Abstract
The ZFP36L3 protein is a rodent-specific, placenta- and yolk sac-specific member of the tristetraprolin (TTP) family of CCCH tandem zinc finger proteins. These proteins bind to AU-rich elements in target mRNAs, and promote their deadenylation and decay. We addressed the hypotheses that the absence of ZFP36L3 would result in the accumulation of target transcripts in placenta and/or yolk sac, and that some of these would be important for female reproductive physiology and overall fecundity. Mice deficient in ZFP36L3 exhibited decreased neonatal survival rates, but no apparent morphological changes in the placenta or surviving offspring. We found Zfp36l3 to be paternally imprinted, with profound parent-of-origin effects on gene expression. The protein was highly expressed in the syncytiotrophoblast cells of the labyrinth layer of the placenta, and the epithelial cells of the yolk sac. RNA-Seq of placental mRNA from Zfp36l3 knockout (KO) mice revealed many significantly upregulated transcripts, whereas there were few changes in KO yolk sacs. Many of the upregulated placental transcripts exhibited decreased decay rates in differentiated trophoblast stem cells derived from KO blastocysts. Several dozen transcripts were deemed high probability targets of ZFP36L3; these include proteins known to be involved in trophoblast and placenta physiology. Type 1 transferrin receptor mRNA was unexpectedly decreased in KO placentas, despite an increase in its stability in KO stem cells. This receptor is crucial for placental iron uptake, and its decrease was accompanied by decreased iron stores in the KO fetus, suggesting that this intrauterine deficiency might have deleterious consequences in later life.
KEY WORDS: Deadenylation, mRNA binding proteins, mRNA decay, Placenta, Zinc finger proteins
Summary: ZFP36L3-deficient mice have fewer offspring than normal and increased placental expression of many potential target genes, including genes essential for placenta function.
INTRODUCTION
The tristetraprolin (TTP) family of CCCH tandem zinc finger proteins consists of mRNA-binding proteins that are thought to regulate gene expression by binding to and promoting the decay of mRNAs containing specific types of AU-rich element-binding sites (Blackshear and Perera, 2014; Brooks and Blackshear, 2013). These sites are generally located in the 3′-untranslated regions (3′UTRs) of mRNAs and have the optimal sequence of UUAUUUAUU (Lai et al., 2005); physiologically relevant confirmed target transcripts often contain multiple binding sites in close proximity to each other (Carballo et al., 1998, 2000). The mechanism of the induced mRNA instability is not entirely understood, but appears to involve initial stimulation of deadenylation, or removal of the poly(A) tail, thought to be the rate-limiting step in mRNA decay in all eukaryotes (Carballo et al., 2000; Fabian et al., 2013; Lai et al., 2003).
In most mammals, the TTP protein family consists of three members, which behave similarly in biochemical assays of mRNA binding and destabilization (Frederick et al., 2008; Lai et al., 2003). However, knockout (KO) mice for these three family members exhibit dramatic physiological specificity. For example, TTP KO mice (Zfp36−/−) develop a systemic inflammatory syndrome that is largely due to chronic elevations in tumor necrosis factor alpha (TNFα); the Tnf mRNA was found to be a direct target of TTP binding and induced mRNA destabilization (Carballo and Blackshear, 2001; Carballo et al., 1998; Taylor et al., 1996). KO of a second family member, Zfp36l1, results in embryonic lethality, apparently due to failure of chorioallantoic fusion, an essential step in the development of the umbilical circulation (Stumpo et al., 2004). The third family member, Zfp36l2, is crucial for definitive hematopoiesis (Stumpo et al., 2009).
A fourth mammalian gene in this family, Zfp36l3, is an X chromosomal gene expressed only in the yolk sac and placenta of certain rodents, including mice and rats (Blackshear et al., 2005; Frederick et al., 2008; Gingerich et al., 2016). The ZFP36L3 protein differs from its other family members in several respects; for example, in contrast to the other family members, which are nucleocytoplasmic shuttling proteins (Phillips et al., 2002), the mouse and rat ZFP36L3 proteins contain long series of carboxyl-terminal repeats that serve to maintain the protein in the cytoplasm (Frederick et al., 2008). We describe here the development of a Zfp36l3 KO mouse, and its phenotypic and biochemical characterization. Our findings highlight the importance of ZFP36L3 in mouse fertility, as well as its influence on post-transcriptional gene expression in placenta.
RESULTS
Expression of ZFP36L3 during normal development
In placenta, Zfp36l3 mRNA was readily detected by embryonic day (E) 9.5, increased to near maximum levels by E14.5, and then remained elevated until E18.5. In the yolk sac, Zfp36l3 mRNA was essentially undetectable at E10.5, and then began accumulating, reaching a peak by E18.5 (Fig. 1A). Transcripts for the other family members were readily detectable and largely constant in both yolk sac and placenta during development (Fig. 1A). Zfp36l3 mRNA was undetectable in the embryo at all time points (Fig. 1A), and was not found in other tissues of the adult mouse (Blackshear et al., 2005).
Fig. 1.
Expression patterns of TTP family member mRNAs during development. (A) Total cellular RNA was obtained from yolk sacs, placentas and embryos at the indicated times of development, and northern blots were probed with cDNAs for Zfp36 (TTP), Zfp36l1, Zfp36l2 and Zfp36l3. Each lane in each blot contains the same amount of total cellular RNA (10 µg), and a single blot was used for each transcript; thus, the expression levels of a given mRNA can be directly compared between tissues and developmental stages. However, possible differences in probe specific activity and affinity mean that these blots cannot be used to compare expression quantitatively among the four different transcripts. The exposure lengths for the autoradiographs were 7, 7, 7 and 5 days for Zfp36, Zfp36l1, Zfp36l2 and Zfp36l3 mRNAs, respectively. Consistency of RNA loading was demonstrated by ethidium bromide (EtBr) staining of the gels. (B) Immunostaining of ZFP36L3 in mouse placenta at E17.5. The brown color indicates ZFP36L3 staining, which was greatest in the syncytiotrophoblast cells of the labyrinth zone of the placenta. There was also good staining of the trophoblast giant cells, much less staining in the spongiotrophoblasts, and no detectable staining either in the maternal decidua or in the allantois. Staining with pre-immune serum under the same conditions was entirely negative at this exposure (data not shown). (C) Immunostaining of ZFP36L3 in yolk sac at E15.5. The specific ZFP36L3 staining is indicated by the brown color (Imm.). Adjacent sections were stained with pre-immune serum under otherwise identical conditions (Pre-imm.). Note the strong staining of the single layer of epithelial cells.
Immunohistochemical staining revealed that within the placenta, ZFP36L3 protein was highly expressed in the syncytiotrophoblast cells of the labyrinth region of the placenta at E17.5; in addition, it was readily detected in the parietal trophoblast giant cells of the junctional zone (Fig. 1B; Fig. S1). Spongiotrophoblast cells of the junctional zone were relatively poorly stained, and immunoreactive protein was not detected either in the allantois or in the maternal decidua (Fig. 1B; Fig. S1). In keeping with the northern blot results, immunoreactive ZFP36L3 was detected in the developing labyrinth zone as early as E9.5, and ZFP36L3-expressing syncytiotrophoblast cells were the major cell type of the mature placenta (Fig. S2).
In the visceral yolk sac at E15.5, there was a high level expression of ZFP36L3 in the single layer of endodermal epithelial cells, but no apparent expression in the neighboring mesenchymal cells (Fig. 1C). Higher power views of both the highly folded yolk sac facing the placenta, and the unfolded yolk sac facing the embryo, confirmed this cellular localization and the lack of staining in the amnion (Fig. S3). Under these conditions, the pre-immune serum did not react with proteins in either the placenta (Fig. S4) or the yolk sac (Fig. 1C).
Phenotype of ZFP36L3-deficient mice
The Zfp36l3 gene was knocked out by conventional procedures in C57Bl/6 mice (Fig. S5A). The mutant allele could be readily detected by PCR (Fig. S5B). Northern blotting of placenta total cellular RNA confirmed the absence of Zfp36l3 mRNA in the KO placentas (Fig. S5C). This was confirmed by immunoblotting (not shown) and immunohistochemistry of the KO placentas (Fig. S4). The histology of the KO placentas, however, appeared to be normal at this age (Fig. S4).
Both male and female KO mice were viable, lived to adulthood, and were fertile. There was, however, a decrease in the total number of surviving, weaned Zfp36l3 KO pups, when 103 litters that were the products of male wild type (WT) versus female heterozygote (Het) matings were analyzed (Table S1A). Male KO mice survived at a frequency of 14% compared with the expected rate of 25% (P<0.0001). Interestingly, surviving female Hets from these matings were also decreased, with 19% surviving compared with the expected 25% (P<0.0001). This suggested the possibility that in some cases there might be X inactivation of the single allele present in the female Het mice.
We also performed timed matings, and examined 12 litters at E15.5; average litter size was higher than expected at 8.6. Viable male KO embryos were present at 28% at E15.5, compared with the expected 25%. These data suggest that the observed fall-off in survival of the male KOs occurred at some time after E15.5.
In matings of KO males versus Het females, KO survivors (male and female) represented 34% of total, compared with the expected 50%. Both male and female KO mice were decreased in frequency, with 16% of males surviving (versus 25% expected; P<0.001), and 18% of females (versus 25% expected; P<0.01) (Table S1B).
Evidence for parent-of-origin effects
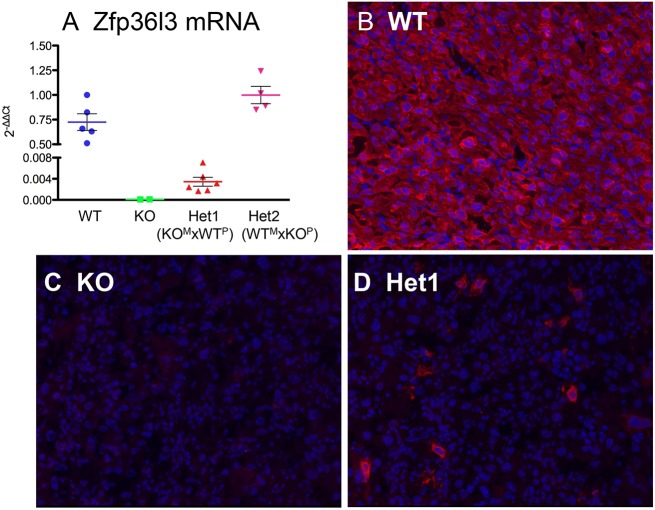
To test for possible parent-of-origin effects on Zfp36l3 expression, we evaluated Zfp36l3 mRNA concentrations in WT, KO and Het placentas at E15.5 by real-time RT-PCR. As expected, there was no detectable Zfp36l3 mRNA in the KO placentas (Fig. 2A). In six Het placentas that were the products of female Het versus male WT matings (Het1), there were profound decreases in mRNA levels compared with WT, to levels less than 1% of WT (Fig. 2A; note the break in the y-axis). These six Het placentas were from three different Het females. By contrast, when the mutant alleles came from the male (Het2), the average levels of mRNA expression were slightly higher than the WT levels (Fig. 2A).
Fig. 2.
Zfp36l3 mRNA expression and ZFP36L3 immunostaining in WT, KO and Het placentas at E15.5. (A) Real-time RT-PCR measurements of Zfp36l3 mRNA from placentas of the indicated genotypes. Each dot represents data from a single placenta. Mean ± s.e.m. are indicated. ‘Het1’ refers to heterozygous animals derived from matings of a Het female (maternal KO allele, or KOM) and a WT male (paternal WT allele, or WTP); ‘Het2’ refers to heterozygous animals derived from matings of a WT female (WTM allele) and a KO male (KOP allele). Note the break in the vertical axis. (B-D) High magnification views of ZFP36L3 immunostained syncytiotrophoblast cells of the labyrinth zone from placentas at E15.5. In this case, the ZFP36L3 protein is stained red and is shown in a WT placenta section (B), and a section from a Het1 placenta (D); the DAPI nuclear stain is blue (B-D). The Het1 sample in D is from a mating between a WT male and a Het female, and shows a few positively stained cells in a field of non-expressing syncytiotrophoblast cells.
By immunohistochemistry, placentas from the female Het versus male WT matings (Het1) exhibited a striking pattern, in which only a few cells expressed ZFP36L3 (Fig. 2D). These cells did not appear syncytial, but instead appeared as single, discrete, nucleated cells, in which the ZFP36L3 protein was expressed in the cytosol. The identity of these rare immunopositive cells is not known. By contrast, ZFP36L3 expression patterns were similar to WT in Het placentas that were the product of WT female versus KO male matings (Het2) (not shown).
These results demonstrated that, when the KO allele comes from the male, the maternal WT allele is expressed normally, whereas when the KO allele comes from the female, the paternal WT allele is inactivated, resulting in Het placentas that resemble KO placentas in terms of Zfp36l3 expression. This means that parental origin of the KO allele determines the resulting phenotype of the otherwise genetically identical ‘Het’ mice. This is, therefore, an interesting example of apparent paternal imprinting, in which the extra-embryonic tissues, the only tissues expressing Zfp36l3, experience persistent inactivation of the paternal X chromosome (Bermejo-Alvarez et al., 2012).
Identification of potential physiological targets of ZFP36L3 by deep sequencing
Our hypothesis was that the complete absence of ZFP36L3 in the placenta and/or yolk sac would cause abnormal accumulation of its direct mRNA targets in those tissues. As there were no apparent morphological or histological abnormalities in the KO placenta, molecular changes in the KO placentas should take place in the apparent absence of placental structural abnormalities. The absence of Zfp36l3 mRNA in the samples used for deep mRNA sequencing (mRNA-Seq) was confirmed by northern blotting (not shown).
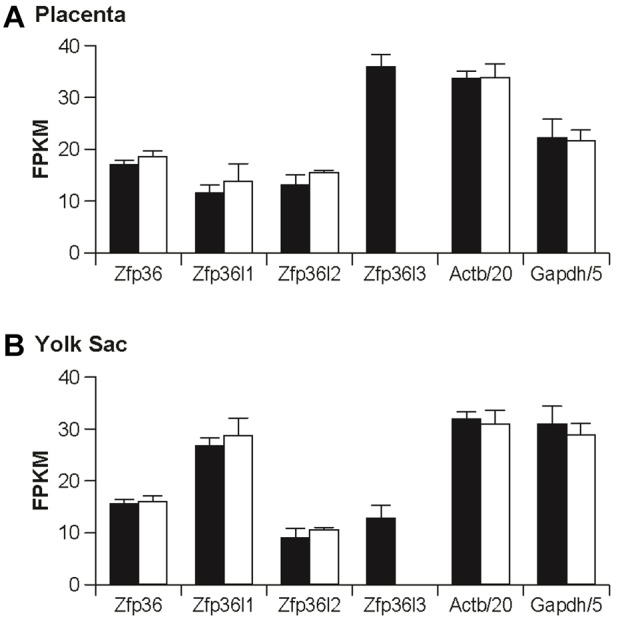
We first compared mRNA levels from whole WT and KO placentas, including maternal decidua, at E15.5, using mRNA-Seq; five samples consisting of individual placentas were used in each group. The complete dataset has been deposited in GEO under accession number GSE66137. A total of 28,886 transcript models were quantified. The range of expression of transcripts in the WT placentas was very large, ranging from an average number of fragments per kilobase of exon per million fragments mapped (FPKM) of 5558 for prolactin family 3, subfamily b, member 1 mRNA (Prl3b1; GenBank accession number NM_008865) and 5285 for trophoblast-specific protein alpha (Tpbpa) mRNA (NM_009411), to <1 FPKM for 15,139 mRNA models.
We also performed a similar analysis with WT and KO yolk sacs at E15.5, using mRNA-Seq; four samples were used in each group, and each sample consisted of a single yolk sac. The complete dataset has been deposited in GEO under accession number GSE66138. A total of 27,713 transcript models were quantified. The levels of expression of transcripts in the WT yolk sacs ranged from an average of 25,228 FPKM for alpha fetoprotein (Afp) mRNA (NM_007423), to <1 for 13,880 mRNA models.
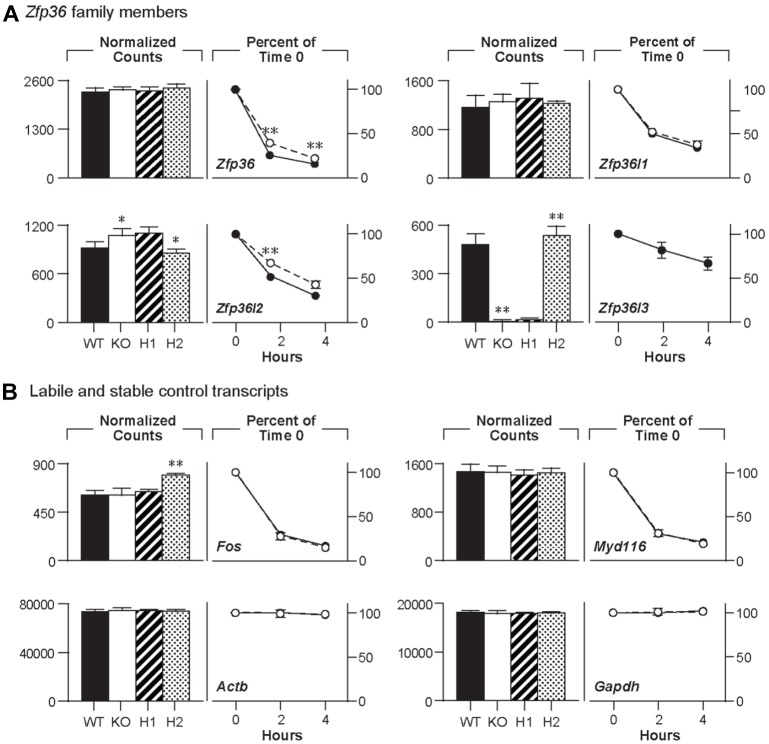
Expression of the four mouse TTP family members is shown in Fig. 3, along with two highly expressed control transcripts, Gapdh and Actb mRNAs. In placenta (Fig. 3A), Zfp36l3 mRNA was the most highly expressed of the family members in the WT tissues, and it was not detected in the KO samples. There were no significant changes in expression of the other TTP family members, or of two internal control transcripts, Gapdh and Actb mRNA, in the KO samples. In the yolk sac, Zfp36l1 mRNA was the most highly expressed family member, followed by Zfp36 mRNA, and then Zfp36l3 and Zfp36l2 mRNA (Fig. 3B). Average mRNA levels for the family members other than Zfp36l3, and for Gapdh and Actb, were not significantly different in the two sets of samples.
Fig. 3.
Expression of TTP family member mRNAs in Zfp36l3 KO placentas. (A) Average transcript levels for the four TTP family members in E15.5 placentas from WT and KO mice, as determined by mRNA-Seq and expressed as FPKM. Each bar represents the average of five values, each determined from an individual placenta, ±s.d. Shown as internal controls are the equivalent values for Actb and Gapdh transcripts, after dividing the individual values by 20 and 5, respectively. With the exception of the Zfp36l3 transcript, the other WT versus KO pairs were not significantly different. (B) Similar results from the mRNA-Seq analysis of yolk sacs from E15.5 conceptuses, using the same format. Each bar represents the average of four determinations, ±s.d. With the exception of the Zfp36l3 transcript, none of the WT versus KO means was significantly different. Note that the vertical axis is the same for both graphs, so that the FPKM values can be compared between genotypes, among genes, and between tissues. Black bars, WT; white bars, KO.
Differentially expressed transcripts between placental WT and KO samples were identified using the EpiCenter tool (Huang et al., 2011). Briefly, transcripts with normalized read counts <100 were filtered out, and then a list of significant transcripts was generated using a cut-off of 5% false discovery rate (FDR). In the placenta, mean values were significantly different from each other using these criteria for a total of 711 transcript models. Of these, after removing duplicates, 91 transcripts were significantly increased in the KO placentas by >1.5-fold, and 72 were decreased by >1.5-fold, not including Zfp36l3 mRNA (Table S2).
In the yolk sacs, when we applied the same selection criteria, we found only five transcripts that were upregulated and one that was downregulated (besides Zfp36l3 mRNA, which was absent in the KO samples). The upregulated transcripts included glycogen synthase 2 mRNA (Gys2) (upregulated 2.41-fold); solute carrier family 39 (zinc transporter), member 4 (Slc39a4) (1.72); lectin, mannose-binding, 1 (Lman1), transcript variant 2 (1.67); centrosomal protein 290 (Cep290) (1.64); and RAP1 GTPase activating protein 2 (Rap1gap2) (1.54). Only one of these, Cep290 mRNA, contained copies of the core 7-mer TTP family member binding site, UAUUUAU, in its 3′UTR (seven copies). The one significantly downregulated transcript was solute carrier family 30 (zinc transporter), member 2 (Slc30a2) mRNA, which was downregulated by 1.77-fold. It also did not contain the core 7-mer of the ideal binding site.
Thus, in the placenta, Zfp36l3 mRNA was the most highly expressed family member in the WT tissue, and its deletion significantly affected the levels of many transcripts. By contrast, Zfp36l3 mRNA expression was lower than that of two other family members in the WT yolk sacs, and its deletion had few effects on mRNA levels in this tissue. It seems likely that the minimal effects of the Zfp36l3 deletion on yolk sac transcripts were due to the higher level expression of the other TTP family members in both the WT and KO tissues (Fig. 3B). For whatever reason, the effect of this genetic manipulation on gene expression in yolk sac appears to be minimal at this stage of development, and will not be discussed in detail further.
A gene ontology (GO) analysis of the significantly affected transcripts in placenta using DAVID (http://david.abcc.ncifcrf.gov/gene2gene.jsp) revealed that only a few functional clusters were significantly elevated with scores of >2. These included clusters related to lysosome vacuole activity (enrichment score 2.31) and to GTPase activity (2.27). For the downregulated transcripts, the GO analysis revealed several clusters that were significantly elevated, including organic ion transport (2.57); intracellular lumens (2.51); metal ion binding (2.25); and tRNA metabolic processes (2.21).
The complete set of transcripts that were significantly increased or decreased by >1.5-fold were then analyzed for the presence or absence of the core sequence of an optimal TTP family member binding site, the 7-mer UAUUUAU (Table S2). Of the 91 upregulated transcripts, 54 (59%) contained at least one 7-mer potential TTP family member binding site, many of which were conserved in other mammals; by contrast, only 5 of 72 (7%) of the downregulated transcripts contained one or more potential binding sites (Table S2).
To test whether some of the most highly affected transcripts could bind directly to ZFP36L3, we performed immunoprecipitations of ZFP36L3 in normal placenta at E15.5 (Keene et al., 2006), and analyzed associated mRNAs by real-time RT-PCR. Immunoprecipitations of this type brought down ZFP36L3 specifically (not shown). Real-time RT-PCR of the immunoprecipitated transcripts showed average enrichment of 156-, 57- and 82-fold for Hbegf, Lipg and Tfrc mRNAs, respectively, whereas Gapdh and Actb mRNAs were enriched three and eightfold, respectively. These data suggest that, at least for the specific mRNAs tested, there was relative enrichment with the immune serum compared with the pre-immune serum, supporting their identification as direct ZFP36L3-binding targets.
Seventy-two transcripts were significantly downregulated by >1.5-fold in the KO placenta (Table S2), not counting Zfp36l3 mRNA. We presume that most of these downregulation events occurred as secondary consequences of one or more of the upregulated transcript-encoded proteins. The most profoundly downregulated transcript (6.37-fold) was the product of the Tfrc gene, which encodes the type 1 transferrin receptor.
NanoString quantification of mRNA levels in WT, KO and Het placentas, and analysis of mRNA decay in trophoblast stem cells
To validate the changes in steady state mRNA levels found in the placentas by mRNA-Seq analysis, we applied NanoString nCounter analysis (Geiss et al., 2008) to four separate new sets of placentas: WT, KO, Het1 (KO allele comes from the female) and Het2 (KO allele comes from the male). In this way, in addition to comparing the conventional WT versus KO samples, we could compare samples from a genetically distinct pair of sample sets that would represent ‘effective’ WT and KO samples, despite their heterozygous states. The set of 160 transcripts analyzed was largely based on the potential targets listed in Table S2, but also included a number of previously identified or suspected targets of TTP (Brooks and Blackshear, 2013), as well as internal controls, controls representing rapidly decaying and stable mRNAs, Tfrc mRNA, and mRNAs of the other TTP family members. The mRNAs analyzed in these experiments are listed in Table S3.
We also wished to determine whether the ZFP36L3 deficiency affected the stability of potential target mRNAs. Because both the syncytiotrophoblast cells and trophoblast giant cells highly express ZFP36L3, we performed experiments on trophoblast stem cells (TSCs), which, when differentiated in culture, express markers of both syncytiotrophoblast and trophoblast giant cells (Simmons et al., 2008). These cells were isolated from Zfp36l3 WT and KO blastocysts, and then differentiated in culture as described in supplementary Materials and Methods. We performed NanoString analyses on the WT and KO cells before and after differentiation (Table S4), and on mRNA decay in these cells after differentiation (Table S5).
In the following sections, the NanoString data from both the new placental samples and the TSC samples will be presented together. We first evaluated mRNAs encoding the four TTP family members. As expected, Zfp36l3 mRNA levels were essentially undetectable in samples from both the KO and Het1 mice, whereas they were indistinguishable from WT mice in the Het2 placentas (Fig. 4A). For Zfp36 mRNA, there were no differences in mRNA levels in the four groups of placentas, but there were modest and significant increases in mRNA stability in the KO TSC actinomycin D decay curves (Fig. 4A). In the case of Zfp36l1 mRNA, there were no steady state changes in levels in the four groups of samples, and the decay curves were superimposable (Fig. 4A). In the case of Zfp36l2 mRNA, there were significant increases in steady state levels in the two ZFP36L3-deficient genotypes (Fig. 4A); these were accompanied by increases in the stability of this mRNA in the decay curves (Fig. 4A). The effect of ZFP36L3 deficiency on its own mRNA decay could not be examined in these experiments because of its absence in the KO cells, but this mRNA decayed rather slowly in the WT cells, with a decay half-life of >4 h (Fig. 4A). There was a striking, ∼20-fold increase in the levels of Zfp36l3 mRNA in the differentiated TSCs compared with their undifferentiated state: 7.0±2.5 (mean±s.d.) versus 143.4±4.2 normalized counts for the undifferentiated WT cells versus the differentiated WT cells (n=4 in each group).
Fig. 4.
Expression of TTP family member mRNAs in WT, KO and Het placentas, and decay in WT and KO TSCs. In this and subsequent figures, the left side of each panel shows the NanoString nCounter quantification of the indicated transcripts, expressed as mean±s.d. of five independent biological samples, from four sets of RNA samples: WT, KO, Het1 (H1; KO allele comes from the mother) and Het2 (H2; KO allele comes from the father). The right side of each panel represents the decay of each transcript, expressed as a percentage of the control mean at time 0, in differentiated TSCs 4 h after actinomycin D treatment. Each point represents the mean of four separate cultures, ±s.d. *P<0.05; **P<0.01, as determined by unpaired Student's t-tests. (A) Data of this type for the four TTP family members expressed in mouse placenta. (B) The same type of data for two transcripts known to be labile in other cell types, Fos and Myd116 mRNAs, and two transcripts known to be stable in other cell types, Actb and Gapdh mRNAs. There were no significant differences in the decay rates of the four transcripts shown in B.
We next evaluated four internal control transcripts, two that were selected for their extreme lability in other cells [Fos and Myd116 (Ppp1r15a) mRNAs], and two that were expected to be stable (Actb and Gapdh mRNAs). As shown in Fig. 4B, with the exception of Fos mRNA levels, which were significantly increased in the Het2 samples compared with the Het1 samples, all of the transcripts were present at similar levels in all four genotypes. In the TSC experiments, Fos and Myd116 mRNAs decayed rapidly, and Actb and Gapdh mRNA remained stable (Fig. 4B), during the 4 h experiment; results from the two genotypes were superimposable in all four cases. The data on the rapidly decaying transcripts suggest that the actinomycin D inhibition of transcription was working as expected in this experimental system.
We then segregated transcripts according to whether they were significantly different in four different comparisons: WT versus KO placenta; Het1 versus Het2 placenta; and WT versus KO TSCs at 2 and 4 h after actinomycin D. The complete datasets for these categorizations are contained in Tables S3 and S5. Several transcripts were removed from these tables because expression in the WT placenta samples was below the limits of detection of the assay, leaving 139 evaluable transcripts. We then divided the results into different categories based on the significance of the comparisons listed above; definitions of the different categories are contained in the legend to Table S3. For example, Category 1 transcripts (n=22) were significantly different in both placenta comparisons and at both the 2 h and 4 h time points of the decay time course.
We view these Category 1 transcripts as the most likely to be direct targets of ZFP36L3 in placenta; their results are summarized graphically in Fig. 5 (and Fig. 4A, showing Zfp36l2 mRNA). Most, if not all, of the 22 mRNAs in this category contained potential TTP family member binding sites, and a few of them were confirmed as direct binders to ZFP36L3 by co-immunoprecipitation. One transcript of interest in this group was Hbegf mRNA, encoding heparin binding epidermal growth factor; this was reasonably well expressed in placenta, and its 3′UTR contained two very well conserved potential TTP family member binding sites (Fig. S6A). Another top candidate was the Lipg mRNA, encoding an endothelial cell lipase. Its mRNA also contained two highly conserved 7-mer potential binding sites (Fig. S6B). Other noteworthy potential ZFP36L3 targets on this list included mRNAs coding for SLC38A3 (solute carrier family 38, member 3), LON peptidase N-terminal domain and ring finger 3 (LONRF3), the mRNA stability factor ELAVL1, steroidogenic acute regulatory protein (STAR), the low density lipoprotein receptor (LDLR), and at least two transcription factors, transcription factor CP2-like 1 (TCFCP2L1; also known as TFCP2L1) and Iroquois related homeobox 2 (IRX2). Despite the increased levels and stability of the STAR transcript, we found no differences in levels of either progesterone or corticosterone in placentas from E15.5 KO mice (data not shown).
Fig. 5.
Category 1 transcripts. The organization of the figure is as described in the legend to Fig. 4. This figure contains data from all the Category 1 transcripts except Zfp36l2, which was described separately in Fig. 4A. Note that although the Tfrc mRNA in the bottom right hand corner is profoundly decreased in both the Zfp36l3 KO and H1 genotypes, its mRNA was nonetheless significantly stabilized in the absence of ZFP36L3. Note: Dos is also known as Cbarp.
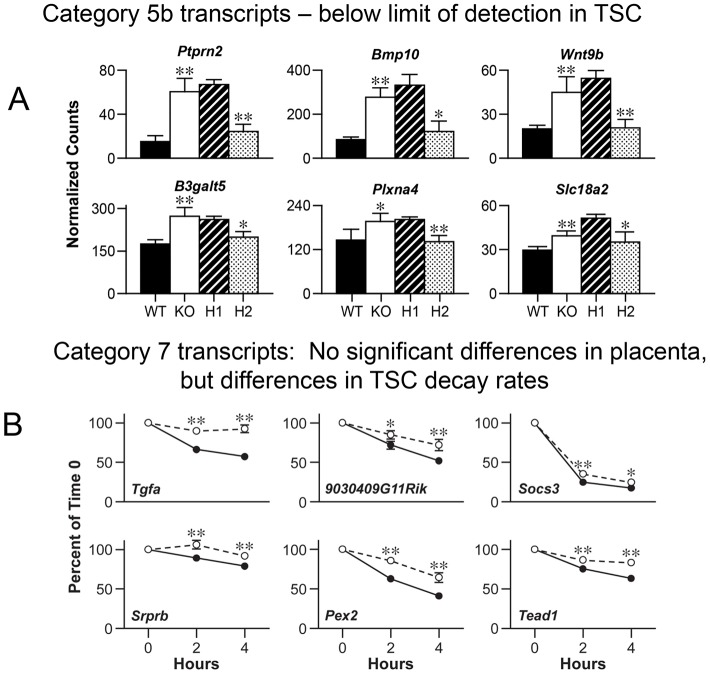
We view the other categories as less likely to contain direct ZFP36L3 targets, but two of the other categories will be highlighted here. In one case (Category 5B), both placenta comparisons were significant, but the expression levels of the transcripts in the TSCs were below the limits of the assay detection (Fig. 6A). This group contained other interesting transcripts, including those encoding protein tyrosine phosphatase, receptor type, N polypeptide 2 (PTPRN2); bone morphogenetic protein 10 (BMP10); the transcription factor wingless-type MMTV integration site family, member 9B (WNT9B); UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 5 (B3GALT5), transcript variant 2; plexin A4 (PLXNA4); and solute carrier family 18 (vesicular monoamine), member 2 (SLC18A2).
Fig. 6.
Category 5b and 7 transcripts. (A,B) The organization of the histograms and decay curves is as described in the legend to Fig. 4. However, this figure contains data from placenta measurements of all of the Category 5b transcripts (A), which were below the limits of the assay detection in the TSCs, and all of the Category 7 transcripts (B), which showed significant differences in decay rates but no significant differences in placenta.
Another interesting category (Category 7) contains transcripts that were stabilized in the TSCs in the absence of ZFP36L3, but were not significantly affected in the ZFP36L3-deficient placentas (Fig. 6B). In one case, that of transforming growth factor alpha mRNA (Tgfa mRNA), the decay rate differences were striking, and significant at both time points; there was an average upregulation of 1.4-fold in both placental comparisons, but these did not achieve statistical significance. In the case of peroxisomal biogenesis factor 2 (Pex2 mRNA), there were significant differences in decay, but these were not reflected in changes in the two placental comparisons (Fig. 6B), suggesting the possibility of compensatory changes in transcription.
Finally, there is the interesting example of the Tfrc mRNA, a member of the Category 1 class of transcripts. As shown in Fig. 5, this transcript was strikingly downregulated in both types of ZFP36L3-deficient placenta, confirming the previous mRNA-Seq results. This was accompanied by parallel changes in the immunoreactive protein (Fig. S7A). Despite these decreases in expression in the placenta, its mRNA was significantly stabilized in the ZFP36L3-deficient TSCs (Fig. 5). The Tfrc transcript was nicely induced in the differentiated TSCs (Fig. S7B), and steady state levels were significantly decreased in both undifferentiated and differentiated ZFP36L3-deficient TSCs, although not to the same extent as in placenta. Its mRNA contained two nearly ideal TTP family member binding sites in its 3′UTR that are highly conserved among mammals (Fig. S7C). As noted above, immunoprecipitation with ZFP36L3 antiserum pulled down more Tfrc mRNA than a pre-immune serum. Thus, despite evident stabilization of the Tfrc mRNA in the absence of ZFP36L3 in the TSCs, the ZFP36L3 deficiency caused profound decreases in Tfrc mRNA and protein levels, possibly by inhibiting transcription. By contrast, in the yolk sac mRNA-Seq experiments, the Tfrc mRNA was increased by 24% in the ZFP36L3-deficient yolk sacs, a difference that was not significant.
Immunohistochemistry of TFRC in the WT placenta showed remarkable colocalization of both TFRC and ZFP36L3 in the syncytiotrophoblast cells of the labyrinth zone (Fig. 7), suggesting an ‘intracrine’ effect.
Fig. 7.
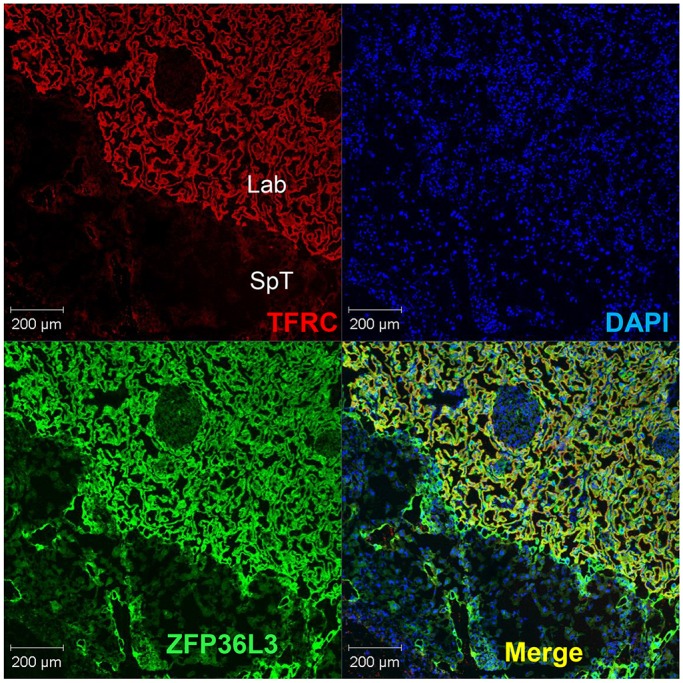
Immunostaining of ZFP36L3 and TFRC in WT placenta at E15.5. Frozen placental sections were stained with rat anti-mouse TFRC (CD71) antibody (1:500) and rabbit anti-ZFP36L3 antibody (1:1000), with secondary antibodies donkey anti-rat IgG Alexa Fluor-594 (red for CD71) and donkey anti-rabbit IgG Alexa Fluor-488 (green for ZFP36L3). Nuclear staining with DAPI is also shown (blue). The figure is focused on a region at the border between the spongiotrophoblast (SpT) and labyrinth (Lab) zones. The panel labeled ‘Merge’ shows the colocalization (yellow) of TFRC and ZFP36L3 in the syncytiotrophoblast cells of the labyrinth zone.
Fetal element analysis
To determine if the decreased placental TFRC expression resulted in decreased iron accumulation in the fetus, we measured various elements in whole KO (n=8) and WT (n=9) fetuses from three litters at E15.5 using inductively coupled plasma atomic emission spectroscopy (ICP-AES). Of the 20 elements measured, only iron, zinc and calcium were significantly changed: iron was decreased by 29% (P=1.8×10−4), zinc by 10% (P=7×10−6), and calcium was decreased by 13% with marginal significance (P=0.044). A second similar experiment was performed at E15.5, in which five littermate fetuses were compared for element concentration (three KO, two WT). In this experiment, iron was decreased by 31% in the KO (P=0.033), whereas zinc was decreased by 7% (not significant) and calcium was unchanged. These data suggest that the decrease in TFRC expression seen in the ZFP36L3-deficient placentas resulted in decreased overall iron accumulation in the fetus.
DISCUSSION
The Zfp36l3 gene is unique within the TTP family of genes in that it is an imprinted X chromosomal gene, expression of which is limited to the placenta and yolk sac of certain rodents, but not other mammals (Blackshear et al., 2005; Frederick et al., 2008; Gingerich et al., 2016). As we show here, its complete deficiency in the mouse leads to significant decreases in the numbers of surviving offspring. If born alive, the KO mice appear to develop normally, without any apparent adult abnormalities, and KO mice of both sexes are fertile. These data suggest that Zfp36l3 in some way promotes peripartum survival, and presumably confers a survival advantage to mice and other rodents that express it.
This relatively mild reproductive phenotype was accompanied by a lack of morphological or histological abnormalities in the placentas of KO mice. This allowed for molecular comparisons of WT and KO placentas from the same litters, in the absence of any gross anatomical abnormalities in the KO placenta.
Our working hypothesis was that ZFP36L3-deficient placentas would exhibit increases in some mRNAs that would be direct targets of ZFP36L3. In addition, there would be changes in steady state levels of many transcripts that could be secondary to the primary increases in direct mRNA targets, as well as tertiary responses, etc. To try to increase the probability that an individual transcript was a direct target, we analyzed potential targets in two types of placenta comparisons, between WT and null or ‘effective null’ animals, as well as in mRNA decay assays in TSCs. These and other analyses allowed us to generate lists of high probability target transcripts for ZFP36L3.
An example of one of the top ranked transcripts was the Hbegf mRNA, which was significantly increased 3.15-fold in the KO placentas in the mRNA-Seq study, and was enriched 156-fold in the immunoprecipitation experiment. It contains three core binding sites for TTP family proteins, which are conserved among several mammalian species. We confirmed by immunohistochemistry that the HBEGF protein was expressed in the same syncytiotrophoblast cells that express the ZFP36L3 protein (data not shown). In the NanoString assays, we confirmed the increase in Hbegf mRNA expression in both the KO versus WT comparison, as well as the Het1 versus Het2 comparison (2.62- and 2.48-fold increases, respectively). Finally, Hbegf mRNA decayed significantly more slowly in TSCs isolated from KO blastocysts. All of these data suggest that this transcript is serving as a direct target of ZFP36L3 in this tissue.
HBEGF is a secreted member of the epidermal growth factor (EGF) family of growth factors, and is well known in placental physiology as a factor that promotes trophoblast invasiveness, and also prevents hypoxemic damage (Imudia et al., 2008; Jessmon et al., 2010, 2009; Leach et al., 2008; Lim and Dey, 2009). To our knowledge, the regulation of its expression by modulated mRNA decay has not been evaluated previously. Interestingly, the mRNA encoding another member of the EGF agonist family, TGFA, was also strikingly stabilized in the ZFP36L3-deficient TSCs. This growth factor is also thought to be involved in trophoblast proliferation and invasion (Lysiak et al., 1993), and both HBEGF and TGFA have been shown to be decreased in placentas from patients with pre-eclampsia (Armant et al., 2015).
A second high probability target was Lipg mRNA, which encodes an endothelial lipase. This was highly expressed in the WT placenta, and its levels were significantly increased 2.59-fold in the KO placenta; it was also enriched by 57-fold in the ZFP36L3 immunoprecipitates. Its 3′UTR contains two highly conserved, closely spaced potential ZFP36L3-binding sites. This lipase has been shown to be preferentially involved in the breakdown of phospholipids, and is important for high-density lipoprotein metabolism (Lindegaard et al., 2005). It is expressed in human syncytiotrophoblast cells, and has been shown to be dysregulated in intrauterine growth retardation and gestational diabetes with obesity (Gauster et al., 2007, 2011). It is thought to be important for the hydrolysis and uptake of lipids in maternal circulation by the placenta to aid in fetal growth and development.
Many other potential ZFP36L3 targets transcripts were identified, as described in detail in the Results section. However, we also identified many transcripts that changed significantly in the KO placentas that did not seem likely to be direct ZFP36L3 targets, particularly the downregulated transcripts. These are most likely to be a result of secondary expression changes, perhaps in some cases from increases in transcription factor mRNAs that were directly stabilized in the absence of ZFP36L3. The most strikingly downregulated transcript was that encoding the type 1 transferrin receptor TFRC. This receptor is the only transferrin receptor expressed in placenta, and as such is solely responsible for iron uptake by the placenta for the developing fetus (Cetin et al., 2011). The Tfrc transcript was downregulated by 6.37-fold in the KO placentas in the mRNA-Seq experiment, with a similar decrease in protein levels confirmed by western blotting and immunohistochemistry. The NanoString assays of completely different placental samples of two genotypes showed that there was a 5.9-fold downregulation in the KO placentas compared with WT, and a 9.1-fold downregulation in the Het1 placentas compared with Het2. The protein was expressed in the same syncytiotrophoblast cells as expressed the ZFP36L3 protein, suggesting the possibility of an effect of ZFP36L3 on the expression of TFRC in the same cells, or an intracrine effect; however, we cannot exclude autocrine or paracrine effects leading to the same result.
Nonetheless, the Tfrc transcript was stabilized in the ZFP36L3-deficient TSCs, its 3′UTR contains two highly conserved and closely spaced 8-mer potential ZFP36L3-binding sites, and it was highly enriched in the ZFP36L3 immunoprecipitates. It was previously identified as a possible direct TTP target in other tissues (Bayeva et al., 2012). These data suggest that the profound decrease in Tfrc mRNA levels might be a secondary effect of ZFP36L3 deficiency resulting in inhibition of transcription of Tfrc in the placenta.
The observed profound decrease of TFRC in the syncytiotrophoblast cells is likely to have an effect on iron uptake by the fetus. These cells are located between the maternal and fetal blood spaces in the labyrinth zone, and express other transporters important for placental nutrient uptake. Decreased in utero iron uptake because of maternal iron deficiency anemia is a common global health problem, and is thought to result in a variety of long-term consequences for the offspring, including cognitive defects (Carter et al., 2010; Congdon et al., 2012). We found that the ZFP36L3-deficient pups contained ∼30% less iron than their WT littermates, a finding confirmed in a second study that compared littermates. It will be of great interest to determine whether this in utero experience of decreased iron uptake will lead to any lasting consequences in surviving ZFP36L3-deficient offspring. In addition, the markedly decreased expression of TFRC in the placental cells from the KO mice could have led to the decreased rates of fetal survival observed, as it has previously been shown that Tfrc−/− mice die at about E12.5 and Tfrc+/− mice exhibit abnormalities in hematopoiesis and iron homeostasis (Levy et al., 1999).
This mouse model is of potential interest in studies of mRNA turnover, regulation of nutrient and element uptake by the placenta, intrauterine programming, imprinting, and others. However, the fact that there is no orthologous gene in non-rodent mammals limits the attractiveness of this model for studies of human placental physiology. One possibility is that one or more of the three remaining TTP family members expressed in the placenta of humans and other non-rodent mammals can assume the role of ZFP36L3 in modulating placental gene expression and physiology during development. We suggest that the TTP family member ZFP36L1 is the best candidate for this position for several reasons. First, it is the only family member (besides ZFP36L3) for which a deficiency is known to cause defects in the mouse placenta, leading to failure of chorioallantoic fusion (Stumpo et al., 2004). Second, deep sequencing experiments have shown that its mRNA levels are the highest of the three family members found in human placenta (Saben et al., 2014). Finally, Zfp36l1 mRNA is the most highly expressed of the family members in the placentas from the rodent Nannospalax galili, a species very close evolutionarily to the rodents that express ZFP36L3, but which apparently does not express ZFP36L3 in the placenta (Gingerich et al., 2016). Although there are no good cellular models available of human syncytiotrophoblast cells, it might be possible to do specific mRNA knockdown experiments in either human trophoblast or choriocarcinoma cell lines that have been used to model placental transport systems in previous experiments (Ikeda et al., 2011; Lager et al., 2011). Alternatively, immunoprecipitation of TTP family members from human placental extracts might point to one or more of the proteins associating with the human version of the transcripts identified here. Attempts to identify possible human ‘functional orthologs’ of ZFP36L3 will be an important future goal of this work.
MATERIALS AND METHODS
Mice
Detailed procedures for the generation and characterization of the Zfp36l3 knockout mice analyzed in this study are contained in supplementary Materials and Methods. The embryonic stem cells (ESCs) and mice used in these experiments were both on 100% C57BL/6 backgrounds. The trophoblast stem cells (TSCs) used in these studies were generated as described (Tanaka et al., 1998), and differentiated as described in supplementary Materials and Methods. All mouse experiments were conducted according to US Public Health Service Policy on the humane care and use of laboratory animals. The National Institute of Environmental Health Sciences Institutional Animal Care and Use Committee approved all animal procedures used in this study.
RNA techniques
Methods for RNA isolation, northern blotting, mRNA-Seq analyses, NanoString n-counter analyses, real-time RT-PCR, and RNA co-immunoprecipitation are described in detail in supplementary Materials and Methods.
Immunohistochemistry
For routine paraffin section immunohistochemistry of ZFP36L3 in yolk sac and placenta, tissues were processed, embedded, sectioned and stained as described previously (Blackshear et al., 2005; Frederick et al., 2008). Detailed methods for the immunostaining of frozen sections for confocal microscopy can be found in supplementary Materials and Methods.
Element analysis
Fetal mice were removed at E15.5 from three pregnant dams, quick frozen in liquid nitrogen and stored at −80°C until genotyping could be performed. At that point, nine WT and eight KO fetuses from three litters were used for total element analysis, using a panel of 20 elements, performed at the University of Georgia Chemical Analysis Laboratory at the Center for Applied Isotope Studies. The samples were ashed and then subjected to inductively coupled plasma atomic emission spectroscopy, according to EPA method 6010C. See http://www.cais.uga.edu/analytical_services/chemical_analysis/services.htm for further details.
Steroid hormone analysis
Placentas from E15.5 mice were frozen and used for analysis of progesterone and corticosterone concentrations at the University of Virginia Ligand Assay and Analysis Core of the Center for Research in Reproduction (https://med.virginia.edu/research-in-reproduction/ligand-assay-analysis-core/).
Acknowledgements
We are grateful to Rebecca Auxier of the University of Georgia for performing the element analysis; Liwen Liu and Grace Kissling of the Biostatics Branch, NIEHS, for help with the Gene Ontology analysis and statistical analysis of breeding data, respectively; Julie Foley for help with the analysis of yolk sac sections; Dee Wenzel for animal husbandry and breeding support; and the staff of the Laboratory of Experimental Pathology for some of the immunohistochemistry. We especially thank Kathleen Caron and Jay Cross for helpful comments on the manuscript.
Footnotes
Competing interests
D. Bortner and K. Kluckman are employed by, have equity ownership in, and serve on the board of directors of TransViragen.
Author contributions
D.J.S., K.K., D.M.B. and P.J.B. designed the experiments; D.J.S., C.S.T., C.J.T., K.K. and D.M.B. performed the experiments; D.J.S., W.H., L.L. and P.J.B. analyzed the data; D.J.S. and P.J.B. wrote the manuscript.
Funding
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Environmental Health Sciences. Deposited in PMC for release after 12 months.
Data availability
mRNA-Seq datasets have been deposited in Gene Expression Omnibus (GEO) under accession numbers GSE66137 and GSE66138. NanoString datasets have been deposited in GEO under accession number GSE79767.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.130369/-/DC1
References
- Armant D. R., Fritz R., Kilburn B. A., Kim Y. M., Nien J. K., Maihle N. J., Romero R. and Leach R. E. (2015). Reduced expression of the epidermal growth factor signaling system in preeclampsia. Placenta 36, 270-278. 10.1016/j.placenta.2014.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayeva M., Khechaduri A., Puig S., Chang H.-C., Patial S., Blackshear P. J. and Ardehali H. (2012). mTOR regulates cellular iron homeostasis through tristetraprolin. Cell Metab. 16, 645-657. 10.1016/j.cmet.2012.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bermejo-Alvarez P., Ramos-Ibeas P. and Gutierrez-Adan A. (2012). Solving the ‘X’ in embryos and stem cells. Stem Cells Dev. 21, 1215-1224. 10.1089/scd.2011.0685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear P. J. and Perera L. (2014). Phylogenetic distribution and evolution of the linked RNA-binding and NOT1-binding domains in the tristetraprolin family of tandem CCCH zinc finger proteins. J. Interferon Cytokine Res. 34, 297-306. 10.1089/jir.2013.0150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackshear P. J., Phillips R. S., Ghosh S., Ramos S. B. V., Richfield E. K. and Lai W. S. (2005). Zfp36l3, a rodent X chromosome gene encoding a placenta-specific member of the Tristetraprolin family of CCCH tandem zinc finger proteins. Biol. Reprod. 73, 297-307. 10.1095/biolreprod.105.040527 [DOI] [PubMed] [Google Scholar]
- Brooks S. A. and Blackshear P. J. (2013). Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim. Biophys. Acta 1829, 666-679. 10.1016/j.bbagrm.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E. and Blackshear P. J. (2001). Roles of tumor necrosis factor-alpha receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 98, 2389-2395. 10.1182/blood.V98.8.2389 [DOI] [PubMed] [Google Scholar]
- Carballo E., Lai W. S. and Blackshear P. J. (1998). Feedback inhibition of macrophage tumor necrosis factor-alpha production by tristetraprolin. Science 281, 1001-1005. 10.1126/science.281.5379.1001 [DOI] [PubMed] [Google Scholar]
- Carballo E., Lai W. S. and Blackshear P. J. (2000). Evidence that tristetraprolin is a physiological regulator of granulocyte-macrophage colony-stimulating factor messenger RNA deadenylation and stability. Blood 95, 1891-1899. [PubMed] [Google Scholar]
- Carter R. C., Jacobson J. L., Burden M. J., Armony-Sivan R., Dodge N. C., Angelilli M. L., Lozoff B. and Jacobson S. W. (2010). Iron deficiency anemia and cognitive function in infancy. Pediatrics 126, e427-e434. 10.1542/peds.2009-2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin I., Berti C., Mandò C. and Parisi F. (2011). Placental iron transport and maternal absorption. Ann. Nutr. Metab. 59, 55-58. 10.1159/000332133 [DOI] [PubMed] [Google Scholar]
- Congdon E. L., Westerlund A., Algarin C. R., Peirano P. D., Gregas M., Lozoff B. and Nelson C. A. (2012). Iron deficiency in infancy is associated with altered neural correlates of recognition memory at 10 years. J. Pediatr. 160, 1027-1033. 10.1016/j.jpeds.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian M. R., Frank F., Rouya C., Siddiqui N., Lai W. S., Karetnikov A., Blackshear P. J., Nagar B. and Sonenberg N. (2013). Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin. Nat. Struct. Mol. Biol. 20, 735-739. 10.1038/nsmb.2572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick E. D., Ramos S. B. V. and Blackshear P. J. (2008). A unique C-terminal repeat domain maintains the cytosolic localization of the placenta-specific tristetraprolin family member ZFP36L3. J. Biol. Chem. 283, 14792-14800. 10.1074/jbc.M801234200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauster M., Hiden U., Blaschitz A., Frank S., Lang U., Alvino G., Cetin I., Desoye G. and Wadsack C. (2007). Dysregulation of placental endothelial lipase and lipoprotein lipase in intrauterine growth-restricted pregnancies. J. Clin. Endocrinol. Metab. 92, 2256-2263. 10.1210/jc.2006-2403 [DOI] [PubMed] [Google Scholar]
- Gauster M., Hiden U., van Poppel M., Frank S., Wadsack C., Hauguel-de Mouzon S. and Desoye G. (2011). Dysregulation of placental endothelial lipase in obese women with gestational diabetes mellitus. Diabetes 60, 2457-2464. 10.2337/db10-1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiss G. K., Bumgarner R. E., Birditt B., Dahl T., Dowidar N., Dunaway D. L., Fell H. P., Ferree S., George R. D., Grogan T. et al. (2008). Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 26, 317-325. 10.1038/nbt1385 [DOI] [PubMed] [Google Scholar]
- Gingerich T. J., Stumpo D. J., Lai W. S., Randall T. A., Steppan S. J. and Blackshear P. J. (2016). Emergence and evolution of Zfp36l3. Mol. Phylogenet. Evol. 94, 518-530. 10.1016/j.ympev.2015.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Umbach D. M., Vincent Jordan N., Abell A. N., Johnson G. L. and Li L. (2011). Efficiently identifying genome-wide changes with next-generation sequencing data. Nucleic Acids Res. 39, e130 10.1093/nar/gkr592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K., Utoguchi N., Tsutsui H., Yamaue S., Homemoto M., Nakao E., Hukunaga Y., Yamasaki K., Myotoku M. and Hirotani Y. (2011). In vitro approaches to evaluate placental drug transport by using differentiating JEG-3 human choriocarcinoma cells. Basic Clin. Pharmacol. Toxicol. 108, 138-145. 10.1111/j.1742-7843.2010.00634.x [DOI] [PubMed] [Google Scholar]
- Imudia A. N., Kilburn B. A., Petkova A., Edwin S. S., Romero R. and Armant D. R. (2008). Expression of heparin-binding EGF-like growth factor in term chorionic villous explants and its role in trophoblast survival. Placenta 29, 784-789. 10.1016/j.placenta.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessmon P., Leach R. E. and Armant D. R. (2009). Diverse functions of HBEGF during pregnancy. Mol. Reprod. Dev. 76, 1116-1127. 10.1002/mrd.21066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessmon P., Kilburn B. A., Romero R., Leach R. E. and Armant D. R. (2010). Function-specific intracellular signaling pathways downstream of heparin-binding EGF-like growth factor utilized by human trophoblasts. Biol. Reprod. 82, 921-929. 10.1095/biolreprod.109.082305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J. D., Komisarow J. M. and Friedersdorf M. B. (2006). RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 1, 302-307. 10.1038/nprot.2006.47 [DOI] [PubMed] [Google Scholar]
- Lager S., Jansson N., Olsson A. L., Wennergren M., Jansson T. and Powell T. L. (2011). Effect of IL-6 and TNF-alpha on fatty acid uptake in cultured human primary trophoblast cells. Placenta 32, 121-127. 10.1016/j.placenta.2010.10.012 [DOI] [PubMed] [Google Scholar]
- Lai W. S., Kennington E. A. and Blackshear P. J. (2003). Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol. Cell. Biol. 23, 3798-3812. 10.1128/MCB.23.11.3798-3812.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W. S., Carrick D. M. and Blackshear P. J. (2005). Influence of nonameric AU-rich tristetraprolin-binding sites on mRNA deadenylation and turnover. J. Biol. Chem. 280, 34365-34377. 10.1074/jbc.M506757200 [DOI] [PubMed] [Google Scholar]
- Leach R. E., Kilburn B. A., Petkova A., Romero R. and Armant D. R. (2008). Diminished survival of human cytotrophoblast cells exposed to hypoxia/reoxygenation injury and associated reduction of heparin-binding epidermal growth factor-like growth factor. Am. J. Obstet. Gynecol. 198, 471.e471-471.e477; discussion 471.e477-478 10.1016/j.ajog.2008.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J. E., Jin O., Fujiwara Y., Kuo F. and Andrews N. (1999). Transferrin receptor is necessary for development of erythrocytes and the nervous system. Nat. Genet. 21, 396-399. 10.1038/7727 [DOI] [PubMed] [Google Scholar]
- Lim H. J. and Dey S. K. (2009). HB-EGF: a unique mediator of embryo-uterine interactions during implantation. Exp. Cell Res. 315, 619-626. 10.1016/j.yexcr.2008.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindegaard M. L. S., Nielsen J. E., Hannibal J. and Nielsen L. B. (2005). Expression of the endothelial lipase gene in murine embryos and reproductive organs. J. Lipid Res. 46, 439-444. 10.1194/jlr.M400417-JLR200 [DOI] [PubMed] [Google Scholar]
- Lysiak J. J., Han V. K. and Lala P. K. (1993). Localization of transforming growth factor alpha in the human placenta and decidua: role in trophoblast growth. Biol. Reprod. 49, 885-894. 10.1095/biolreprod49.5.885 [DOI] [PubMed] [Google Scholar]
- Phillips R. S., Ramos S. B. V. and Blackshear P. J. (2002). Members of the tristetraprolin family of tandem CCCH zinc finger proteins exhibit CRM1-dependent nucleocytoplasmic shuttling. J. Biol. Chem. 277, 11606-11613. 10.1074/jbc.M111457200 [DOI] [PubMed] [Google Scholar]
- Saben J., Zhong Y., McKelvey S., Dajani N. K., Andres A., Badger T. M., Gomez-Acevedo H. and Shankar K. (2014). A comprehensive analysis of the human placenta transcriptome. Placenta 35, 125-131. 10.1016/j.placenta.2013.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons D. G., Natale D. R. C., Begay V., Hughes M., Leutz A. and Cross J. C. (2008). Early patterning of the chorion leads to the trilaminar trophoblast cell structure in the placental labyrinth. Development 135, 2083-2091. 10.1242/dev.020099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo D. J., Byrd N. A., Phillips R. S., Ghosh S., Maronpot R. R., Castranio T., Meyers E. N., Mishina Y. and Blackshear P. J. (2004). Chorioallantoic fusion defects and embryonic lethality resulting from disruption of Zfp36L1, a gene encoding a CCCH tandem zinc finger protein of the Tristetraprolin family. Mol. Cell. Biol. 24, 6445-6455. 10.1128/MCB.24.14.6445-6455.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpo D. J., Broxmeyer H. E., Ward T., Cooper S., Hangoc G., Chung Y. J., Shelley W. C., Richfield E. K., Ray M. K., Yoder M. C. et al. (2009). Targeted disruption of Zfp36l2, encoding a CCCH tandem zinc finger RNA-binding protein, results in defective hematopoiesis. Blood 114, 2401-2410. 10.1182/blood-2009-04-214619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Kunath T., Hadjantonakis A.-K., Nagy A. and Rossant J. (1998). Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072-2075. 10.1126/science.282.5396.2072 [DOI] [PubMed] [Google Scholar]
- Taylor G. A., Carballo E., Lee D. M., Lai W. S., Thompson M. J., Patel D. D., Schenkman D. I., Gilkeson G. S., Broxmeyer H. E., Haynes B. F. et al. (1996). A pathogenetic role for TNF alpha in the syndrome of cachexia, arthritis, and autoimmunity resulting from tristetraprolin (TTP) deficiency. Immunity 4, 445-454. 10.1016/S1074-7613(00)80411-2 [DOI] [PubMed] [Google Scholar]