Abstract
Purpose
To develop a technique to generate on-board volumetric-cine MRI (VC-MRI) using patient prior images, motion modeling and on-board 2D-cine MRI.
Methods
One phase of a 4D-MRI acquired during patient simulation is used as patient prior images. 3 major respiratory deformation patterns of the patient are extracted from 4D-MRI based on principal-component-analysis. The on-board VC-MRI at any instant is considered as a deformation of the prior MRI. The deformation field is represented as a linear combination of the 3 major deformation patterns. The coefficients of the deformation patterns are solved by the data fidelity constraint using the acquired on-board single 2D-cine MRI. The method was evaluated using both XCAT simulation of lung cancer patients and MRI data from four real liver cancer patients. The accuracy of the estimated VC-MRI was quantitatively evaluated using Volume-Percent-Difference(VPD), Center-of-Mass-Shift(COMS), and target tracking errors. Effects of acquisition orientation, region-of-interest(ROI) selection, patient breathing pattern change and noise on the estimation accuracy were also evaluated.
Results
Image subtraction of ground-truth with estimated on-board VC-MRI shows fewer differences than image subtraction of ground-truth with prior image. Agreement between profiles in the estimated and ground-truth VC-MRI was achieved with less than 6% error for both XCAT and patient data. Among all XCAT scenarios, the VPD between ground-truth and estimated lesion volumes was on average 8.43±1.52% and the COMS was on average 0.93±0.58mm across all time-steps for estimation based on the ROI region in the sagittal cine images. Matching to ROI in the sagittal view achieved better accuracy when there was substantial breathing pattern change. The technique was robust against noise levels up to SNR=20. For patient data, average tracking errors were less than 2 mm in all directions for all patients.
Conclusions
Preliminary studies demonstrated the feasibility to generate real-time VC-MRI for on-board localization of moving targets in radiotherapy.
I. INTRODUCTION
Reducing target localization errors is crucial for radiation therapy treatments, as previous studies showed that it was correlated with improved tumor control and reduced normal tissue toxicity. [24,29] This is especially important for lung and liver cancer treatments due to the uncertainties caused by tumor motions.[3,10] Stereotactic body radiation therapy (SBRT) is becoming an emerging and effective treatment paradigm in radiation therapy to treat early stage non-small cell lung cancer and liver cancer patients with promising early clinical outcome.[6,14,16,17] Compared to traditional fractionated radiotherapy, SBRT delivers much higher radiation dose per fraction in one to five fractions. On-board 4D or real time volumetric verification of the target location before and during the treatment is critical for lung and liver SBRT treatments due to its tight PTV margin, high fractional dose and long treatment time.
Cone-beam CT (CBCT) has been developed for on-board target localization; however, its applications are limited by the long scanning time, high imaging dose and poor soft tissue contrast.[9,22,23] Previous work has been done to use patient prior knowledge and deformation models for fast estimation of 4D-CBCT with low dose.[11-13,18,20,25,30].However, MRI is still needed for better soft tissue contrast. Currently, commercial MRI-Radiotherapy systems[4] are only capable of generating 2D cine images without volumetric information for real-time verification of moving targets. 4D MRI has been proposed and researched in many studies through either a prospective or a retrospective approach. However, due to limitations of hardware and software, prospective 4D-MRI suffers from poor temporal resolution (~1s) and poor spatial resolution (4-5mm).[2,5,8] Retrospective 4D-MRI suffers from long acquisition time (5-30min), and poor plane-to-plane resolution (3-5mm slice thickness).[1,2,19,27,28] No real time volumetric cine MRI has been developed due to limitation of the MR data acquisition speed.
In this work, we developed a novel algorithm to generate volumetric cine MRI (VC-MRI) images based on prior images and motion models for real time volumetric target localization in radiotherapy. Both 4D Digital Extended-Cardiac Torso (XCAT) [21] simulation of lung cancer patients and real liver cancer patients’ data were used to evaluate the clinical efficacy of the method.
II. METHODS
In VC-MRI, each time step of the VC-MRI images is considered a deformation of the prior MRI images acquired during patient simulation. In our study, one phase of the prior 4D MRI was used as the prior image, MRIprior. The new on-board VC-MRI at any time-step can be expressed as a function of the Deformation Field Map (DFM), D, and MRIprior as shown in Eq. (1)
| (1) |
Dx, Dy, Dz represent the deformation fields along the three canonical directions of the Cartesian coordinate system. The deformation field D in Eq. (1) has a large number of variables, which makes the searching of the optimal D inefficient and prone to be trapped at locally optimal values. To solve this problem, we use a Principal Component Analysis (PCA) based motion modeling method to reduce the number of variables in the deformation field D.
II.A. Calculate PCA Motion Modes from Prior Knowledge
Deformation fields were obtained by registering the one phase used as MRIprior with all other phases of the prior 4D-MRI images. PCA analysis was used to extract three principal deformation modes of the patient. Details about the PCA analysis can be found in previous publications. [20,30]
On-board DFM D to be solved in Equation 1 can be expressed as a linear combination of the three principal deformation modes
| (2) |
D0ave is the average of the original DFMs from the prior 4D MRI. wj (j=1,2,3) are the weightings corresponding to each principal motion mode.
II.B. VC-MRI Estimation Using PCA Motion Model
On-board 2D cine MRI images are used for VC-MRI estimation. The weighting coefficients, wj (j=1,2,3) in Eq. (2), can be solved by using a data fidelity constraint which states that the corresponding 2D slice of the estimated VC-MRI matches with the on-board 2D-cine slice
| (3) |
Where S is the operator to extract the corresponding 2D slice from the estimated VC-MRI images. This data fidelity constraint is met by minimizing the following objective function:
| (4) |
A gradient descent method was used to find the wj that minimizes f(w).
Once is solved, the DFM D can be built according to Eq (2). Then, the DFM D can be applied to the prior image to obtain the VC-MRI based on Eq (1).
II.C Effect of acquisition orientation
Axial, sagittal and coronal 2D cine MR images were all used for VC-MRI estimation separately for both XCAT and patient data. The accuracies of the estimated VC-MRI based on different cine images were compared to investigate the effect of acquisition orientation of 2D cine MR images on the VC-MRI estimation.
II.D Effect of region of interest (ROI) selection
In XCAT study, the VC-MRI was estimated using both the entire image and the ROI surrounding the tumor region in the 2D-cine MR image to evaluate the effect of ROI selection on the estimation accuracy. In the patient studies, the entire 2D cine image was used for image estimation.
II.E. Simulation study using XCAT Phantom
We used a digital anthropomorphic phantom, XCAT, to simulate the prior 4D-MRI set, onboard VC-MRIs and 2D-cine MRIs. XCAT uses nonuniform rational B-spline surfaces to model detailed human anatomy based on databases from the National Library of Medicine and patient datasets.[21] The 4D XCAT image generation contains two parts: the body volume and the lesion. The respiratory motion of the body volume and lesion can be controlled separately. Each respiratory motion is controlled by two respiratory curves: the diaphragm curve and the chest wall curve. The diaphragm curve mainly determines the motion in the superior-inferior (SI) direction, and the chest wall curve mainly controls the motion in the anterior-posterior (AP) direction.
II.E.1. Prior 4D MRI simulation
A spherical lesion of 30 mm diameter was simulated in the middle of the lung in XCAT. Both the body volume and lesion volume were simulated to move according to the same diaphragm and chest wall curves with a respiratory cycle of 5 seconds. The peak-to-peak amplitudes of the diaphragm curve and the chest wall curve were set to 3 and 2 cm, respectively. A ten-phase 4D MRI was then simulated as the prior 4D MRI. The MRI volume of each phase was composed of 256 × 256 × 100 voxels, with each voxel measuring 1.875× 1.875 × 3 mm in dimension. The XCAT phantom was generated in the activity mode in order to produce MRI-like images. Signal intensities of organs and tissues were assigned using values derived from FIESTA/TrueFISP MR images [15]. The end-expiration phase of the prior 4D MRI was selected as MRIprior.
II.E.2. On-board 2D Cine MR Simulation
To simulate on-board 2D-cine MR acquisition with a frame rate of 3-5 frames/s as it is typically achievable in a MRI scanner, 20 frames of on-board volumetric MRI images were generated within a respiratory cycle of 5 seconds, with a frame rate of 4 frames/s. These 20 sets of on-board volumetric MRI images were used as the “ground-truth” on-board VC-MRI images. 2D slices were extracted from the ground-truth VC-MRI at the location corresponding to the central slice of the lesion in the MRIprior images to simulate the on-board 2D-cine MR images acquired.
To evaluate the effects of potential patient breathing pattern change from simulation to treatment, four patient scenarios were simulated for the on-board volumetric cine MRI sets:
No breathing pattern change.
For both body and lesion volume, the peak-to-peak amplitude of the diaphragm curve changes to 2 cm, and that of the chest wall curve changes to 1.2 cm.
Body and lesion move according to different diaphragm and chest wall curves: the peak-to-peak amplitudes of the diaphragm curve for body volume and lesion are 4 and 2 cm, respectively; and those of the chest wall curve for body volume and lesion are 3 and 1.2 cm, respectively.
Based on scenario 2, but with the lesion having 20% phases shift relative to the body volume respiratory cycle.
II.E.3. Noise Study
To investigate the effects of noise on the estimation accuracy, noise was incorporated into the 4D-MRI and on-board 2D sagittal cine images for the study using XCAT scenario 2 as an example. The noise was added based on the typical assumption that the MRI signal is governed by a Rayleigh distribution in the presence of noise[7]. To evaluate how the VC-MRI reconstruction method will behave under different magnitudes of noise, SNR levels of 500, 100, 50, and 20 within the lesion were tested.
II.F. Patient study using liver cancer patient data
The VC-MRI method was evaluated using four liver cancer patients. The patient studies were conducted under an IRB-approved protocol. Details of image acquisition and 4D MRI reconstruction can be found in our previous publications [2,15]. In brief, 4D MRI image data were acquired using a fast imaging employing steady state (FIESTA) sequence on a 1.5T GE scanner. The images were retrospectively reconstructed for 10 phases using our in-house developed method which uses body area of either axial or sagittal MR images as an internal respiratory surrogate for 4D sorting. Each phase consisted of volumetric images of 256 × 256 × 40 voxels, with each voxel measuring 1.875 × 1.875 × 5 mm. The end-inspiration phase was used as MRIprior. Single-slice cine MR was acquired separately in axial, coronal and sagittal planes across the center of the tumor for 30 s at about 3 frames/s.
II.G. Evaluation methods
The estimation accuracy for lesion location and volume in the on-board VC-MRI was evaluated at every cine time-step, and results were reported as averages across the time steps with corresponding standard deviations. For the XCAT simulations, the lesions were automatically contoured in an in-house MATLAB code based on a threshold voxel value in both the estimated images and the “ground-truth” VC-MRI images for comparison. Two metrics were defined to quantify the accuracy of the estimated lesion volume: volume percentage-difference (VPD) (Eq. 5) and center-of-mass-shift (COMS) (Eq. 6).
| (5) |
V is the lesion volume contoured in the estimated image and V0 is that contoured in the “ground-truth” image.
| (6) |
Δx, Δy, Δz are center-of mass distances from V to V0.
In addition, the estimation results were evaluated by comparing the image differences and normalized profiles between the estimated and ground-truth VC-MRI.
For the four patient cases, a previously developed ROI feature-based motion tracking method [2] was used to calculate and compare the target tracking based on VC-MRI and the 2D cine images acquired along different directions.
III. RESULTS
III.A. XCAT Results
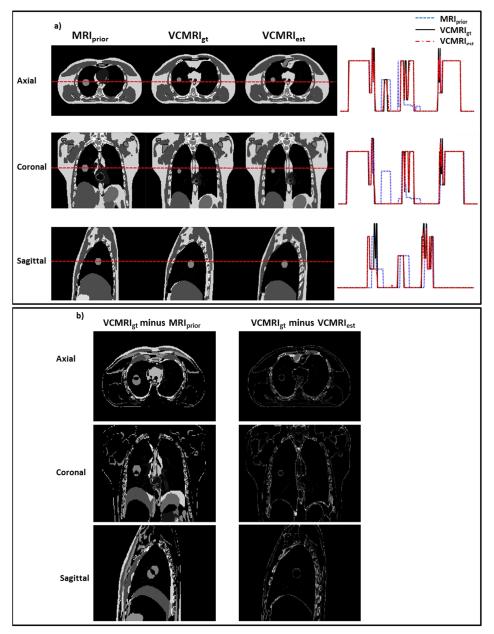
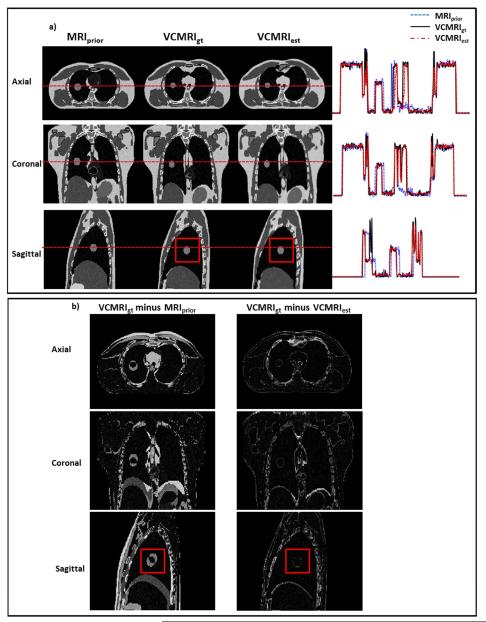
Figure 1 shows a) the prior MRI image (end expiration phase of 4D MRI), ground truth VC-MRI, estimated VC-MRI, as well as their profile comparisons and b) the corresponding subtraction images for XCAT scenario 1. The VC-MRI images were estimated by globally matching with the entire on-board axial 2D cine MRI image. The profile errors between ground-truth VC-MRI and estimated VC-MRI shown in Figure 1a) are on average 2.31%, 1.92%, and 3.04% for axial, coronal and sagittal views, respectively. Figure 2 shows a) the prior image, ground truth VC-MRI and estimated VC-MRI, as well as their profile comparisons and b) the subtraction images for XCAT scenario 2 with simulated noise. The VC-MRI images were estimated by matching with the ROI region of the sagittal 2D cine MRI image with a noise level of SNR = 20. The profile errors between ground-truth VC-MRI and estimated VC-MRI shown in Figure 2a) are on average 5.52%, 4.44%, and 3.48% for axial, coronal and sagittal views, respectively. Note that the ground truth and estimated images of the VC MRIs in both Figures 1 and 2 were taken from the end-inspiration phase as it has the most deformation from the prior image. Ten iterations were performed before the algorithm reached convergence, which has also been shown in previous PCA studies.[11,30]
Figure 1.
a) Comparison of prior MRI (MRIprior) at end-expiration phase, ground-truth on-board VC-MRI (VCMRIgt) at end-inspiration phase and estimated on-board VC-MRI (VCMRIest) for XCAT scenario 1. The VC-MRI was estimated using the entire 2D cine-MRI in the axial view. The horizontal red dotted line corresponds to the location of the profile curves shown to the right of the images. b) Subtraction images for the axial, coronal and sagittal images shown in Fig. 1a).
Figure 2.
a) Comparison of MRIprior at end-expiration phase, ground-truth VC-MRI (VCMRIgt) at end-inspiration phase and estimated VC-MRI (VCMRIest) for XCAT patient scenario 2. VC-MRI was estimated using the ROI around the lesion (the red box) in the 2D cine image in the sagittal view with added noise (SNR = 20). The horizontal red dotted line corresponds to the location of the profile curves shown to the right of the images. b) Subtraction images for the axial, coronal and sagittal images shown in Fig. 2a). The boxes in images signify the ROI regions used for VC-MRI estimation.
The VPD of the prior image compared with the ground-truth VC-MRI was 66.67±91.95% and COMS was 6.13±8.35 mm on average across all four XCAT scenarios and cine time steps. Table 1a shows the average VPD and COMS across all cine time-steps for different scenarios, acquisition orientations and estimation schemes (matching globally or matching ROI). Table 1b shows the VPD and COMS for the noise study performed on XCAT scenario 2.
Table 1.
Table 1a shows VPD and COMS for XCAT scenarios for the six different estimation schemes. Table 1b shows VPD and COMS for XCAT scenario 2 with various noise levels. The estimation scheme was to match an ROI in the sagittal cine images.
VPD and COMS for XCAT Scenarios
| a. | ||||
|---|---|---|---|---|
| XCAT Scenarios | 1 | 2 | 3 | 4 |
| VPD1(%) | ||||
| Axial, Global | 9.48±2.83 | 10.74±3.22 | 52.58±30.89 | 23.90±11.07 |
| Axial, ROI | 11.22±5.07 | 10.83±4.54 | 14.09±4.40 | 10.75±4.25 |
| Coronal, Global | 7.89±1.48 | 10.65±4.54 | 56.97±29.56 | 23.50±12.28 |
| Coronal, ROI | 15.22±8.69 | 15.98±10.56 | 15.97±10.07 | 15.61±10.24 |
| Sagittal, Global | 8.17±1.08 | 11.14±3.62 | 56.06±33.68 | 21.46±13.60 |
| Sagittal, ROI | 8.93±1.72 | 8.26±1.45 | 8.25±1.44 | 8.27±1.46 |
|
| ||||
| COMS2(mm) | ||||
| Axial, Global | 1.22±0.90 | 1.24±0.64 | 5.82±3.42 | 2.58±1.46 |
| Axial, ROI | 1.32±0.48 | 1.65±0.89 | 1.82±1.07 | 1.33±1.00 |
| Coronal, Global | 1.31±0.80 | 0.98±0.98 | 6.27±3.59 | 2.37±1.16 |
| Coronal, ROI | 2.14±1.00 | 2.03±1.14 | 2.18±1.28 | 1.91±1.20 |
| Sagittal, Global | 1.38±0.60 | 1.03±0.53 | 6.15±3.66 | 2.27±1.43 |
| Sagittal, ROI | 1.01±0.54 | 0.87±0.58 | 0.92±0.60 | 0.92±0.60 |
|
| ||||
| b. | ||||
| SNR | 500 | 100 | 50 | 20 |
|
| ||||
| VPD (%) |
8.22±1.43 | 8.27±1.47 | 8.15±1.32 | 7.95±1.61 |
| COMS (mm) | 0.89±0.54 | 0.89±0.54 | 0.99±0.55 | 0.75±0.55 |
Volume Percent Error
Center of Mass Shift
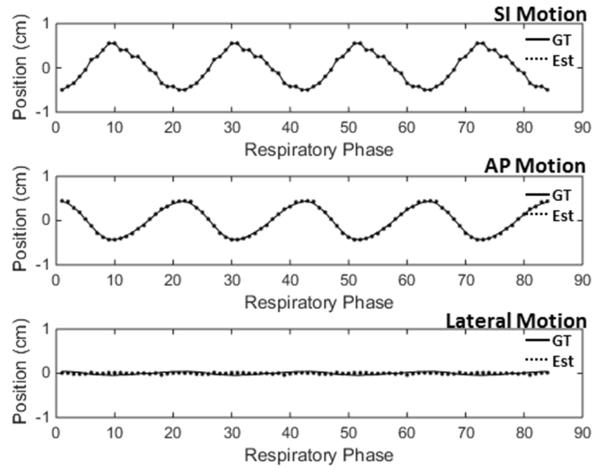
Figure 3 shows the centroid locations of the lesion along three axes over four respiratory cycles in the ground-truth images and the estimated VC-MRI for XCAT scenario 2. The VC-MRI estimation scheme used was to match with the ROI around the lesion in the sagittal cine image.
Figure 3.
Lesion centroid position curve along different axes for all time steps for XCAT scenario 2. The VC-MRI was estimated using the ROI around the lesion in the sagittal on-board 2D cine image. The solid line corresponds to the ground-truth lesion positon, and the dotted line corresponds to the estimated lesion position in VC-MRI.
III.B. Patient Results
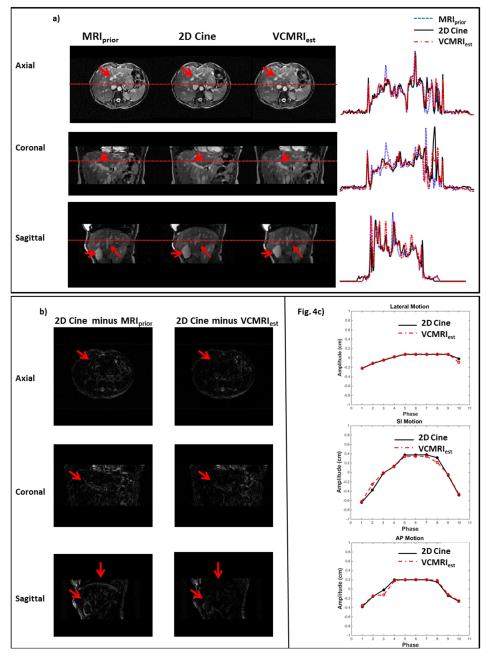
Figure 4a) shows the prior MRI image at end-inspiration phase, 2D cine, estimated VC-MRI, as well as their profile comparisons for Patient 1. Figure 4b) shows the subtraction images between estimated VC-MRI and 2D cine for Patient 1. Figure 4c) shows the tumor tracking based on estimated VC-MRI and 2D cine for average cycles in the lateral, SI and AP directions. Note that the VC-MRI was estimated based on sagittal cine images, and the tracking along SI and lateral directions were based on the coronal slice of VC-MRI and coronal cine images. The profile errors between 2D cine and estimated VC-MRI shown in Figure 4a) are on average 3.87%, 5.88%, and 5.41% for axial, coronal and sagittal views, respectively. Table 2 shows the mean, standard deviation and max tracking errors based on VC-MRI for each patient.
Figure 4.
a) prior MRI (MRIprior), 2D cine MRI and estimated VC-MRI for Patient 1. VC-MRI was estimated by matching to the axial, coronal and sagittal cine images, respectively. The horizontal red dotted line corresponds to the location of the profile curves shown to the right of the images. Fig4b) shows subtraction images for the axial, coronal and sagittal images in Fig4a). Fig 4c) shows the tracking curves between 2D cine and VCMRI for an average cycle. Note that the VCMRI was generated by matching to sagittal cine images. The tracking along Lat and SI directions were based on the coronal slice of the VCMRI and coronal cine, while tracking along AP direction was based on the sagittal slice of VCMRI and sagittal cine.
Table 2.
Tumor tracking errors (mean, standard deviation and max errors) based on VC-MRI in SI, AP and lateral directions in the patient study. VC-MRI was estimated based on sagittal cine images.
Motion Tracking Errors
| SI | AP | Lateral | ||||
|---|---|---|---|---|---|---|
| μ±σ (mm) | errormax (mm) | μ±σ (mm) | errormax (mm) |
μ±σ (mm) | errormax (mm) |
|
| Patient 1 | 0.40±0.40 | 1.26 | 0.26±0.30 | 1.07 | 0.15±0.21 | 0.75 |
| Patient 2 | 0.57±0.37 | 1.35 | 0.03±0.04 | 0.14 | 0.37±0.21 | 0.88 |
| Patient 3 | 1.30±0.94 | 2.64 | 1.11±0.58 | 2.30 | 1.16±0.56 | 1.99 |
| Patient 4 | 0.55±0.51 | 1.52 | 0.87±0.67 | 2.21 | 0.28±0.40 | 1.41 |
IV. DISCUSSIONS
To our knowledge, VC-MRI is the first technique capable of generating real time volumetric MRI images for target localization in radiation therapy treatments. Different from 2D cine MRI and 4D-MRI, VC-MRI can potentially achieve both high temporal resolution of 3-5 frames/s and high 3D spatial resolution of ~1mm in-plane, 1-3mm plane-to-plane resolution depending on the quality of the prior images. With these significant improvements, VC-MRI allows us for the first time to localize the full 3D target volume continuously in real time to improve the localization accuracy of SBRT both before and during the actual treatment. The real time volumetric localization by VC-MRI also make it feasible to perform real time 4D gating or target tracking during treatments, which paves the road to further margin reduction and dose escalation for lung and liver SBRT.
Results in Table 1 showed that the VC-MRI estimation based on the ROI region of the cine MR image achieved better accuracy for tumor localization than that based on the entire cine MR image when there was substantial breathing pattern change from prior to on-board images. This is because the deformation models from prior images were not accurate to model the entire body motion, but were still accurate enough to model the local tumor motion under these scenarios. For the VC-MRI estimation based on the ROI region, sagittal MR cine images produced better estimation accuracy than coronal and axial cine images. This is due to the fact that the sagittal image captured most of the prominent tumor motion along the superior-inferior and anterior-posterior directions, which provided more motion information for the VC-MRI estimation.
Note that the PCA model in this study was built from the deformation fields obtained from the prior 4D-MRI data. In the situation where no 4D-MRI is available, one possible solution is to build the PCA model from deformations obtained from 4D-CT data, as reported in previous studies.[30]
This manuscript mainly presents a pilot study of the feasibility of the VC-MRI technique. Further investigations are warranted to fully evaluate or improve the VC-MRI in the following aspects: (1). Patient breathing irregularities during prior 4D-MRI scan may affect the image quality of the 4D-MRI and the deformation field derived for motion modeling. The robustness of VC-MRI against breathing irregularities needs to be evaluated through either phantom or patient studies. (2). Effects of patient breathing pattern changes from prior to on-board imaging on VC-MRI need to be further studied. To make the technique more robust against such change, we can potentially incorporate free form deformation model to correct any errors induced by the motion modeling.[20,30] (3). Acceleration of VC-MRI. Currently, VC-MRI reconstruction takes ~2min. We can potentially accelerate the reconstruction using the graphics card and parallel computing to achieve real time imaging. (4). Comparison with 2D cine techniques. A method was reported before to use interleaved orthogonal 2D-cine images for target tracking.[26] A comparison between VC-MRI and the 2D cine technique is warranted to fully understand the advantages and limitation of each technique.
V. Conclusion
A novel VC-MRI technique has been developed to use patient prior images, deformation models and real-time on-board 2D cine images to generate volumetric cine MRI images. Preliminary studies using both XCAT simulation of lung cancer patients and real liver cancer patients data demonstrated the feasibility of the VC-MRI technique, which can potentially become a valuable tool for both inter-and intra-fraction target localization in lung and liver SBRT treatments.
SUMMARY.
A novel technique has been developed to generate volumetric cine MRI (VC-MRI) using patient prior information. The VC-MRI was generated by deforming the prior MRI images based on the on-board 2D cine MRI and patient respiratory breathing model. The technique was evaluated using both anthropomorphic digital phantom and patient data. Results demonstrated the feasibility of generating VC-MRI for both inter- and intra-fraction verification of moving targets in radiotherapy.
Acknowledgments
The authors would like to thank Dr. Paul Segars for use of his XCAT digital phantom and Chunhao Wang for thought-provoking discussions and suggestions involving this work.
Conflict of Interests: This work was supported by the National Institutes of Health under Grant No. R01-CA184173 and a research grant from Varian Medical Systems.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Bjerre T, et al. Three-dimensional mri-linac intra-fraction guidance using multiple orthogonal cine-mri planes. Physics in medicine and biology. 2013;58:4943–4950. doi: 10.1088/0031-9155/58/14/4943. [DOI] [PubMed] [Google Scholar]
- [2].Cai J, et al. Four-dimensional magnetic resonance imaging (4d-mri) using image-based respiratory surrogate: A feasibility study. Medical physics. 2011;38:6384–6394. doi: 10.1118/1.3658737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Cervino LI, et al. The diaphragm as an anatomic surrogate for lung tumor motion. Physics in medicine and biology. 2009;54:3529–3541. doi: 10.1088/0031-9155/54/11/017. [DOI] [PubMed] [Google Scholar]
- [4].Dempsey J, et al. We-e-vala-06: A real-time mri guided external beam radiotherapy delivery system. Medical physics. 2006;33:2254–2254. [Google Scholar]
- [5].Dinkel J, et al. 4d-mri analysis of lung tumor motion in patients with hemidiaphragmatic paralysis. Radiotherapy and oncology : journal of the European Society for Therapeutic Radiology and Oncology. 2009;91:449–454. doi: 10.1016/j.radonc.2009.03.021. [DOI] [PubMed] [Google Scholar]
- [6].Fakiris AJ, et al. Stereotactic body radiation therapy for early-stage non-small-cell lung carcinoma: Four-year results of a prospective phase ii study. International journal of radiation oncology, biology, physics. 2009;75:677–682. doi: 10.1016/j.ijrobp.2008.11.042. [DOI] [PubMed] [Google Scholar]
- [7].Gudbjartsson H, Patz S. The rician distribution of noisy mri data. Magnetic resonance in medicine. 1995;34:910–914. doi: 10.1002/mrm.1910340618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hu Y, et al. Respiratory amplitude guided 4-dimensional magnetic resonance imaging. International journal of radiation oncology, biology, physics. 2013;86:198–204. doi: 10.1016/j.ijrobp.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jaffray DA, Siewerdsen JH. Cone-beam computed tomography with a flat-panel imager: Initial performance characterization. Medical physics. 2000;27:1311–1323. doi: 10.1118/1.599009. [DOI] [PubMed] [Google Scholar]
- [10].Keall PJ, et al. The management of respiratory motion in radiation oncology report of aapm task group 76. Medical physics. 2006;33:3874–3900. doi: 10.1118/1.2349696. [DOI] [PubMed] [Google Scholar]
- [11].Li R, et al. Real-time volumetric image reconstruction and 3d tumor localization based on a single x-ray projection image for lung cancer radiotherapy. Medical physics. 2010;37:2822–2826. doi: 10.1118/1.3426002. [DOI] [PubMed] [Google Scholar]
- [12].Li R, et al. Single-projection based volumetric image reconstruction and 3d tumor localization in real time for lung cancer radiotherapy. Medical image computing and computer-assisted intervention : MICCAI ... International Conference on Medical Image Computing and Computer-Assisted Intervention. 2010;13:449–456. doi: 10.1007/978-3-642-15711-0_56. [DOI] [PubMed] [Google Scholar]
- [13].Li R, et al. On a pca-based lung motion model. Physics in medicine and biology. 2011;56:6009–6030. doi: 10.1088/0031-9155/56/18/015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Liu E, et al. Stereotactic body radiation therapy for primary and metastatic liver tumors. Translational oncology. 2013;6:442–446. doi: 10.1593/tlo.12448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liu Y, et al. Investigation of sagittal image acquisition for 4d-mri with body area as respiratory surrogate. Medical physics. 2014;41:101902. doi: 10.1118/1.4894726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Mendez Romero A, et al. Stereotactic body radiation therapy for primary and metastatic liver tumors: A single institution phase i-ii study. Acta oncologica. 2006;45:831–837. doi: 10.1080/02841860600897934. [DOI] [PubMed] [Google Scholar]
- [17].Mendez Romero A, et al. Quality of life after stereotactic body radiation therapy for primary and metastatic liver tumors. International journal of radiation oncology, biology, physics. 2008;70:1447–1452. doi: 10.1016/j.ijrobp.2007.08.058. [DOI] [PubMed] [Google Scholar]
- [18].Mishra P, et al. Evaluation of 3d fluoroscopic image generation from a single planar treatment image on patient data with a modified xcat phantom. Physics in medicine and biology. 2013;58:841–858. doi: 10.1088/0031-9155/58/4/841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Remmert G, et al. Four-dimensional magnetic resonance imaging for the determination of tumour movement and its evaluation using a dynamic porcine lung phantom. Physics in medicine and biology. 2007;52:N401–415. doi: 10.1088/0031-9155/52/18/N02. [DOI] [PubMed] [Google Scholar]
- [20].Ren L, Zhang Y, Yin FF. A limited-angle intrafraction verification (live) system for radiation therapy. Medical physics. 2014;41:020701. doi: 10.1118/1.4861820. [DOI] [PubMed] [Google Scholar]
- [21].Segars WP, et al. 4d xcat phantom for multimodality imaging research. Medical physics. 2010;37:4902–4915. doi: 10.1118/1.3480985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Siewerdsen JH, Jaffray DA. Cone-beam computed tomography with a flat-panel imager: Effects of image lag. Medical physics. 1999;26:2635–2647. doi: 10.1118/1.598803. [DOI] [PubMed] [Google Scholar]
- [23].Siewerdsen JH, Jaffray DA. Cone-beam computed tomography with a flat-panel imager: Magnitude and effects of x-ray scatter. Medical physics. 2001;28:220–231. doi: 10.1118/1.1339879. [DOI] [PubMed] [Google Scholar]
- [24].Soike M, et al. Image guided radiation therapy results in improved local control in lung cancer patients treated with fractionated radiation therapy for stage iib-iiib disease. International journal of radiation oncology, biology, physics. 2013;87:S547–548. [Google Scholar]
- [25].Staub D, et al. 4d cone-beam ct reconstruction using a motion model based on principal component analysis. Medical physics. 2011;38:6697–6709. doi: 10.1118/1.3662895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Tryggestad E, et al. 4d tumor centroid tracking using orthogonal 2d dynamic mri: Implications for radiotherapy planning. Medical physics. 2013;40:091712. doi: 10.1118/1.4818656. [DOI] [PubMed] [Google Scholar]
- [27].Tryggestad E, et al. Respiration-based sorting of dynamic mri to derive representative 4d-mri for radiotherapy planning. Medical physics. 2013;40:051909. doi: 10.1118/1.4800808. [DOI] [PubMed] [Google Scholar]
- [28].von Siebenthal M, et al. 4d mr imaging of respiratory organ motion and its variability. Physics in medicine and biology. 2007;52:1547–1564. doi: 10.1088/0031-9155/52/6/001. [DOI] [PubMed] [Google Scholar]
- [29].Zelefsky MJ, et al. Improved clinical outcomes with high-dose image guided radiotherapy compared with non-igrt for the treatment of clinically localized prostate cancer. International journal of radiation oncology, biology, physics. 2012;84:125–129. doi: 10.1016/j.ijrobp.2011.11.047. [DOI] [PubMed] [Google Scholar]
- [30].Zhang Y, et al. A technique for estimating 4d-cbct using prior knowledge and limited-angle projections. Medical physics. 2013;40:121701. doi: 10.1118/1.4825097. [DOI] [PubMed] [Google Scholar]