Abstract
The persistence of certain strains of Listeria monocytogenes, even after the food processing environment has been cleaned and disinfected, suggests that this may be related to phenomena that reduce the concentration of the disinfectants to subinhibitory levels. This includes (i) the existence of environmental niches or reservoirs that are difficult for disinfectants to reach, (ii) microorganisms that form biofilms and create microenvironments in which adequate concentrations of disinfectants cannot be attained, and (iii) the acquisition of resistance mechanisms in L. monocytogenes, including those that lead to a reduction in the intracellular concentration of the disinfectants. The only available data with regard to the resistance of L. monocytogenes to disinfectants applied in food production environments refer to genotypic resistance to quaternary ammonium compounds (QACs). Although there are several well-characterized efflux pumps that confer resistance to QACs, it is a low-level resistance that does not generate resistance to QACs at the concentrations applied in the food industry. However, dilution in the environment and biodegradation result in QAC concentration gradients. As a result, the microorganisms are frequently exposed to subinhibitory concentrations of QACs. Therefore, the low-level resistance to QACs in L. monocytogenes may contribute to its environmental adaptation and persistence. In fact, in certain cases, the relationship between low-level resistance and the environmental persistence of L. monocytogenes in different food production chains has been previously established. The resistant strains would have survival advantages in these environments over sensitive strains, such as the ability to form biofilms in the presence of increased biocide concentrations.
Keywords: Listeria monocytogenes, food processing, bacterial persistence, quaternary ammonium disinfectants, resistance
Introduction
Listeria monocytogenes is a Gram-positive foodborne pathogen that can cause listeriosis, a relatively uncommon illness with a 20–30% case fatality rate (Silk et al., 2012; CDC, 2014; de Noordhout et al., 2014; EFSA and ECDC, 2014). The majority of cases of human listeriosis are caused by contaminated processed foods (McLauchlin et al., 2004; Lianou and Sofos, 2007). Although the original source of the contamination may be the raw food materials used in the processing plants, the strains of L. monocytogenes present in the final food products are usually different from the strains in the raw materials (Tompkin, 2002; Thévenot et al., 2006). In addition, when listeriosis outbreaks have been investigated, contamination is usually traced to the processing environment and equipment (Orsi et al., 2008; Nakari et al., 2014). This suggests that the contamination mainly occurs during the processing of food and is primarily due to strains from the processing plant environment (Malley et al., 2015; Garner and Kathariou, 2016).
Environmental Persistence
The molecular characterization of L. monocytogenes isolates from the food processing environment regularly shows the presence of a reduced number of molecular subtypes and the long-term persistence of specific strains that can contaminate food and cause foodborne listeriosis (Ortiz et al., 2010; Ferreira et al., 2011; Morganti et al., 2015). The detection of highly similar isolates from different areas inside single establishments, and their environmental persistence, is a matter of concern for the hygienic management of food establishments (Cramer, 2006; Ferreira et al., 2014).
Carpentier and Cerf (2011) assert that L. monocytogenes persistence is primarily a random process given that “there are no strains with unique properties that lead to persistence but harborage sites in food industry premises and equipment where L. monocytogenes can persist.” Persistent strains of L. monocytogenes have occasionally been isolated in food processing environments after cleaning and disinfection (Lundén et al., 2003b; Soumet et al., 2005; Thévenot et al., 2006; Bērziņš et al., 2010; Ortiz et al., 2016). As demonstrated by the findings of studies reviewed here, the persistence of certain strains of L. monocytogenes after cleaning and disinfecting is not a completely random process, but rather an event related to different situations that result in subinhibitory concentrations of disinfectants at different scales. This includes: (i) local environmental conditions that can lead to the formation of niches or reservoirs that are difficult for disinfectants to reach; (ii) microorganisms that form biofilms and create microenvironments in which adequate concentrations of disinfectants cannot be attained; and (iii) the acquisition of resistance mechanisms in L. monocytogenes, including those that lead to a reduction in the intracellular concentration of the disinfectants.
There are numerous factors in food processing plants, such as insufficient cleaning before disinfection, disinfection of wet surfaces, and dosage failure that can lead to subinhibitory concentrations of disinfectants, thus reducing their efficiency. This reduction may be especially significant in certain niches or reservoirs in which water and organic matter are abundant, creating an environment where bacteria can survive and multiply readily (Cramer, 2006; Carpentier and Cerf, 2011).
Failure to effectively clean and disinfect the processing plant environment may contribute to biofilm formation in certain niches, leading to bacterial persistence (Gandhi and Chikindas, 2007; Renier et al., 2011). Cells embedded in the biofilm matrix display an increased resistance to biocide treatments (Bridier et al., 2011). However, the resistance to disinfectants in sessile cells (biofilms) and in planktonic cells (free-floating) are clearly different phenomena (Fatemi and Frank, 1999; Norwood and Gilmour, 2000; Stopforth et al., 2002; Kastbjerg and Gram, 2009). Resistance of biofilms to disinfectants is considered to be a form of “phenotypic resistance” since bacterial resistance is mainly induced by a physiological adaptation to the biofilm mode of life and can be lost or markedly reduced when biofilm cells revert to the planktonic state (Pan et al., 2006; Bridier et al., 2011). In contrast, in the case of planktonic cells, a bacterial strain is defined as being resistant to a biocide if it is not inhibited by a specific concentration that usually inhibit the majority of other strains (Bridier et al., 2011); that is, planktonic cell resistance depends on intrinsic cellular attributes such as intra-species (i.e., strain) variability in the MIC of a given disinfectant (Kastbjerg and Gram, 2012).
Resistance To Disinfectants
The only available data with regard to the resistance of L. monocytogenes to disinfectants applied in food production environments refer to genotypic resistance to QACs (Hegstad et al., 2010; Gerba, 2015). BAC is typically used in studies assessing in vitro the activity of QACs. Increased MICs of BAC have been found in L. monocytogenes strains from different food production chains in different countries (Table 1). BAC resistance in L. monocytogenes is a low-level resistance. This means that the resistant strains only have a two to eight-fold increase in the MIC compared to the rest of the strains (Table 1). This resistance does not lead to QAC resistance at the concentrations that are normally used in the food industry (typically 200–1000 mg L-1) (SCENIHR, 2009; Ferreira et al., 2014; Tezel and Pavlostathis, 2015). Therefore, QACs are considered to be an effective means of eliminating the resistant L. monocytogenes strains (Kastbjerg and Gram, 2009, 2012).
Table 1.
Reports of resistance to BAC in planktonic cells of Listeria monocytogenes.
| No. assayed isolates | No. resistant isolates (%) | MIC (mg L-1) of BAC |
Susceptibility testing medium | Main origin of strains | Reference | |
|---|---|---|---|---|---|---|
| Resistant isolates | Susceptible isolates | |||||
| 132 | 12 (9%) | 16.0 | 2.0–8.0 | Mueller Hinton agar (MHA) with blood | Poultry (France) | Lemaitre et al., 1998 |
| 200 | 20 (10%) | 4.0–7.0 | ≤2.0 | Tryptic soy broth (TSB) | Fish (Norway) | Aase et al., 2000 |
| 97 | 7 (7%) | ≥8.0 | ≤4.0 | MHA | Food and others (France) | Mereghetti et al., 2000 |
| 19 | 5 (26%) | ≥5.0 | ≤1.25 | TSB | Meat, poultry (USA and Canada) | Romanova et al., 2002 |
| 112 | 17 (15%) | 4.0–8.0 | 2.0–3.0 | TSB | Meat (Norway) | Heir et al., 2004 |
| 254 | 108 (42%) | >7.5 | ≤7.5 | MHA | Fish (France) | Soumet et al., 2005 |
| 114 | 9 (8%) | 16.0–32.0 | 4.0 | MHA with blood | Retail food (Denmark) | Aarestrup et al., 2007 |
| 123 | 57 (46%) | >10.0 | ≤10.0 | MHA with blood | Poultry (turkey) (USA) | Mullapudi et al., 2008 |
| 138 | 19 (14%) | >10.0 | ≤10.0 | MHA with blood | Different foods (USA) | Ratani et al., 2012 |
| 91 | 15 (16%) | 28.0 | 14.0 | MHA with blood | Foods and others (Austria) | Müller et al., 2013 |
| 29 | 3 (10%) | ≥10.0 | ≤2.5 | MHA | Pork (Spain) | Ortiz et al., 2014a |
| 71 | 19 (27%) | ≥16.0 | <16.0 | MHA with blood | Retail foods (China) | Xu et al., 2014 |
| 59 | 13 (22%) | ≥12.0 | ≤10.0 | Brain heart infussion (BHI) broth | Retail foods (China) | Jiang et al., 2015 |
| 142 | 25 (18%) | ≥10.0 | <10.0 | MHA with blood | Different foods (Switzerland) | Ebner et al., 2015 |
| 20 | 3 (15%) | ≥10.0 | <10.0 | MHA with blood | Different foods (China) | Zhang et al., 2015 |
| 14a | 11 (79%) | ≥10.0 | ≤2.5 | MHA | Pork (Spain) | Ortiz et al., 2016 |
aAll from disinfected surfaces.
Some of these studies have been conducted with a large number of strains, and they report a variable frequency of resistant strains due to the diversity of the selection criteria for the strains studied. For example, the study of Ortiz et al. (2016) reports a very high frequency of resistant isolates because only samples of disinfected surfaces are included (Table 1). In some cases, selection for L. monocytogenes isolates resistant to QACs has been associated with the repeated use of this class of disinfectants (Heir et al., 2004; Ortiz et al., 2014a, 2016).
In food processing environments, L. monocytogenes is exposed to different disinfectants and sanitizers, and sometimes at subinhibitory cocentrations. This is particularly true for disinfectants that are not fully biodegradable which may persist in sewage for long periods. For example, QACs are biodegradable only under aerobic conditions, resulting in continuously fluctuating concentration gradients (Tezel and Pavlostathis, 2015). As a result, the microorganisms are frequently exposed to subinhibitory concentrations of QACs. Repeated exposure to subinhibitory concentrations of QACs and prolonged environmental persistence of certain strains may facilitate the development of resistance over time (Ortiz et al., 2014a). Subinhibitory concentrations of antimicrobials may cause genetic changes in the bacteria by means of different pathways, including an increase in free radicals inside the cell or oxidative stress (Liu et al., 2016). This can trigger the SOS response which can promote either the expression of genes involved in horizontal gene transfer or mutagenesis through induction of the error-prone DNA polymerases (Shapiro, 2015).
Strains of L. monocytogenes with genotypic resistance to QACs may have mutations that lead to a reduction in cell permeability (Mereghetti et al., 2000; To et al., 2002). For instance, resistant strains may have modifications in membrane fatty acids and phospholipids (Fox et al., 2011), which can lead to a more anionic and hydrophobic cell surface. This makes it difficult for QACs to pass through the membrane and enter the cell (To et al., 2002).
In other cases, resistance to QACs in L. monocytogenes may be due to the acquisition of QAC-specific efflux pumps through recombinant elements and mobile genetic elements. Different genetic markers have been identified that confer L. monocytogenes with a low-level resistance to QACs, including the resistance determinant bcrABC (Elhanafi et al., 2010) and the qacH gene of transposon Tn6188 (Müller et al., 2013), as well as various qac determinants originally identified in staphylococci (Xu et al., 2014). All of these genes encode components of the efflux system in the small multidrug resistance (SMR) protein family group (Blair et al., 2015). Plasmids are also associated with L. monocytogenes resistance to BAC (Elhanafi et al., 2010). Furthermore, plasmids with genes that confer resistance to BAC can be transferred between different pathogenic and non-pathogenic species of Listeria in the presence of QACs. This process also leads to the co-selection of resistance against heavy metals (Katharios-Lanwermeyer et al., 2012). In several screening studies of resistant strains, the qacH gene and the determinant bcrABC have been found (Müller et al., 2013). In one study, for example, resistance to BAC has been associated with the qacH gene in the majority (80%) of the tested strains, and a minority of the strains (12%) have been associated with the determinant bcrABC (Ebner et al., 2015). Nevertheless, in other studies, only the qacH gene has been detected (Ortiz et al., 2014b, 2016), while in others a clear predominance of the determinant bcrABC has been reported (Dutta et al., 2013).
On the other hand, in some studies, the aforementioned determinants are not detected in certain resistant strains (Ortiz et al., 2014b, 2016; Ebner et al., 2015). In these cases, resistance may be due to the overexpression of endogenous efflux pumps due to mutations in regulatory elements. This can occur due to exposure to QACs or the stress induced by these compounds (Tezel and Pavlostathis, 2015). These pumps are usually chromosomally encoded and affect a broad spectrum of antimicrobial compounds (Buffet-Bataillon et al., 2012; Ortiz et al., 2016). In certain strains of L. monocytogenes that are resistant to BAC, QAC resistance has been associated with the overexpression of efflux pumps in the MFS (major facilitator superfamily) group (Blair et al., 2015), such as MdrL (multidrug resistant Listeria) and Lde (Listeria drug efflux) (Romanova et al., 2006; Rakic-Martinez et al., 2011). Recently, the overexpression of a new L. monocytogenes efflux pump (encoded by emrE) has been implicated in the resistance to QACs (Kovacevic et al., 2015).
The Problem of Defining and Detecting Resistance
For various reasons, data on bacterial resistance to biocides is often difficult to interpret and compare (Buffet-Bataillon et al., 2012). A potential reason for the difficulty in interpreting data on bacterial resistance to biocides is the absence of clear criteria for defining a microorganism as resistant to disinfectants. An ecological concept of resistance to biocides has been proposed based on the “natural” susceptibility of a given species (Morrissey et al., 2014). According to this criterion, an isolate is defined as resistant when it is not inhibited by a concentration that would inhibit most of the strains of that particular species. The resistant isolate is typically phenotypically different from the wild type because it has acquired a resistance mechanism through mutation or horizontal gene transfer (EFSA, 2008; SCENIHR, 2009; Buffet-Bataillon et al., 2012; Morrissey et al., 2014). This commonly happens to L. monocytogenes strains that are resistant to BAC (Mereghetti et al., 2000; Elhanafi et al., 2010; Müller et al., 2013).
Another reason for the difficulty in comparing results on bacterial resistance is the lack of standardized tests for examining in vitro susceptibility to disinfectants (Buffet-Bataillon et al., 2012). Susceptibility to disinfectants is often determined through the MIC technique (usually employing agar dilution or broth dilution methods) (Table 1). However, in some cases, the MICs obtained through different methods can produce different results, which is why standardization is necessary. The influence of the culture media is especially important to consider when detecting low-level resistance (Table 1). Therefore, we recommend the use of standardized clinical protocols that are designed to evaluate the inhibitory activity of antibiotics (EUCAST, 2000; CLSI, 2009). Consequently, changes in the MIC of the disinfectant of several “test” strains can be measured and the resistant strains can be detected (Ortiz et al., 2014a). In contrast, current standardized tests for the evaluation and comparison of commercial disinfectants measure the bactericidal activity of the products by using one specific “reference” strain (European Committee for Standardisation, 2010; Buffet-Bataillon et al., 2012); the different tests use planktonic cells (suspension test) or cells attached to stainless steel (carrier test).
Despite the low MIC values, L. monocytogenes strains with low-level resistance to QACs have efflux pumps that can reduce the intracellular concentration of the biocides to subinhibitory levels, “such that the bacterium can survive longer than may have been predicted from the MIC for that organism” (Piddock, 2006). For quinolone-resistant Staphylococcus aureus, for example, the MICs afforded by mutations in the gene encoding the target topoisomerase are too low to allow survival, and it has been proposed that efflux gives rise to low intracellular concentrations of the drugs (Piddock, 2006). In addition, microorganisms in the environment may be exposed to enormously variable concentrations of QACs. For example, in domestic wastewater the average concentration of QACs is approximately 0.5 mg L-1, and in wastewater treatment plant effluents the concentration decreases to 0.05 mg L-1 (Tezel and Pavlostathis, 2015). As a result, low-level resistance to QACs may contribute to the persistence of L. monocytogenes in the environment.
Is Persistence Linked to Resistance?
The hypothesis that the persistence of certain subtypes of L. monocytogenes is linked to resistance to disinfectants has been investigated in numerous studies (revised by Ferreira et al., 2014). However, an association between low-level resistance to QACs and persistence of the pathogen in different food processing environments has been demonstrated only in a few cases (Aase et al., 2000; Lundén et al., 2003a; Fox et al., 2011; Ortiz et al., 2014a, 2016).
The susceptibility of 200 strains of L. monocytogenes to BAC has been investigated in a fish production plant in Norway. In this study, 10% of the strains are resistant strains, and all of these are previously identified persistent strains (Aase et al., 2000; Table 1). The relationship between persistence and resistance to two QACs has also been analyzed in four strains of poultry originating in Finland. In one of the persistent strains, the MIC values are higher (2.5–5.0 mg L-1) than the MICs for the rest of the strains (0.63–1.25 mg L-1) (Lundén et al., 2003a). Additionally, the MICs of a different QAC, benzethonium chloride, have been analyzed in 11 strains of L. monocytogenes from the cheese industry in Ireland. Two of the persistent strains have MIC values of 1.5–4.0 mg L-1, compared with a MIC of 0.5 mg L-1 for the rest of the strains (Fox et al., 2011). In an Iberian pork processing plant in Spain, 29 different subtypes of L. monocytogenes have been identified over 3 years, and the resistance to BAC has been associated with the persistence of three subtypes of the molecular serotype 1/2a (Ortiz et al., 2014a). Subsequently, in a similar plant, five different molecular subtypes have been detected in environmental samples from clean, disinfected surfaces (Ortiz et al., 2016). In these eight persistent subtypes, a low-level resistance to QACs has been confirmed. In both of these studies, the MIC values of BAC are 10–20 mg L-1 in the resistant strains and 1.25–2.5 mg L-1 in the sensitive strains (Table 1). To improve the control of L. monocytogenes in food processing environments, additional research is needed to evaluate attributes specific to resistant and persistent strains of this pathogen.
Advantages of Resistant Strains for Persisting in the Environment
The L. monocytogenes strains that have acquired mechanisms of resistance to QACs may have specific advantages over sensitive strains with regard to persisting in food processing environments. For example, the influence of residual QACs may vary in sensitive strains and resistant strains, as an optimum concentration (inhibitory) for the former may be suboptimum (subinhibitory) for the latter. This has been confirmed by Ortiz et al. (2014b) in strains of L. monocytogenes resistant to BAC. When biofilm formation in the presence of BAC has been studied, only three concentrations of BAC (1.25, 2.5, and 5 mg L-1) have been identified that show different effects on the biofilm formation by the group of strains of L. monocytogenes included in the study. It was observed that certain resistant strains are able to form biofilm at 5 mg L-1, a concentration of BAC that is inhibitory for most strains (Ortiz et al., 2014b). However, biofilm formation in the presence of increased concentrations of BAC may differ between resistant strains with similar MICs but different genetic determinants of BAC resistance. For example, biofilm production in the presence of BAC differs between persistent L. monocytogenes strains belonging to sequence type ST121 that have transposon Tn6188 with the gene qacH, and prfA mutants belonging to sequence type ST31 that are also persistent and resistant (Ortiz et al., 2016). Figure 1 shows the percentage of the biofilm formed in the presence of BAC relative to the biofilm formed without BAC by two representative ST31 and ST121 resistant strains. Thus, ST121 strains are able to form biofilm in the presence of BAC concentrations (5 mg L-1) higher than the MIC of sensitive strains (1.25–2.5 mg L-1) (Ortiz et al., 2016; Figure 1). These results may help explain the persistence of the resistant ST121 strains (Ortiz et al., 2014b, 2016; Schmitz-Esser et al., 2015).
FIGURE 1.
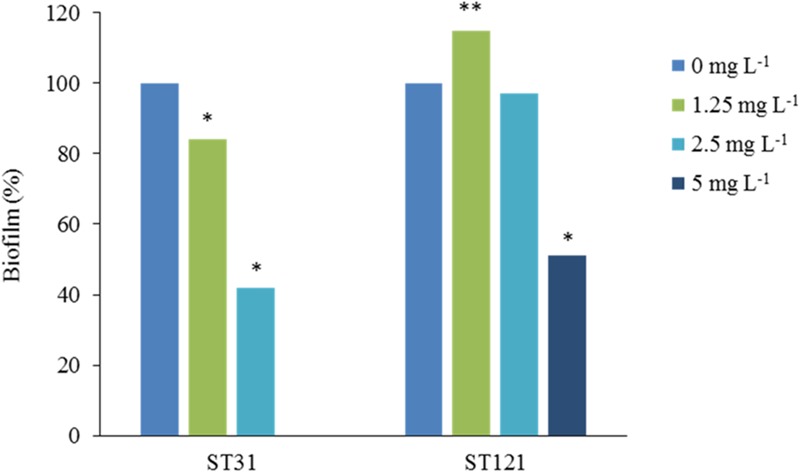
Effect of BAC on biofilm formation by representative sequence type (ST) 31 and ST121 Listeria monocytogenes strains. The results are presented as the average percentages of three measurements of biofilm formed with the addition of BAC concentrations of 1.25 mg L-1, 2.5 mg L-1 and 5 mg L-1, compared with the control without the addition of BAC (0 mg L-1, 100%). Reduced (∗) or increased (∗∗) percentages of biofilm differed significantly from the controls without BAC (p < 0.05, student’s t-test), adapted from Ortiz (2015).
Additionally, studies have shown that L. monocytogenes strains that are resistant to QACs can form biofilms faster than sensitive strains, which increases the likelihood of their survival (Nakamura et al., 2013). The expression of certain genes associated with the stress response or its regulation may also lead to an increased formation of biofilms by resistant strains (van der Veen and Abee, 2010). Therefore, the exposure of L. monocytogenes to subinhibitory concentrations of QACs and the consequent selection of resistant microorganisms may increase the ability of these bacteria to form biofilms and survive future treatment with high concentrations of the same compounds (Tezel and Pavlostathis, 2015).
Concluding Remarks
In recent years, the evaluation of the relationship between resistance to disinfectants and persistence, have allowed researchers to identify L. monocytogenes persistent subtypes that are resistant to disinfectants. The low-level resistance to QACs that has been detected in these subtypes is not a phenotypic adaptation, but rather, it is a stable genotypic resistance that is relatively frequent in certain environments and has important implications. For example, the formation of biofilms by resistant strains in the presence of biocide concentrations that are inhibitory for the sensitive strains may undoubtedly have an effect on their environmental survival. Therefore, a better understanding of the ecological and genetic characteristics of the strains resistant to QACs is needed, as well as standardization and consensus of the techniques used to detect the resistant strains.
Author Contributions
All authors listed, have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors were supported by grants RTA2011-00098-C02 and RTA2014-00045-C03 (Ministry of Economy and Competitiveness, Spain, Plan Nacional I+D+i co-financed by INIA and FEDER) during the preparation of this manuscript.
Abbreviations
- BAC
benzalkonium chloride
- MIC
minimum inhibitory concentration
- QACs
quaternary ammonium compounds
References
- Aarestrup F. M., Knöchel S., Hasman H. (2007). Antimicrobial susceptibility of Listeria monocytogenes from food products. Foodborne Pathog. Dis. 4 216–221. 10.1089/fpd.2006.0078 [DOI] [PubMed] [Google Scholar]
- Aase B., Sundheim G., Langsrud S., Rørvik L. M. (2000). Occurrence of and a possible mechanism for resistance to a quaternary ammonium compound in Listeria monocytogenes. Int. J. Food Microbiol. 62 57–63. 10.1016/S0168-1605(00)00357-3 [DOI] [PubMed] [Google Scholar]
- Bērziņš A., Hellström S., Siliņš I., Korkeala H. (2010). Contamination patterns of Listeria monocytogenes in cold-smoked pork processing. J. Food Prot. 73 2103–2109. [DOI] [PubMed] [Google Scholar]
- Blair J. M., Webber M. A., Baylay A. J., Ogbolu D. O., Piddock L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13 42–51. 10.1038/nrmicro3380 [DOI] [PubMed] [Google Scholar]
- Bridier A., Briandet R., Thomas V., Dubois-Brissonnet F. (2011). Resistance of bacterial biofilms to disinfectants: a review. Biofouling 27 1017–1032. 10.1080/08927014.2011.626899 [DOI] [PubMed] [Google Scholar]
- Buffet-Bataillon S., Tattevin P., Bonnaure-Mallet M., Jolivet-Gougeon A. (2012). Emergence of resistance to antibacterial agents: the role of quaternary ammonium compounds-a critical review. Int. J. Antimicrob. Agents 39 381–389. 10.1016/j.ijantimicag.2012.01.011 [DOI] [PubMed] [Google Scholar]
- Carpentier B., Cerf O. (2011). Review–persistence of Listeria monocytogenes in food industry equipment and premises. Int. J. Food Microbiol. 145 1–8. 10.1016/j.ijfoodmicro.2011.01.005 [DOI] [PubMed] [Google Scholar]
- CDC (2014). Incidence and trends of infection with pathogens transmitted commonly through food — foodborne diseases active surveillance network, 10 U.S. sites, 2006–2013. Morb. Mortal. Wkly. Rep. 63 328–332. [PMC free article] [PubMed] [Google Scholar]
- CLSI (2009). Clinical Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically. Approved Standard, 8th ed. M7-A8. Wayne, PA: CLSI. [Google Scholar]
- Cramer M. M. (2006). Food Plant Sanitation. Design, Maintenance, and Good Manufacturing Practices. Boca Raton, FL: CRC Press. [Google Scholar]
- de Noordhout C. M., Devleesschauwer B., Angulo F. J., Verbeke G., Haagsma J., Kirk M., et al. (2014). The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect. Dis. 14 1073–1082. 10.1016/S1473-3099(14)70870-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta V., Elhanafi D., Kathariou S. (2013). Conservation and distribution of the benzalkonium chloride resistance cassette bcrABC in Listeria monocytogenes. Appl. Environ. Microbiol. 79 6067–6074. 10.1128/AEM.01751-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebner R., Stephan R., Althaus D., Brisse S., Maury M., Tasara T. (2015). Phenotypic and genotypic characteristics of Listeria monocytogenes strains isolated during 2011-2014 from different food matrices in Switzerland. Food Control 57 321–326. 10.1016/j.foodcont.2015.04.030 [DOI] [Google Scholar]
- EFSA (2008). Scientific opinion of the panel on biological hazards on a request from the European Food Safety Authority on foodborne antimicrobial resistance as a biological hazard. EFSA J. 765 1–87. [Google Scholar]
- EFSA and ECDC (2014). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2012. EFSA J. 12 3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhanafi D., Dutta V., Kathariou S. (2010). Genetic characterisation of plasmid associated benzalkonium chloride resistance determinants in a strain of Listeria monocytogenes from the 1998–1999 outbreak. Appl. Environ. Microbiol. 76 8231–8238. 10.1128/AEM.02056-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2000). European committee on antimicrobial susceptibility testing. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by agar dilution. Definitive document E.DEF 3.1. Clin. Microbiol. Infect. 6 509–515. [DOI] [PubMed] [Google Scholar]
- European Committee for Standardisation (2010). European Standard EN 1276:2009/AC:2010. Chemical Disinfectants and Antiseptics–Quantitative Suspension Test for the Evaluation of Bactericidal Activity of Chemical Disinfectants and Antiseptics used in Food, Industrial, Domestic and Institutional Areas - Test Method and Requirements (phase 2, step 1). Brussels: European Committee for Standardisation. [Google Scholar]
- Fatemi P., Frank J. F. (1999). Inactivation of Listeria monocytogenes/Pseudomonas biofilms by peracid sanitizers. J. Food Prot. 62 761–765. [DOI] [PubMed] [Google Scholar]
- Ferreira V., Barbosa J., Stasiewicz M., Vongkamjan K., Moreno Switt A., Hogg T., et al. (2011). Diverse geno- and phenotypes of persistent Listeria monocytogenes isolates from fermented meat sausage production facilities in Portugal. Appl. Environ. Microbiol. 77 2701–2715. 10.1128/AEM.02553-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira V., Wiedmann M., Teixeira P., Stasiewicz M. J. (2014). Listeria monocytogenes persistence in food-associated environments: epidemiology, strain characteristics, and implications for public health. J. Food Prot. 77 150–170. 10.4315/0362-028X.JFP-13-150 [DOI] [PubMed] [Google Scholar]
- Fox E. M., Leonard N., Jordan K. (2011). Physiological and transcriptional characterization of persistent and nonpersistent Listeria monocytogenes isolates. Appl. Environ. Microbiol. 77 6559–6569. 10.1128/AEM.05529-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandhi M., Chikindas M. L. (2007). Listeria: a foodborne pathogen that knows how to survive. Int. J. Food Microbiol. 113 1–15. 10.1016/j.ijfoodmicro.2006.07.008 [DOI] [PubMed] [Google Scholar]
- Garner D., Kathariou S. (2016). Fresh produce-associated listeriosis outbreaks, sources of concern, teachable moments, and insights. J. Food Prot. 79 337–344. 10.4315/0362-028X.JFP-15-387 [DOI] [PubMed] [Google Scholar]
- Gerba C. P. (2015). Quaternary ammonium biocides: efficacy in application. Appl. Environ. Microbiol. 81 464–469. 10.1128/AEM.02633-2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegstad K., Langsrud S., Lunestad B. T., Scheie A. A., Sunde M., Yazdankhah S. P. (2010). Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health? Microb. Drug Resist. 16 91–104. 10.1089/mdr.2009.0120 [DOI] [PubMed] [Google Scholar]
- Heir E., Lindstedt B. A., Røtterud O. J., Vardund T., Kapperud G., Nesbakken T. (2004). Molecular epidemiology and disinfectant susceptibility of Listeria monocytogenes from meat processing plants and human infections. Int. J. Food Microbiol. 96 85–96. 10.1016/j.ijfoodmicro.2004.03.014 [DOI] [PubMed] [Google Scholar]
- Jiang X., Yu T., Liang Y., Ji S., Guo X., Ma J., et al. (2015). Efflux pump-mediated benzalkonium chloride resistance in Listeria monocytogenes isolated from retail food. Int. J. Food Microbiol. 217 141–145. 10.1016/j.ijfoodmicro.2015.10.022 [DOI] [PubMed] [Google Scholar]
- Kastbjerg V. G., Gram L. (2009). Model systems allowing quantification of sensitivity to disinfectants and comparison of disinfectant susceptibility of persistent and presumed nonpersistent Listeria monocytogenes. J. Appl. Microbiol. 106 1667–1681. 10.1111/j.1365-2672.2008.04134.x [DOI] [PubMed] [Google Scholar]
- Kastbjerg V. G., Gram L. (2012). Industrial disinfectants do not select for resistance in Listeria monocytogenes following long term exposure. Int. J. Food Microbiol. 160 11–15. 10.1016/j.ijfoodmicro.2012.09.009 [DOI] [PubMed] [Google Scholar]
- Katharios-Lanwermeyer S., Rakic-Martinez M., Elhanafi D., Ratani S., Tiedje J. M., Kathariou S. (2012). Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other Listeriae. Appl. Environ. Microbiol. 78 7549–7556. 10.1128/AEM.02245-2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacevic J., Ziegler J., Wałecka-Zacharska E., Reimer A., Kitts D. D., Gilmour M. W. (2015). Tolerance of Listeria monocytogenes to quaternary ammonium sanitizers is mediated by a novel efflux pump encoded by emrE. Appl. Environ. Microbiol. 82 939–953. 10.1128/AEM.03741-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre J. P., Echchannaoui H., Michaut G., Divies C., Rousset A. (1998). Plasmid-mediated resistance to antimicrobial agents among listeriae. J. Food Protect. 61 1459–1464. [DOI] [PubMed] [Google Scholar]
- Lianou A., Sofos J. N. (2007). A review of the incidence and transmission of Listeria monocytogenes in ready-to-eat products in retail and food service environments. J. Food Prot. 70 2172–2198. [DOI] [PubMed] [Google Scholar]
- Liu X., Marrakchi M., Jahne M., Rogers S., Andreescu S. (2016). Real-time investigation of antibiotics-induced oxidative stress and superoxide release in bacteria using an electrochemical biosensor. Free Radic. Biol. Med. 91 25–33. 10.1016/j.freeradbiomed.2015.12.001 [DOI] [PubMed] [Google Scholar]
- Lundén J. M., Autio T. J., Markkula A., Hellström S., Korkeala H. (2003a). Adaptive and cross-adaptive responses of persistent and nonpersistent Listeria monocytogenes strains to disinfectants. Int. J. Food Microbiol. 82 265–272. 10.1016/S0168-1605(02)00312-4 [DOI] [PubMed] [Google Scholar]
- Lundén J. M., Autio T. J., Sjoberg A. M., Korkeala H. J. (2003b). Persistent and nonpersistent Listeria monocytogenes contamination in meat and poultry processing plants. J. Food Prot. 66 2062–2069. [DOI] [PubMed] [Google Scholar]
- Malley T. J., Butts J., Wiedmann M. (2015). Seek and destroy process: Listeria monocytogenes process controls in the ready-to-eat meat and poultry industry. J. Food Prot. 78 436–445. 10.4315/0362-028X.JFP-13-507 [DOI] [PubMed] [Google Scholar]
- McLauchlin J., Mitchell R. T., Smerdon W. J., Jewell K. (2004). Listeria monocytogenes and listeriosis: a review of hazard characterisation for use in microbiological risk assessment of foods. Int. J. Food Microbiol. 92 15–33. 10.1016/S0168-1605(03)00326-X [DOI] [PubMed] [Google Scholar]
- Mereghetti L., Quentin R., Mee N., Audurier A. (2000). Low sensitivity of Listeria monocytogenes to quaternary ammonium compounds. Appl. Environ. Microbiol. 66 5083–5086. 10.1128/AEM.66.11.5083-5086.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morganti M., Scaltriti E., Cozzolino P., Bolzoni L., Casadei G., Pierantoni M., et al. (2015). Processing-dependent and clonal contamination patterns of Listeria monocytogenes in the cured ham food chain revealed by genetic analysis. Appl. Environ. Microbiol. 82 822–831. 10.1128/AEM.03103-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey I., Oggioni M. R., Knight D., Curiao T., Coque T., Kalkanci A., et al. (2014). Evaluation of epidemiological cut-off values indicates that biocide resistant subpopulations are uncommon in natural isolates of clinically-relevant microorganisms. PLoS ONE 9:e86669 10.1371/journal.pone.0086669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullapudi S., Siletzky R. M., Kathariou S. (2008). Heavy-metal and benzalkonium chloride resistance of Listeria monocytogenes isolates from the environment of turkey-processing plants. Appl. Environ. Microbiol. 74 1464–1468. 10.1128/AEM.02426-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller A., Rychli K., Muhterem-Uyar M., Zaiser A., Stessl B., Guinane C. M., et al. (2013). Tn6188 - a novel transposon in Listeria monocytogenes responsible for tolerance to benzalkonium chloride. PLoS ONE 8:e76835 10.1371/journal.pone.0076835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Takakura K., Sone Y., Itano Y., Nishikawa Y. (2013). Biofilm formation and resistance to benzalkonium chloride in Listeria monocytogenes isolated from a fish processing plant. J. Food Prot. 76 1179–1186. 10.4315/0362-028X.JFP-12-225 [DOI] [PubMed] [Google Scholar]
- Nakari U. M., Rantala L., Pihlajasaari A., Toikkanen S., Johansson T., Hellsten C., et al. (2014). Investigation of increased listeriosis revealed two fishery production plants with persistent Listeria contamination in Finland in 2010. Epidemiol. Infect. 142 2261–2269. 10.1017/S095026881300349X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwood D., Gilmour A. (2000). The growth and resistance to sodium hypochlorite of Listeria monocytogenes in a steady-state multispecies biofilm. J. Appl. Microbiol. 88 512–520. 10.1046/j.1365-2672.2000.00990.x [DOI] [PubMed] [Google Scholar]
- Orsi R. H., Borowsky M. L., Lauer P., Young S. K., Nusbaum C., Galagan J. E., et al. (2008). Short-term genome evolution of Listeria monocytogenes in a non-controlled environment. BMC Genomics 9:539 10.1186/1471-2164-9-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz S. (2015). Diversidad Genética y Persistencia Ambiental de Listeria monocytogenes En Dos Plantas de Procesado de Carne de Cerdo Ibérico: Influencia de la Resistencia a Desinfectantes de Amonio Cuaternario. Ph.D. thesis, Universidad Complutense de Madrid; Madrid. [Google Scholar]
- Ortiz S., López V., Martínez-Suárez J. V. (2014a). Control of Listeria monocytogenes contamination in an Iberian pork processing plant and selection of benzalkonium chloride-resistant strains. Food Microbiol. 39 81–88. 10.1016/j.fm.2013.11.007 [DOI] [PubMed] [Google Scholar]
- Ortiz S., López V., Martínez-Suárez J. V. (2014b). The influence of subminimal inhibitory concentrations of benzalkonium chloride on biofilm formation by Listeria monocytogenes. Int. J. Food Microbiol. 189 106–112. 10.1016/j.ijfoodmicro.2014.08.007 [DOI] [PubMed] [Google Scholar]
- Ortiz S., López V., Villatoro D., López P., Dávila J. C., Martínez-Suárez J. V. (2010). A 3-year surveillance of the genetic diversity and persistence of Listeria monocytogenes in an Iberian pig slaughterhouse and processing plant. Foodborne Pathog. Dis. 7 1177–1184. 10.1089/fpd.2010.0535 [DOI] [PubMed] [Google Scholar]
- Ortiz S., López-Alonso V., Rodríguez P., Martínez-Suárez J. V. (2016). The connection between persistent, disinfectant-resistant Listeria monocytogenes strains from two geographically separate Iberian pork processing plants: evidence from comparative genome analysis. Appl. Environ. Microbiol. 82 308–317. 10.1128/AEM.02824-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Breidt F., Jr., Kathariou S. (2006). Resistance of Listeria monocytogenes biofilms to sanitizing agents in a simulated food processing environment. Appl. Environ. Microbiol. 72 7711–7717. 10.1128/AEM.01065-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock L. J. (2006). Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 19 382–402. 10.1128/CMR.19.2.382-402.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakic-Martinez M., Drevets D. A., Dutta V., Katic V., Kathariou S. (2011). Listeria monocytogenes strains selected on ciprofloxacin or the disinfectant benzalkonium chloride exhibit reduced susceptibility to ciprofloxacin, gentamicin, benzalkonium chloride, and other toxic compounds. Appl. Environ. Microbiol. 77 8714–8721. 10.1128/AEM.05941-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratani S. S., Siletzky R. M., Dutta V., Yildirim S., Osborne J. A., Lin W., et al. (2012). Heavy metal and disinfectant resistance of Listeria monocytogenes from foods and food processing plants. Appl. Environ. Microbiol. 78 6938–6945. 10.1128/AEM.01553-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renier S., Hébraud M., Desvaux M. (2011). Molecular biology of surface colonization by Listeria monocytogenes: an additional facet of an opportunistic Gram-positive foodborne pathogen. Environ. Microbiol. 13 835–850. 10.1111/j.1462-2920.2010.02378.x [DOI] [PubMed] [Google Scholar]
- Romanova N., Favrin S., Griffiths M. W. (2002). Sensitivity of Listeria monocytogenes to sanitizers used in the meat processing industry. Appl. Environ. Microbiol. 68 6405–6409. 10.1128/AEM.68.12.6405-6409.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanova N. A., Wolffs P. F., Brovko L. Y., Griffiths M. W. (2006). Role of efflux pumps in adaptation and resistance of Listeria monocytogenes to benzalkonium chloride. Appl. Environ. Microbiol. 72 3498–3503. 10.1128/AEM.72.5.3498-3503.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCENIHR (2009). Scientific Committee on Emerging and Newly Identified Health Risks. Assessment of the Antibiotic Resistance Effects of Biocides. Brussels: European Commission. Health & Consumer Protection DG. [Google Scholar]
- Schmitz-Esser S., Müller A., Stessl B., Wagner M. (2015). Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 6:380 10.3389/fmicb.2015.00380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro R. S. (2015). Antimicrobial-induced DNA damage and genomic instability in microbial pathogens. PLoS Pathog. 11:e1004678 10.1371/journal.ppat.1004678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk B. J., Date K. A., Jackson K. A., Pouillot R., Holt K. G., Graves L. M., et al. (2012). Invasive listeriosis in the foodborne diseases active surveillance network (FoodNet), 2004-2009: further targeted prevention needed for higher-risk groups. Clin. Infect. Dis. 54(Suppl. 5) S396–S404. 10.1093/cid/cis268 [DOI] [PubMed] [Google Scholar]
- Soumet C., Ragimbeau C., Maris P. (2005). Screening of benzalkonium chloride resistance in Listeria monocytogenes strains isolated during cold smoked fish production. Lett. Appl. Microbiol. 41 291–296. 10.1111/j.1472-765X.2005.01763.x [DOI] [PubMed] [Google Scholar]
- Stopforth J. D., Samelis J., Sofos J. N., Kendall P. A., Smith G. C. (2002). Biofilm formation by acid-adapted and nonadapted Listeria monocytogenes in fresh beef decontamination washings and its subsequent inactivation with sanitizers. J. Food Prot. 65 1717–1727. [DOI] [PubMed] [Google Scholar]
- Tezel U., Pavlostathis S. G. (2015). Quaternary ammonium disinfectants: microbial adaptation, degradation and ecology. Curr. Opin. Biotechnol. 33 296–304. 10.1016/j.copbio.2015.03.018 [DOI] [PubMed] [Google Scholar]
- Thévenot D., Dernburg A., Vernozy-Rozand C. (2006). An updated review of Listeria monocytogenes in the pork meat industry and its products. J. Appl. Microbiol. 101 7–17. 10.1111/j.1365-2672.2006.02962.x [DOI] [PubMed] [Google Scholar]
- To M. S., Favrin S., Romanova N., Griffiths M. W. (2002). Postadaptational resistance to benzalkonium chloride and subsequent physicochemical modifications of Listeria monocytogenes. Appl. Environ. Microbiol. 68 5258–5264. 10.1128/AEM.68.11.5258-5264.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompkin R. B. (2002). Control of Listeria monocytogenes in the food-processing environment. J. Food Prot. 65 709–725. [DOI] [PubMed] [Google Scholar]
- van der Veen S., Abee T. (2010). Importance of SigB for Listeria monocytogenes static and continuous-flow biofilm formation and disinfectant resistance. Appl. Environ. Microbiol. 76 7854–7860. 10.1128/AEM.01519-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., Li Y., Zahid M. S., Yamasaki S., Shi L., Li J. R., et al. (2014). Benzalkonium chloride and heavy-metal tolerance in Listeria monocytogenes from retail foods. Int. J. Food Microbiol. 190 24–30. 10.1016/j.ijfoodmicro.2014.08.017 [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhou Y., Bao H., Zhang L., Wang R., Zhou X. (2015). Plasmid-borne cadmium resistant determinants are associated with the susceptibility of Listeria monocytogenes to bacteriophage. Microbiol. Res. 172 1–6. 10.1016/j.micres.2015.01.008 [DOI] [PubMed] [Google Scholar]


