Abstract
Background
Propionic acidemia (PA) is a disorder of intermediary metabolism with defects in the alpha or beta subunits of propionyl CoA carboxylase (PCCA and PCCB respectively) enzyme. We previously described a liver culture system that uses liver-derived hemodynamic blood flow and transport parameters to restore and maintain primary human hepatocyte biology and metabolism utilizing physiologically relevant milieu concentrations.
Methods
In this study, primary hepatocytes isolated from the explanted liver of an 8-year-old PA patient were cultured in the liver system for 10 days and evaluated for retention of differentiated polarized morphology. The expression of PCCA and PCCB was assessed at a gene and protein level relative to healthy donor controls. Ammonia and urea levels were measured in the presence and absence of amino acid supplements to assess the metabolic consequences of branched-chain amino acid metabolism in this disease.
Results
Primary hepatocytes from the PA patient maintained a differentiated polarized morphology (peripheral actin staining) over 10 days of culture in the system. We noted lower levels of PCCA and PCCB relative to normal healthy controls at the mRNA and protein level. Supplementation of branched-chain amino acids, isoleucine (5 mM) and valine (5 mM) in the medium, resulted in increased ammonia and decreased urea in the PA patient hepatocyte system, but no such response was seen in healthy hepatocytes or patient-derived fibroblasts.
Conclusions
We demonstrate for the first time the successful culture of PA patient-derived primary hepatocytes in a differentiated state, that stably retain the PCCA and PCCB enzyme defects at a gene and protein level. Phenotypic response of the system to an increased load of branched-chain amino acids, not possible with fibroblasts, underscores the utility of this system in the better understanding of the molecular pathophysiology of PA and examining the effectiveness of potential therapeutic agents in the most relevant tissue.
Keywords: Organic acidemia, Propionic acidemia, Ammonia, Urea, Hepatocyte, Hemodynamic flow
1. Introduction
Propionic acidemia (PA, OMIM #606054) is an inborn error of metabolism that affects 1 in 100,000 individuals in the US, and the prevalence can be as high as 1 in 1000–2000 in specific populations that are genetically at higher risk (e.g. Inuit, Greenland and Saudi Arabia) [1–3]. The disease is caused by a dysfunction of the propionyl CoA carboxylase (EC 6.4.1.3) enzyme, which blocks the conversion of propionyl CoA to methylmalonyl CoA resulting in the accumulation of metabolic intermediates 3-hydroxypropionic acid and methylcitrate in the urine, and propionylcarnitine in the blood [4]. Inhibition of the urea cycle (assumed to be by propionic acid or propionyl CoA) results in clinically significant elevations in blood ammonia, contributing to both morbidity and mortality [5]. Patients with PA often present acutely with metabolic acidosis, cardiac arrhythmias and hyperammonemia causing severe central nervous system dysfunction. Long term complications, include seizures, cardiomyopathies, metabolic stroke like episodes, cardiac arrhythmias, impaired consciousness, ketosis, pancreatitis and optic atrophy, which severely impact the quality of life and cause progressive deterioration, sometimes ending in sudden death, [6–8]. There are no current definitive treatments for PA. Therapeutic options focus on the dietary restriction of the precursors of propionyl CoA, such as odd-chain fatty acids and branched-chain amino acids (valine, methionine, isoleucine and threonine), while trying to maintain normal growth, scavenging excess propionic acid using carnitine, and symptomatically treating complications as they occur [9,10]. Some patients with PA receive liver transplants to ameliorate symptoms primarily due to hyperammonemia. However, in spite of the symptomatic relief, many of these patients still progress to the long-term sequelae of the disease. This highlights the unmet need to develop better therapies that would improve quality of life and lifespan of these patients.
While small patient populations represent a major challenge in the discovery and development of new therapies for rare diseases like PA, the most significant issue is the lack of adequate pre-clinical models in which to study the disease and test new drugs. Existing genetic models in mice do not adequately recapitulate disease biology due to inherent species-related differences in metabolism and protein turnover rates [11–14]. Even within a species, there are metabolic differences dependent on the tissue type [15–17]. Current in vitro models use fibroblasts derived from PA patients [18–20]. However, there are significant quantitative and qualitative differences between fibroblasts and more highly metabolic cells such as hepatocytes [21,22]. Additionally, certain metabolic pathways are either restricted or active only in certain tissues, e.g. only the liver expresses the complete urea cycle enzymes converting ammonia into urea. The liver is the most metabolically versatile organ in the body. Hepatocytes, the epithelial cells of the liver, are the primary cell type responsible for converting nitrogenous waste arising from the breakdown of proteins and amino acids into urea, prior to excretion by the kidney. However, the use of traditional cultured hepatocytes has been limited by the rapid loss of the liver-like phenotype known to occur due to dedifferentiation in vitro [23–25]. Approaches to preserve liver-specific function and response of hepatocytes by modifications to culture medium, three dimensional aggregation into spheroids, co-culturing with supporting cell types and/or introducing flow are associated with varying levels of functional restoration. This said, the non-physiological nature and responsiveness of these cultured cells are demonstrated by the fact that baseline maintenance media to support hepatocyte survival still requires extremely high glucose concentrations and super-physiological levels of growth factors and hormones like insulin, up to 20,000 times normal circulating levels [26,27], further undermining the usefulness of these systems for studying metabolic diseases.
The role of physiological parameters such as biomechanical effects of flow and the nutrient and oxygen gradients in supporting liver biology has long been appreciated [28–30]. We previously demonstrated that primary hepatocytes cultured in a three dimensional context with the restoration of sinusoidal hemodynamic flow patterns and transport, retain an in vivo-like, stable, mature differentiated phenotype [29]. This is not only reflected by stable polarized morphology and in vivo-like transcriptomic signatures distinct from static cultures, but also by consistently higher liver-specific functions such as albumin and urea production and drug and xenobiotic metabolic activity over 2 weeks [31]. More importantly, this results in response to drugs at levels that match therapeutic concentrations, and the hepatocytes are cultured in a milieu that is reflective of normal glucose and near physiological insulin levels [32–34]. We hypothesized that we could apply this model to build a stable system using hepatocytes isolated from patients with specific rare diseases like PA, creating new opportunities for research and therapeutic evaluation. Explanted livers from PA patients undergoing liver transplantation provide a potential source of primary hepatocytes that are otherwise wasted. These cells typically possess a specific genetic biochemical defect, but are otherwise healthy, and could be used to create a disease model that retains the functionality of the tissue while preserving the inherent defect in a system which can be utilized for research. Here, for the first time, we describe the recapitulation of PA disease biology, including hepatocyte morphology, organization and function, and metabolic defects associated with PA, in an organotypic model that deploys primary hepatocytes from a patient with PA. Innovative approaches like this provide a means to understand the biochemistry and function of cells from an individual with an inborn error of metabolism while maintaining its biochemical function (or in this case, dysfunction). This approach will also allow the evaluation of potential therapeutic interventions and their direct effects in a highly relevant system.
2. Materials and methods
2.1. Liver tissue procurement and isolation of hepatic cells
Fresh hepatocytes were procured from liver explanted from an individual with propionic acidemia whose mutation is c.937C > T/c.937C > T; pArg313Stop/p.Arg313Stop (Fig. 1A). The individual was an eight year old female patient with neonatal presentation of PA followed by repeated hospitalizations for hyperammonemia resulting in the decision to proceed with transplantation. The subject was consented to the Children’s National Bio-repository for cells, tissues and DNA (CNHS IRB Pro0004911). Most of the explanted liver was prepared as hepatocytes (below). Part of the caudate lobe was prepared as clinical specimens (formalin imbedded) and sent to pathology for staining and EM analysis. The remainder of the caudate lobe was flash frozen and entered into Children National Medical Center’s bio-repository storage. Protocol H&E stains were performed on formalin imbedded liver tissue. Hepatocyte isolation from liver tissue obtained from the propionic acidemia patient was performed at QPS Hepatic Biosciences using a protocol previously described by Lecluyse et al. [35]. Cryopreserved control hepatocytes from healthy controls were procured from an age and sex-matched healthy donor from QPS Hepatic Biosciences.
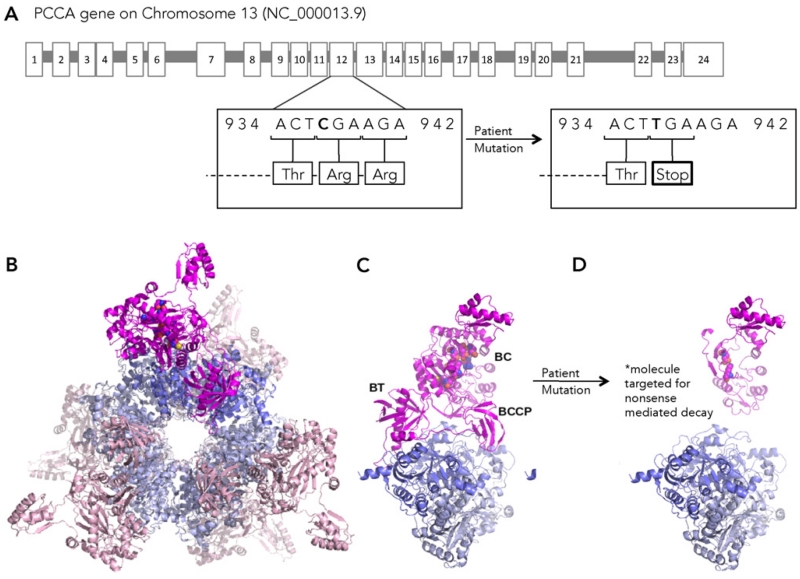
Fig. 1.
A. The PCCA gene mutation in the patient used in this study (c.937C > T/c.937C > T; pArg313Stop/p.Arg313Stop) is a C > T transition located at nucleotide 973 on exon 11, resulting in the replacement of the arginine at position 313 with a stop codon. This mutation results in the loss of the PCCA active site and loss of the domains responsible for PCCA interaction with PCCB. B. A homology model of the wild-type human PCC α6β6 holoenzyme, created by the program PyMOL, viewed along the three-fold axis of the dodecamer. PCCA subunits are shown in magenta and pink with active site residues shown as spheres; PCCB subunits are shown in slate blue and cobalt. C. A single wild-type PCCA subunit showing the domain arrangement and interactions with a PCCB dimer. D. Hypothetical reconstruction illustrating the impact of the deletion of residues 313–728 from PCCA. Although the premature stop codon targets the molecule for nonsense mediated decay, the model illustrates that the Arg313stop mutation results in the complete loss of the biotin carboxyl carrier protein (BCCP) and carboxyltransferase (CT) domains that enable binding of PCCA to PCCB, and loss of all but two of the PCCA active site residues from the biotin carboxylase (BC).
2.2. Cell culture and device operating conditions
Primary hepatocytes obtained as described above were plated in a collagen gel sandwich configuration on the undersurface of the membranes of 75 mm polycarbonate transwells (Corning) using previously described protocols [29]. The cultures were left overnight in a maintenance medium (MM) that consisted of DMEM/F-12 supplemented with fetal bovine serum (10% at the time of plating). Additionally, the medium contained gentamicin (50 μg/mL), 0.2% ITS (Fisher/MediaTech MT-25–800CR), and dexamethasone (Cat# D4902, Sigma Aldrich, St. Louis, MO, 1 μM at plating and 250 nM thereafter). On the 2nd day, the transwells were set up within HemoShear devices in a configuration to allow for control of hemodynamics and transport as described previously [29]. A proprietary hepatocyte flow medium (HFM), modified from the MM but with significantly lower levels of key hormones and growth factors, was continuously perfused on both sides while shear stress was applied on the top surface based on the calculations described below. The devices were housed in a controlled environment at 37 °C with 5% CO2 mixed with air. The maintenance medium was replaced every 48 h in non-flow cultures in a standard CO2 incubator. For all the flow experiments described in this study the shear stress of 0.6 dyn/cm2 was used derived from reference values for the pressure gradient across the sinusoid (ΔP), the radius of sinusoids (r), and the length of the sinusoids (l) obtained from the literature [36,37]. The hepatocytes were cultured for 10 days.
Fibroblast lines (GM03590 and GM00057) from patients with deficient Propionyl CoA carboxylase activity were identified in the NIGMS Human Genetic Cell Repository and purchased from the Coriell Institute for Medical Research. They were cultured according to the recommended protocol in Eagle’s Minimum Essential Medium with Earle’s salts and non-essential amino acids along with 15% FBS under standard tissue culture conditions.
2.3. Drug preparation and exposure
Primary hepatocytes from individual donors were plated in a collagen gel sandwich configuration on 24-well plates (Corning) using previously described protocols. The cultures were incubated in normal or maintenance media that consisted of DMEM/F-12 supplemented with fetal bovine serum (10% at the time of plating). Additionally, the medium contained gentamicin (50 μg/mL), 0.2% ITS (Fisher/MediaTech MT-25–800CR), and dexamethasone (Cat# D4902, Sigma Aldrich, St. Louis, MO, 1 μM at plating and 250 nM thereafter). To recreate a milieu representative of protein loading, we prepared amino acid supplemented media with the addition of L-isoleucine and L-valine (Cat# I7403, and V0513, Sigma Aldrich, St. Louis, MO). The amino acids were dissolved in the maintenance media as a 10× stock and further diluted to a final concentration of 5 mM each for the experiments. Cells were incubated in either maintenance or amino acid supplemented media after 4 days of cell culture until the end of the experiment.
2.4. Homology model
Homology models of human PCC α and β subunits were downloaded from MODBASE (doi: 10.1093/nar/gkj059). Each subunit was aligned to a single corresponding protomer of a bacterial PCC α6β6 holoenzyme X-ray crystal structure (PDB code: 3N6R, doi:10.1038/nature09302) in the program PyMOL (The PyMOL Molecular Graphics System, Version 1.7.4 Schrödinger, LLC). The aligned PCC α and β subunits were combined as the PCC heterodimer, which was then copied and aligned to the remaining heterodimers of the 3N6R structure to generate a model of the human PCC α6β6 dodecamer. The human PCC homology model was analyzed with MolProbity (doi: 10.1107/S0907444909042073) and the quality of the model was similar to that of the 3N6R structure. Figures were created in PyMOL.
2.5. H & E of tissue
Frozen liver sections were cut on a cryostat and prepared for staining with Hematoxylin and Eosin using standard protocols established at Georgetown University Hospital for the processing of clinical tissue samples. Phase contrast images of the histology specimens were obtained with a Nikon Eclipse TS100 microscope.
2.6. Immunostaining
At the prescribed time points in the experimental design, the device transwells were removed and washed gently with PBS followed by fixation of islands with 4% paraformaldehyde for 20 min. The samples were first permeabilized with 0.1% Triton-X for 20 min then washed with PBS and incubated with AlexaFluor-488 Phalloidin (Cat# A12379, Life Technologies), and TOPRO-3 (Cat# T3605, Life Technologies) stains for 1 h to label F-actin and nuclei, respectively. The samples were washed with PBS and then mounted for imaging on a Nikon C1 + Confocal System microscope.
2.7. RNA preparation and RT-PCR
Hepatocyte cell pellets were lysed in Trizol and RNA isolated using the Invitrogen Purelink RNA Mini kit (Cat# 12183018A) according to the manufacturer’s instructions. RNA concentrations were determined with the Nanodrop.
After total RNA was isolated, RNA was reverse transcribed to cDNA using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Taqman FAM-MGB Probes against PCCA and PCCB (Applied Biosystems) were used for determining RNA expression by RT-PCR using Taqman® Universal PCR Master Mix (Applied Biosystems) and a CFX96 Real-Time System (with C1000 Thermal Cycler; BioRad). RNA data were normalized to endogenous expression of β2-microglobulin.
2.8. Protein and Western blots
Samples from 1 well from 24-well plates were harvested using 100 uL of 2× Laemmli sample buffer (BioRad 161-0737) containing 2-beta-mercaptoethanol (BioRad). Samples were boiled and sonicated on ice. Total protein lysates were resolved on a 10% SDS-PAGE gel and transferred to nitrocellulose. Blots were probed overnight with primary antibody and 30 min at room temperature with secondary antibody in blocking buffer (LI-COR 927-40000). Primary antibodies (1:1000) include: βactin (Sigma A1978), PCCA (Bethyl Laboratories), and PCCB (Origene). Secondary antibodies (1:15,000) include: IRDye 800CW Donkey Anti-Mouse (LI-COR 926-32212), IRDye 680LT Donkey Anti-Rabbit (LI-COR 926-68,023), and IRDye 800CW Donkey Anti-Rabbit (LI-COR 926-32,213). The LI-COR Odyssey infrared imager was used for image acquisition and the LI-COR Odyssey Image Studio software used for densitometry analysis.
2.9. Urea
The medium collected from static cultures at various time points was assayed for urea using a standard colorimetric assay (QuantiChrom Urea Assay Kit, DIUR-500; BioAssay Systems). DNA content of the samples was measured using Quant-IT PicoGreen (Life Technologies) as normalization means. ATP content of samples was measured using Cell Titer-Glo Luminescent Cell Viability Assay (Promega) and also used for normalization.
2.10. Ammonia extraction assay
Sodium acetate was utilized to extract ammonia from PBS following a modified protocol developed by Ayyub et al. [38]. Briefly, hepatocytes were incubated in PBS for 2 h. Following incubation, 100 μL of PBS is loaded into the bisected wells of an ammonia sensor and incubated for 20 min with 1 M sodium acetate to allow ion exchange to extract ammonia. 40 μL of ammonium enriched sodium acetate was then removed and incubated in a 96-well plate for 10 min with 10 μL of 2-phenylphenol, 10 μL of NaOH, 10 μL 0.75% hypochlorite and 10 μL of sodium nitroprusside. Absorbance values were recorded at 635 nm and compared to a standard curve to determine ammonia concentration.
2.11. Statistics
For PCR data, mixed-effects linear models were fit to the B2M-normalized Cq values for each gene. In these models, donor identity was included as a random effect. Differences between the normal and PA states were assessed by fitting nested models; i.e., models including/excluding disease state as a fixed effect. These models were compared using the likelihood ratio test, from which the reported p-values were derived. The same approach was used to assess differences in the ammonia and urea assay results in the system — in this case including either donor or assay batch as random effects where appropriate. Finally, in the case of protein abundance, p-values were calculated using a two-sided Student’s t-test, where the input values were the log2 ratios of the protein of interest to its control (either B-actin or tubulin).
3. Results
3.1. PCCA mutation and the impact on protein structure
Our 8-year-old female patient possesses a homozygous PCCA gene mutation (Fig. 1A). The mutation (c.937C > T/c.937C > T; pArg313Stop/p.Arg313Stop) is a C > T transition located at nucleotide 937 on exon 12, resulting in the replacement of the arginine at position 313 with a stop codon. This mutation results in presumptive nonsense mediated decay of mRNA for PCCA with resulting lack of production of PCCA protein. In healthy individuals, propionyl CoA carboxylase is a 750 kDa alpha(6)-beta(6)-dodecamer with the alpha subunits arranged as monomers, around the central beta-6 hexameric core as shown in our homology model (Fig. 1B). The PCCA subunit interacts with a PCCB dimer along active site residues (Fig. 1C). Due to its significant impact on the primary structure of the protein (Fig. 1D), the Arg313stop mutation would result in RNA degradation and absence of PCCA protein.
3.2. Primary hepatocytes from the PA patient stably maintain differentiated morphology and function in a model with physiological hemodynamics and transport
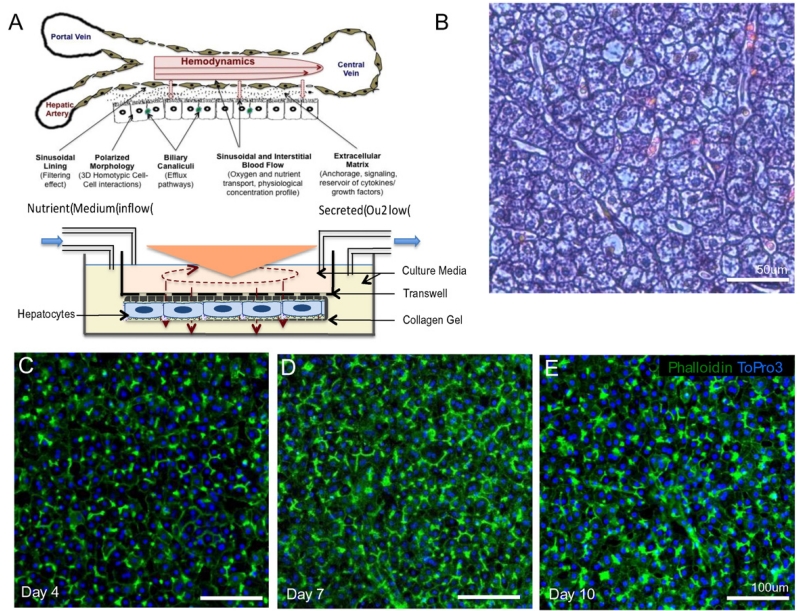
Hepatocytes isolated from the liver of an 8 year old PA patient were cultured with physiological hemodynamics and transport using a previously described system modeled on the sinusoidal configuration [29] (Fig. 2A). After 10 days of culture in the system, the hepatocytes from the PA patient maintained a normal differentiated and polarized morphology as evidenced by the peripheral actin staining (green) (Fig. 2C–E). Hematoxylin and eosin staining of paraffin imbedded sections of the liver from the PA patient exhibited normal morphology (Fig. 2B). To ensure the viability and differentiated function at the end of the 10 day duration of the experiment, we measured the levels of albumin in the effluent and performed MTT assay to assess the cellular metabolic activity. The levels of albumin (406.4 +/− 53.4 ng/mL) were within the range that we see with healthy human hepatocytes in the system as were the MTT readouts.
Fig. 2.
A. Primary hepatocytes were maintained in a system model based on the sinusoidal configuration [29] under conditions that maintain the physiological hemodynamics and transport, and shown to retain and restore liver like phenotype, morphology, function and responses. B. An H&E stain of the liver from the propionic acidemia patient used in this study, reveals normal in vivo morphology. Hepatocytes isolated from the propionic acidemia patient and cultured in the system demonstrate retention of polarized morphology (Green — Phalloidin, Blue — Draq 5) that is stable over time (C. — 4 days, D. — 7 days, E. — 10 days).
3.3. Primary hepatocytes from the PA patient cultured in the system exhibit decreased expression of PCCA and PCCB at the gene and protein level
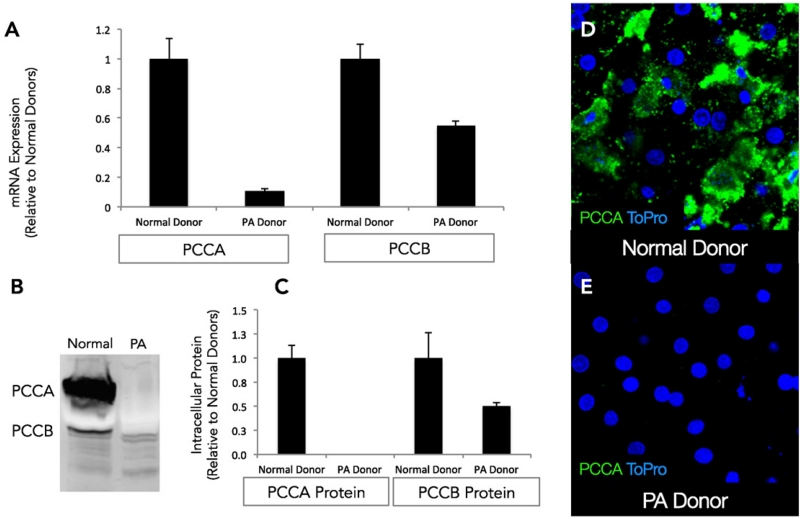
The expression of PCCA and PCCB was assessed at the gene and protein level and compared to hepatocytes obtained from healthy donors cultured in a similar manner. There was a decreased level of both PCCA RNA (~9-fold) and PCCB RNA (~2-fold) relative to normal healthy controls (Fig. 3A). At the protein level, PCCA expression was completely absent while PCCB expression was reduced by 50% in the PA patient-derived hepatocytes relative to healthy donors, as shown in Fig. 3B and C. Detection of PCCA by immunofluorescence microscopy showed punctate intracellular staining in normal healthy hepatocytes (Fig. 3D), whereas it could not be detected in hepatocytes from the PA patient (Fig. 3E).
Fig. 3.
A. RT-PCR of RNA obtained from propionic acidemia patient hepatocytes following 10 days in culture reveals significantly lower levels of mRNA expression of the PCCA and PCCB genes relative to time matched healthy normal controls. B. Western blots using antibodies directed against PCCA and PCCB proteins demonstrate complete absence of PCCA and very low levels of PCCB. C. Quantification of the protein levels reveals significant differences from time matched normal controls. D. Immunostaining with PCCA antibody indicates cytoplasmic distribution of PCCA in hepatocytes cultured from normal donors (Green — PCCA, Blue — ToPro). E. Expression of PCCA is completely lost in hepatocytes cultured from the propionic acidemia donor.
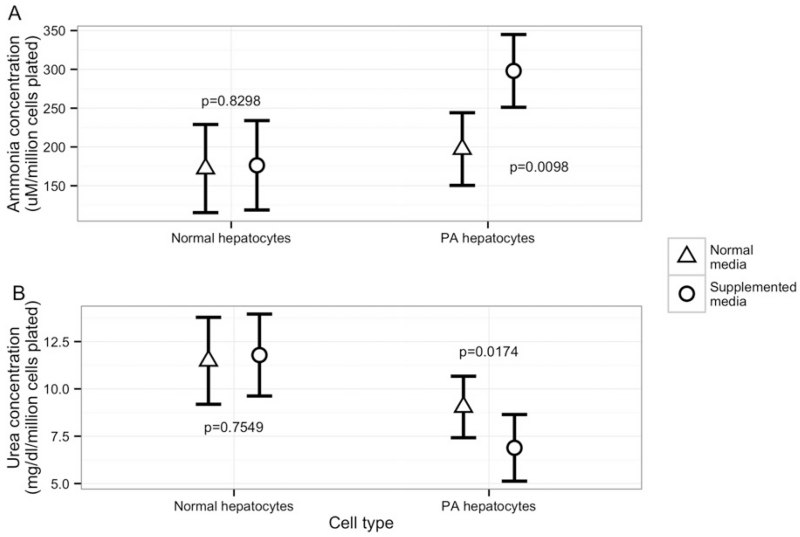
3.4. Primary hepatocytes from the PA patient produce increased ammonia and decreased urea
To mimic a clinical metabolic decompensation event, the medium was supplemented with branched-chain amino acids, isoleucine (5 mM) and valine (5 mM). Hepatocytes from the PA patient produced a significantly higher amount of ammonia (measured in supernatant) when pre-conditioned with supplemented media (p = 0.0098), relative to normal cell culture media (Fig. 4A). However, a similar response to supplemented media was not observed in corresponding experiments using healthy donor hepatocytes (Fig. 4A). Urea levels in the supernatants of hepatocytes from the PA patient were significantly decreased (p = 0.017) in the presence of supplemented media relative to normal media conditions while healthy donor hepatocytes did not show a significant change in response to media supplementation (Fig. 4B). Supernatants of PA patient derived dermal fibroblasts demonstrated overall lower levels of ammonia that was not significantly changed by media supplementation, and urea levels were undetectable (data not shown).
Fig. 4.
To recreate a milieu similar to the clinical situation of high protein diet, the medium was supplemented with branched-chain amino acids, isoleucine (5 mM) and valine (5 mM). Ammonia levels in supernatants of day 10 hepatocytes from the PA patient were significantly increased in supplemented media relative to culture in normal media (Fig. 4A) unlike healthy donor hepatocytes. Urea levels in the supernatants of day 10 PA patient hepatocytes were significantly decreased in the presence of supplemented media relative to normal media conditions (Fig. 4B) unlike healthy donor hepatocytes.
4. Discussion
The purpose of this study was to create a patient-based organotypic model that preserved the clinical phenotype associated with PA, in order to explore the disease biology and metabolic responses associated with this rare inborn error of metabolism. A number of novel results are presented. First, hepatocytes from the liver of a PA patient retained differentiated polarized morphology over 10 days when maintained in a three-dimensional environment that restored hemodynamics and transport representative of the sinusoidal space. Second, stable retention of the key defect of the disease was reflected by the absence or significantly lowered levels of the isoforms of the propionyl CoA carboxylase enzyme at the mRNA and protein level. Third, when PA hepatocytes were metabolically challenged with an increased load of branched-chain amino acids, we quantified an increased production of ammonia and a decreased production of urea, recapitulating the PA-associated metabolic phenotype of hyperammonemia. This response was not observed in fibroblasts from PA patients or hepatocytes from healthy controls. Taken together, such approaches can be applied to further our understanding of the basic disease biology of PA, and other IEMs, and ultimately provide insight for new therapeutic interventions.
4.1. Approaches to experimental modeling of genetic disease
Drug discovery research in the rare disease arena is challenging due to limited patient populations, as well as the lack of robust metabolically complex and responsive experimental models. Several approaches are possible to recreate an experimental liver disease system for PA: First, preservation of the altered functional phenotype of the existing defect using patient-derived tissue as described here; second, introduction of the altered functional phenotype in healthy tissue using a range of genetic manipulation tools e.g. silencing RNA (siRNA) and clustered regularly interspaced short palindromic repeats (CRISPR/Cas9), that can allow one to knock out or edit the disease gene involved [39,40]. We chose the former approach, as it is the most faithful representation of the patient’s original genotype. This approach depends on access to patient tissue with isolation of high quality hepatocytes, and the ability to maintain the hepatocytes in a functionally differentiated state in culture. The latter point is critical as primary human hepatocytes are known to rapidly dedifferentiate in traditional static cell cultures. In this study, the tissue was procured from a PA patient at the time of liver transplant that would have been otherwise discarded as medical waste. The timely isolation and ability to keep the cells differentiated within the system with controlled hemodynamics and transport were important to preserve the functionality as well as the metabolic defect. The alternate approach of using genetic manipulation tools to create a disease phenotype would be particularly useful in the absence of the availability of patient tissue. The described system provides an excellent test-bed to further develop introduced genetic alteration/suppression models. Another hybrid approach would be to use hepatocytes derived from inducible pluripotent stem cells (iPSC) created from fibroblasts obtained from the skin or peripheral blood of patients. These cells stably possess the original patient’s genetic defect making them relevant from a genotypic standpoint. However, the existing iPSC-derived hepatocytes are reported to still retain some aspects of fetal phenotype and lack the complete metabolic maturity of primary human hepatocytes, limiting their use for disease modeling in juvenile or adult livers [41].
4.2. Variable disease spectrum and need to understand biochemical consequences of defects
Organic acidemias are a diverse group of genetic metabolic disorders with disrupted branched-chain amino acid metabolism. Though there are commonalities in the buildup of certain metabolites, e.g. ammonia, there is extensive variation in the severity of the disease phenotypes associated with the specific enzyme defects. Even in the case of PA, depending on the specific mutations in PCCA or PCCB, the severity of the disease can vary greatly [42,43]. This highlights the need to generate a deeper understanding of specific underlying defects and correlate it with resulting biochemical perturbations that could offer insights into possible therapeutic options. In this particular study, the hepatocytes used were from a patient with homozygous non-sense mutations in alleles for subunit alpha of PCC. This mutation results in nonsense-mediated decay of messenger RNA before translation which is reflected in the low levels of detectable RNA and absence of PCCA protein. The absence of subunit A protein would be expected to cause a complete loss of enzyme function which correlates well with the clinically severe phenotype seen in this patient. In addition, our data indicate that both the mRNA and protein expression of subunit B is compromised. Using an in silico modeling approach, we hypothesize that the absence of the A subunit, and consequent loss of the B subunit interaction site, provide a possible reason for the decrease in PCCB protein expression. Despite stable PCCB gene expression at the RNA level, there is a decrease in subunit B protein level due to reduced stability of the protein in the absence of the subunit A/B interaction. This mechanism has yet to be proven.
4.3. Recreating disease relevance: milieu, endpoints and tissue
A critical component that contributes to the physiological relevance of a disease model is a dynamic milieu reflective of changes in the pathologic state. This is crucial to capture the adaptive responses and down-stream metabolic fluxes occurring as a result of nutrient loads that can impose stress upon the compromised cellular metabolic machinery. For instance, in healthy primary hepatocytes maintained in a differentiated state under controlled hemodynamics and transport, we have previously demonstrated progressive pathologic changes reminiscent of non-alcoholic steatohepatitis (NASH) by increasing the glucose load in the presence of high levels of insulin [33]. Clinically, in patients with PA who have been stabilized, conditions that cause increased circulatory nitrogen load, e.g. catabolism or dietary increases in protein and propiogenic amino acids, can precipitate acute crises characterized by hyperammonemia that require hospitalization [44].
In this study, we supplemented the culture medium with increased levels of propiogenic branched-chain amino acids to recreate a milieu analogous to a clinical situation of decompensation associated with increased propionic acid burden. Interestingly, while we noted a significant increase in ammonia accumulation in the PA liver hepatocytes exposed to supplemented branched-chain amino acid conditions, control liver hepatocytes isolated from healthy donors showed no such trend. This most likely reflects the ability of normal cells to process propionyl CoA without the resultant effects on urea cycle metabolism. Our choice of ammonia as a measure of functional response was designed to establish a clinically relevant phenotypic biomarker in the PA patient population. It is important to note that dermal fibroblasts from the liver of PA patients exhibited a lack of change in ammonia accumulation under supplemented amino acid media conditions unlike differentiated patient hepatocytes in culture. Similarly, only patient hepatocytes showed a decrease in urea levels under supplemented media conditions unlike healthy donor hepatocytes, and urea levels from fibroblasts were undetectable. This reflects the lack of urea cycle function, nitrogen processing, and limited propionic acid pathway function in these non-hepatic cells. Secondary dysfunction with inability to process ammonia at early steps of the urea cycle resulting in the accumulation of ammonia and decreased urea production has long been the proposed cause for hyperammonemia seen in patients with PA (Fig. 5) [5,45,46]. Here, we recapitulate the patient phenomenon resulting in the elevation of ammonia, but also a decrease in urea levels in the cell lines from PA patients treated with propiogenic amino acids. Taken together, these results highlight the necessity to use metabolically relevant tissue, in this case, hepatocytes, while creating a PA disease model. The use of primary hepatocytes will likely apply to additional models for studying diseases of inborn errors of metabolism such as methylmalonic acidemia, maple syrup urine disease, alpha-1 antitrypsin deficiency, phenylketonuria, galactosemia and urea cycle disorders.
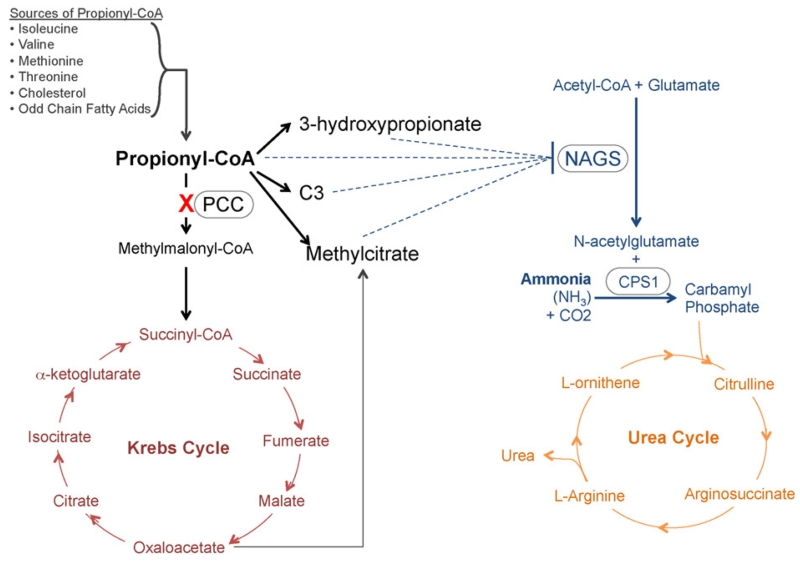
Fig. 5.
Postulated relationship between propionyl CoA carboxylase (PCC) defect and levels of ammonia and urea. Accumulation of Propionyl CoA and its metabolites inhibits N-acetylglutamate synthase (NAGS) and consequently lower levels of N-acetylglutamate, resulting in decreased conversion of ammonia to urea. As a result, under conditions of high protein in the patient’s hepatocytes, we observe an increase of ammonia and decrease in urea levels.
We have presented direct evidence that some aspects of the propionic acidemia disease phenotype such as enzymatic defects at the gene and protein level, and hyperammonemia in response to increased branched chain amino acid loads, can be retained in primary human hepatocytes from a patient with PA, when placed into the appropriate physiological context. We feel that further use of this model system as in the present studies might have utility for elucidating candidate factors and regulatory mechanisms that contribute to metabolic decompensation in PA. We strongly believe that the system as described has great potential for identifying new therapeutic options. Finally, the paradigm deployed herein directly applies to other rare diseases with primary defects in hepatocytes and where mouse models are deficient.
Acknowledgments
The authors would like to express thanks to Metin Ozdemirli, Christin Hamilton, Andrew Pryor, Nathan Day, Diana Berry, Morgan Donovan, Crystal Passmore, Banumati Cole and Joshua Thomas for technical assistance. This study was funded by a NIH SBIR grant R44GM109539 to HemoShear Therapeutics, Charlottesville, VA. Additionally it was funded through the Division of Genetics & Metabolism at Children’s National Health System, Washington, DC, by the Margaret O’Malley Chair in Molecular Genetics, and via a NIH K08 grant (DK105233).
Footnotes
Conflicts: None.
References
- [1].Ravn K, Chloupkova M, Christensen E, Brandt NJ, Simonsen H, Kraus JP, et al. High incidence of propionic acidemia in greenland is due to a prevalent mutation, 1540insCCC, in the gene for the beta-subunit of propionyl CoA carboxylase. Am. J. Hum. Genet. 2000;67(1):203–206. doi: 10.1086/302971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Carrillo-Carrasco N, Venditti C. In: Propionic acidemia. Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, LJH B, et al., editors. GeneReviews(R), University of Washington; Seattle (WA): 1993. Seattle University of Washington, Seattle. All rights reserved. [Google Scholar]
- [3].Ozand PT, Rashed M, Gascon GG, Youssef NG, Harfi H, Rahbeeni Z, et al. Unusual presentations of propionic acidemia. Brain Dev. 1994;16(Suppl):46–57. doi: 10.1016/0387-7604(94)90096-5. [DOI] [PubMed] [Google Scholar]
- [4].Fenton WA, Gravel RA, Rosenblatt DS. Disorders of propionate and methylmalonate metabolism. In: Scriver CRBALSWSVD, editor. The Metabolic & Molecular Basis of Inherited Disease. McGraw-Hill; New York: 2001. pp. 2165–2193. [Google Scholar]
- [5].Coude FX, Sweetman L, Nyhan WL. Inhibition by propionyl-coenzyme A of N-acetylglutamate synthetase in rat liver mitochondria. A possible explanation for hyperammonemia in propionic and methylmalonic acidemia. J. Clin. Invest. 1979;64(6):1544–1551. doi: 10.1172/JCI109614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Deodato F, Boenzi S, Santorelli FM, Dionisi-Vici C. Methylmalonic and propionic aciduria. Am. J. Med. Genet. C: Semin. Med. Genet. 2006;142C(2):104–112. doi: 10.1002/ajmg.c.30090. [DOI] [PubMed] [Google Scholar]
- [7].Pena L, Burton BK. Survey of health status and complications among propionic acidemia patients. Am. J. Med. Genet. A. 2012;158A:1641–1646. doi: 10.1002/ajmg.a.35387. [DOI] [PubMed] [Google Scholar]
- [8].Pena L, Franks J, Chapman KA, Gropman A, Ah Mew N, Chakrapani A, et al. Natural history of propionic acidemia. Mol. Genet. Metab. 2012;105(1):5–9. doi: 10.1016/j.ymgme.2011.09.022. [DOI] [PubMed] [Google Scholar]
- [9].Baumgartner MR, Horster F, Dionisi-Vici C, Haliloglu G, Karall D, Chapman KA, et al. Proposed guidelines for the diagnosis and management of methylmalonic and propionic acidemia. Orphanet J. Rare Dis. 2014;9:130. doi: 10.1186/s13023-014-0130-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sutton VR, Chapman KA, Gropman AL, MacLeod E, Stagni K, Summar ML, et al. Chronic management and health supervision of individuals with propionic acidemia. Mol. Genet. Metab. 2012;105(1):26–33. doi: 10.1016/j.ymgme.2011.08.034. [DOI] [PubMed] [Google Scholar]
- [11].Graham MJ, Lake BG. Induction of drug metabolism: species differences and toxicological relevance. Toxicology. 2008;254(3):184–191. doi: 10.1016/j.tox.2008.09.002. [DOI] [PubMed] [Google Scholar]
- [12].Martignoni M, Groothuis GM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin. Drug Metab. Toxicol. 2006;2(6):875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- [13].Ball RO, Urschel KL, Pencharz PB. Nutritional consequences of interspecies differences in arginine and lysine metabolism. J. Nutr. 2007;137(6 Suppl. 2):1626S–1641S. doi: 10.1093/jn/137.6.1626S. [DOI] [PubMed] [Google Scholar]
- [14].Waterlow JC. Protein turnover with special reference to man. Q. J. Exp. Physiol. 1984;69(3):409–438. doi: 10.1113/expphysiol.1984.sp002829. [DOI] [PubMed] [Google Scholar]
- [15].Kun E, Loh HH, Volfin P. Tissue specific differences in rate limiting factors of glutamate metabolism in enzyme systems of kidney and liver. Biochem. Biophys. Res. Commun. 1966;23(5):702–706. doi: 10.1016/0006-291x(66)90457-8. [DOI] [PubMed] [Google Scholar]
- [16].Kun E, Volfin P. Metabolic differences between mitochondria isolated from various tissues. Biochem. Biophys. Res. Commun. 1966;23(5):696–701. doi: 10.1016/0006-291x(66)90456-6. [DOI] [PubMed] [Google Scholar]
- [17].Cigolini M, Bonora E, Querena M, Moghetti P, Cacciatori V, Zancanaro C, et al. Differences in glucose metabolic enzyme activities in human adipose tissue from abdominal and gluteal regions. Metabolism. 1988;37(9):820–823. doi: 10.1016/0026-0495(88)90114-x. [DOI] [PubMed] [Google Scholar]
- [18].Barash V, Elpeleg O, Amit R, Gottfried S, Yatziv S, Gutman A. Propionic acidemia—biochemical studies. Isr. J. Med. Sci. 1989;25(2):103–106. [PubMed] [Google Scholar]
- [19].Perez-Cerda C, Clavero S, Perez B, Rodriguez-Pombo P, Desviat LR, Ugarte M. Functional analysis of PCCB mutations causing propionic acidemia based on expression studies in deficient human skin fibroblasts. Biochim. Biophys. Acta. 2003;1638(1):43–49. doi: 10.1016/s0925-4439(03)00039-5. [DOI] [PubMed] [Google Scholar]
- [20].Ugarte M, Perez-Cerda C, Rodriguez-Pombo P, Desviat LR, Perez B, Richard E, et al. Overview of mutations in the PCCA and PCCB genes causing propionic acidemia. Hum. Mutat. 1999;14(4):275–282. doi: 10.1002/(SICI)1098-1004(199910)14:4<275::AID-HUMU1>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- [21].Varani J, Dame M, Rediske J, Beals TF, Hillegas W. Substrate-dependent differences in growth and biological properties of fibroblasts and epithelial cells grown in microcarrier culture. J. Biol. Stand. 1985;13(1):67–76. doi: 10.1016/s0092-1157(85)80035-4. [DOI] [PubMed] [Google Scholar]
- [22].Berman JJ, Tong C, Williams GM. Differences between rat liver epithelial and fibroblast cells in metabolism of purines. J. Cell. Physiol. 1980;103(3):393–398. doi: 10.1002/jcp.1041030304. [DOI] [PubMed] [Google Scholar]
- [23].Baker TK, Carfagna MA, Gao H, Dow ER, Li Q, Searfoss GH, et al. Temporal gene expression analysis of monolayer cultured rat hepatocytes. Chem. Res. Toxicol. 2001;14(9):1218–1231. doi: 10.1021/tx015518a. [DOI] [PubMed] [Google Scholar]
- [24].Boess F, Kamber M, Romer S, Gasser R, Muller D, Albertini S, et al. Gene expression in two hepatic cell lines, cultured primary hepatocytes, and liver slices compared to the in vivo liver gene expression in rats: possible implications for toxicogenomics use of in vitro systems. Toxicol. Sci. 2003;73(2):386–402. doi: 10.1093/toxsci/kfg064. [DOI] [PubMed] [Google Scholar]
- [25].Rodriguez-Antona C, Donato MT, Boobis A, Edwards RJ, Watts PS, Castell JV, et al. Cytochrome P450 expression in human hepatocytes and hepatoma cell lines: molecular mechanisms that determine lower expression in cultured cells. Xenobiotica. 2002;32(6):505–520. doi: 10.1080/00498250210128675. [DOI] [PubMed] [Google Scholar]
- [26].Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26(1):120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- [27].Nelson LJ, Treskes P, Howie AF, Walker SW, Hayes PC, Plevris JN. Profiling the impact of medium formulation on morphology and functionality of primary hepatocytes in vitro. Sci. Rep. 2013;3:2735. doi: 10.1038/srep02735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Dash A, Inman W, Hoffmaster K, Sevidal S, Kelly J, Obach RS, et al. Liver tissue engineering in the evaluation of drug safety. Expert Opin. Drug Metab. Toxicol. 2009;5(10):1159–1174. doi: 10.1517/17425250903160664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dash A, Simmers MB, Deering TG, Berry DJ, Feaver RE, Hastings NE, et al. Hemodynamic flow improves rat hepatocyte morphology, function, and metabolic activity in vitro. Am. J. Physiol. Cell Physiol. 2013;304(11):C1053–C1063. doi: 10.1152/ajpcell.00331.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Domansky K, Inman W, Serdy J, Dash A, Lim MH, Griffith LG. Perfused multiwell plate for 3D liver tissue engineering. Lab Chip. 2010;10(1):51–58. doi: 10.1039/b913221j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Dash A, Deering TG, Marukian S, Thomas J, Desbans C, Alexandre E, et al., editors. Human Primary Hepatocytes Under Controlled Hemodynamics Elicit Induction Responses to Drugs at Clinical Cmax Concentrations; 52nd Annual Meeting of the Society of Toxicology; San Antonio, Texas. 2013. [Google Scholar]
- [32].Dash A, Deering TG, Marukian S, Thomas J, Wamhoff BR, Blackman BR, editors. Physiological Hemodynamics And Transport Restore Insulin and Glucagon Responses in a Normal Glucose Milieu in Hepatocytes In Vitro; 73rd Scientific Sessions of the American Diabetes Association; Chicago. 2013. [Google Scholar]
- [33].Deering TGTJT, Blackman BR, Wamhoff BR, Dash A, editors. Development of an In-vitro Model of Hepatic Steatosis Using Rat Hepatocytes Under Controlled Hemodynamics in a Diabetic Milieu. American Association for the Study of Liver Diseases (AASLD); Boston: 2012. [Google Scholar]
- [34].Terelius Y, Figler RA, Marukian S, Collado MS, Lawson MJ, Mackey AJ, et al. Transcriptional profiling suggests that nevirapine and ritonavir cause drug induced liver injury through distinct mechanisms in primary human hepatocytes. Chem. Biol. Interact. 2015 doi: 10.1016/j.cbi.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lecluyse EL, Alexandre E. Isolation and culture of primary hepatocytes from resected human liver tissue. Methods Mol. Biol. 2010;640:57–82. doi: 10.1007/978-1-60761-688-7_3. [DOI] [PubMed] [Google Scholar]
- [36].Maass-Moreno R, Rothe CF. Distribution of pressure gradients along hepatic vasculature. Am. J. Phys. 1997;272(6 Pt 2):H2826–H2832. doi: 10.1152/ajpheart.1997.272.6.H2826. [DOI] [PubMed] [Google Scholar]
- [37].Wambaugh J, Shah I. Simulating microdosimetry in a virtual hepatic lobule. PLoS Comput. Biol. 2010;6(4):e1000756. doi: 10.1371/journal.pcbi.1000756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Ayyub OB, Behrens AM, Heligman BT, Natoli ME, Ayoub JJ, Cunningham G, et al. Simple and inexpensive quantification of ammonia in whole blood. Mol. Genet. Metab. 2015;115(2–3):95–100. doi: 10.1016/j.ymgme.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dorsett Y, Tuschl T. siRNAs: applications in functional genomics and potential as therapeutics. Nat. Rev. Drug Discov. 2004;3(4):318–329. doi: 10.1038/nrd1345. [DOI] [PubMed] [Google Scholar]
- [40].Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157(6):1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Baxter M, Withey S, Harrison S, Segeritz CP, Zhang F, Atkinson-Dell R, et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015;62(3):581–589. doi: 10.1016/j.jhep.2014.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Chapman KA, Bush WS, Zhang Z. Gene expression in cell lines from propionic acidemia patients, carrier parents, and controls. Mol. Genet. Metab. 2015;115(4):174–179. doi: 10.1016/j.ymgme.2015.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kolker S, Valayannopoulos V, Burlina AB, Sykut-Cegielska J, Wijburg FA, Teles EL, et al. The phenotypic spectrum of organic acidurias and urea cycle disorders. Part 2: the evolving clinical phenotype. J. Inherit. Metab. Dis. 2015 doi: 10.1007/s10545-015-9840-x. [DOI] [PubMed] [Google Scholar]
- [44].Vockley J, Chapman KA, Arnold GL. Development of clinical guidelines for inborn errors of metabolism: commentary. Mol. Genet. Metab. 2013;108(4):203–205. doi: 10.1016/j.ymgme.2013.01.013. [DOI] [PubMed] [Google Scholar]
- [45].Ah MN, McCarter R, Daikhin Y, Nissim I, Yudkoff M, Tuchman M. N-carbamylglutamate augments ureagenesis and reduces ammonia and glutamine in propionic acidemia. Pediatrics. 2010;126(1):e208–e214. doi: 10.1542/peds.2010-0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Dercksen M, IJ L, Duran M, Mienie LJ, van Cruchten A, van der Westhuizen FH, et al. Inhibition of N-acetylglutamate synthase by various monocarboxylic and dicarboxylic short-chain coenzyme A esters and the production of alternative glutamate esters. Biochim. Biophys. Acta. 2014;1842(12 Pt A):2510–2516. doi: 10.1016/j.bbadis.2013.04.027. [DOI] [PubMed] [Google Scholar]