Abstract
Object
To establish and compare normative metabolite concentrations in 2nd and 3rd trimester human amniotic fluid samples in an effort to reveal metabolic biomarkers of fetal health and development.
Materials and methods
Twenty-one metabolite concentrations were compared between 2nd (15–27 weeks gestation, N = 23) and 3rd (29–39 weeks gestation, N = 27) trimester amniotic fluid samples using 1H high resolution magic angle spinning (HR-MAS) spectroscopy. Data were acquired using the electronic reference to access in vivo concentrations method and quantified using a modified semi-parametric quantum estimation algorithm modified for high-resolution ex vivo data.
Results
Sixteen of 21 metabolite concentrations differed significantly between 2nd and 3rd trimester groups. Betaine (0.00846±0.00206 mmol/kg vs. 0.0133±0.0058 mmol/kg, P <0.002) and creatinine (0.0124±0.0058 mmol/kg vs. 0.247±0.011 mmol/kg, P <0.001) concentrations increased significantly, while glucose (5.96±1.66 mmol/kg vs. 2.41±1.69 mmol/kg, P <0.001), citrate (0.740±0.217 mmol/kg vs. 0.399±0.137 mmol/kg, P <0.001), pyruvate (0.0659±0.0103 mmol/kg vs. 0.0299±0.286 mmol/kg, P <0.001), and numerous amino acid (e.g. alanine, glutamate, isoleucine, leucine, lysine, and valine) concentrations decreased significantly with advancing gestation. A stepwise multiple linear regression model applied to 50 samples showed that gestational age can be accurately predicted using combinations of alanine, glucose and creatinine concentrations.
Conclusion
These results provide key normative data for 2nd and 3rd trimester amniotic fluid metabolite concentrations and provide the foundation for future development of magnetic resonance spectroscopy (MRS) biomarkers to evaluate fetal health and development.
Keywords: Electronic reference to access in vivo concentrations (ERETIC), Gestational age, High-resolution magic angle spinning (HR-MAS), Spectroscopy, High-resolution quantum estimation (HR-QUEST), Human amniotic fluid
Introduction
High-resolution magnetic resonance spectroscopy (MRS) of human amniotic fluid has the potential to become a valuable technique for the evaluation of fetal health and development [1–3]. Although numerous clinical assays utilize amniotic fluid to evaluate chromosomal abnormalities, metabolic errors, and fetal lung maturity [4], none currently uses MRS to exploit the complex metabolic information contained within amniotic fluid. Many of these clinical assays require time-consuming laboratory procedures and provide only a limited amount of information. MRS requires minimal sample preparation and can be used to quantify several metabolites simultaneously. However, before MRS-detectable biomarkers can be used clinically, the general relationships between amniotic fluid metabolites, gestational age, and the health status of the mother and fetus must be characterized.
Several metabolites associated with fetal developmental abnormalities have previously been identified by MRS. 31P MRS studies of human amniotic fluid detected various phospholipid analytes related to fetal respiratory distress syndrome (RDS) [5–7]. Subsequent 1H MRS investigations identified several organic acids, amino acids, sugars, choline-containing compounds and developmental catabolites [1,4,8,9]. Other studies employing a variety of methods searched for biomarkers of renal function [1], diabetes [8,10], pre-eclampsia [8,11], perinatal infection [12], spina bifida [8,13], trisomy 21 [8,14], cystic fibrosis [14] and fetal lung maturity [2,3,15,16]. These studies described the variance of specific compounds with respect to disease state, but often lacked robust quantitative analysis of the data. Recent high field studies performed at 800MHz have identified several additional amniotic fluid metabolites, determined the effects of freeze-thawing, and evaluated long term sample stability at room temperature, −20, and −70°C [17].
Because water suppression can be challenging in 1HMRS studies of amniotic fluid, previous investigators typically centrifuged or filtered the samples to remove insoluble matter or lyophilized and redissolved the samples in deuterium oxide (D2O) [1,9,13]. These procedures require additional sample preparation steps, which could lead to quantitation errors. Furthermore, the freeze-drying process can cause sample degradation or other spectral changes, particularly if the samples are not adjusted to neutral pH prior to freeze-drying [17]. We previously applied high resolution magic angle spinning (HR-MAS) spectroscopy to whole amniotic fluid to investigate surfactant molecules and other metabolites as potential markers of fetal lung maturity [3,16]. Using this approach, high quality MR spectra were obtained with good water suppression and minimal sample preparation.
Quantification of amniotic fluid metabolites is complicated due to the poor reproducibility of internal quantitation standards and severe spectral overlap. For example, the commonly used standard trimethylsilyltetradeuteriopropionate (TSP) can bind to macromolecules [18] and may also overlap with other signals of interest [19]. The Electronic REference To access In Vivo Concentrations (ERETIC) method [20] provides a synthetic radiofrequency signal and has been shown to be more robust than TSP for HR-MAS spectroscopy quantification [21].
Simple integration and Lorentzian–Gaussian peak-fitting cannot accurately quantify metabolites that co-resonate, and the problem is compounded when the overlapping peaks are multiplets. Semi-parametric quantum estimation (QUEST) is a time domain quantification algorithm that can model several metabolites simultaneously using prior knowledge of chemical shift and coupling constant information obtained from basis set spectra [22,23]. In the present study, metabolite concentrations in 2nd and 3rd trimester amniotic fluid samples were determined from 1H HR-MAS spectroscopy data acquired using ERETIC and quantified using a modified QUEST algorithm for high-resolution ex vivo MRS data (HR-QUEST). The purpose of this study was to establish and compare normative metabolite concentrations in 2nd and 3rd trimester human amniotic fluid samples.
Materials and methods
Sample acquisition
All samples were collected for clinical indications and a waiver of consent that satisfies federal regulations for Protection of Human Subjects and Standards for Privacy of Individually Identifiable Health Information (HIPAA) for analyses of existing biofluids was granted for this study. This protocol was reviewed and approved by our Institutional Review Board. Fifty amniotic fluid specimens (mean: 27 weeks gestation, range: 15–39 weeks) were retrospectively identified, transferred from their initial storage locations to −80°C freezers, and analyzed within 2 months of amniocentesis. Twenty-three samples were collected during the 2nd trimester (range: 15–27 weeks) and twenty-seven samples during the 3rd trimester (range: 29–39 weeks). The 2nd trimester samples were obtained for karyotype analysis in patients at risk for genetic disorders, however, the samples included in this study were karyotypically normal. The 3rd trimester samples were acquired from patients in pre-term labor or at risk for premature delivery to assess fetal lung maturity, as is the standard of care. While many of the patients were noted to be diabetic; each patient was undergoing medical management for their condition and was euglycemic at the time of amniocentesis. No first trimester samples were included because amniocentesis is not typically performed until at least 14 weeks of gestation at our institution. Gestational age was determined by date of last menstrual period or crown-rump length measurement at time of first trimester ultrasound per the standard of practice within our obstetric clinics. Demographic information is detailed in Table 1.
Table 1.
Patient demographic data for 2nd versus 3rd trimester amniotic fluid samples
| 2nd Trimester | 3rd Trimester | Significance | |
|---|---|---|---|
| Total (n) | 23 | 27 | |
| Gestational age (weeks) | |||
| n | 23 | 27 | |
| Mean | 18.28 ± 2.45 | 36.35 ± 1.97 | 0.226 |
| Maternal age (years) | |||
| n | 23 | 27 | |
| Mean | 33.00 ± 6.78 | 32.30 ± 7.71 | 0.256 |
| Gravidity | |||
| n | 14 | 16 | |
| Mean | 2.00 ± 1.41 | 3.65 ± 1.98 | 0.039 |
| Parity | |||
| n | 14 | 16 | |
| Mean | 0.714 ± 0.914 | 1.69 ± 1.35 | 0.131 |
| Ethnicity | |||
| African American/Black | 1 | 4 | |
| Asian/Pacific Islander | 5 | 8 | |
| Caucasian | 2 | 3 | |
| Hispanic/Latina | 5 | 10 | |
| Undisclosed | 10 | 3 |
Significance determined by 2-tailed t-test for nominal data sets
Reference metabolite spectra
A reference database was created containing quantitative 1D spectra of the most common metabolites in amniotic fluid and urine as well as metabolites associated with fetal maturity [4,17,24]. All chemicals were purchased from Sigma–Aldrich (Sigma, Aldrich, Fluka and Supelco, St Louis, MO). Stock solutions of each compound were prepared with 1× phosphate buffered saline solution and pH adjusted (to between 7.0 and 8.0) using concentrated hydrochloric acid or sodium hydroxide to match the physiologic pH of amniotic fluid [25]. Diluted samples of known concentrations (between 0.01 and 100.0 mM) were prepared to approximate physiologic concentrations of metabolites in human urine or amniotic fluid. Reference solutions were kept frozen at −80°C when not in use and were analyzed as described below.
1H HR-MAS spectroscopy protocol
1H HR-MAS spectroscopy was performed at 11.7 T (500MHz for 1H) using a Varian INOVA spectrometer equipped with a 4 mm gHX nanoprobe. Data were acquired at 1°C to minimize degradation and a 2,250 Hz spin rate in order to place the spinning sideband of water between the frequency reference TSP (0 ppm) and the first metabolites (~0.8 ppm). Samples were analyzed using custom designed 35-µl volume zirconium oxide rotors, which can be purchased from Varian, Inc (Palo Alto, CA). The rotors were designed to be easy to assemble, and are effectively leak proof when properly tightened [26]. If leakage occurred during sample preparation, as evidenced by the presence of liquid on the outside of the rotor or the loss of lock signal, the rotors were cleaned and dried, and the samples were remade. The sealed rotors were also routinely weighed before and after analysis to ensure that no leakage occurred during spectral analysis.
At HR-MAS analysis, the reference solutions or amniotic fluid samples were thawed at room temperature for up to 45 min and briefly vortexed for ~5–10 s to mix the contents. To provide a frequency and lock reference, 3.0 µl of D2O containing 0.75% TSP was pipetted into the rotor and weighed to ±0.01 mg, after which a sample aliquot was pipetted into the rotor and weighed (mean: 22.53 mg, range: 19.01–26.15 mg). The rotor was then assembled and placed into the probe for analysis. Quantitative 1D spectra were acquired with a 90°pulse, 4 s water presaturation, 2 s acquisition, 256 transients, 40,000 points, and 20,000 Hz spectral width. The total repetition rate was 6 s for amniotic fluid samples and 10 s for reference solutions, which was 4× and 5× the longest T1 (1.5 s for lactate) of the metabolites of interest at 1°C, respectively. Water suppression was achieved by continuous wave presaturation using the standard Varian “presat” sequence. During experimental optimization the power level and duration of the presaturation pulses were optimized to minimize saturation of nearby resonances (e.g., glucose). These parameters were then used for all samples.
The ERETIC signal was created using PBox (Varian Inc, Palo Alto, CA) and transmitted during the acquisition period using 0 dB of power, a full width at half height of 3.5 Hz, and an offset frequency equivalent to −0.5 ppm. The ERETIC signal was phase cycled in synchronization with the receiver to maintain phase coherence across scans. At the beginning of each acquisition, the phase of the ERETIC signal was manually adjusted to match the other signals in the spectrum. The ERETIC signal was calibrated daily using a standard amount of D2O+TSP. Detailed information on the implementation of ERETIC on our spectrometer has been reported elsewhere [21]. The total time required for data acquisition, including sample preparation, rotor assembly, tuning, shimming, pulse width calibrations, phasing of the ERETIC signal and data collection was ≤45 min.
Quantification
Spectra were processed and displayed using the Advanced Chemistry Development (ACD) 1D NMR processor (ACD/Labs, Toronto), the Java-based graphical user interface for the Magnetic Resonance User Interface (jMRUI) quantitation package, and MATLAB (The MathWorks, Natick, MA) and quantified using a custom version of the semiparametric quantum estimation program developed by Ratiney and co-workers which was adapted for analysis of short-echo time HR-MAS spectra containing 40,000 points (HR-QUEST) [22]. The first two points of each free-induction decay (FID) were backward linear predicted in ACD using base points 3–1,000 and 32 coefficients. Each spectrum was then saved as a text file and imported into jMRUI. The spectra were reversed, phased, and referenced to TSP. Spectral regions downfield of 4.75 ppm (including residual water) and upfield of −1 ppm were deleted. The HR-QUEST program estimated the macromolecule signals using the Hankel-Lanczos singular value decomposition (HLSVD) algorithm and iterated between fitting the metabolites and modeling the macromolecules 12 times. Minor frequency shifts, which could be caused by differences in pH between samples, were also accounted for during the iterative process. HR-QUEST results were displayed in MATLAB and metabolite peak areas were then exported into an excel spreadsheet and converted to concentrations using a conversion factor for the ERETIC signal as previously described [21].
Statistical analysis
Signal not accounted for by the HR-QUEST modeling algorithm was represented as a residual spectrum (the difference between the measured and HR-QUEST modeled spectrum). Model error was calculated using MATLAB as the area under the residual spectrum divided by the area under the measured spectrum. Errors for the complete spectrum and the spectral range of interest (from 0.2 to 4.5 ppm) were both calculated.
Differences between 2nd trimester (N = 23) and 3rd trimester (N = 27) amniotic fluid samples were assessed using the 2-sample Wilcoxon/Mann–Whitney test. The Wilcoxon test is non-parameteric and therefore avoids the need to make distributional assumptions about the data. In addition, an analysis was performed to determine whether metabolites provided complementary (i.e. additive) information. Five pre-selected metabolites (alanine, betaine, choline, creatinine and glucose) were entered as explanatory variables in a stepwise multiple linear regression model with gestational age as the outcome variable. These five metabolites were selected to test the hypothesis that members of each of their representative groups (amino acids, surfactant catabolites, surfactant components, indicators of renal function and sugars, respectively) would contribute to the prediction of gestational age. Terms were retained or dropped at a significance level of P = 0.01. Finally, the stepwise linear regression modeling procedure was repeated with all 21 metabolite concentrations included as explanatory variables to account for all possible predictors of gestational age.
Results
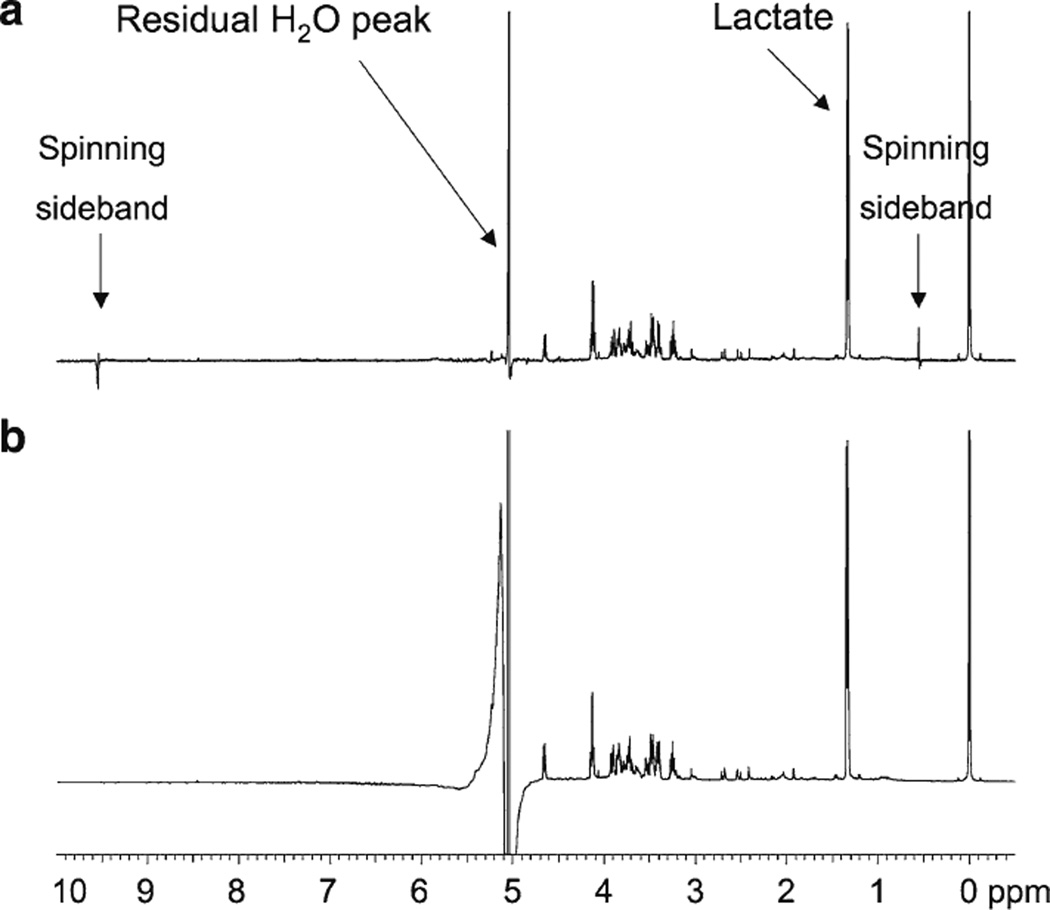
Figure 1 shows 1H MR spectra of the same amniotic fluid sample obtained using (a) a 4-mm HR-MAS probe and (b) a conventional 5-mm inverse probe. For the HR-MAS spectrum, the sample tube was briefly vortexed and then an aliquot was pipetted into the rotor; whereas for the inverse probe, the sample was centrifuged and an aliquot of the supernatant was pipetted into a 5-mm NMR tube. It is important to note that both samples were 90% aqueous with 10% D2O added to provide a lock signal. As demonstrated in the figure, the water suppression achieved with the HR-MAS probe was superior to that achieved with the inverse probe, both in terms of the degree of water suppression and baseline distortion. Note that the α-glucose doublet (δ = 5.22 ppm), which resonates very close to water, can be observed in the HR-MAS spectrum but not the conventional NMR spectrum. Also note in the HR-MAS spectrum that the spinning side bands of water near 0.5 and 9.5 ppm are very small and do not have a significant effect on the baseline or nearby resonances. During preliminary investigations, some samples were also centrifuged and/or filtered or were lyophilized and reconstituted in 100% D2O prior to analysis. However, these approaches required additional sample preparation steps and degradation is known to occur if the samples are not closely adjusted to pH 7 prior to freeze drying. Since high quality spectra were achieved with no additional sample preparation steps using the HR-MAS probe, we elected to use this probe rather than a liquids probe for this study.
Fig. 1.
500 MHz 1H MR spectra of the same amniotic fluid sample obtained using a a 4 mm HR-MAS probe and b a conventional 5 mm inverse probe. Both samples were 90% aqueous with 10% D2O added to provide a lock signal. For HR-MAS, the sample was vortexed and an aliquot was analyzed; for the inverse probe, the sample was centrifuged and the supernatant was analyzed
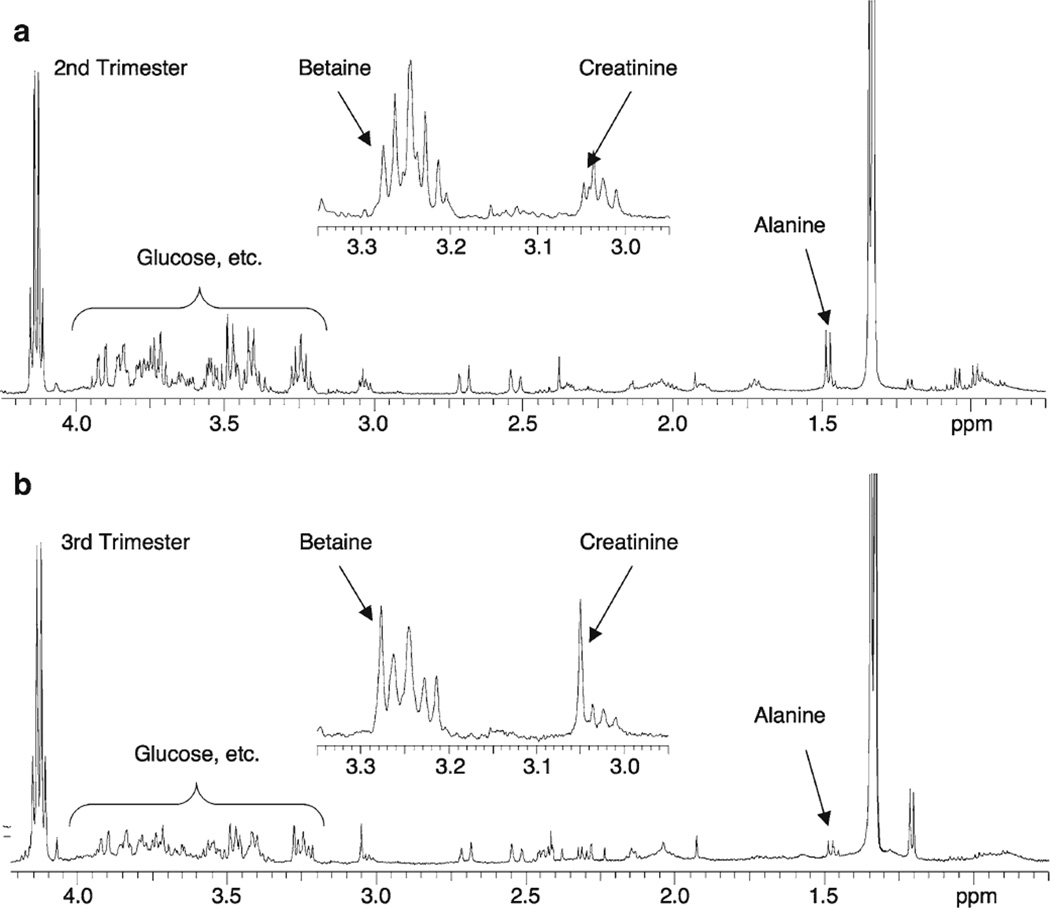
Figure 2 shows representative 1H HR-MAS spectra of (a) 2nd (16 weeks 6 days gestation) and (b) 3rd trimester (36 weeks) amniotic fluid specimens. Both spectra are scaled relative to lactate and were chosen arbitrarily; however, the metabolic changes illustrated in this figure reflect the overall trends that were observed between 2nd and 3rd trimester samples. For example, note the visually apparent decreases in glucose and alanine between 2nd and 3rd trimester samples. The region between 2.95 and 3.35 ppm is shown as an inset to illustrate the significant increase in betaine and creatinine between 2nd and 3rd trimester samples.
Fig. 2.
Representative 500 MHz 1H HR-MAS spectra of the 0.75–4.25 ppm region of a 2nd and b 3rd trimester human amniotic fluid samples. The 2.95–3.35 ppm region of each spectrum is shown in inset. Note the significant decrease in glucose and alanine and concurrent increase in creatinine and betaine with advancing gestational age
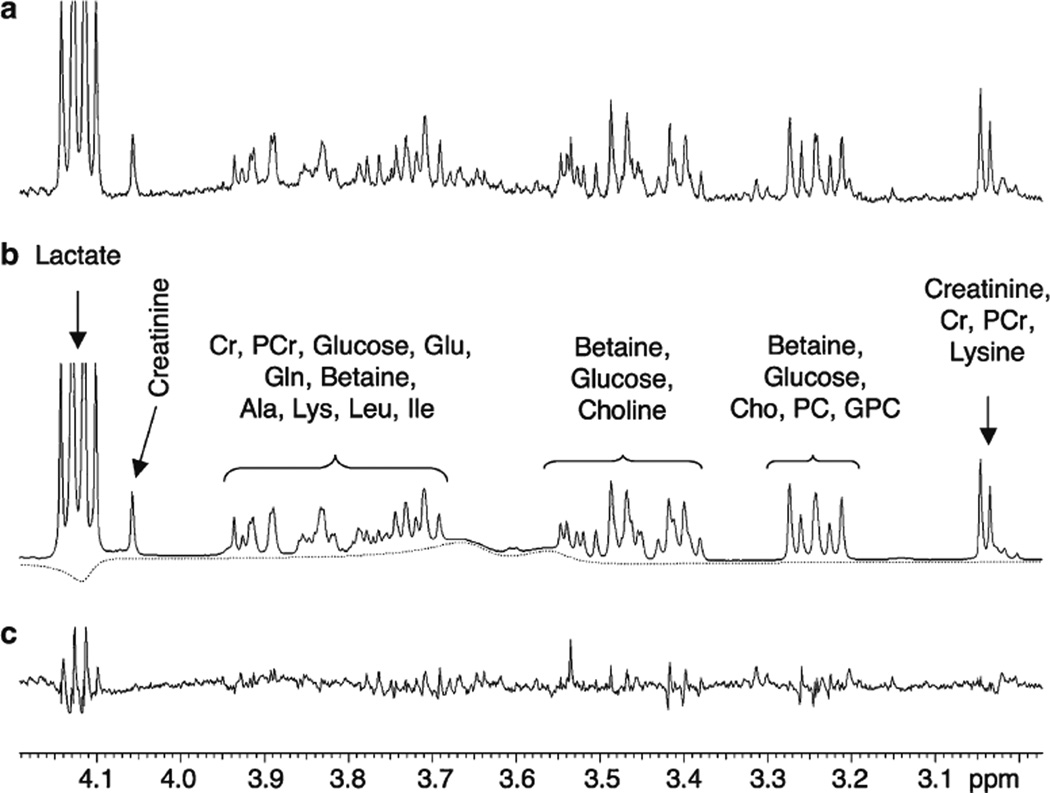
Figure 3 shows the ~3.0 to 4.2 ppm region of (a) 3rd trimester amniotic fluid sample, (b) the HR-QUEST modeled spectrum, and (c) the residual or difference spectrum. The modeled spectrum is composed only of the metabolites contained in the reference spectral database and a baseline correction (dotted line). Twenty-one reference metabolites were identified and quantified in amniotic fluid, accounting for 99.7% of all resonances in the spectrum by area. Ethanolamine was quantified in less than half of the spectra and thus was excluded from statistical analyses. Three reference metabolites (phosphatidylcholine (lecithin), phosphatidylglycerol, and sphingomyelin) were not observed in any of the amniotic fluid samples. The mean spectral fitting error was −0.27 ± 0.9% for all samples over the entire spectral range. The mean spectral fitting error over the range of interest between 0.2 and 4.5 ppm was nearly zero (0.07 ± 0.7%).
Fig. 3.
Example of HR-QUEST fitting of a 3rd trimester amniotic fluid sample. The figure demonstrates the ~3.0–4.2 ppm region of a the measured spectrum, b the modeled spectrum and c the residual spectrum. The modeled spectrum is composed only of metabolites contained in reference spectral database and a baseline correction (dotted line)
Table 2 lists the mean concentrations (mmol/kg), standard deviations, and percent change of the significantly different metabolites in 2nd versus 3rd trimester amniotic fluid samples. Sixteen of 21 metabolites were significantly different between 2nd and 3rd trimester amniotic fluid samples (P < 0.05, Wilcoxon test), without adjusting significance thresholds to account for multiple comparisons. Applying a strict Bonferroni correction (i.e. a significance threshold of P < 0.05/21 = 0.00238), 12 metabolites (alanine, betaine, citrate, creatinine, glutamate, glucose, GPC, lactate, leucine, lysine, pyruvate and valine) were still significantly different. These metabolites reflect diverse chemical classes, including sugars, amino acids, components of the Krebs cycle and biomarkers for renal function.
Table 2.
Average metabolite concentrations (mmol/kg) and percent change of significantly different metabolites (16 of 21) in 2nd vs. 3rd trimester amniotic fluid samples
| Metabolite | 2nd Trimester (Mean ± SD) | 3rd Trimester (Mean ± SD) | Percent change | P (Wilcoxon) |
|---|---|---|---|---|
| Alanine | 0.860 ± 0.229 | 0.228 ± 0.132 | −73.5 | <0.001 |
| Betaine | 0.00846 ± 0.00206 | 0.0133 ± 0.0058 | +56.7 | <0.002 |
| Citrate | 0.740 ± 0.217 | 0.399 ± 0.137 | −46.0 | <0.001 |
| Creatine | 0.154 ± 0.084 | 0.097 ± 0.053 | −37.2 | 0.008 |
| Creatinine | 0.0124 ± 0.0058 | 0.0247 ± 0.011 | +99.5 | <0.001 |
| GPC | 0.0424 ± 0.0407 | 0.0181 ± 0.0178 | −57.4 | <0.001 |
| Glutamine | 0.219 ± 0.106 | 0.310 ± 0.166 | +41.6 | 0.029 |
| Glutamate | 1.10 ± 0.26 | 0.319 ± 0.305 | −71.0 | <0.001 |
| Glucose | 5.96 ± 1.66 | 2.41 ± 1.69 | −59.4 | <0.001 |
| Isoleucine | 0.0847 ± 0.0889 | 0.0409 ± 0.0512 | −51.7 | 0.043 |
| Lactate | 20.0 ± 5.1 | 15.4 ± 4.1 | −23.3 | 0.001 |
| Leucine | 0.0787 ± 0.0318 | 0.0240 ± 0.0230 | −69.5 | <0.001 |
| Lysine | 0.719 ± 0.196 | 0.270 ± 0.209 | −62.4 | <0.001 |
| Pyruvate | 0.0659 ± 0.0103 | 0.0299 ± 0.0286 | −54.6 | <0.001 |
| Succinate | 0.0562 ± 0.0624 | 0.0842 ± 0.0663 | +49.9 | 0.007 |
| Valine | 0.828 ± 0.288 | 0.323 ± 0.179 | −60.9 | <0.001 |
GPC glycerophosphocholine
Table 3 displays results from a stepwise multiple linear regression model, which assessed the joint capability of five selected metabolites (alanine, betaine, choline, creatinine and glucose) to predict gestational age. These metabolites were selected a priori as representative compounds that may be related to fetal lung and renal maturity. Terms were retained or dropped at a significance level of 0.01. This model yielded a significant additive contribution from 3 of the 5 metabolites. When applied sequentially, the combination of alanine, creatinine and glucose created a linear prediction model that fit the data with an R2 value of 0.90. When this model “fitting procedure” was repeated for all 21 metabolites, these same three compounds were part of the final model (with a minor added contribution from 3-hydroxybutyrate).
Table 3.
Metabolites most predictive of gestational age based on stepwise linear regression model
| Variable | R2 | Cp | Prob F |
|---|---|---|---|
| Alanine | 0.77 | 54.18 | <0.0001 |
| Creatinine | 0.83 | 29.79 | 0.0002 |
| Glucose | 0.90 | 5.84 | <0.0001 |
Discussion
In this study, 2nd and 3rd trimester amniotic fluid metabolite concentrations were determined using 1H HR-MAS spectroscopy by incorporating an improved external quantitation reference (ERETIC) into the experiment and a semi-automated spectral modeling algorithm (HR-QUEST) for data analysis. The combination of HR-MAS spectroscopy with ERETIC and HR-QUEST provided several improvements over previous MRS studies of amniotic fluid. As shown in Fig. 1, improved water suppression was achieved using a HR-MAS probe compared to a conventional inverse probe designed for liquids. The improved water suppression is likely due to several factors. First, greater B0 homogeneity can be obtained by reducing the sample volume [27]. The volume of sample typically used for a 5-mm NMR tube is 500–600 µl, whereas the volume typically used for HR-MAS is ≤40 µl. However, it should be noted that small volume microtubes could also be used in a conventional NMR probe without the need for water suppression. Secondly, the HR-MA Srotors used in this study were designed to have an oblate spheroid geometry, which provides improved homogeneity compared to cylindrical designs [26,28]. Additionally, in a HR-MAS probe, the coil is slightly larger than the sample chamber [21], whereas in an inverse probe the coil is smaller than the sample length. Therefore, in an inverse probe, unsuppressed water can diffuse in and out of the region of detection resulting in a residual water signal [29], whereas in an HR-MAS probe this does not significantly occur. Although HR-MAS probes are less sensitive than conventional inverse probes, HR-MAS spectroscopy can be used as an alternative when the analysis of intact biofluid samples is desired. It should also be noted that although the samples in this study were briefly vortexed prior to analysis, any solid matter would be effectively centrifuged to the bottom of the rotor because of the high-spin rate used for HR-MAS.
The determination of metabolite concentrations using the ERETIC method also represents a major advance. Some earlier studies reported metabolite ratios (based on integrals or intensities) [5–7,14], which are not ideal because they do not illustrate how individual metabolites change independently of each other. Consequently, ratios can yield ambiguously high or low values when either the numerator or the denominator approaches zero. The use of external or internal standards (e.g., TSP, solvent signals, standard addition) [1,9,10,13] can yield absolute concentrations, however, errors associated with the use of integrals or peak intensities still remain. Although ERETIC has been shown to be more reliable than TSP for HR-MAS applications in tissue [21], PUlse Length based CONcentration determination (PULCON) is another external reference technique—based upon the principle of reciprocity—has recently been reported [19]. Both ERETIC [30] and PULCON are now commercially available on Bruker spectrometers and future studies will be needed to compare the results obtained using these two techniques.
Previous studies often used integration [1,9,10,13], peak intensities [8,14], or peak fitting [6] to quantify amniotic fluid metabolites. The use of integration and peak intensities is only accurate for completely resolved resonances, while peak fitting is also limited in cases of extreme spectral overlap and prone to user-bias. HR-QUEST is much better suited to fit overlapping metabolites because it uses a priori knowledge of all of the frequencies, multiplicities, and linewidths of each metabolite in the library. This is particularly useful for metabolites such as glucose, which have complex splitting patterns and also co-resonate with several metabolites of interest. In this study, HR-QUEST modeled amniotic fluid spectra with a fitting error of less than 1%, representing a significant improvement over manual techniques [31]. Moreover, although the reference metabolite solutions were adjusted to physiological pH, no efforts were made to adjust the pH of the amniotic fluid samples. The frequencies of protons located near ionizable groups (e.g., phosphate) are known to shift with changes in pH; however, HR-QUEST is able to compensate for minor frequency differences during the iterative process. Although HR-QUEST is very robust, several other approaches including principal component analysis [32], linear combination models (LCModel) [33], and various time-domain and frequency-domain fitting algorithms [34] have been reported that may be equally effective.
Consistent with previous studies, metabolite profiles for 2nd and 3rd trimester amniotic fluid samples showed significant differences with gestational age [8,9,11,13]. As illustrated in Fig. 2 and shown in Table 2, the concentrations of glucose, creatine, and various amino acids decreased while the concentrations of creatinine and betaine increased with advancing gestational age. These data provide excellent metabolomic evidence of fetal maturation. In addition to general fetal maturation, the detection and quantification of creatinine may provide a biomarker for assessing fetal renal status. While prior reports have suggested that creatinine would be a poor MRS biomarker for renal maturation because it co-resonates with multiple glucose resonances, the present technique reliably identified and quantified creatinine within this region of the 1H spectrum. Prior authors have investigated surrogate markers of renal function with limited success. Given the inherent difficulties reported in quantifying surrogate markers of renal maturity (urea, indoxyl-sulfate, formate) [1], the accurate quantification of creatinine presents a significant development in the MR assessment of fetal renal function.
This work elaborated upon similar studies in human urine [35,36], blood, serum and plasma [37], which vetted the ability of rapid metabolic profiling techniques to provide accurate quantitation of biofluid constituents. As reported in Table 2, there was a much greater change in betaine than choline between 2nd and 3rd trimester fetuses. Betaine is thought to be produced in the liver by oxidation of choline, an essential nutrient that functions in multiple physiological pathways, and is a component of phosphatidylcholine and sphingomyelin [38]. Choline is transported across the placenta, and is higher in fetal than maternal plasma [39,40] [41], but no published studies indicate whether betaine is transferred across the human placenta. If betaine is not freely transported, it may be a more accurate marker of fetal choline metabolism than choline proper. Therefore, betaine may prove to be a more specific marker of fetal lung maturity than choline. In addition, recent case–control studies have provided evidence that low dietary intake of betaine and choline are associated with increased risk of neural defects in humans, offering an additional indication for the monitoring of these metabolites using 1HMRS [42].
Stepwise linear regression models performed on the present data suggest that gestational age can be accurately predicted using combinations of alanine, glucose and creatinine concentrations. At present, the clinical estimation of gestational age is based upon a woman’s last menstrual period (LMP) or early ultrasound (<20 weeks) findings. However, as gestation advances into the late second and third trimesters, the ability of existing techniques to accurately assess fetal age decreases markedly [43,44]. While it is not our contention that this technique would replace existing dating techniques early in gestation, this model may provide concurrent physiologic evidence to complement existing tests (i.e. biometric ultrasound studies), which rely on anatomic measurements. In this case, a metabolite-based method for estimating gestational age may prove more reliable and is the topic of ongoing investigation.
In any NMR study involving biological fluid or tissue, sample stability and degradation are critical issues. Graca et al. [17] previously showed that amniotic fluid samples were stable at room temperature for 4–5 h and for up to 6 months at −70°C. In the current study, data were acquired at 1°C to minimize degradation, and the total time allowed for sample thawing, sample preparation, rotor assembly, tuning, shimming, pulse width calibrations, and finally data collection was ≤90 min. Additionally, all of the samples included in this paper were analyzed within 1–2 months of amniocentesis. During preliminary studies, serial 1D experiments were performed over 12 h and found that there were negligible spectroscopic changes over this time period at this temperature. This is in contrast to tissue studies where significant spectroscopic (and histologic) changes occur even over 2 h [26]. Additionally, prior experiments using a temperature standard (methanol) demonstrated negligible sample heating using the conventional pulse-acquire sequence with presaturation turned on. While phase separation is an issue for HR-MAS studies of cells and tissue, it does not pose a major problem in amniotic fluid, which is acellular and closely resembles urine.
Finally, sample leakage is an issue which must also be addressed in HR-MAS studies of biofluids. Sample leakage can occur during either rotor assembly or analysis and results in a uniform overestimation of metabolite concentrations. When leakage occurs during rotor assembly it is often visually apparent as the liquid is displaced from the rotor. If leakage occurs during HR-MAS analysis (e.g., due to inadequate tightening of the rotor), the lock signal will usually be lost and the sample will be pushed out of the rotor because of the high spin rate. Weighing rotors before and after analysis is a simple method to determine if leakage has occurred. The use of specially designed rotors can also be helpful. For example, the rotors used in the current study have a simple plunger that gently slides into the rotor and requires almost no force at all to assemble. At the top of the plunger is a rubber O-ring, which creates a seal when the rotor is tightened. Early designs included inserts to constrict the sample volume, however, fluids become trapped underneath them resulting in residual signals. Instead, the current plungers have fixed lengths corresponding to specific rotor volumes, and the volume between the side of the rotor and the plunger is factored into the concentration calculations. These rotors have proven to be very robust and have resulted in fewer problems with sample leakage compared to other rotor designs.
Conclusion
Normative metabolite concentrations were established and compared for 2nd and 3rd trimester amniotic fluid samples. The simultaneous assessment of multiple metabolic markers with advancing gestational age has the potential to improve the evaluation of fetal health and development.
Acknowledgments
The authors acknowledge and thank Drs. Hélène Ratiney and Herald Rabeson of Creatis-Lrmn, University of Lyon, for providing the HR-QUEST algorithm used in this research. We also thank Drs. Mark J. Albers, Andrew S. Zektzer, and Daina Avizonis for technical support and helpful discussions. This work was supported in part by grants from the RSNA Research and Education Foundation (Berlex Laboratories/RSNA Research Scholar Grant), the UCSF Research Evaluation and Allocation Committee, and the UCSF Academic Senate Committee on Research. In addition, this publication was made possible by Grant Number: 1 TL1 RR 024129 from the National Center for Research Resources (NCRR), a component of the National Institutes of Health (NIH), and NIH Roadmap for Medical Research. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NCRR of NIH. Information on NCRR is available at http://www.ncrr.nih.gov/. Information on Re-engineering the Clinical Research Enterprise can be obtained from http://nihroadmap.nih.gov/clinicalresearch/overview-translational.asp.
Contributor Information
Brad R. Cohn, Email: brad.cohn@ucsf.edu, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA.
Bonnie N. Joe, Email: bonnie.joe@radiology.ucsf.edu, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA.
Shoujun Zhao, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA.
John Kornak, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA; Department of Epidemiology & Biostatistics, University of California, San Francisco, CA, USA.
Vickie Y. Zhang, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA
Rahwa Iman, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA.
John Kurhanewicz, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA.
Kiarash Vahidi, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA.
Jingwei Yu, Department of Laboratory Medicine, University of California, San Francisco, CA, USA.
Aaron B. Caughey, Department of Obstetrics & Gynecology, University of California, San Francisco, CA, USA
Mark G. Swanson, Department of Radiology & Biomedical Imaging, University of California, 1600 Divisadero Street, Room C-250, Box 1667, San Francisco, CA 94115, USA
References
- 1.McGowan PE, Reglinski J, Wilson R, Walker JJ, Wisdoms S, McKillop JH. Quantitative 1H-NMR analysis of amniotic fluid. J Pharm Biomed Anal. 1993;11(8):629–632. doi: 10.1016/0731-7085(93)80167-y. [DOI] [PubMed] [Google Scholar]
- 2.Fenton BW, Lin CS, Ascher S, Macedonia C. Magnetic resonance spectroscopy to detect lecithin in amniotic fluid and fetal lung. Obstet Gynecol. 2000;95(3):457–460. doi: 10.1016/s0029-7844(99)00566-9. [DOI] [PubMed] [Google Scholar]
- 3.Clifton MS, Joe BN, Zektzer AS, Kurhanewicz J, Vigneron DB, Coakley FV, Nobuhara KK, Swanson MG. Feasibility of magnetic resonance spectroscopy for evaluating fetal lung maturity. J Pediatr Surg. 2006;41(4):768–773. doi: 10.1016/j.jpedsurg.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 4.Nelson TR, Gillies RJ, Powell DA, Schrader MC, Manchester DK, Pretorius DH. High resolution proton NMR spectroscopy of human amniotic fluid. Prenat Diagn. 1987;7(5):363–372. doi: 10.1002/pd.1970070511. [DOI] [PubMed] [Google Scholar]
- 5.Pearce JM, Komoroski RA. Resolution of phospholipid molecular species by 31P NMR. Magn Reson Med. 1993;29(6):724–731. doi: 10.1002/mrm.1910290603. [DOI] [PubMed] [Google Scholar]
- 6.Pearce JM, Krone JT, Pappas AA, Komoroski RA. Analysis of saturated phosphatidylcholine in amniotic fluid by 31P NMR. Magn Reson Med. 1993;30(4):476–484. doi: 10.1002/mrm.1910300410. [DOI] [PubMed] [Google Scholar]
- 7.Pearce JM, Shifman MA, Pappas AA, Komoroski RA. Analysis of phospholipids in human amniotic fluid by 31P NMR. Magn Reson Med. 1991;21(1):107–116. doi: 10.1002/mrm.1910210113. [DOI] [PubMed] [Google Scholar]
- 8.Bock JL. Metabolic profiling of amniotic fluid by proton nuclear magnetic resonance spectroscopy: correlation with fetal maturation and other clinical variables. Clin Chem. 1994;40(1):56–61. [PubMed] [Google Scholar]
- 9.Sims CJ, Fujito DT, Burholt DR, Dadok J, Giles HR, Wilkinson DA. Quantification of human amniotic fluid constituents by high resolution proton nuclear magnetic resonance (NMR) spectroscopy. Prenat Diagn. 1993;13(6):473–480. doi: 10.1002/pd.1970130609. [DOI] [PubMed] [Google Scholar]
- 10.McGowan PE, Lawrie WC, Reglinski J, Spickett CM, Wilson R, Walker JJ, Wisdom S, Maclean MA. 1H NMR as a non-invasive probe of amniotic fluid in insulin dependent diabetes mellitus. J Perinat Med. 1999;27(5):404–408. doi: 10.1515/JPM.1999.056. [DOI] [PubMed] [Google Scholar]
- 11.Roopnarinesingh S, Morris D. Amniotic fluid urea and creatinine in normal pregnancy and in pre-eclampsia. J Obstet Gynaecol Br Commonw. 1971;78(1):29–33. doi: 10.1111/j.1471-0528.1971.tb00187.x. [DOI] [PubMed] [Google Scholar]
- 12.Romero R, Jimenez C, Lohda AK, Nores J, Hanaoka S, Avila C, Callahan R, Mazor M, Hobbins JC, Diamond MP. Amniotic fluid glucose concentration: a rapid and simple method for the detection of intraamniotic infection in preterm labor. Am J Obstet Gynecol. 1990;163(3):968–974. doi: 10.1016/0002-9378(90)91106-m. [DOI] [PubMed] [Google Scholar]
- 13.Groenen PM, Engelke UF, Wevers RA, Hendriks JC, Eskes TK, Merkus HM, Steegers-Theunissen RP. High-resolution 1H NMR spectroscopy of amniotic fluids from spina bifida fetuses and controls. Eur J Obstet Gynecol Reprod Biol. 2004;112(1):16–23. doi: 10.1016/s0301-2115(03)00279-3. [DOI] [PubMed] [Google Scholar]
- 14.Le Moyec L, Muller F, Eugene M, Spraul M. Proton magnetic resonance spectroscopy of human amniotic fluids sampled at 17–18 weeks of pregnancy in cases of decreased digestive enzyme activities and detected cystic fibrosis. Clin Biochem. 1994;27(6):475–483. doi: 10.1016/0009-9120(94)00051-v. [DOI] [PubMed] [Google Scholar]
- 15.Joe BN, Swanson MG, Zektzer AS, Kurhanewicz J, Qayyum A, Coakley FV, Yeh BM. Non-invasive evaluation of fetal lung maturity by MR spectroscopy: a preliminary ex-vivo investigation. Am J Roentgenol. 2005;184(4):16–17. [Google Scholar]
- 16.Joe BN, Vahidi K, Zektzer A, Chen MH, Clifton MS, Butler T, Keshari K, Kurhanewicz J, Coakley F, Swanson MG. H-1 HR-MAS spectroscopy for quantitative measurement of choline concentration in amniotic fluid as a marker of fetal lung maturity: inter-and intraobserver reproducibility study. J Magn Reson Imaging. 2008;28(6):1540–1545. doi: 10.1002/jmri.21592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Graca G, Duarte IF, Goodfellow BJ, Barros AS, Carreira IM, Couceiro AB, Spraul M, Gil AM. Potential of NMR spectroscopy for the study of human amniotic fluid. Anal Chem. 2007;79(21):8367–8375. doi: 10.1021/ac071278d. [DOI] [PubMed] [Google Scholar]
- 18.Kriat M, Confortgouny S, Viondury J, Sciaky M, Viout P, Cozzone PJ. Quantitation of metabolites in human blood serum by proton magnetic resonance spectroscopy—a comparative study of the use of formate and TSP as concentration standards. NMR Biomed. 1992;5(4):179–184. doi: 10.1002/nbm.1940050404. [DOI] [PubMed] [Google Scholar]
- 19.Wider G, Dreier L. Measuring protein concentrations by NMR spectroscopy. J Am Chem Soc. 2006;128(8):2571–2576. doi: 10.1021/ja055336t. [DOI] [PubMed] [Google Scholar]
- 20.Akoka S, Barantin L, Trierweiler M. Concentration measurement by proton NMR using the ERETIC method. Anal Chem. 1999;71(13):2554–2557. doi: 10.1021/ac981422i. [DOI] [PubMed] [Google Scholar]
- 21.Albers MJ, Butler TN, Rahwa I, Bao N, Keshari KR, Swanson MG, Kurhanewicz J. Evaluation of the ERETIC method as an improved quantitative reference for H-1 HR-MAS spectroscopy of prostate tissue. Magn Reson Med. 2009;61(3):525–532. doi: 10.1002/mrm.21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ratiney H, Sdika M, Coenradie Y, Cavassila S, van Ormondt D, Graveron-Demilly D. Time-domain semi-parametric estimation based on a metabolite basis set. NMR Biomed. 2005;18(1):1–13. doi: 10.1002/nbm.895. [DOI] [PubMed] [Google Scholar]
- 23.Rabeson H, Fauvelle F, Testylier G, Foquin A, Carpentier P, Dorandeu F, van Ormondt D, Graveron-Demilly D. Quantitation with QUEST of brain HRMAS-NMR signals: application to metabolic disorders in experimental epileptic seizures. Magn Reson Med. 2008;59(6):1266–1273. doi: 10.1002/mrm.21610. [DOI] [PubMed] [Google Scholar]
- 24.Holmes E, Foxall PJ, Spraul M, Farrant RD, Nicholson JK, Lindon JC. 750 MHz 1H NMR spectroscopy characterisation of the complex metabolic pattern of urine from patients with inborn errors of metabolism: 2-hydroxyglutaric aciduria and maple syrup urine disease. J Pharm Biomed Anal. 1997;15(11):1647–1659. doi: 10.1016/s0731-7085(97)00066-6. [DOI] [PubMed] [Google Scholar]
- 25.Uyeno D. The physical properties and chemical composition of human amniotic fluid. J Biol Chem. 1919;37(1):77–103. [Google Scholar]
- 26.Swanson MG, Zektzer AS, Tabatabai ZL, Simko J, Jarso S, Keshari KR, Schmitt L, Carroll PR, Shinohara K, Vigneron DB, Kurhanewicz J. Quantitative analysis of prostate metabolites using 1H HR-MAS spectroscopy. Magn Reson Med. 2006;55(6):1257–1264. doi: 10.1002/mrm.20909. [DOI] [PubMed] [Google Scholar]
- 27.Becker ED. High resolution NMR: theory and chemical applications. San Diego: Academic Press; 2000. [Google Scholar]
- 28.Kuchel PW, Chapman BE, Bubb WA, Hansen PE, Durrant CJ, Hertzberg MP. Magnetic susceptibility: solutions, emulsions, and cells. Concepts Magn Reson A. 2003;18(1):56–71. [Google Scholar]
- 29.Mo HP, Raftery D. Improved residual water suppression: WET180. J Biomol NMR. 2008;41(2):105–111. doi: 10.1007/s10858-008-9246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remaud GS, Silvestre V, Akoka S. Traceability in quantitative NMR using an electronic signal as working standard. Accredit Qual Assur. 2005;10(8):415–420. [Google Scholar]
- 31.Rabenstein DL, Keire DA. Quantitative chemical analysis by NMR. In: Popov IA, Halenga K, editors. Modern NMR techniques and their application in chemistry. New York: Marcel Dekker; 1991. pp. 323–369. [Google Scholar]
- 32.Griffin JL, Bollard M, Nicholson JK, Bhakoo K. Spectral profiles of cultured neuronal and glial cells derived from HRMAS H-1 NMR spectroscopy. NMR Biomed. 2002;15(6):375–384. doi: 10.1002/nbm.792. [DOI] [PubMed] [Google Scholar]
- 33.Provencher SW. Automatic quantitation of localized in vivo 1H spectra with LCModel. NMR Biomed. 2001;14:260–264. doi: 10.1002/nbm.698. [DOI] [PubMed] [Google Scholar]
- 34.Poullet JB, Sima DM, Van Huffel S. MRS signal quantitation: a review of time- and frequency-domain methods. J Magn Reson. 2008;195(2):134–144. doi: 10.1016/j.jmr.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Brown JC, Mills GA, Sadler PJ, Walker V. 1H NMR studies of urine from premature and sick babies. Magn Reson Med. 1989;11(2):193–201. doi: 10.1002/mrm.1910110207. [DOI] [PubMed] [Google Scholar]
- 36.Lauridsen M, Hansen SH, Jaroszewski JW, Cornett C. Human urine as test material in 1H NMR-based metabonomics: recommendations for sample preparation and storage. Anal Chem. 2007;79(3):1181–1186. doi: 10.1021/ac061354x. [DOI] [PubMed] [Google Scholar]
- 37.Teahan O, Gamble S, Holmes E, Waxman J, Nicholson JK, Bevan C, Keun HC. Impact of analytical bias in metabonomic studies of human blood serum and plasma. Anal Chem. 2006;78(13):4307–4318. doi: 10.1021/ac051972y. [DOI] [PubMed] [Google Scholar]
- 38.Friesen RW, Novak EM, Hasman D, Innis SM. Relationship of dimethylglycine, choline, and betaine with oxoproline in plasma of pregnant women and their newborn infants. J Nutr. 2007;137(12):2641–2646. doi: 10.1093/jn/137.12.2641. [DOI] [PubMed] [Google Scholar]
- 39.Grassl SM. Choline transport in human placental brush-border membrane vesicles. Biochim Biophys Acta. 1994;1194(1):203–213. doi: 10.1016/0005-2736(94)90221-6. [DOI] [PubMed] [Google Scholar]
- 40.van der Aa EM, Wouterse AC, Peereboom-Stegeman JH, Russel FG. Uptake of choline into syncytial microvillus membrane vesicles of human term placenta. Biochem Pharmacol. 1994;47(3):453–456. doi: 10.1016/0006-2952(94)90175-9. [DOI] [PubMed] [Google Scholar]
- 41.Eaton BM, Sooranna SR. Regulation of the choline transport system in superfused microcarrier cultures of BeWo cells. Placenta. 1998;19(8):663–669. doi: 10.1016/s0143-4004(98)90028-5. [DOI] [PubMed] [Google Scholar]
- 42.Shaw GM, Carmichael SL, Yang W, Selvin S, Schaffer DM. Periconceptional dietary intake of choline and betaine and neural tube defects in offspring. Am J Epidemiol. 2004;160(2):102–109. doi: 10.1093/aje/kwh187. [DOI] [PubMed] [Google Scholar]
- 43.Johnsen SL, Rasmussen S, Sollien R, Kiserud T. Fetal age assessment based on femur length at 10–25 weeks of gestation, and reference ranges for femur length to head circumference ratios. Acta Obstet Gynecol Scan. 2005;84(8):725–733. doi: 10.1111/j.0001-6349.2005.00691.x. [DOI] [PubMed] [Google Scholar]
- 44.Konje JC, Abrams KR, Bell SC, Taylor DJ. Determination of gestational age after the 24th week of gestation from fetal kidney length measurements. Ultrasound Obstet Gynecol. 2002;19(6):592–597. doi: 10.1046/j.1469-0705.2002.00704.x. [DOI] [PubMed] [Google Scholar]