Abstract
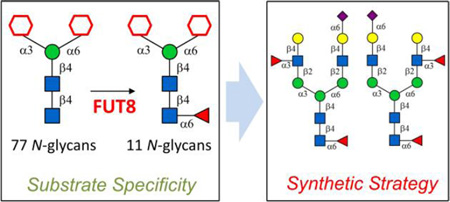
Substrate specificity studies of human FUT8 using 77 structurally-defined N-glycans as acceptors showed a strict requirement towards the α1,3-mannose branch, but a great promiscuity towards the α1,6-mannose branch. Accordingly, a chemoenzymatic strategy was developed for efficient synthesis of core-fucosylated asymmetric N-glycans.
TOC
Based on substrate specificity of human FUT8, an efficient strategy was developed for the synthesis of core-fucosylated asymmetric N-glycans.
Graphical abstract
Asparagine-linked glycosylation (N-glycosylation) represents the most prevalent and structurally varied post-translational modification of eukaryotic proteins. Tremendous advances in glycobiology have demonstrated that asparagine-linked oligosaccharides (N-glycans) can modulate protein’s structure and function in many ways. For example, they were found to play roles in a variety of biological processes, including cell adhesion, pathogen infections, immune responses and tumor metastasis.1 On the other hand, N-glycans can influence proteins biosynthesis, folding, stability, antigenicity and immunogenicity.1c, 2 N-Glycans found in nature possesses inherent complexity and diversity, due to variable connectivity of monosaccharide building blocks as well as additional modifications on the basic scaffolds. Core-fucosylation is a major N-glycan modification in eukaryotic glycomes.
In mammalian cells, core-fucosylation is exclusively found as α1,6-fucosylation of the innermost N-acetyl-glucosamine (GlcNAc) residue of N-glycans. Many glycoproteins are known to be core-fucosylated, and accumulating data indicate that this type of modification regulates the functions of certain glycoproteins. For example, depletion of N-glycan core-fucosylation in human IgG1 enhanced its antibody-dependent cytotoxicity (ADCC) activity to 50 – 100 folds.3 It was shown that core-fucosylation regulates the functions of immunoglobulin by altering its physicochemical properties.4 Another most recent example is that in humoral immune response, core-fucosylation of IgG B cell receptor was shown to mediate antigen recognition and cell signal transduction.5 Alteration of core-fucosylation is frequently found to be closely related to human diseases including liver cancer, pancreatic cancer, lung cancer, breast cancer, etc.6 A well-known example is α-fetoprotein, which was found highly core-fucosylated specifically in hepatocellular carcinoma, but not other liver diseases.7 In addition, E-cadherin, a glycoprotein correlated with cancer metastasis, was found to be core-fucosylated in highly metastatic lung cancer cells but absent in low metastatic ones.8 Compare to controls, increased core-fucosylated N-glycans and decreased non-fucosylated ones were also detected in the clinical ovarian cancer biomarker CA125.9 Moreover, core-fucosylation is pivotal in development, and the loss of such N-glycan modification leads to severe phenotypes such as growth retardation, emphysema-like changes in lung, even death.10 All these evidences strongly suggest that core-fucosylation plays vital roles in various biological processes and events.
FUT8 is the only enzyme responsible for core-fucosylation in mammals, which catalyzes the transfer of an l-fucose residue from GDP-β-l-fucose (GDP-Fuc) onto the innermost GlcNAc of N-glycan to form an α1,6-linkage (Fig. 1).10–11 It has been reported that FUT8 is highly expressed in brain tissue (where core-fucosylated glycoproteins were found abundant), human blood platelets, cancer tissues, and other tissues during certain pathological processes. Previous substrate specificity study suggested that FUT8 requires both α1,3-mannose branch terminal GlcNAc residue and reducing end β-configuration of the acceptor.12 In addition, galactosylation of the GlcNAc residue at β1,3-mannose branch and bisecting of N-glycans prevent FUT8 activity.12a, 12b Most recently, we and others showed that both human and Caenorhabditis elegans FUT8 can fucosylate asymmetric N-glycans (Fig. 1, N211 and N212), indicating that they may have a much relaxed acceptor substrate preference towards the α1,6-mannose branch.13 A comprehensive study is necessary to confirm such specificity, and structurally defined N-glycans that we synthesized previously13b represent perfect substrate candidates for test.
Figure 1.
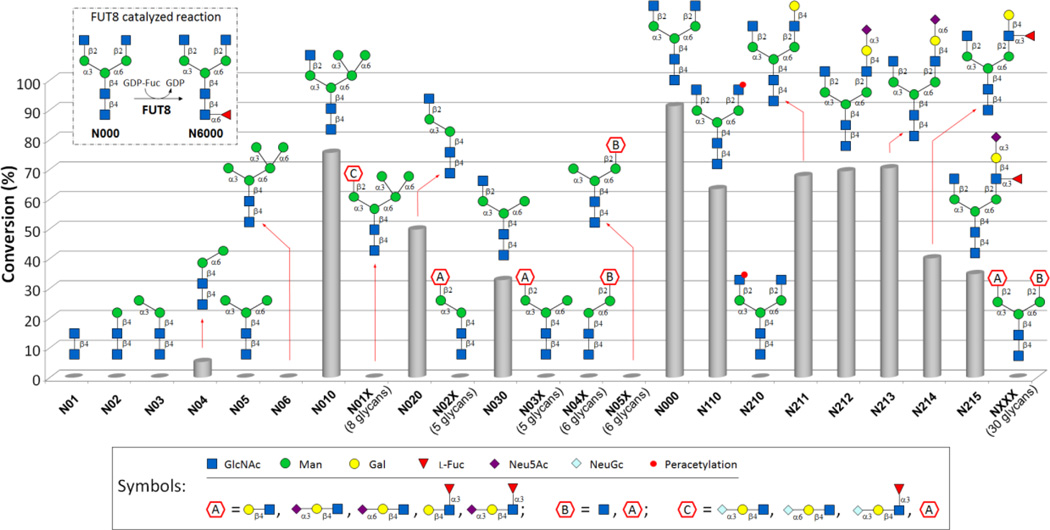
FUT8-catalyzed reaction and substrate specificity of human FUT8 using 77 N-glycans as potential acceptors (see Supplementary Figure S2 for the full list of N-glycans, Supplementary Table S1 for conversion percentage towards each glycan). The average conversion percentage of three replicates for each acceptor was shown.
Given the significance, FUT8 and core-fucosylation have been extensively investigated in terms of glycomics and in vivo biological functions, while in depth molecular elucidation is largely limited by the unavailability of structurally well-defined N-glycans with core-fucosylation. In the past decade, a few chemical and enzymatic methodologies were developed for the preparation of such molecules. Unverzagt, used a modular chemical synthesis strategy where the α1,6-fucose was attached in the last step.14 On the other hand, Huang applied a pre-activation-based one-pot chemical strategy to quickly assemble a dodecasaccharide N-glycan structure.15 Enzymatically, Reichardt used C. elegans FUT8 to core-fucosylate chemically prepared simple N-glycan structures (Fig. 1, N000 and N010),16 while Wang applied glycosynthases to conjugate chemically prepared glycan oxazolines and Fucα1,6-GlcNAc to form homogeneous core-fucosylated N-glycans and glycoproteins.17 In all cases, only simple or symmetric core-fucosylated N-glycans were prepared, and an efficient strategy for rapid access of more complicated and asymmetric ones is yet to be developed. In this work, human FUT8 was overexpressed and purified, detailed acceptor substrate specificity was investigated towards 77 structurally defined N-glycans and derivatives. Accordingly, a chemoenzymatic strategy was developed for efficient synthesis of core-fucosylated asymmetric N-glycans.
Human FUT8 was chosen for substrate specificity study and synthetic purpose. Heterogeneous expression was achieved via a Titerless Infected-cell Preservation and Scale-up (TIPS) approach18 (Supplementary Information II) using a baculorvirus system (see http://glycoenzymes.ccrc.uga.edu for system and construction details) provided by Dr. Jarvis from the University of Wyoming. After one-step Ni-Sepharose Excell (GE Healthcare) affinity purification, recombinant FUT8 with a purity of over 90% (Supplementary Figure S1) was obtained in a scale of 0.42 mg/L of cultures. Substrate specificity study was then performed in reactions with a total volume of 20 µL containing an acceptor N-glycan (0.3 mM), donor GDP-Fuc (1 mM), MES buffer (100 mM, pH 7.0) and 50 µg/mL of the recombinant human FUT8. After 4 h of incubation at 37 °C, the reactions were quenched by the addition of equal volumes of ice cold ethanol, followed by analysis using mass spectrometry (MS) and high performance liquid chromatography (HPLC) monitored by an evaporative light scattering detector (ELSD) (Supplementary Information III). Consistent with previous reports12 as shown in Figure 1, FUT8 could core-fucosylate N-glycans with an agalactosylated and unprotected GlcNAc residue on the α1,3-mannose branch (N000, N010, N020, N030, N110, and N211–N215), but not those either galactosylated or with protected GlcNAc residue on the branch. Among these N-glycans, FUT8 showed the highest activity towards biantennary complex N-glycan structures terminated with GlcNAc at the non-reducing end of both α1,3-and α1,6-mannose branches N000 (91.3% conversion) and slightly decreased activities towards N110 (63.4% conversion) and N211–N213 (67.8% to 70.4% conversion), indicating that galactosylation with or without additional sialylation or chemical protection of GlcNAc on the α1,6-mannose branch can be well tolerated. The addition of an α1,3-linked fucose to the terminal N-acetyllactosamine (LacNAc) or sialyl LacNAc structures on N211 and N212 to form the Lewis x and sialyl Lewis x structures on the α1,6-mannose branch of N214 (40.1%) and N215 (34.8%), respectively, further decrease the efficiency for FUT8-catalyzed core fucosylation. Quite interestingly, FUT8 was found to be able to glycosylate N04, an N-glycan without a α1,3-mannose branch, even though with a neglectable percentage conversion (5.26%). The result was confirmed by milligram scale synthesis and product N604 purification followed by mass spectrometry (MS) and proton nuclear magnetic resonance (1H NMR) characterization (Supplementary Information VI). Overall, our substrate specificity study using a library of 77 structurally defined N-glycans confirmed that FUT8 has a quite strict requirement towards the α1,3-mannose branch, and suggested a very relaxed preference towards the α1,6-mannose branch.
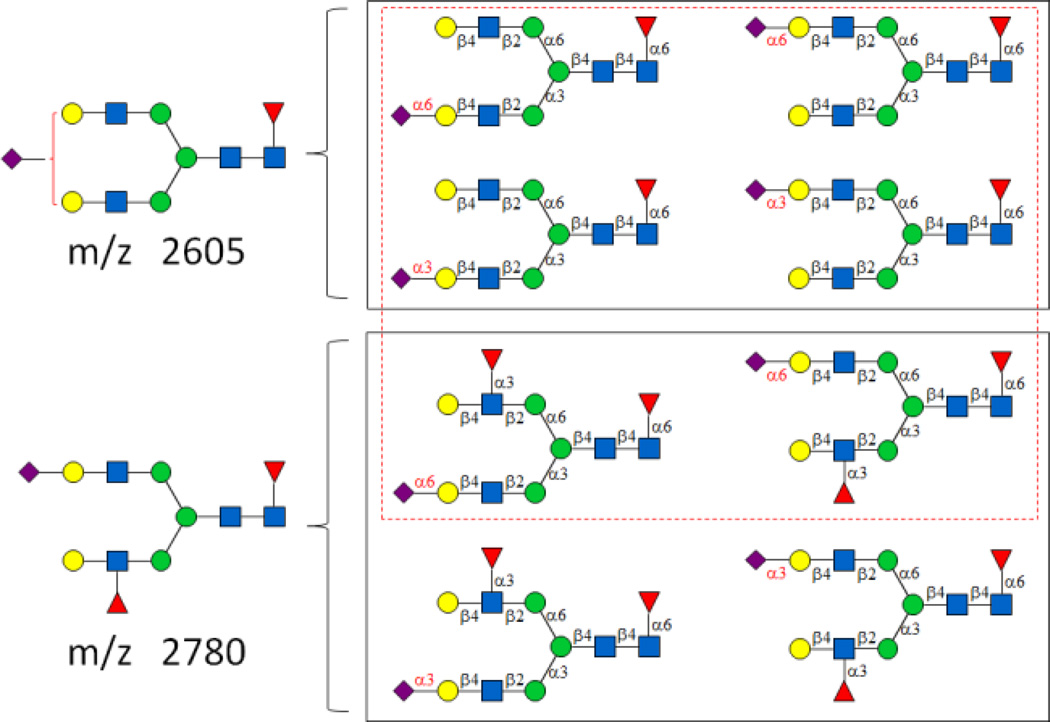
With the information about substrate specificity of FUT8 in hand, we were ready to design effective routes for synthesizing asymmetric N-glycans with core-fucosylation which have been frequently found in the glycomes of mammalian cells, especially on disease related proteins.19 For example, putative structures of m/z 2605 and 2780 found on human ovarian biomarker glycoprotein CA125 were determined by matrix-assisted laser desorption/ionization-mass spectrometry (MALDI-MS) and collision activated dissociation-tandem mass spectrometry (CAD-MS/MS).19b Nevertheless, a few possible isomers (Fig. 2) were not ruled out. To access such asymmetric N-glycans with core-fucosylation which can be used as glycan standards, a facile chemoenzymatic synthetic strategy was developed (Fig. 3 & 4).
Figure 2.
Two N-glycans (m/z) identified from CA125 and possible N-glycans (black squared). Red dash squared N-glycan structures were synthesized in this study.
Figure 3.
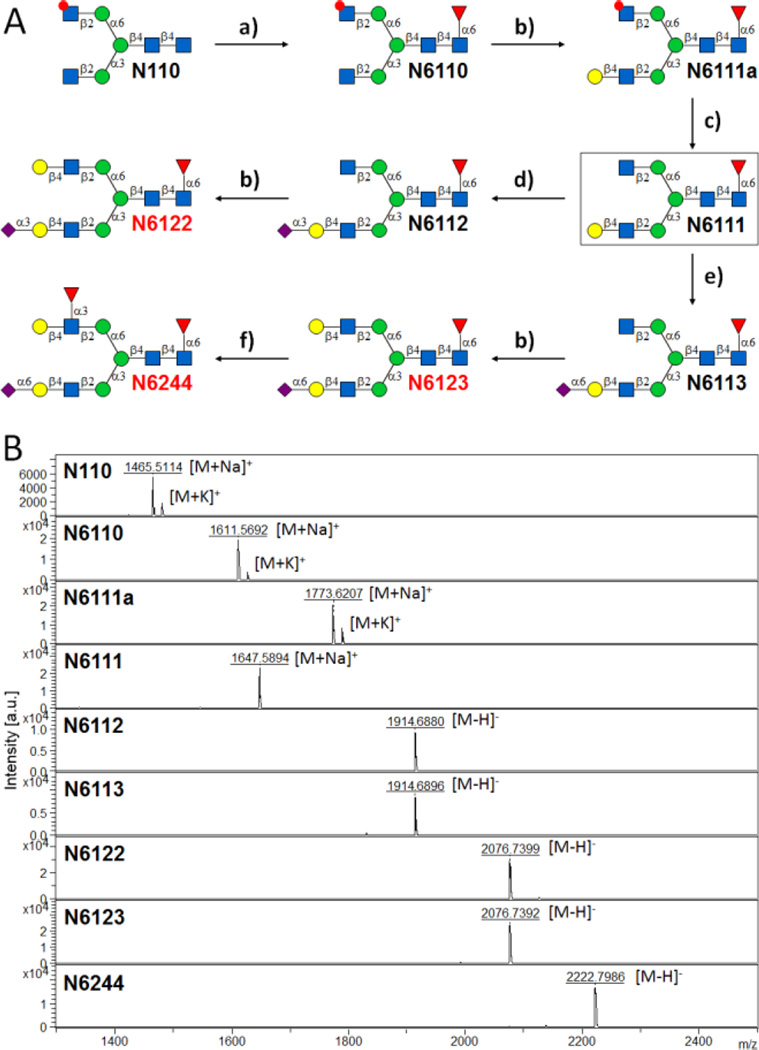
Chemoenzymatic synthesis of core-fucosylated asymmetric N-glycans from N110 (A), and MALDI-MS analysis of purified glycans (B). Reaction conditions: a) FUT8, GDP-Fuc, FastAP; b) β4GALT1, UDP-Gal, FastAP, Mn2+; c) 30% ammonium hydroxide : H2O (1 : 10), 6 h; d) PmST1m, CMP-Neu5Ac; e) Pd2,6ST, CMP-Neu5Ac; f) Hpα1,3FT, GDP-Fuc, FastAP, Mn2+. FUT8, human α1,6-fucosyltransferase; B4GALT1, bovine β1,4-galactosyltransferase; PmST1m, Pasteurella multocida α2,3-sialyltransferase 1 mutant E271F/R313Y; Pd2,6ST, Photobacterium damselae α 2,6-sialyltransferase; Hpα1,3FT, c-terminal 66 amino acids truncated Helicobacter pylori α1,3-fucosyltransferase; FastAP, thermo-sensitive alkaline phosphatase.
Figure 4.
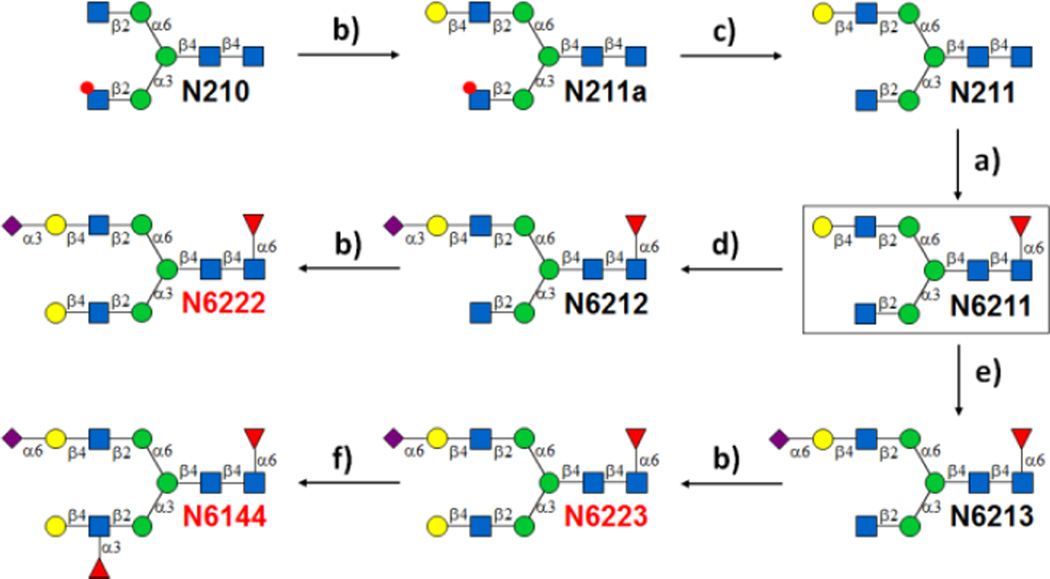
Enzymatic synthesis of core-fucosylated N-glycans starting with N210. See Figure 3A for detailed description of each step.
Similar to our core synthesis/enzymatic extension strategy (CSEE) for the preparation of asymmetric N-glycans,13b we envisaged N6111 and N6211 as two key intermediates for the synthesis of core-fucosylated ones. As illustrated in Figure 3A, to obtain intermediate N6111, we chose chemically prepared N-glycan N11013b as a starting point, which was shown to be an well tolerated substrate of human FUT8 in the substrate specificity study described above. Briefly, in a 2.5 mL reaction system containing 100 mM MES buffer (pH 7.0), 7.2 mg (5 µmole, 2 mM) of N110 was incubated at 37 °C with GDP-Fuc (4 mM), recombinant FUT8 (0.2 mg/mL), and 10 U/mL of thermosensitive alkaline phosphatase (FastAP, ThermoFisher) to drive the reaction forward by digesting byproduct GDP. Every other hour, 2 µL of the reaction mixture was withdrawn for analysis. MALDI-MS analysis showed a peak at m/z = 1611.5695, corresponding to N6110 [M + H]+. Meanwhile, on the HPLC-ELSD profile, a new peak (TR = 6.99 min) was observed, of which the area increased while that of the peak corresponding to N110 (TR = 6.06 min) decreased. After N110 was completely fucosylated (8 h incubation), the reaction was freeze-quenched at −80 °C for 30 min, and the mixture was concentrated to 300 mL for HPLC purification with a water/acetonitrile gradient elution (see Supplementary Information IV for details) to provide 6.9 mg of N6110 (87% yield). The purified N6110 was then galactosylated to afford N6111a by bovine β1,4-galactosyltransferase (B4GALT1) as described previously.13b In this case, only the GlcNAc residue on the α1,3-mannose branch was galactosylated, as the other one on the α1,3-mannose branch was protected by peracetylation. After HPLC purification, the GlcNAc residue on the α1,6-mannose branch of N6111a was de-acetylated with 3% of ammonium hydroxide to afford 6.5 mg (after HPLC purification) of key intermediate N6111 (80% yield over 3 steps).
With N6111 in hand, core-fucosylated asymmetric N-glycans N6122, N6123 and N6244 were prepared in a step-wise manner similar to that of the non-core-fucosylated ones.13b Besides B4GALT1, 3 bacterial glycosyltransferases (GTs) were employed in the synthesis, including a sialidase activity reduced mutant (E271F/R313Y) of Pasteurella multocida α2,3-sialyltransferase 1 (PmST1m)20 to catalyze the formation of α2,3-sialosides, a Photobacterium damselae α2,6-sialyltransferase (Pd2,6ST)21 to catalyze the formation of α2,6-sialosides, and a C-terminal 66 amino acids truncated version of Helicobacter pylori α1,3-fucosyltransferase (Hpα1,3FT)13b to fucosylate branched GlcNAc residues (Fig. 3A). All glycans were purified by HPLC after each glycosylation step and characterized by MALDI-MS (Fig. 3B). N-Glycan purity was confirmed by HPLC-ELSD (Supplementary Information V). It is worth to mention that none of the 4 GTs showed altered activity towards core-fucosylated N-glycan substrates compare to that for non-core-fucosylated ones, indicating that the α1,6-core fucose has negligible effect on acceptor glycan interaction with the GTs. More core-fucosylated asymmetric N-glycans can thus be readily synthesized from N6111 following previously reported routes.13b
In contrast to N110 with a per-acetylated GlcNAc at the α1,6-mannose branch which is a suitable acceptor for FUT8, N210 with a per-acetylated GlcNAc at the α1,3-mannose branch is not a suitable substrate for FUT8. Therefore, a key intermediate N6211 was needed for further enzymatic extension. It was synthesized from N210 by β1,4-galactosylation of the non-protected GlcNAc on the α1,6-mannose branch for the formation of N211 followed by deprotection of the GlcNAc on the α1,3-mannose branch and FUT8-catalyzed core-fucosylation of the asymmetric N-glycan (Fig. 4). Starting from N6211, desired N-glycans N6222, N6223 and N6144 were then enzymatically synthesized in a step-wise manner as described above (Fig. 4). The purified products were characterized by MALDI-MS, 1H NMR (Supplementary Information VII), and HPLC-ELSD (Supplementary Information V).
In conclusion, the available of previously synthesized 77 structurally-defined N-glycans allowed detailed substrate specificity study of a recombinant human FUT8. Results showed a quite strict requirement of FUT8 towards the α1,3-mannose branch but a great promiscuity towards the α1,6-mannose branch. Interestingly, human FUT8 can also core-fucosylated an N-glycan without the α1,3-mannose branch, which was not observed before. In addition, a facile chemoenzymatic strategy was developed and employed for the efficient synthesis of a panel of core-fucosylated asymmetric N-glycans. These well-defined structures are valuable standards and probes for glycomics analysis and elucidating functions of glycan-binding protein.
Supplementary Material
Acknowledgments
We are grateful to Dr. Donald Jarvis from the University of Wyoming for kindly providing the Sf9 cells and FUT8 baculovirus stock. This work was supported by National Institutes of Health (U01GM0116263 to P.G. Wang and L. Li) and National Cancer Institute (Contract SBIR: HHSN261201500021C to Chemily, LLC).
Footnotes
Electronic Supplementary Information (ESI) available: Experimental details for expression and purification of FUT8, substrate specificity study, N-glycan synthesis, purification and characterization. See DOI: 10.1039/x0xx00000x
Contributor Information
Lei Li, Email: lli22@gsu.edu.
Peng G. Wang, Email: pwang11@gsu.edu.
Notes and References
- 1.(a) Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. Cold Spring Harbor (NY): 2009. [PubMed] [Google Scholar]; (b) Zhao YY, Takahashi M, Gu JG, Miyoshi E, Matsumoto A, Kitazume S, Taniguchi N. Cancer Sci. 2008;99:1304. doi: 10.1111/j.1349-7006.2008.00839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Dwek RA. Chem Rev. 1996;96:683. doi: 10.1021/cr940283b. [DOI] [PubMed] [Google Scholar]
- 2.(a) Imperiali B, O'Connor SE. Curr Opin Chem Biol. 1999;3:643. doi: 10.1016/s1367-5931(99)00021-6. [DOI] [PubMed] [Google Scholar]; (b) Helenius A, Aebi M. Science. 2001;291:2364. doi: 10.1126/science.291.5512.2364. [DOI] [PubMed] [Google Scholar]; (c) Annu Rev Biochem. 2004;73:1019. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- 3.(a) Shields RL, Lai J, Keck R, O'Connell LY, Hong K, Meng YG, Weikert SH, Presta LG. J Biol Chem. 2002;277:26733. doi: 10.1074/jbc.M202069200. [DOI] [PubMed] [Google Scholar]; (b) Shinkawa T, Nakamura K, Yamane N, Shoji-Hosaka E, Kanda Y, Sakurada M, Uchida K, Anazawa H, Satoh M, Yamasaki M, Hanai N, Shitara K. J Biol Chem. 2003;278:3466. doi: 10.1074/jbc.M210665200. [DOI] [PubMed] [Google Scholar]
- 4.Okazaki A, Shoji-Hosaka E, Nakamura K, Wakitani M, Uchida K, Kakita S, Tsumoto K, Kumagai I, Shitara K. J Mol Biol. 2004;336:1239. doi: 10.1016/j.jmb.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Li W, Yu R, Ma B, Yang Y, Jiao X, Liu Y, Cao H, Dong W, Liu L, Ma K, Fukuda T, Liu Q, Ma T, Wang Z, Gu J, Zhang J, Taniguchi N. J Immunol. 2015;194:2596. doi: 10.4049/jimmunol.1402678. [DOI] [PubMed] [Google Scholar]
- 6.(a) Taniguchi N, Kizuka Y. Adv Cancer Res. 2015;126:11. doi: 10.1016/bs.acr.2014.11.001. [DOI] [PubMed] [Google Scholar]; (b) Pinho SS, Reis CA. Nat Rev Cancer. 2015;15:540. doi: 10.1038/nrc3982. [DOI] [PubMed] [Google Scholar]
- 7.(a) Aoyagi Y, Isokawa O, Suda T, Watanabe M, Suzuki Y, Asakura H. Cancer. 1998;83:2076. [PubMed] [Google Scholar]; (b) Aoyagi Y, Suzuki Y, Isemura M, Nomoto M, Sekine C, Igarashi K, Ichida F. Cancer. 1988;61:769. doi: 10.1002/1097-0142(19880215)61:4<769::aid-cncr2820610422>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]; (c) Taketa K, Endo Y, Sekiya C, Tanikawa K, Koji T, Taga H, Satomura S, Matsuura S, Kawai T, Hirai H. Cancer Res. 1993;53:5419. [PubMed] [Google Scholar]; (d) Miyoshi E, Noda K, Yamaguchi Y, Inoue S, Ikeda Y, Wang W, Ko JH, Uozumi N, Li W, Taniguchi N. Biochim Biophys Acta. 1999;1473:9. doi: 10.1016/s0304-4165(99)00166-x. [DOI] [PubMed] [Google Scholar]
- 8.Geng F, Shi BZ, Yuan YF, Wu XZ. Cell Res. 2004;14:423. doi: 10.1038/sj.cr.7290243. [DOI] [PubMed] [Google Scholar]
- 9.Saldova R, Struwe WB, Wynne K, Elia G, Duffy MJ, Rudd PM. Int J Mol Sci. 2013;14:15636. doi: 10.3390/ijms140815636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Inoue S, Gu J, Miyoshi E, Noda K, Li W, Mizuno-Horikawa Y, Nakano M, Asahi M, Takahashi M, Uozumi N, Ihara S, Lee SH, Ikeda Y, Yamaguchi Y, Aze Y, Tomiyama Y, Fujii J, Suzuki K, Kondo A, Shapiro SD, Lopez-Otin C, Kuwaki T, Okabe M, Honke K, Taniguchi N. Proc Natl Acad Sci U S A. 2005;102:15791. doi: 10.1073/pnas.0507375102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson JR, Williams D, Schachter H. Biochem Biophys Res Commun. 1976;72:909. doi: 10.1016/s0006-291x(76)80218-5. [DOI] [PubMed] [Google Scholar]
- 12.(a) Longmore GD, Schachter H. Carbohydr Res. 1982;100:365. doi: 10.1016/s0008-6215(00)81049-6. [DOI] [PubMed] [Google Scholar]; (b) Voynow JA, Kaiser RS, Scanlin TF, Glick MC. J Biol Chem. 1991;266:21572. [PubMed] [Google Scholar]; (c) Kaminska J, Glick MC, Koscielak J. Glycoconj J. 1998;15:783. doi: 10.1023/a:1006959915435. [DOI] [PubMed] [Google Scholar]; (d) Paschinger K, Staudacher E, Stemmer U, Fabini G, Wilson IB. Glycobiology. 2005;15:463. doi: 10.1093/glycob/cwi028. [DOI] [PubMed] [Google Scholar]
- 13.(a) Echeverria B, Etxebarria J, Ruiz N, Hernandez A, Calvo J, Haberger M, Reusch D, Reichardt NC. Anal Chem. 2015;87:11460. doi: 10.1021/acs.analchem.5b03135. [DOI] [PubMed] [Google Scholar]; (b) Li L, Liu Y, Ma C, Qu J, Calderon AD, Wu B, Wei N, Wang X, Guo Y, Xiao Z, Song J, Sugiarto G, Li Y, Yu H, Chen X, Wang PG. Chem Sci. 2015;6:5652. doi: 10.1039/c5sc02025e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.(a) Prahl I, Unverzagt C. Tetrahedron Letters. 2000;41:10189. [Google Scholar]; (b) Eller S, Schuberth R, Gundel G, Seifert J, Unverzagt C. Angew Chem Int Ed Engl. 2007;46:4173. doi: 10.1002/anie.200604788. [DOI] [PubMed] [Google Scholar]; (c) Ott D, Seifert J, Prahl I, Niemietz M, Hoffman J, Guder J, Mönnich M, Unverzagt C. Eur J Org Chem. 2012;2012:5054. [Google Scholar]
- 15.Sun B, Srinivasan B, Huang X. Chemistry. 2008;14:7072. doi: 10.1002/chem.200800757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serna S, Yan S, Martin-Lomas M, Wilson IB, Reichardt NC. J Am Chem Soc. 2011;133:16495. doi: 10.1021/ja205392z. [DOI] [PubMed] [Google Scholar]
- 17.(a) Fan SQ, Huang W, Wang LX. J Biol Chem. 2012;287:11272. doi: 10.1074/jbc.M112.340497. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Huang W, Giddens J, Fan SQ, Toonstra C, Wang LX. J Am Chem Soc. 2012;134:12308. doi: 10.1021/ja3051266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wasilko DJ, Lee SE, Stutzman-Engwall KJ, Reitz BA, Emmons TL, Mathis KJ, Bienkowski MJ, Tomasselli AG, Fischer HD. Protein Expr Purif. 2009;65:122. doi: 10.1016/j.pep.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 19.(a) Martin K, Talukder R, Hay FC, Axford JS. J Rheumatol. 2001;28:1531. [PubMed] [Google Scholar]; (b) O'Riordan CR, Lachapelle AL, Marshall J, Higgins EA, Cheng SH. Glycobiology. 2000;10:1225. doi: 10.1093/glycob/10.11.1225. [DOI] [PubMed] [Google Scholar]; (c) Kratz EM, Ferens-Sieczkowska M, Faundez R, Katnik-Prastowska I. Glycoconj J. 2014;31:51. doi: 10.1007/s10719-013-9501-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugiarto G, Lau K, Li Y, Khedri Z, Yu H, Le DT, Chen X. Mol Biosyst. 2011;7:3021. doi: 10.1039/c1mb05182b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu H, Huang S, Chokhawala H, Sun M, Zheng H, Chen X. Angew Chem Int Ed Engl. 2006;45:3938. doi: 10.1002/anie.200600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.