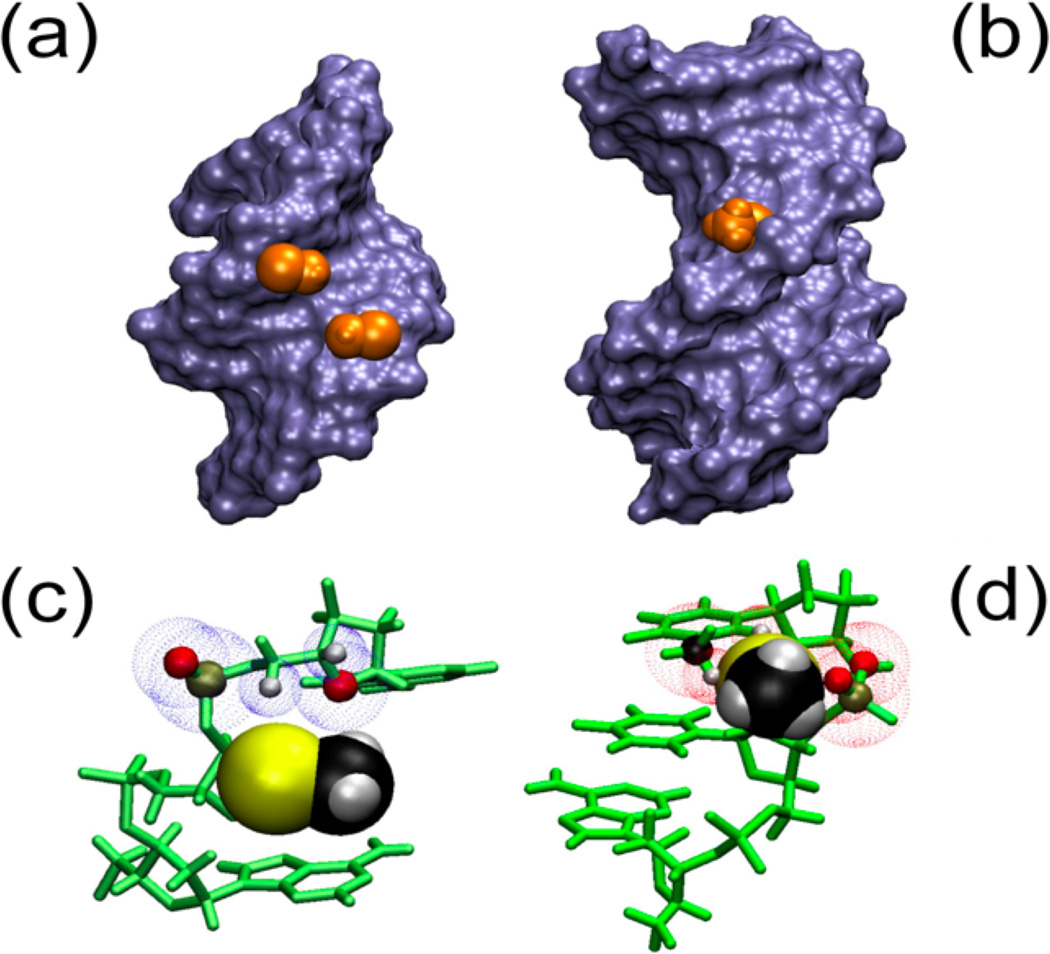
Taken in context, this demonstrates that a 2’-methylseleno group destabilizes a B-type DNA helix in solution, even though this modification has an intrinsic preference for a southern sugar conformation for a free nucleoside. To obtain further insight into why 2’-methylseleno-uridine (6) adopts a northern conformation when becoming a part of a DNA duplex as seen in crystal structures, standard A- and B- type DNA helical models with the appropriate modifications (sequence III) were investigated. As evident from Figure 6, the 2’-methylseleno group is readily accommodated in a standard A-helix and fits snuggly into the minor groove. There are no steric clashes with the backbone, neighboring bases, or deoxyribose ring. In contrast, in a B-helical model the modification is situated in the major groove, but the 3' phosphate and the base on the 3' side of the modification clash with –SeCH3 group. This is especially apparent when the 3’ neighboring base is thymine, whose methyl group is also in the major groove. Therefore, a base with a smaller footprint in the major groove would be expected to be less perturbed, which agrees with our previous NMR data where the 2’ modified residue was flanked by cytosines (Salon et al., 2010).
Figure 6.
2’-SeCH3 groups implanted in A and B helical structures. Panels A and B represent Connelly surfaces with the Se-CH3 group depicted as Van der Waals (VdW) representations in orange. In panel C and D the bases and sugars are shown in green and Se-CH3 is depicted as VdW spheres. A) The group is nestled comfortably in the minor groove of an A-type helix (pdb: 1MA8). C) No clashes are apparent between the 3’ neighboring residue and –SeCH3. The blue dotted spheres depict VdW spheres of close atoms. B) In a B-type helical model (dGCGAAUSeMeTCGC), the Se-CH3 group points away from the major groove but experiences significant clashes with the backbone as well as the base on the 3’ side of the modification, which would disrupt base stacking. D) Predicted clashes for the B helix model are indicated with red VdW spheres for backbone (O and P) as well as the base (CH3 and H6).
To further explore this concept, we used unrestrained AMBER molecular dynamics (MD) to gain a qualitative insight into the impact that the –SeCH3 moieties may be having on overall duplex formation and stability. Models of sequence III were subjected to a melting and annealing process and then assessed for global and local helical integrity. This approach can give a qualitative impression for how the non-canonical moiety could be incorporated. The control duplex (sequence II) was built in an overall B type conformation, and upon heating and cooling annealed back into a B-type duplex with intact base pairing. However, the –SeCH3 containing duplex (sequence III) when built in an overall B-type duplex with all sugars in S conformations produced structures with extrahelical bases and poor base stacking. When the same duplex was built with the –SeCH3 nucleotides in an N sugar conformation, annealing produces a base paired duplex. In these duplexes, the –SeCH3 groups are positioned in the minor groove. (SI Figure 1) All simulations were conducted in duplicate yielding similar results. Taken together this shows that a 2’-SeCH3 modified nucleotide destabilizes a B-type helix.
The notion that the southern sugar conformation of 2’-methylseleno modified nucleotides is not tolerated well in a B-helix because of the steric interactions could also rationalize the enhanced crystal growth. Recent data has shown that the nature of the base of the modified nucleotide is not a determining factor for crystal growth, as enhanced crystal growth was also observed for other 2’-methylseleno-modified nucleotides (Salon et al., 2010; Sheng & Huang, 2010; Sheng, Salon, Gan, & Huang, 2010). As exhibited by DNA oligonucleotides containing embedded ribose moieties, localized A-type perturbations in overall B-type DNA oligonucleotides can be observed in solution structures, yet this mixed conformation is rarely seen in crystal structures, which favor longer-lived species. As compared to overall A or B type global geometries, mixed A / B type helical conformations, which contain destabilizing junctions, are energetically unfavored and thus transiently present (DeRose, Perera, Murray, Kunkel, & London, 2012).
The embedded 2’-methylseleno nucleotide cannot tolerate a localized B-type geometry due to steric clashes with neighboring residues. These results are not unexpected; 2’-methoxy substituents will preorganize the ribose moiety into an N type sugar pucker (Teplova et al., 2002). The MD data supports the requirement of an N type sugar puckering for the 2’-methylseleno nucleotide (SI figure 1); this would destabilize a B-type duplex and serve as a nucleation point for A helix formation. Our empirical data indicate a low population of stable duplex formation as exhibited by weak ethidium bromide staining, a shifted, shallow Tm profile, and weak base pair formation demonstrating the absence of a stable helical structure in solution. The absence of an A-type duplex is expected given the aqueous environment and predominant DNA sequence context. However, under conditions of low hydration, i.e. crystallization, A-helix formation is inherently favored and the 2’-methylseleno modification tips the balance by providing a nucleation center for A-helix formation. It should also be noted that short DNA oligonucleotides have frequently been reported to crystalize in an A-form helix (Kennard, 1987). For DNA, the 2’-methylseleno modification introduces a bias sufficient to drive A-helix formation in crystallization. Other studies have utilized the 2’-methylselno modified uracil for RNA oligonucleotides; these resulting structures yielded canonical A-form RNA conformations with anticipated structures and minimal perturbations resulting from the 2’ substitution, yet contain the benefits of the selenium addition (Freisz, Lang, Micura, Dumas, & Ennifar, 2008; Serganov et al., 2005).
Thus we provide the following rationale: A single 2’-methylseleno group narrows the conformational space by destabilizing the B-helical form while promoting A-helix formation. It is noted that the free nucleoside with the 2’ modification still populates the northern conformational space (Table 2, −SeCH3 84% S and thus 16% N). Moreover, the 2’-methylseleno group fits snuggly into the minor groove of an A-helix and can serve as the origin for a B- to A- conversion, which is also aided by dehydration during crystallization. In addition, the 2’-methylseleno group locally dehydrates the minor groove, which further facilitates the crystallization process.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health [AI/GM47459]; National Science Foundation [MCB-0824837], and the Georgia Cancer Coalition; AMS is supported by the Georgia State University Molecular Basis of Disease program.
Footnotes
Supplemental material consists of a figure detailing the ending molecular dynamics models for –SeCH3 containing duplexes (sequence III).
REFERENCES
- Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. Journal of the American Chemical Society. 1972;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Altona C, Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. Improved method for the interpretation of proton magnetic resonance coupling constants. Journal of the American Chemical Society. 1973;95(7):2333–2344. doi: 10.1021/ja00788a038. [DOI] [PubMed] [Google Scholar]
- Altona C, Ippel JH, Hoekzema AJAW, Erkelens C, Groesbeek M, Donders LA. Relationship between proton-proton NMR coupling constants and substituent electronegativities. V—Empirical substituent constants deduced from ethanes and propanes. Organic Magnetic Resonance. 1989;27(6):564–576. [Google Scholar]
- Aramini JM, Kalisch BW, Pon RT, van de Sande JH, Germann MW. Structure of a DNA duplex that contains alpha-anomeric nucleotides and 3'-3' and 5“-5” phosphodiester linkages: coexistence of parallel and antiparallel DNA. Biochemistry. 1996;35(29):9355–9365. doi: 10.1021/bi960612p. [DOI] [PubMed] [Google Scholar]
- Aramini JM, Mujeeb A, Germann MW. NMR solution structures of [d(GCGAAT-3'-3'-alphaT-5“-5-”CGC)2] and its unmodified control. Nucleic Acids Research. 1998;26(24):5644–5654. doi: 10.1093/nar/26.24.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beecher JF. Microwave spectrum, dipole moment, structure, and internal rotation of dimethyl selenide. Journal of Molecular Spectroscopy. 1966;21(1–4):414–424. [Google Scholar]
- Case DA, Meyer T, editors. modified bases appearing in tRNA-phe from yeast. (n.d.) Retrieved April 29, 2013, from http://www.pharmacy.manchester.ac.uk/bryce/amber#nuc. [Google Scholar]
- Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, et al. AMBER 9.0. 2006. [Google Scholar]
- Cheatham TE, Cieplak P, Kollman PA. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. Journal of biomolecular structure & dynamics. 1999;16(4):845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- Davies DB, Danyluk SS. Nuclear magnetic resonance studies of 5'-ribo- and deoxyribonucleotide structures in solution. Biochemistry. 1974;13(21):4417–4434. doi: 10.1021/bi00718a027. [DOI] [PubMed] [Google Scholar]
- De Leeuw Frank AAM, Altona C. Computer-assisted pseudorotation analysis of five-membered rings by means of proton spin-spin coupling constants: ProgramPSEUROT. Journal of Computational Chemistry. 1983;4(3):428–437. [Google Scholar]
- DeRose EF, Perera L, Murray MS, Kunkel TA, London RE. Solution structure of the Dickerson DNA dodecamer containing a single ribonucleotide. Biochemistry. 2012;51(12):2407–2416. doi: 10.1021/bi201710q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donders LA, De Leeuw FAAM, Altona C. Relationship between proton—proton NMR coupling constants and substituent electronegativities. IV—An extended karplus equation accounting for interactions between substituents and its application to coupling constant data calculated by the Extended Hückel method. Organic Magnetic Resonance. 1989;27(6):556–563. [Google Scholar]
- Du Q, Carrasco N, Teplova M, Wilds CJ, Egli M, Huang Z. Internal derivatization of oligonucleotides with selenium for X-ray crystallography using MAD. Journal of the American Chemical Society. 2002;124(1):24–25. doi: 10.1021/ja0171097. [DOI] [PubMed] [Google Scholar]
- Dupradeau F-Y, Pigache A, Zaffran T, Savineau C, Lelong R, Grivel N, et al. The R.E.D. tools: advances in RESP and ESP charge derivation and force field library building. Physical chemistry chemical physics : PCCP. 2010;12(28):7821–7839. doi: 10.1039/c0cp00111b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A, Wheeler P, Dan Cook P, Sanghvi YS. Synthesis and conformational properties of 2′-deoxy-2′-methylthio-pyrimidine and -purine nucleosides: Potential antisense applications. Journal of Heterocyclic Chemistry. 1993;30(5):1277–1287. [Google Scholar]
- Freisz S, Lang K, Micura R, Dumas P, Ennifar E. Binding of aminoglycoside antibiotics to the duplex form of the HIV-1 genomic RNA dimerization initiation site. Angewandte Chemie (International ed. in English) 2008;47(22):4110–4113. doi: 10.1002/anie.200800726. [DOI] [PubMed] [Google Scholar]
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 03 C.02. (n.d.) [Google Scholar]
- Germann MW, Kalisch BW, Lundberg P, Vogel HJ, van de Sande JH. Perturbation of DNA hairpins containing the EcoRI recognition site by hairpin loops of varying size and composition: physical (NMR and UV) and enzymatic (EcoRI) studies. Nucleic Acids Research. 1990;18(6):1489–1498. doi: 10.1093/nar/18.6.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschlbauer W, Jankowski K. Nucleoside conformation is determined by the electronegativity of the sugar substituent. Nucleic Acids Research. 1980;8(6):1421–1433. doi: 10.1093/nar/8.6.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haasnoot CAG, de Leeuw FAAM, de Leeuw HPM, Altona C. The relationship between proton-proton NMR coupling constants and substituent electronegativities. II—conformational analysis of the sugar ring in nucleosides and nucleotides in solution using a generalized Karplus equation. Organic Magnetic Resonance. 1981;15(1):43–52. [Google Scholar]
- Hendrickx PMS, Martins JC. A user-friendly Matlab program and GUI for the pseudorotation analysis of saturated five-membered ring systems based on scalar coupling constants. Chemistry Central journal. 2008;2:20. doi: 10.1186/1752-153X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseknecht JB, Altona C, Hadad CM, Lowary TL. Conformational analysis of furanose rings with PSEUROT: parametrization for rings possessing the arabino, lyxo, ribo, and xylo stereochemistry and application to arabinofuranosides. The Journal of organic chemistry. 2002;67(14):4647–4651. doi: 10.1021/jo025635q. [DOI] [PubMed] [Google Scholar]
- Höbartner C, Micura R. Chemical Synthesis of Selenium-Modified Oligoribonucleotides and Their Enzymatic Ligation Leading to an U6 SnRNA Stem−Loop Segment. Journal of the American Chemical Society. 2004;126(4):1141–1149. doi: 10.1021/ja038481k. [DOI] [PubMed] [Google Scholar]
- Jiang J, Sheng J, Carrasco N, Huang Z. Selenium derivatization of nucleic acids for crystallography. Nucleic Acids Research. 2007;35(2):477–485. doi: 10.1093/nar/gkl1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joecks A, K ppel H, Schleinitz KD, Cech D. NMR-spektroskopische Untersuchungen zum Konformations-verhalten von 2?- und 3?-halogensubstituierten Pyrimidinnucleosiden. Journal für Praktische Chemie. 1983;325(6):881–892. [Google Scholar]
- Johnson CN, Spring AM, Sergueev D, Shaw BR, Germann MW. Structural basis of the RNase H1 activity on stereo regular borano phosphonate DNA/RNA hybrids. Biochemistry. 2011;50(19):3903–3912. doi: 10.1021/bi200083d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karplus M. Vicinal Proton Coupling in Nuclear Magnetic Resonance. Journal of the American Chemical Society. 1963;85(18):2870–2871. [Google Scholar]
- Kennard O. DNA—Ligand Interactions. Boston, MA: Springer US; 1987. DNA Structure: Current Results from Single Crystal X-Ray Diffraction Studies; pp. 1–21. [Google Scholar]
- Marenich AV, Olson RM, Kelly CP, Cramer CJ, Truhlar DG. Self-Consistent Reaction Field Model for Aqueous and Nonaqueous Solutions Based on Accurate Polarized Partial Charges. Journal of Chemical Theory and Computation. 2007;3(6):2011–2033. doi: 10.1021/ct7001418. [DOI] [PubMed] [Google Scholar]
- Pigache A, Cieplak P, Dupradeau FY. Automatic and highly reproducible RESP and ESP charge derivation: Application to the Development of Programs RED and X RED. 227th ACS National Meeting.2004. [Google Scholar]
- Plateau P, Gueron M. Exchangeable proton NMR without base-line distorsion, using new strong-pulse sequences. Journal of the American Chemical Society. 1982;104(25):7310–7311. [Google Scholar]
- Rinkel LJ, Altona C. Conformational analysis of the deoxyribofuranose ring in DNA by means of sums of proton-proton coupling constants: a graphical method. Journal of biomolecular structure & dynamics. 1987;4(4):621–649. doi: 10.1080/07391102.1987.10507665. [DOI] [PubMed] [Google Scholar]
- Robins MJ, Mullah KB, Wnuk SF, Dalley NK. Nucleic acid related compounds. 73. Fluorination of uridine 2'-thioethers with xenon difluoride or (diethylamino)sulfur trifluoride. Synthesis of stable 2'-[alkyl(or aryl)sulfonyl]-2“-deoxy-2-”fluorouridines. The Journal of organic chemistry. 1992;57(8):2357–2364. [Google Scholar]
- Rosemeyer H, Zulauf M, Ramzaeva N, Becher G, Feiling E, Mühlegger K, et al. Stereoelectronic Effects of Modified Purines on the Sugar Conformation of Nucleosides and Fluorescence Properties. Nucleosides and Nucleotides. 1997;16(5–6):821–828. [Google Scholar]
- Rozners E. Carbohydrate Chemistry for RNA Interference: Synthesis and Properties of RNA Analogues Modified in Sugar-Phosphate Backbone. Current Organic Chemistry. 2006;10(6):675–692. [Google Scholar]
- Salon J, Sheng J, Gan J, Huang Z. Synthesis and crystal structure of 2'-se-modified guanosine containing DNA. The Journal of organic chemistry. 2010;75(3):637–641. doi: 10.1021/jo902190c. [DOI] [PubMed] [Google Scholar]
- Serganov A, Keiper S, Malinina L, Tereshko V, Skripkin E, Höbartner C, et al. Structural basis for Diels-Alder ribozyme-catalyzed carbon-carbon bond formation. Nature Structural & Molecular Biology. 2005;12(3):218–224. doi: 10.1038/nsmb906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Huang Z. Selenium derivatization of nucleic acids for X-ray crystal-structure and function studies. Chemistry & biodiversity. 2010;7(4):753–785. doi: 10.1002/cbdv.200900200. [DOI] [PubMed] [Google Scholar]
- Sheng J, Jiang J, Salon J, Huang Z. Synthesis of a 2'-Se-thymidine phosphoramidite and its incorporation into oligonucleotides for crystal structure study. Organic letters. 2007;9(5):749–752. doi: 10.1021/ol062937w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng J, Salon J, Gan J, Huang Z. Synthesis and crystal structure study of 2′-Se-adenosine-derivatized DNA. Science China Chemistry. 2010;53(1):78–85. [Google Scholar]
- Teplova M, Wilds CJ, Wawrzak Z, Tereshko V, Du Q, Carrasco N, et al. Covalent incorporation of selenium into oligonucleotides for X-ray crystal structure determination via MAD: proof of principle. Multiwavelength anomalous dispersion. Biochimie. 2002;84(9):849–858. doi: 10.1016/s0300-9084(02)01440-2. [DOI] [PubMed] [Google Scholar]
- Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. Journal of Computational Chemistry. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- Watts JK, Sadalapure K, Choubdar N, Pinto BM, Damha MJ. Synthesis and conformational analysis of 2'-fluoro-5-methyl-4‘-thioarabinouridine (4’S-FMAU) The Journal of organic chemistry. 2006;71(3):921–925. doi: 10.1021/jo051844+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.