Abstract
Cancer is a dreadful disease constituting abnormal growth and proliferation of malignant cells in the body. Next to lung cancer, breast cancer is the most common form of cancer affecting women. The apoptotic pathway regulators, B cell lymphoma family of protein, play a key role in various malignancies defining cancer and their constitutive expression plays an integral role in breast cancer chemotherapy. The research work discusses the identification and molecular cloning of a B cell lymphoma like gene from human breast cancer cell line. The open reading frame of the gene consisted of 965 nucleotides, encoding a protein of 380 amino acids with a predicted molecular weight of 42.5 kilodalton. The predicted physiochemical properties of the gene were as follows: Isoelectric point - 9.49, molecular formula - C1893H3004N534O548S16, total number of negatively charged residues, (Aspartate+Glutamate) - 26, total number of positively charged residues, (Arginine+Lysine)-39, instability index-42.08 (unstable protein) and grand average of hydropathicity is -0.202. Additionally, phobius prediction suggested non-cytoplasmic localization of the putative protein. The presence of secondary structure in the protein was determined by Memsat program. A 3 dimensional protein homology model was generated using threading based method of protein modeling for structural and functional annotation of the putative protein. Future prospects accounts for the biochemical characterization of the enzyme including in vitro assays on breast cancer cell line would establish the functional characteristics of the protein and its physiological mechanisms in breast cancer development and its therapeutic-target role in future.
Keywords: Apoptosis, breast cancer, Bcl-2, molecular cloning, MCF-7, phylogenetics, protein homology modeling
Cancer, defined as malignant neoplasm of cells and tissues, affect millions and is one of the leading causes of death worldwide. According to published report of International Agency for Research on Cancer (IARC) of the World Health Organization/IARC GLOBOCAN database (2013), 14.1 million new cancer cases were registered and approximately 8.2 million cancer-related deaths were reported worldwide in 2013. The prevalence rate indicated that 32.6 million people were diagnosed with cancer in the past five years. The percentage statistics of the diagnosed cancer types are estimated according to the frequency of occurrence: Lung cancer (1.8 million, 13.0% of the total), breast cancer (1.7 million, 11.9%) and colorectal cancer (1.4 million, 9.7%), respectively. Additionally, the future projection rates of global occurrence of cancer are estimated to be 19.3 million new cases per year by 2025, with a higher frequency in developing countries. On the physiological platform, the disease is characterized by uncontrolled growth and proliferation of abnormal cells, gradually affecting major organs of the body. The disease may result from internal factors (mutations in genome, hormones) and external factors (chemicals, tobacco, radiation). All cancers are caused by mutations or malfunctioning of genes that control cell growth and division. About 5% of all cancers are inherited while most of the cancers are caused due to somatic gene mutations[1]. Furthermore, the spread of malignant cells to other organs, a process known as metastasis, is responsible for 90% cancer deaths in the world[2].
Considering the rising frequency of occurrence, breast cancer is a leading cause of mortality in women leading to 5.22 lakh deaths in 2012 worldwide. According to a survey, approximately 1.7 million women were diagnosed and 6.3 million women are living with breast cancer[3]. Past decades have witnessed the prevalence of breast cancer by more than 20% and mortality rate by 14%[4]. Breast cancer is the second major cancer occurring in women (after lung cancer)[5] and is a global cause of concern. According to a survey, the global mortality rate of breast cancer was 438,000 women in 2010 and estimated to reach 747,802 women in 2030[6,7]. Several reasons can be attributed for the development of breast cancer, from genetic predispositions like obesity, genetic defects in breast cancer susceptible genes namely BRCA 1 and BRCA 2 and immunity to environmental agents like harmful radiation as well as exposures to hormonal therapies including progestin and estrogen hormones. Use of cancer cell lines is a prerequisite requirement for development of in vitro systems in research pertaining to breast cancer studies and drug development. MCF-7 is a breast cancer cell line isolated in 1970 from a 69 year old Caucasian woman from a pleural effusion[8] and named after Michigan Cancer Foundation; the cell line is used as cellular model for breast cancer studies. An alteration in normal course of programmed cell death mechanisms plays a critical role in the development and progression of breast cancer. The Bcl-2 family of proteins acts as the key regulators of apoptotic pathway in cancer. The Bcl-2 gene family was first identified through its involvement in B-cell lymphomas and Bcl-2 expression is altered by a chromosomal translocation between 14th and 18th chromosome in humans[9]. Bcl-2 protein family plays a key role in regulation of apoptosis including necrosis and autophagy[10,11]. The overexpression of antiapoptotic gene of the Bcl-2 family namely Bcl-2 and Bcl-xL is responsible for resistance to breast cancer chemotherapy while proapoptotic proteins like Bax promote apoptosis[12]. Studies have reported that Bcl-2 can prevent apoptosis initiation by a wide variety of stimuli, namely (i) neurotrophic factor withdrawal from neurons; (ii) chemotherapeutic drugs and gamma irradiation in cancer cells; (iii) cytotoxic cytokines such as tumor necrosis factor-α, Fas-ligand, and transforming growth factor-β (iv) chemicals that induce oxidative injury (v) heat shock; and (vi) calcium ionophores[13,14]. It has been suggested that Bcl-2 protein might regulate a final event in the common pathway involved in programmed and apoptotic cell death. Bcl-2 gene expression in mammary epithelial cells and in estrogen receptor (ER)-positive breast cancer cell lines is regulated by estrogens[15]. It is interesting to note that statistical analysis had revealed that breast cancer patients with positive expression of Bcl-2 show better survival rate as compared to Bcl-2 negative patients[16]. Several members of the gene family have been studied from animal species namely chicken[17], mouse[18], yeast artificial chromosome (YAC) clones, human T lymphoid cell line[19], rat[20,21] and chicken lymphoid cells[22].
The research discusses the isolation and molecular cloning of a Bcl-2 like gene from breast cancer cell line, MCF-7. In silico characterization of the putative protein predicted the physiochemical properties and protein homology modeling was employed to elucidate three dimensional structure of the putative protein. Future prospects of the study focusing on the biochemical characterization of enzyme through in vitro assays would help to understand the molecular mechanism underlying breast cancer and in the development of target-based therapeutic interventions against the disease.
MATERIALS AND METHODS
Cryopreservation and maintenance of cell lines:
The breast cancer cell line, MCF-7 was obtained from King George Medical University, Lucknow and maintained and stored in a cryopreservation cylinder at -196° (Nalgene). General chemicals and reagents were purchased from Sigma-Aldrich, Sisco Research Lab Ltd. and Gibco/Life Technologies. Molecular biology kits were obtained from Qiagen, Promega and Invitrogen while the restriction enzymes used were from Fermentas. Antibiotics namely penicillin and streptomycin were products of Sigma-Aldrich. The cell lines were stored in a cryopreservation cylinder at -196° and sub-cultured every week in Dulbecco modified Eagle's Medium (dMEM) according to growth of cancer cells (confluency is 50% of the culture flask) in laminar air flow chamber (Heraeus Instruments). The cells were given a quick washing with phosphate buffer saline (PBS) and proteinized with trypsin. Cells were counted in haemocytometer slide and cell density (1×105 cells/25 cm2 flask approximately) were inoculated in fresh dMEM media. The culture flask was incubated in CO2 incubator at 37°.
Genomic DNA isolation and PCR amplification:
Genomic DNA was isolated from breast cancer cells, MCF-7 by Axygen DNA Isolation Kit as per manufacturer's instructions. Nanodrop spectrophotometer was used for the qualitative and quantitative estimation of the genomic DNA by measuring relative absorbance values at 260 and 280 nm (A260/A280) and agarose gel (1.2%) electrophoresis. The PCR conditions were initial denaturation at 94° for 5 min, followed by 45 cycles at 94° for 1 min, annealing at 59.8° for 1 min, extension at 72° for 2 min and final extension at 72° for 5 min. The RT-PCR products were subjected to gel electrophoresis on 1.2% agarose gel.
Molecular cloning of Bcl-2 gene:
The resolved fragments were purified through Qiagen columns and cloned in pGEMT vector (Promega) according to manufacturer's instructions[23]. The recombinant plasmid containing the cloned gene was subjected to automated sequencing using universal sequencing primers (M-13F and M-13R). The purified PCR products were then analyzed on automated capillary based DNA sequencer (3130 excel applied biosystems, USA) for validation of the respective gene.
Phylogenetic studies:
Phylogenetic analysis of the Bcl-2 like gene was performed utilizing NCBI database (http://www.ncbi.nlm.nih.gov) and Expasy proteomics server (http://www.expasy.org). Phylogenetic tree was constructed by Phylogeny. fr at Expasy bioinformatics resource portal to establish the evolutionary relationship between Bcl-2 like gene and members of Bcl-2 gene family from different animal species[24]. Protparam and Protscale tools at Expasy Proteomics server were employed to predict the various physiochemical properties of the cloned gene such as molecular weight, molecular formula, isoelectric point (PI), amino acid composition, instability index, molar extinction coefficient, estimated half-life, aliphatic index, total number of positively and negatively charged residues and grand average of hydropathicity (GRAVY) respectively.
Protein homology modeling:
Protein modeling of the putative protein was performed through I-TASSER online program (zhanglab.ccmb.med.umich.edu/I-TASSER/)[25] for structural and functional predictions on default parameters. Structural genomics studies have revealed that many proteins have disordered regions which may interfere with their purification and crystallization. Thus, it is important to determine such regions in proteins from their amino acid sequences. The prediction of disordered residues in protein and presence of transmembrane segment was performed employing memsat program (www.sacs.ucsf.edu/cgi-bin/memsat.py) for functional annotation of protein. Studies had reported that such regions may be involved in many biological processes namely cell signaling, regulation and control of cell cycle[26]. Such factors play a key role in the determination of functional aspects of protein.
RESULTS
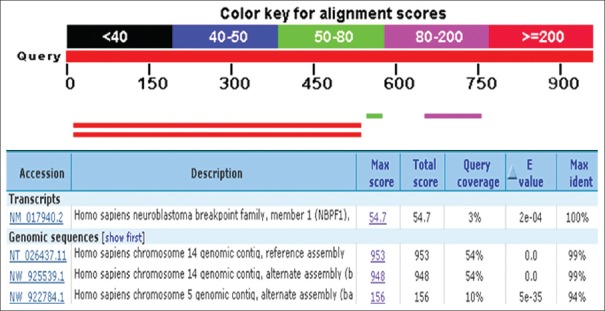
A PCR based homology screening approach was used for the identification and isolation of Bcl-2 gene from human breast cancer cell line, MCF-7. PCR amplification resulted in Bcl-2 amplicon (965 bp) from an enriched library of MCF-7 cancer cell line. Molecular cloning of the gene in pGEMT vector and automated sequencing of the cloned fragment (965 bp) established its homology with Homo sapiens neuroblastoma breakpoint family and Homo sapiens chromosome 14 genomic contig through BLAST search (BLASTn and BLASTp) and related member of Bcl-2 gene family (fig. 1). The Bcl-2 gene consisted of 965 nucleotides encoding a protein of 380 amino acids with a predicted molecular weight of 42.5 KDa. The various physiochemical properties predicted through protparam and protscale servers at expasy proteomics server (http://www.expasy.org) are listed in Table 1. The evolutionary relationship of Bcl-2 like gene and those reported in NCBI database was elucidated by maximum likelihood method of tree construction employing Phylogeny. Fr at Expasy proteomics server (http://www.expasy.org, fig. 2).
Fig. 1.
BLASTn analysis of the Bcl-2 like gene with those reported in NCBI Database.
TABLE 1.
PHYSIOCHEMICAL PROPERTIES OF THE BCL-2 LIKE GENE GENERATED BY PROTPARAM

Fig. 2.
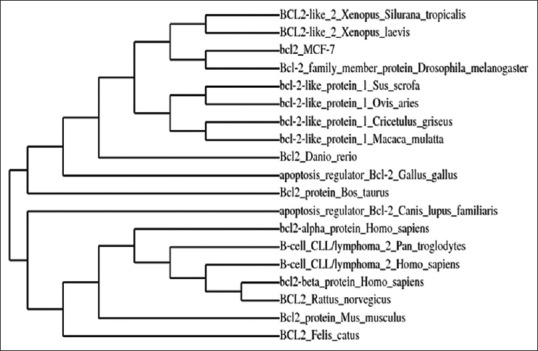
Phylogenetic tree was constructed using Phylogeny. fr tool at Expasy Proteomics sever.
The evolutionary relationship between Bcl-2 like protein and different members of Bcl-2 gene family was established by maximum likelihood method of tree construction (http://www.expasy.org).
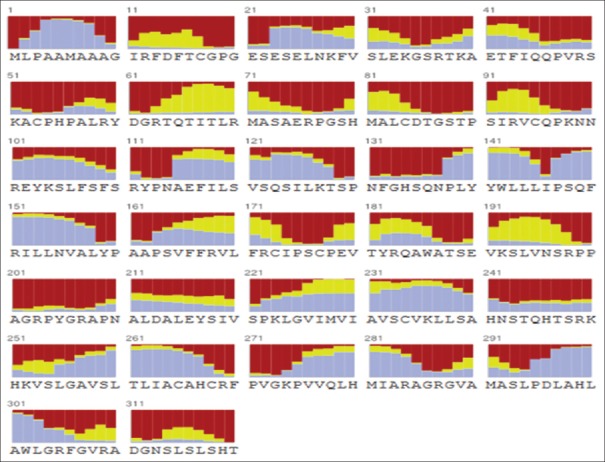
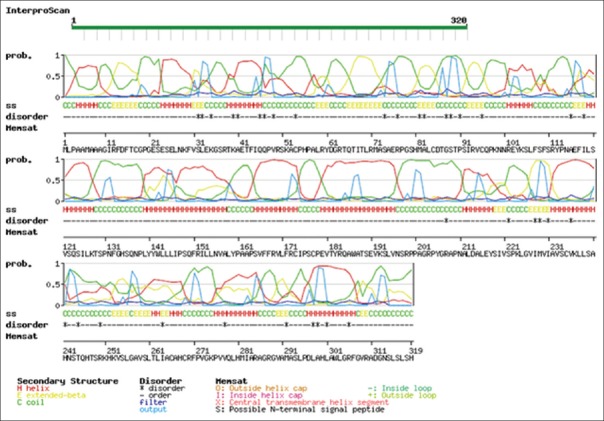
For structural elucidation of the putative protein, threading based protein modeling approach was followed for construction of a 3D protein model via I-TASSER online tool. The active binding sites in the Bcl-2 protein model were predicted and analyzed on default parameters following standard protocol. The secondary structure of the protein was determined by Raptor X prediction method (raptorx.uchicago.edu/, fig. 3). Furthermore, phobius prediction suggested the non-cytoplasmic localization of Bcl-2 like gene (fig. 4). The presence of transmembrane segment and disordered state of Bcl-2 gene was determined by memsat tool (www.sacs.ucsf.edu/cgi-bin/memsat.py, fig. 5).
Fig. 3.
Secondary structure of the Bcl-2 was determined by Raptor X prediction method.
Secondary structure of the Bcl-2 was determined by Raptor X prediction method from raptorx.uchicago.edu/. The color diagram shows the distribution of secondary structures and represented by various color combination as follows: Alpha helix (blue), Beta (yellow) and Loop/irregular region (red), respectively.
Fig. 4.
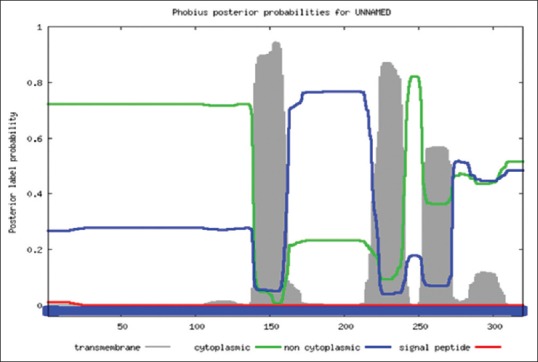
Plot of phobius prediction method.
The localization of Bcl-2 like gene and the presence of transmembrane segment were determined by Phobius prediction method (phobius.sbc.su.se/).
Fig. 5.
Putative protein prediction through Memsat online tool.
The state of disorderliness in the Bcl-2 like putative protein was predicted through Memsat online tool (www.sacs.ucsf.edu/cgi-bin/memsat.py). The presence of secondary structures was represented in various colors as follows: Alpha helix (red), extended beta (yellow) and coil (green) respectively. The disorderliness state in the protein is represented by a star (black) in the diagram.
DISCUSSION
The Bcl-2 families of proteins are the key regulators of apoptotic pathway in cancer. These are involved in various forms of malignancies and play a crucial role in breast cancer chemotherapy. The overexpression of antiapoptotic Bcl-2 members namely Bcl-2 and Bcl-xL has chemoresistance implications in cancer whereas pro-apoptotic proteins such as Bax enhance tumor cells sensitization to anticancer therapies. Moreover, the Bcl-2 family acts diversely in apoptotic regulation, governing the key conclusive step of activation or silencing of the caspases for cell survival.
The research investigation discusses the identification, molecular cloning and bioinformatics analysis of a member of the apoptotic pathway regulator, Bcl-2 gene family. A Bcl-2 homology was isolated from MCF-7 cancer cell line employing PCR based screening approach. Automated sequencing of the cloned gene revealed it to be closely related to Homo sapiens neuroblastoma breakpoint family and Homo sapiens chromosome 14 genomic contig, respectively. Phylogenetic analysis predicted the physiochemical properties of Bcl-2 like gene namely molecular weight of 42.5 KDa, isoelectric point (pI)-9.49, molecular formula- C1893H3004N534O548S16, total number of negatively charged residues, (Asp+Glu)-26, total number of positively charged residues, (Arg+Lys)-39, instability index- 42.08 (unstable protein), total number of atoms-5995, extinction coefficients-35910, 0.844, assuming all pairs of cysteine residues form cystines and 35410, 0.833, assuming all cysteine residues are reduced, aliphatic index-75.00 and grand average of hydropathicity (GRAVY) is -0.202 (Table 1). Further, phobius prediction suggested non cytoplasmic localization of Bcl-2 like gene (fig. 4) in agreement with the literature which suggest the intracellular localization of the Bcl-2 gene including the nuclear membrane and outer mitochondrial membrane and nuclear pore complexes, mitochondrial complexes, and some parts of endoplasmic reticulum[27]. The expression of Bcl-2 gene in breast cancer patients is controlled by estrogen in mammary epithelial cells and estrogen receptor positive breast cancer cell lines[28,29]. Studies have suggested better survival and prognosis rate in Bcl-2 positive patients compared to the Bcl-2 negative patients[30,31]. Although the exact mechanism is unknown, the antiproliferative effect of the gene is responsible for this phenomenon, is a prospective hypothesis. Moreover, the prediction of protein homology model of Bcl-2 gene is significant for structural and functional annotation of the protein (fig. 6). The presence of disordered region and transmembrane segment in Bcl-2 gene suggests its role in biological processes namely cell signaling, regulation and control of cell cycle as reported in literature[26].
Fig. 6.
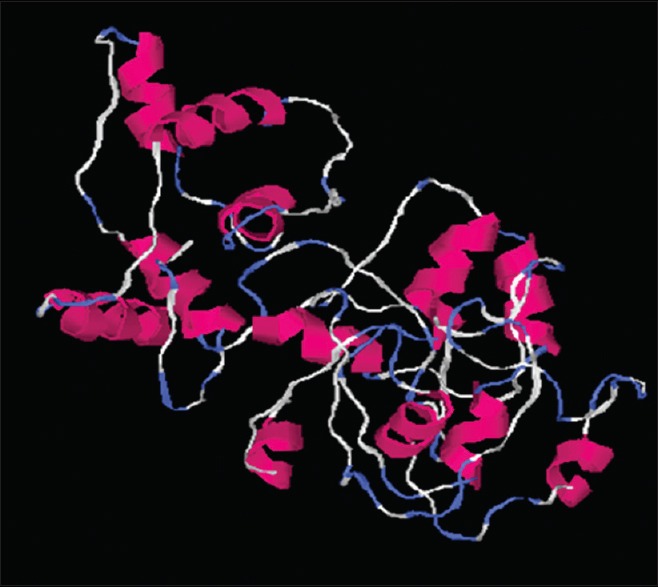
The 3 dimensional structure of the putative Bcl-2 protein.
The 3 dimensional structure of the putative Bcl-2 protein was determined via protein homology modeling (I-TASSER protein modeling suite) (zhanglab.ccmb.med.umich.edu/I-TASSER/).
The future prospects in the study highlight the requirement of biochemical characterization of the cloned gene through in vitro assays for defining the functional characteristics of the putative Bcl-2 like protein. Furthermore, protein docking studies would decipher the role of key amino acids in the protein's active site and would help to establish the functional properties of the protein. Such studies in future would help in target based approach for drug discovery and management in breast cancer studies. Moreover, the estrogen receptor regulated expression of the Bcl-2 gene in mammary epithelial cell suggests the expression of Bcl-2 gene as a prospective biomarker for tumors, showing the functional aspects of hormone receptors in breast cancer patients. Another significant area correlates with Bcl-2 expression and higher survival rate suggesting better treatment possibilities in breast cancer patients showing positive expression. The future studies should focus on the research on apoptotic proteins and their prognostic effect on a larger group of patients.
Financial support and sponsorship:
Nil.
Conflicts of interest:
There are no conflicts of interest.
Acknowledgements:
The author PT is thankful to the Director, Central Institute of Medicinal and Aromatic Plants, Lucknow for providing research facilities.
Footnotes
Tiwari and Khan: Molecular Cloning of Bcl-2 gene from MCF-7
REFERENCES
- 1.Cancer Facts and Figures. Atlanta: American Cancer Society; 2015. American Cancer Society. [Google Scholar]
- 2.Spano D, Heck C, De Antonellis P, Christofori G, Zollo M. Molecular networks that regulate cancer metastasis. Semin Cancer Biol. 2012;22:234–49. doi: 10.1016/j.semcancer.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 3.World Health Organization. International Agency for Research on Cancer. Geneva: World Health Organization; 2013. [Google Scholar]
- 4.Ferlay J, Steliarova FE, Lortet TJ, Rosso S, Coebergh JW, Comber H, et al. GLOBOCAN 2008 v2.0, cancer incidence and mortality worldwide: IARC CancerBase Suppl 10. Lyon, France: International Agency for Research on Cancer; 2010. [Last accessed on 2015 Nov 29]. Available from: http://www.globocan.iarc.fr. [2013] [Google Scholar]
- 5.Hortobagyi GN, de la Garza Salazar J, Pritchard K, Amadori D, Haidinger R, Hudis CA, et al. The global breast cancer burden: Variations in epidemiology and survival. Clin Breast Cancer. 2005;6:391–401. doi: 10.3816/cbc.2005.n.043. [DOI] [PubMed] [Google Scholar]
- 6.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: A systematic analysis for the global burden of disease study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127:2893–917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 8.Soule HD, Vazguez J, Long A, Albert S, Brennan M. A human cell line from a pleural effusion derived from a breast carcinoma. J Natl Cancer Inst. 1973;51:1409–16. doi: 10.1093/jnci/51.5.1409. [DOI] [PubMed] [Google Scholar]
- 9.Brown R. The Bcl-2 family of proteins. Br Med Bull. 1997;53:466–77. doi: 10.1093/oxfordjournals.bmb.a011624. [DOI] [PubMed] [Google Scholar]
- 10.Levine B, Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed JC. Bcl-2-family proteins and hematologic malignancies: History and future prospects. Blood. 2008;111:3322–30. doi: 10.1182/blood-2007-09-078162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krajewski S, Krajewska M, Turner BC, Pratt C, Howard B, Zapata JM, et al. Prognostic significance of apoptosis regulators in breast cancer. Endocr Relat Cancer. 1999;6:29–40. doi: 10.1677/erc.0.0060029. [DOI] [PubMed] [Google Scholar]
- 13.Reed JC. Bcl-2 and the regulation of programmed cell death. J Cell Biol. 1994;124:1–6. doi: 10.1083/jcb.124.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zamzami N, Brenner C, Marzo I, Susin SA, Kroemer G. Subcellular and submitochondrial mode of action of Bcl-2-like oncoproteins. Oncogene. 1998;16:2265–82. doi: 10.1038/sj.onc.1201989. [DOI] [PubMed] [Google Scholar]
- 15.Zapata JM, Krajewska M, Krajewski S, Huang RP, Takayama S, Wang HG, et al. Expression of multiple apoptosis-regulatory genes in human breast cancer cell lines and primary tumors. Breast Cancer Res Treat. 1998;47:129–40. doi: 10.1023/a:1005940832123. [DOI] [PubMed] [Google Scholar]
- 16.Zhang GJ, Kimijima I, Abe R, Watanabe T, Kanno M, Hara K, et al. Apoptotic index correlates to Bcl-2 and p53 protein expression, histological grade and prognosis in invasive breast cancers. Anticancer Res. 1998;18:1989–98. [PubMed] [Google Scholar]
- 17.Eguchi Y, Ewert DL, Tsujimoto Y. Isolation and characterization of the chicken Bcl-2 gene: Expression in a variety of tissues including lymphoid and neuronal organs in adult and embryo. Nucleic Acids Res. 1992;20:4187–92. doi: 10.1093/nar/20.16.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hatakeyama S, Hamasaki A, Negishi I, Loh DY, Sendo F, Nakayama K, et al. Multiple gene duplication and expression of mouse Bcl-2-related genes, A1. Int Immunol. 1998;10:631–7. doi: 10.1093/intimm/10.5.631. [DOI] [PubMed] [Google Scholar]
- 19.Gong F, Sun L, Wang Z, Shi J, Li W, Wang S, et al. The Bcl-2 gene is regulated by a special AT-rich sequence binding protein 1-mediated long range chromosomal interaction between the promoter and the distal element located within the 3'-UTR. Nucleic Acids Res. 2011;39:4640–52. doi: 10.1093/nar/gkr023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DeoCampo ND, Wilson MR, Trosko JE. Cooperation of Bcl-2 and myc in the neoplastic transformation of normal rat liver epithelial cells is related to the down-regulation of gap junction-mediated intercellular communication. Carcinogenesis. 2000;21:1501–6. [PubMed] [Google Scholar]
- 21.Clark RS, Chen J, Watkins SC, Kochanek PM, Chen M, Stetler RA, et al. Apoptosis-suppressor gene Bcl-2 expression after traumatic brain injury in rats. J Neurosci. 1997;17:9172–82. doi: 10.1523/JNEUROSCI.17-23-09172.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boise LH, González-García M, Postema CE, Ding L, Lindsten T, Turka LA, et al. Bcl-x, a Bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell. 1993;74:597–608. doi: 10.1016/0092-8674(93)90508-n. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbour; 1989. [Google Scholar]
- 24.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F, et al. Phylogeny. fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36:W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Michels J, Kepp O, Senovilla L, Lissa D, Castedo M, Kroemer G, et al. Functions of Bcl-XL at the interface between cell death and metabolism. Int J Cell Biol. 2013;2013:705294. doi: 10.1155/2013/705294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krajewski S, Tanaka S, Takayama S, Schibler MJ, Fenton W, Reed JC. Investigation of the subcellular distribution of the Bcl-2 oncoprotein: Residence in the nuclear envelope, endoplasmic reticulum, and outer mitochondrial membranes. Cancer Res. 1993;53:4701–14. [PubMed] [Google Scholar]
- 28.Johnston SR, MacLennan KA, Sacks NP, Salter J, Smith IE, Dowsett M. Modulation of Bcl-2 and Ki-67 expression in oestrogen receptor-positive human breast cancer by tamoxifen. Eur J Cancer. 1994;30A:1663–9. doi: 10.1016/0959-8049(94)00327-2. [DOI] [PubMed] [Google Scholar]
- 29.Barbareschi M, Caffo O, Veronese S, Leek RD, Fina P, Fox S, et al. Bcl-2 and p53 expression in node-negative breast carcinoma: A study with long-term follow-up. Hum Pathol. 1996;27:1149–55. doi: 10.1016/s0046-8177(96)90307-x. [DOI] [PubMed] [Google Scholar]
- 30.Joensuu H, Pylkkänen L, Toikkanen S. Bcl-2 protein expression and long-term survival in breast cancer. Am J Pathol. 1994;145:1191–8. [PMC free article] [PubMed] [Google Scholar]
- 31.Silvestrini R, Benini E, Veneroni S, Daidone MG, Tomasic G, Squicciarini P, et al. p53 and Bcl-2 expression correlates with clinical outcome in a series of node-positive breast cancer patients. J Clin Oncol. 1996;14:1604–10. doi: 10.1200/JCO.1996.14.5.1604. [DOI] [PubMed] [Google Scholar]