Abstract
Most proteins have multiple functions. Obviously, conventional methods of manipulating the level of the protein of interest in the cell, such as over-expression, knockout or knockdown, affect all of its functions simultaneously. The key advantage of these methods is that over-expression, knockout or knockdown does not require any knowledge of the molecular mechanisms of the function(s) of the protein of interest. The disadvantage is that these approaches are inadequate to elucidate the role of an individual function of the protein in a particular cellular process. An alternative is the use of re-engineered proteins, in which a single function is eliminated or enhanced. The use of mono-functional elements of a multi-functional protein can also yield cleaner answers. This approach requires detailed knowledge of the structural basis of each function of the protein in question. Thus, a lot of preliminary structure-function work is necessary to make it possible. However, when this information is available, replacing the protein of interest with a mutant in which individual functions are modified can shed light on the biological role of those particular functions. Here we illustrate this point using the example of protein kinases, most of which have additional non-enzymatic functions, as well as arrestins, known multi-functional signaling regulators in the cell.
Keywords: knockout, knockdown, overexpression, arrestin, GPCR, GRK, signaling, functional bias
1. Introduction
In the world of proteins, multi-functionality is the rule rather than the exception. It is hard to name a mono-functional protein: only extremely specialized ones, like casein in milk, come to mind. Virtually every protein in the cell interacts with numerous partners and plays a role in a number of biological processes. Although signaling regulators, such as scaffolding proteins, are the prime example, close examination shows that even something as basic as cytochrome C is not a “one-trick-pony” but simultaneously participates in such disparate processes as mitochondrial energy production (Sun et al., 2013) and apoptotic cell death (Danial and Korsmeyer, 2004; Hüttemann et al., 2011).
To determine the role of a particular protein in cell biology, it is customary to manipulate its expression level. However, this approach is rather crude. It should be obvious that when the protein is knocked out or knocked down, all of its functions are suppressed, whereas over-expression augments every function it has. Here we discuss the advantages and limitations of conventional methods of studying protein functions, as well as less conventional approaches that make it possible to sort out exactly how the protein works and what role it plays in a particular signaling circuit.
We would like to emphasize upfront that the approach we discuss is very different from the one taken when disease-causing mutations are discovered in a protein. A good example of the latter is parkin, an E3 ubiquitin ligase with dozens of known mutations that have been shown to cause Parkinson’s disease (Kitada et al., 1998). In cases like this, it is not known what function(s) a particular mutation affects and in what way: Does it affect one protein function or several? Does it suppress or enhance any of them? Since the research objective is to uncover the mechanism of the disease pathogenesis, these disease-associated mutations are examined for their functional characteristics in order to gain insight into the role of that particular protein in the disease. In contrast, the mutations we discuss below are introduced on the basis of a molecular understanding of the protein’s functions. Since these are specially designed mutations, it is known what is affected and in which way, and the aim is to select the mutation(s) affecting only one function out of many. Naturally occurring mutants can be used for mechanistic and/or therapeutic purposes only after the functional consequences of the mutations are unambiguously elucidated.
To illustrate these points and make the discussion more substantive, we use protein kinases and non-visual arrestins as examples of multi-functional proteins. In both cases, simple manipulation of expression, whether up or down, is too crude to elucidate their role in particular cellular functions and can yield misleading results. We believe that the considerations discussed below are applicable to the majority of cellular proteins.
2. Conventional approaches to study protein function: pros and cons
The most common approaches to establishing the function of an individual protein involve manipulation of its expression level. These include knockout, the results of which can be studied in intact animals, when they are viable, or in mouse embryonic fibroblasts (MEFs) lacking a particular protein, when the knockout is lethal. The key advantage of genetic knockout is that the protein is eliminated completely. The downside is that it is missing from the very beginning, so that animals developing in its absence usually compensate in various ways, some of which are hard to detect. Thus, functional changes could be the result either of the knockout per se or of compensational processes, and in many cases it is impossible to determine which effect is prevalent. Furthermore, constitutive general knockout eliminates the protein from all cells in the organism, making it hard to determine the site of action. The advent of inducible and cell-specific knockout technologies has largely circumvented these problems (Feil, Valtcheva, and R., 2009; Lewandoski, 2001). These techniques have their own limitations, such as possible side effects of the inducer when applied chronically, incomplete recombination, which to some extent negates the advantage of knockout, and the “leaky” system, which may result in undesired knockouts. A relatively easy and inexpensive way to obtain definitive data with constitutive knockout animals is the rescue approach that utilizes virus-mediated expression of the missing protein in a particular cell type or organ. When the rescue is successful, one can be certain that the phenotype is the result of the knockout per se, rather than the compensational changes it induces. Furthermore, the rescue could be limited to a specific cell type in the body as well as timed to a specific functional condition, thereby demonstrating that the absence of the protein in question in these cells at this time is critical for the function under investigation. Similar logic applies to cells: without the demonstration of successful rescue by exogenous expression of the missing protein, the phenotype cannot be unambiguously ascribed to the absence of the protein of interest. Unfortunately, rescue by restoration of the missing protein at a particular place and time has rarely been demonstrated, which makes many conclusions based on the constitutive whole body knockout ambiguous.
Although the total absence of the protein of interest following the knockout is generally perceived as an advantage of this technique and does indeed make for a “cleaner” experimental condition, the situation itself is hardly physiological. When a functionally important protein is missing, the cell might switch into an emergency mode, functioning completely out of the normal “box” and overcompensating in a way that it would not normally do. If compensation cannot be achieved, at least in some cells, then the knockout is lethal, which is hardly informative. These considerations might explain why so many knockout lines have a remarkably mild phenotype or no detectable phenotype at all, even when there is every reason to believe that the knocked out protein is highly functionally important and does not seem to have obvious backups. The knockout is often used to recapitulate the effects of the loss-of-function mutations associated with diseases. Indeed, in some cases, the protein is simply lost due to, for example, a genomic rearrangement, as is often the case with parkin, the protein associated with autosomal recessive juvenile parkinsonism (Kitada et al., 1998). However, many mutations, particularly missense mutations, cause alterations in the protein functions or only partial loss of some function(s) but not of others. Parkin is again a case in point, with a large number of missense mutations associated with familial as well as sporadic early onset parkinsonism (Farrer et al., 2001; Khan et al., 2005; Lücking et al., 2000; Pramstaller et al., 2005). Many of these mutations result in the loss of E3 ligase activity, whereas others yield normal or even enhanced enzymatic activity accompanied by an increased propensity to aggregate and/or an altered subcellular distribution (Sriram et al., 2005; Wang et al., 2005).
Knockdown works similarly to knockout in cultured cells (Caplen, 2003) or even in vivo (Ahmed et al., 2010; Gurevich, Ahmed, and Carl, 2014). The advantage of knockdown is that animals/cells develop in the presence of the protein of interest and are therefore less likely to turn on compensational mechanisms, since the protein is only eliminated for a limited period of time. There are several drawbacks, though. First, in contrast to knockout, the knockdown is never complete, so that a certain proportion (up to ~20%) of the targeted protein still remains. Second, all the tools used for knockdown- morpholinos, siRNA, shRNA, or miRNA (the latter three tools are discussed in detail in (Gurevich, Ahmed, and Carl, 2014)) - target the mRNA of the protein of interest. Thus, knockdown is fairly effective in the case of proteins with a relatively short half-life, in the range of minutes to hours, but very ineffective in the case of proteins that live for many hours or days. Finally, despite all controls (usually oligos and RNAs with a scrambled sequence), there is always a chance that the knockdown construct affects other proteins. For example, recently siRNA knockdown of the arrestin domain-containing protein ARRDC3 suggested that this protein recruits ubiquitin ligase Nedd4 to activated β2AR (Nabhan, Pan, and Lu). However, the group that originally proposed that arrestin-3 plays this role (Shenoy et al., 2008) found that the siRNA used in that study also knocks down both non-visual arrestins (Han, Kommaddi, and Shenoy, 2013). Thus, the effect of that siRNA could be ascribed to the intended knockdown of ARRDC3 as well as the unintended reduction of arrestin-2/3 (Han, Kommaddi, and Shenoy, 2013). Upon knockdown, the expression of closely related proteins is usually tested, but as most cell types express >10,000 different proteins (Manteniotis et al., 2013; Pronin et al., 2014; Yu et al., 2010), comprehensive testing for off-target effects of knockdown constructs is simply impossible. Thus, only successful rescue with a knockdown-resistant version of the protein (e.g., from a different species or carrying silent mutations that make knockdown ineffective) constitutes proof that the observed phenotype is caused by knockdown of the targeted protein, rather than off-target effects of the tools used (Jonchere and Bennett, 2013).
Unlike knockout or knockdown, over-expression of a particular protein has not been reported to affect the levels of other proteins. However, this possibility cannot be discounted, particularly when the protein in question is a modifying enzyme, such as a protein kinase, phosphatase, ubiquitin ligase, etc. Thus, although the phenotype observed upon over-expression of a particular protein is more likely to be associated with an increase in the intracellular level of this protein, other mechanisms cannot be excluded. Besides, in many cases it is easy to express a protein at a level many times higher than physiological. These supra-physiological intracellular concentrations can force its interactions with partners that it would not bind at normal expression levels, producing artifacts.
Thus, the main dangers of methods designed to affect the levels of proteins of interest are different. In the case of knockout and knockdown, these lie in compensational changes or off-target effects due to unanticipated changes in the expression of other proteins. Additionally, complete removal of the protein by knockout could bring about a significant rearrangement of the cellular molecular network in a non-physiological manner. In the case of over-expression, off-target effects are less likely, whereas artifacts produced by supra-physiological levels of a protein come to the fore. However, even when off-target effects or non-physiological interactions can be excluded, in both cases there is a problem that is rarely appreciated: changes of expression level, whether up or down, simultaneously affect every function of the protein in question. Since most proteins can do many things, and each aspect of cellular functions is regulated by numerous inputs, this creates ambiguity in the interpretation of the data to a degree that is often unacceptable.
3. Investigation of multi-functional proteins: changing one function at a time
The great majority of proteins have more than one function. To define the biological role of an individual function, it would be beneficial to selectively eliminate this particular one, leaving all other functions, known and unknown, intact. Ideally, the expression of a separated domain of a multi-functional protein that has only one function would yield complementary evidence, allowing the identification of the biological role of that particular function. Below we discuss attempts to do just that, all of which require a deep understanding of the molecular mechanisms involved.
3.1. Enzymes with well-defined activity: protein kinases
Most kinases, in addition to their ability to phosphorylate target proteins, bind numerous partners. Protein-protein interactions not only change kinase localization in the cell, but often play additional roles. The kinase often serves as a scaffold and/or allosteric modulator of other kinases, and these functions do not require catalytic activity, as evidenced by the abundance of pseudokinases (i.e., proteins with a recognizable kinase domain but no catalytic function) in the genome (Shaw et al., 2014). In most cases, the catalytic activity of the kinase can be blocked via two distinct mechanisms. First, a kinase-dead version can be generated by the very conservative replacement of a lysine in the active center by an arginine (Gibbs and Zoller, 1991; Kong, Penn, and Benovic, 1994; Ohno et al., 1990). The resulting protein folds normally and can engage its partners, but cannot transfer phosphate. Second, changing a conserved alanine into phenylalanine, which likely fills the cavity where the adenine ring would normally bind, precludes ATP binding and therefore abolishes kinase activity (Taylor et al., 2013). Such mutants are widely used in cell culture studies to determine the role of kinase activity in cellular functions. Using kinase-dead mutants, it was shown that in some cases the kinase can effectively fulfill its scaffolding/allosteric function without enzymatic activity. For example, kinase-dead B-Raf binds WT B-Raf and C-Raf, and its dimerization with WT Raf family members leads to the activation of the downstream MEK-ERK cascade independently of Ras (Taylor et al., 2013). A scaffolding function has also been suggested for leucine-rich repeat kinase 2 (LRRK2), a large protein kinase containing, in addition to the kinase and GTPase domains, multiple protein-protein interaction domains (reviewed in (Cookson, 2010)).
G protein-coupled receptor kinases (GRKs) represent another example of multidomain kinases, with different domains having distinct well-defined functions (Fig. 1). GRKs are key players in homologous (activation-dependent) desensitization of G protein-coupled receptors (GPCRs) (Carman and Benovic, 1998). The widely accepted model of homologous desensitization posits that it is a two-step process. First, agonist-activated receptors are selectively phosphorylated by GRKs (Gurevich et al., 2012). This reduces, but does not completely stop, G protein coupling. Second, arrestins specifically recognize active, phosphorylated receptors (Gurevich and Gurevich, 2004) and bind them with high affinity, precluding further G protein activation by competing for overlapping receptor sites (Gurevich and Gurevich, 2006). Vertebrates express hundreds of different GPCRs (Bockaert and Pin, 1999), but only seven GRKs (Gurevich et al., 2012) and four arrestin subtypes (Gurevich and Gurevich, 2006), suggesting that each GRK and arrestin participates in the regulation of multiple GPCRs.
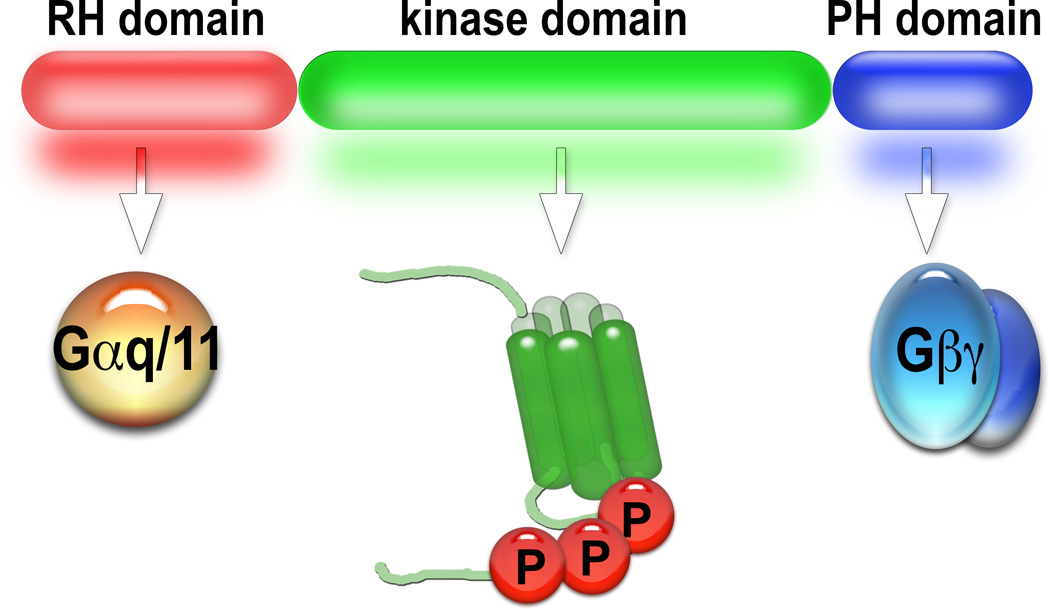
Fig. 1. Domain structure of GRK2/3.
All GRKs have the N-terminal RGS-homology (RH) domain, the central kinase domain, two additional α-helices of the RH domain downstream of the kinase domain (not shown here for simplicity), and C-terminal elements that mediate their membrane localization (Gurevich et al., 2012). The RH domain of the GRK2/3 subfamily has the ability to bind active GTP-liganded α-subunits of Gq/11, sequestering them away from their effectors, thereby suppressing Gq/11-mediated signaling. The kinase domain of all GRKs contains a conserved catalytic site that allows them to phosphorylate GPCRs. At the C-terminus, GRK2/3 have a plekstrin homology (PH) domain that binds the lipid bilayer and G protein βγ-dimers, which are released from heterotrimeric G proteins upon their activation by GPCRs. The PH domain mediates the recruitment of GRK2/3 to the membrane upon receptor activation. The PH domain can be expressed separately (historically termed βARKct), and in that case it acts as a scavenger of Gβγ-dimers, preventing the recruitment to the membrane of endogenous GRK2/3 (Akhter et al., 1999; Salazar et al., 2013) and suppressing βγ-mediated signaling (Raveh et al., 2010). Color version of this figure is available online.
GRKs have a RGS-homology (RH) domain in the N-terminus, followed by a kinase domain, whereas their C-termini are quite diverse and carry a variety of membrane-targeting sequences (Gurevich et al., 2012). In particular, GRK2/3 have a pleckstrin homology (PH) domain that binds lipids in the bilayer and Gβγ (Fig. 1). The PH domain is critical for the recruitment of GRK2/3 to the plasma membrane, and thus for GRK2/3-dependent phosphorylation of GPCRs (Daaka et al., 1997). This has been proven conclusively using the isolated PH domain of GRK2, βARKct (C-terminus of the β-adrenergic receptor kinase, the historic name of GRK2 (Benovic et al., 1989)), that inhibits GRK2-dependent receptor phosphorylation by competing with GRK2 for Gβγ and thus preventing GRK2 recruitment to the membrane. Separately expressed βARKct has been successfully used to elucidate the contribution of the elevated expression of GRK2 in the heart to congestive heart failure. It has been shown that excessive phosphorylation by GRK2 and desensitization of β-adrenergic receptors reduces heart responsiveness to catecholamines, underlying congestive heart failure (Akhter et al., 1999). Heart-specific over-expression of WT GRK2 reduces heart sensitivity to catecholamines (Akhter et al., 1999). In contrast, expression of βARKct, which is the separated GRK2 PH domain that serves as a Gβγ scavenger, inhibits GRK2 recruitment to the receptor, and therefore its phosphorylation, protecting the heart (Akhter et al., 1999; Salazar et al., 2013). This is one of relatively few examples of the use of mutant proteins or their individual domains with a defined function in living animals to elucidate the mechanism of the protein function or to achieve a therapeutically relevant effect. Since the PH domain in the C-terminus of GRK2/3 binds Gβγ, it can sequester it away from its effectors, suppressing every Gβγ-dependent function, known and unknown. Via this mechanism, GRK2, independently of its kinase activity, rapidly suppresses the opening of GPCR-activated potassium channels in cultured cells (Raveh et al., 2010).
The N-terminal RH domain of GRKs 2 and 3, which in contrast to most RGS proteins does not act as a GTPase activator, was shown to bind active (GTP-liganded) α-subunits of the Gq/11 subfamily of G proteins, sequestering them away from their effectors (Carman et al., 1999) (Fig. 1). Using kinase-dead GRK2 as well as the isolated RH domain and RH-dead constructs with key Gαq binding residues inactivated, it has been demonstrated that GRK2 suppresses signaling of mGluR1/5 in cultured cells via its RH domain (Day et al., 2003; Day, Wedegaertner, and Benovic, 2004; Sterne-Marr et al., 2004; Sterne-Marr et al., 2003). A similar strategy was recently employed to tease apart the mechanism of GRK3 action in vivo (Ahmed et al., 2015). However, some background information is needed to appreciate those results. Parkinson’s disease (PD), the second most common neurodegenerative disorder after Alzheimer’s, is caused by the death of dopamine-producing neurons in the substantia nigra and the consequent deficit of dopamine in the striatum. The most effective treatment of PD is dopamine replacement therapy with the dopamine precursor L-DOPA. Problems begin within a few years of L-DOPA therapy: loss of effectiveness necessitates increasing doses, which eventually lead to L-DOPA-induced dyskinesia (LID), i.e. uncontrolled involuntary movements, greatly reducing the quality of life of PD patients. Although the exact pathophysiology of LID remains poorly understood, supersensitivity of the striatal dopamine receptors to dopamine is believed to be a contributing factor (Gurevich and Gurevich, 2010). All five dopamine receptors are G protein-coupled receptors (GPCRs). GRKs play a key role in the signaling termination process for most GPCRs: GRK-dependent receptor phosphorylation is a prerequisite for high-affinity arrestin binding, which blocks further G protein activation (Gurevich and Gurevich, 2004). Thus, it seemed reasonable to hypothesize that the normal desensitization machinery does not have enough capacity to handle the supra-physiological increase in dopamine concentrations upon each L-DOPA dose. Since GRKs tend to be rate-limiting in this process (Violin et al., 2008), the ability of over-expression of GRK6 and GRK3 in the striatum to suppress LID was tested (Ahmed et al., 2010; Ahmed et al., 2015). Both reduced LID in vivo, but in a different manner. Whereas the enzymatic activity of GRK6 was critical for its anti-LID action, it was found to be dispensable in the case of GRK3, as was determined using kinase-dead versions of both GRK isoforms (Ahmed et al., 2015). In contrast, the ability of its RH domain to bind Gαq/11 was found to be critical for the anti-LID activity of GRK3, since a GRK3 mutant with the Gαq/11 binding disabled was ineffective, whereas kinase-dead GRK3 was fully functional (Ahmed et al., 2015). Furthermore, the separated GRK3 RH domain was just as effective in suppressing LID as full-length GRK3 (Ahmed et al., 2015), indicating that only sequestration of Gαq/11 and the resulting suppression of this branch of signaling was necessary. This example demonstrates that the strategy of employing functionally defined mutants can be successfully used in living animals and can yield unambiguous results.
Thus, elimination of certain capabilities of multi-functional proteins by targeted mutations enables identification of their biological role. Moreover, the expression of mono-functional protein elements, such as the PH domain of GRK2 or the RH domain of GRK3, demonstrates what is physiologically important. It should be noted that mono-functionality of the Gβγ-binding PH domain or the Gαq/11-binding RH domain does not mean that each of these elements affects only one signaling event in the cell. In fact, by scavenging Gβγ, the PH domain suppresses all of its effects. Similarly, by preventing GTP-liganded Gαq/11 from interacting with other effectors, the RH domain of GRK2/3 suppresses every branch of signaling initiated by active Gαq/11. However, as far as GRK3 is concerned, this is a way to clarify which function/functional domain is involved, since full-length GRK2/3 has all the functions of these separated domains plus kinase activity towards GPCRs and other substrates (Gurevich et al., 2012). Therefore, this approach helps to elucidate the molecular mechanisms of physiological or pathological processes more precisely. The findings that the expression of βARKct protects against congestive heart failure (Akhter et al., 1999; Salazar et al., 2013), whereas the RH domain of GRK3 has anti-LID activity (Ahmed et al., 2015) suggest that gene therapy based on the expression of these molecular tools is more promising than that based on the expression of full-length GRK2/3. Thus, mutant proteins and/or their mono-functional domains can produce desired changes in cell signaling without affecting most other aspects of the functions of the targeted protein, reducing the chances of unwanted side effects (Gurevich and Gurevich, 2012).
3.2. The ultimate scaffolds: non-visual arrestins
The two non-visual arrestins expressed in virtually every cell in all mammals, arrestin-2 and -31, appear to bind hundreds of different GPCR subtypes (Gurevich and Gurevich, 2006) as well as dozens of non-receptor partners (Xiao et al., 2007), regulating various aspects of cell signaling (DeWire et al., 2007). Since arrestins are such versatile scaffolds, it is particularly important to use mutants designed to enhance or suppress individual functions and/or mono-functional elements of the protein both for mechanistic studies and possible therapeutic applications..
3.2.1. Arrestin functions
The first member of the arrestin family, arrestin-1, was discovered as a protein that selectively binds light-activated, phosphorylated rhodopsin (Kuhn, Hall, and Wilden, 1984), reducing rhodopsin-induced signaling (Wilden, Hall, and Kühn, 1986), apparently by directly blocking the coupling of the cognate G protein, transducin, to rhodopsin via simple competition for overlapping binding sites (Krupnick, Gurevich, and Benovic, 1997; Wilden, 1995). The first non-visual subtype, β-arrestin, was discovered as a protein that plays the same role in the case of the β2-adrenergic receptor (β2AR), clearly preferring this receptor over rhodopsin (Lohse et al., 1992; Lohse et al., 1990). Soon it became apparent that “β-arrestin” is a misnomer, as this non-visual arrestin was shown to interact with a variety of GPCRs in addition to β2AR (Gurevich et al., 1995; Gurevich et al., 1993). Nonetheless, the name is still in use, although the original β-arrestin was retroactively renamed β-arrestin1 after the discovery of another non-visual subtype, which was named β-arrestin2 (Attramadal et al., 1992). More systematic names, arrestin-1 through -4, where the number after the dash indicates the order of cloning, have also been proposed (Sterne-Marr et al., 1993), and are used in parallel with the historic nomenclature. In this system, which we are using here, β-arrestin1 is called arrestin-2 and β-arrestin2 – arrestin-3 (Sterne-Marr et al., 1993).
For a few years, it appeared that binding to the active phosphorylated forms of GPCRs, which induces homologous desensitization, was the only function of arrestin proteins. However, fairly soon evidence emerged that both non-visual subtypes, arrestin-2 and -3, directly interact with non-GPCR partners. The first of these, clathrin, was reported in 1996 (Goodman et al., 1996), followed by the second, the clathrin adaptor AP2 (Laporte et al., 1999), with many others following in rapid succession (reviewed in (DeWire et al., 2007; Gurevich and Gurevich, 2006)). Several years ago, over 150 proteins were reported to bind arrestin-2 and more than 200 interacted with arrestin-3 (Xiao et al., 2007). New partners that were not on these extensive lists continue to be reported (Ahmed et al., 2011; Bhandari et al., 2007; Kook et al., 2013; Zhan et al., 2013). Even an over-simplified picture looks pretty complicated: non-visual arrestins bind the trafficking proteins clathrin, AP2, NSF (Goodman et al., 1996; Laporte et al., 1999; McDonald et al., 1999), as well as Src family kinases (Imamura et al., 2001; Luttrell et al., 1999), MAP kinases belonging to the cascades leading to the activation of JNK3 (McDonald et al., 2000; Song et al., 2009; Zhan et al., 2013), JNK1/2 (Kook et al., 2013), ERK1/2 (Coffa et al., 2011; Coffa et al., 2011; DeFea et al., 2000; Luttrell et al., 2001; Meng et al., 2009; Song et al., 2009), and p38 (Bruchas et al., 2006), the second messenger-eliminating enzymes cAMP phosphodiesterase (Baillie et al., 2007; Perry et al., 2002) and diacylglycerol kinase (Nelson et al., 2007), several E3 ubiquitin ligases, including Mdm2 (Shenoy et al., 2001), AIP (Bhandari et al., 2007), and parkin (Ahmed et al., 2011), deubiquitinating enzymes (Shenoy et al., 2009), and many others (reviewed in (DeWire et al., 2007; Gurevich and Gurevich, 2006). The biological importance of arrestins is underscored by the fact that the absence of visual arrestin-1 leads to night blindness in humans (Fuchs et al., 1995) and retinal degeneration in mice (Xu et al., 1997), simultaneous knockout of the two non-visual subtypes is embryonic lethal in mice (Kohout et al., 2001), which is also the case for knockout of the only non-visual subtype Drosophila has, the product of the gene kurtz (Roman, He, and Davis, 2000). Thus, many functions of arrestins appear to be critical, which makes it harder to establish the role of individual ones in particular cellular processes.
Functionally defined “single function” arrestin mutants could be quite helpful in elucidating the role of arrestins in cellular processes. It is important to note, however, that “single function” is defined in the context of a specific protein and it does not follow that in the cell only one function will be affected, which is obviously not the case. In the case of arrestins, it could mean that an arrestin mutant is competent to mediate receptor desensitization via biding to GPCRs but is unable to support arrestin-mediated signaling. There have been successful attempts to create “biased” receptor agonists that engage only one pathway, either G protein or arrestin-dependent (Violin et al., 2014). Alternatively, GPCR mutants capable of supporting only one line of signaling, G protein or arrestin-dependent, are also being produced (Nakajima and Wess, 2012; Peterson et al., 2015). It is quite obvious, however, that initiation of either pathway will bring about multiple signaling events in the cell. Nevertheless, such an approach would allow assignment of a specific physiological effect to arrestin’s role in receptor desensitization or to its signaling function. Further analysis employing arrestin mutants deficient in particular signaling functions might uncover which of numerous signaling pathways regulated via arrestin-dependent scaffolding is involved, as the examples below illustrate.
3.2.2.Structural elements mediating arrestin functions
Ideally, to investigate an individual function of a protein, one needs to replace it with a mutant where this particular function is disabled, whereas all others remain normal. Comparing the results of replacement of the knocked out protein with the WT form, which should completely rescue the phenotype, and this mutant with a single disabled function would clearly show the role of that particular function. The results would be definitive: if the mutant does not rescue, the function in question is critical for the phenotype; if it rescues like WT, this function does not contribute to the observed changes. The interpretation even of partial rescue with the mutant, compared to full rescue with WT, is fairly straightforward: the function affected by the mutation contributes to the knockout phenotype, but is not the only one doing so.
Although this level of perfection has not been achieved so far, it appears feasible if two conditions can be met. First, we need to know the molecular mechanisms involved in the function of a particular protein in sufficient detail to be able to generate such a mutant. Second, there should be a way of selectively disabling a single function by mutagenesis without significantly affecting others. This approach is best described using as an example a type of protein known to do many things, where we know enough about the structural basis of at least some functions. Arrestins fit the bill: they are versatile signaling regulators that organize multi-protein complexes and target them to particular sub-cellular compartments (Gurevich and Gurevich, 2006; Gurevich and Gurevich, 2014). Structure-function studies of arrestins have generated a lot of information about the elements involved in particular functions (Gurevich and Gurevich, 2012). Several arrestin functions have been localized with sufficient precision to generate selective mutants (Fig. 2)(Table 1). 1. Localization of the phosphate sensor enabled arrestin activation by appropriate mutations, thereby generating “enhanced” versions of different arrestins that bind unphosphorylated active receptors with high affinity (Celver et al., 2002; Gurevich, 1998; Gurevich and Benovic, 1995; Gurevich and Benovic, 1997; Gurevich et al., 1997; Kovoor et al., 1999; Pan, Gurevich, and Gurevich, 2003). 2. Localization of the binding sites for clathrin (Goodman et al., 1996) and the clathrin adaptor AP2 (Laporte et al., 1999) (Fig. 2B) enabled the construction of non-visual arrestins where one or both of these sites are disabled (Goodman et al., 1996; Kim and Benovic, 2002; Laporte et al., 1999). 3. Identification of key residues that determine the receptor preference of arrestins (Vishnivetskiy et al., 2011) (Fig. 2A) allowed the generation of non-visual arrestins with high receptor specificity (Gimenez et al., 2014; Gimenez et al., 2012). 4. Identification of Asp26 and Asp29 as critical MEK1-binding residues in arrestin-2 (Fig. 2C), interacting with Arg47 and Arg49 in the N-terminus of MEK1, enabled the construction of arrestin mutants deficient in MEK1 binding, which cannot promote ERK1/2 activation due to this defect (Meng et al., 2009). In agreement with previous findings that ERK1/2 phosphorylation of arrestin-2 impairs its ability to recruit GPCRs to the coated pit (Lin et al., 1997), the expression of arrestin-2 mutant with impaired MEK1 binding, as well as the peptide mimicking the MEK1 interaction site on arrestin-2, reduced the phosphorylation of arrestin-2 by ERK1/2 at the C-terminal Ser-412 and facilitated agonist-induced internalization of β2AR. 5. The identification of the key c-Raf1 binding residue in arrestin-2 (Fig. 2C) yielded the R307A mutant that has normal ERK1/2 and MEK1 binding, but impaired c-Raf1 interaction, and therefore cannot promote ERK1/2 activation (Coffa et al., 2011). 6. The identification of cAMP phosphodiesterase binding sites on arrestin (Fig. 2B) allowed construction of mutants (R26A, R286A, or a mutant with substitution of an alanine cassette for Leu215-His220) that do not bind PDE4D5, but still interact with GPCRs normally (Baillie et al., 2007). Many of these mutants with a single function disabled by targeted mutagenesis were functionally tested in cells or even in animals. While in many cases the results turned out to be complex, often revealing unintended consequences, this line of experimentation has the potential to yield the almost “perfect” single-defect mutants necessary for analyzing the biological roles of individual protein functions. Below we describe the results and the lessons that were learned from using these mutants.
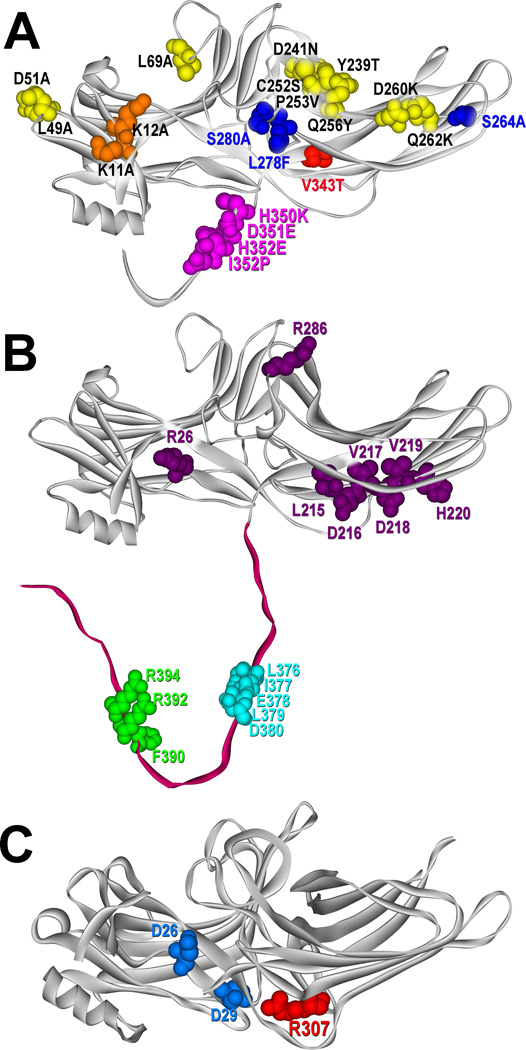
Fig. 2. Arrestin elements implicated in binding different partners.
A. The structure of arrestin-3 (PDB ID: 3P2D (Zhan et al., 2011)), showing the key lysines that bind receptor-attached phosphates (K11, K12), residues that bind other parts of receptors and determine arrestin preference for particular GPCRs (L49, D51, L69, Y239, D241, C252, P253, Q256, D260, and Q262) (Gimenez et al., 2012; Vishnivetskiy et al., 2011), and the elements responsible for arrestin-3’s ability to facilitate JNK3 activation (S264, L278, S280, V343, H350, D351, H352, and I353) (Seo et al., 2011). B. The structure of arrestin-3 (PDB ID: 3P2D (Zhan et al., 2011)), showing the key residues involved in PDE4D binding (R26, L215, D216, V217, D218, V219, H220, and R286) (Baillie et al., 2007), residues involved in clathrin binding (L376, I377, E378, L379, and D380) (Kim and Benovic, 2002; Krupnick et al., 1997), and residues engaged by the clathrin adaptor AP2 (F390, R392, and R394) (Kim and Benovic, 2002; Laporte et al., 1999). C. The structure of arrestin-2 (PDB ID: 1G4M (Han et al., 2001)), showing critical residues involved in MEK1 binding (D26 and D29) (Meng et al., 2009) and Arg-307, which is critical for cRaf1 binding (Coffa et al., 2011). All structures are rendered in ViewerPro; all highlighted residues are shown as CPK models. Parts of the C-tails not resolved in crystal structures were added manually. Color version of this figure is available online.
Table 1.
Arrestin mutations and their functional consequences.
| Arrestin subtype, species |
Mutation | Intended functional effect |
Known collateral effects |
Biological effects in cells and in vivo |
References |
|---|---|---|---|---|---|
| Arrestin- 1, bovine |
R175E, D296R, 3A1, E379Ter |
Binding to active unphosphorylated rhodopsin |
Reduced self- association; enhanced microtubule binding |
? | (Gray-Keller et al., 1997; Gurevich, 1998; Gurevich and Benovic, 1997; Hanson et al., 2006; Nair et al., 2005) |
| Arrestin- 1, mouse |
R176E, 3A1, E378Ter |
Binding to active unphosphorylated rhodopsin |
Reduced thermal stability and self- association; enhanced microtubule binding |
Faster shutoff of unphosphorylated rhodopsin; improved rod survival; cytotoxicity at supra- physiological expression |
(Nair et al., 2005; Song et al., 2013; Song et al., 2009) |
| Arrestin- 2, bovine |
R169E, 3A1 | Binding to active unphosphorylated GPCRs |
Increased flexibility; enhanced clathrin and AP-2 binding |
Shutoff of unphosphorylated GPCRs; faster GPCR cycling; reduced receptor down-regulation |
(Carter et al., 2005; Kim and Benovic, 2002; Kovoor et al., 1999; Pan, Gurevich, and Gurevich, 2003) |
| Arrestin- 3, bovine |
R170E, 3A1 | Binding to active unphosphorylated GPCRs |
Increased flexibility; enhanced clathrin and AP-2 binding |
Shutoff of unphosphorylated GPCRs |
(Celver et al., 2002) |
| Arrestin- 1, bovine |
K14A,K15A | Loss of binding to receptor-attached phosphates |
? | Loss of phosphorylation- induced increase in GPCR binding |
(Gimenez et al., 2012; Vishnivetskiy et al., 2000) |
| Arrestin- 2, bovine |
K12A,K15A | Loss of binding to receptor-attached phosphates |
? | Loss of phosphorylation- induced increase in GPCR binding |
(Gimenez et al., 2012) |
| Arrestin- 3, bovine |
K11A,K12A | Loss of binding to receptor-attached phosphates |
? | Loss of phosphorylation- induced increase in GPCR binding |
(Gimenez et al., 2012) |
| Arrestin- 3, bovine |
NCA2 | Loss of binding to unphosphorylated elements of many GPCRs |
Loss of GPCR binding |
(Gimenez et al., 2014; Gimenez et al., 2012) |
|
| Arrestin- 3, bovine |
KNC3 | Loss of interactions with all GPCRs tested |
Loss of ability to facilitate JNK3 activation |
Loss of interactions with all GPCRs tested; suppression of JNK3 activation by other scaffolds |
(Breitman et al., 2012; Gimenez et al., 2014; Gimenez et al., 2012; Gimenez et al., 2012) |
| Arrestin- 3, bovine |
Individual mutations of receptor- discriminator residues4 |
Changed receptor preference among β2AR, M2R, D1R, D2R, Y1R, and Y2R5 |
? | Changed receptor preference in cells among β2AR, M2R, D1R, D2R, Y1R, and Y2R5 |
(Gimenez et al., 2014; Gimenez et al., 2012) |
| Arrestin- 3, bovine |
V343T | Suppresses the ability to facilitate JNK3 activation |
? | Suppresses the ability to facilitate JNK3 activation |
(Seo et al., 2011) |
| Arrestin- 3, bovine |
R26A, R286A, L215-H220A |
Blocks PDE4D5 binding |
? | Blocks PDE4D5 binding, reduces arrestin- dependent decrease of PKA activity |
(Baillie et al., 2007) |
| Arrestin- 2, bovine |
Deletion of LIELD (residues 376–380); LIEL(376– 379)-AAEA |
Blocks clathrin binding |
? | Blocks clathrin binding; reduces arrestin- dependent GPCR internalization; reduces arrestin- dependent focal adhesion disassembly |
(Cleghorn et al., 2015; Kim and Benovic, 2002) |
| Arrestin- 3, bovine |
F391A, R395A |
Blocks AP-2 binding |
? | Blocks AP-2 binding; reduces arrestin- dependent GPCR internalization |
(Kim and Benovic, 2002; Laporte et al., 1999) |
| Arrestin- 3, bovine |
LIEF(373– 376)-AAEA |
Blocks clathrin binding |
? | Blocks clathrin binding; reduces arrestin- dependent focal adhesion disassembly |
(Cleghorn et al., 2015) |
| Arrestin- 2, bovine |
D26A, D29A | Blocks MEK1 binding |
? | Prevents arrestin- dependent ERK1/2 activation; enhances arrestin- dependent GPCR internalization |
(Meng et al., 2009) |
| Arrestin- 2, bovine |
R307A | Blocks cRaf1 binding |
? | Prevents arrestin- dependent ERK1/2 activation |
(Coffa et al., 2011) |
3A – triple alanine substitution that detaches the C-tail from the body of the N-domain of arrestin. The following residues were replaced with alanines: bovine arrestin-1, F375, V376, F377; mouse arrestin-1, L374, V375, F376; bovine arrestin-2, I386, V387, F388; bovine arrestin-5, I385, V386, F387.
NCA – alanine replacement of receptor-discriminator residues: L49A, D51A, R52A, L69A, Y239A, D241A, C252A, P253A, Q256A, D260A, Q262A.
KNC – a combination of NCA with alanine substitutions of lysines 11 and 12.
Mutations of receptor-discriminator residues tested: D51G, D51QN, L69A, Y231T, D241N, D241E, C252S+P253V, C252K+P253T, Q256Y, D260K, D260G, Q262K, Q262P, DDQ(260–262)REGCP, DDQ(260–262)NEGFP.
Receptor abbreviations: β2-adrenergic receptor, β2AR; M2 muscarinic receptor M2R; D1 and D2 dopamine receptors, D1R and D2R; Y1 and Y2 neuropeptide Y receptors, Y1R and Y2R.
3.2.2.1. Phosphorylation-independent arrestin mutants
Enhanced arrestins were tested in vitro (Gray-Keller et al., 1997), in Xenopus oocytes (Celver et al., 2002; Kovoor et al., 1999), in cultured cells (Pan, Gurevich, and Gurevich, 2003), and in vivo in living mice (Song et al., 2009). While the tests in simpler systems just confirmed the ability of these mutants to quench signaling by unphosphorylated GPCRs (Celver et al., 2002; Gray-Keller et al., 1997; Kovoor et al., 1999), the experiments in cultured cells revealed that the complex of unphosphorylated receptor with enhanced arrestin-2 internalizes normally, but then the receptor recycles back to the plasma membrane much faster than in the presence of WT arrestin-2 (Pan, Gurevich, and Gurevich, 2003). Interestingly, this rapid cycling apparently prevented receptor degradation even after 24 h of continuous agonist treatment (Pan, Gurevich, and Gurevich, 2003). Thus, the ability of arrestin-2 to bind unphosphorylated GPCRs protected the receptor from down-regulation.
Disease-causing mutations in various proteins, including GPCRs (Schoneberg et al., 2004), fall into two categories, gain-of-function and loss-of-function. The latter are usually recessive, because the second correct allele supplies the normal protein, so that the disease only develops when both alleles are damaged. In this case, the delivery of DNA driving expression of WT protein is likely to be sufficient to cure the disease. In contrast, gain-of-function mutations are dominant. For example, when a mutant GPCR signals too much due to constitutive activity or cannot be shut down by the normal GRK-arrestin-mediated mechanism because it has lost its phosphorylation sites, the presence of a perfectly good receptor encoded by the other allele does not alleviate the problem. There are no commonly accepted strategies to deal with gain-of-function mutations. One possibility is compensational gene therapy: to counteract excessive GPCR signaling with an arrestin mutant that suppresses it more effectively then WT protein.
Testing of enhanced mouse arrestin-1 mutant for the ability to compensate for defects of rhodopsin phosphorylation yielded some expected and a lot of unexpected results (Song et al., 2009). It turned out that polar core mutant R176E (Table 1) was not stable enough for in vivo expression, so two more stable mutants were chosen: 1) arrestin-1-3A, where the C-tail was forcibly detached by triple alanine substitution, removing bulky hydrophobic residues that anchored it to the N-domain (Hirsch et al., 1999), mimicking its detachment upon receptor binding (Hanson et al., 2006; Vishnivetskiy et al., 2010; Zhuo et al., 2014), and 2) arrestin-1-(1–377), where the C-tail was deleted (Song et al., 2009). The expression of mouse arrestin-1-3A instead of WT arrestin-1 in rods lacking rhodopsin kinase (RK KO) improved retinal morphology, prolonged photoreceptor survival, and improved the functional performance of mouse rods (Song et al., 2009), demonstrating the feasibility of functional compensation. This was an expected result, and it is quite encouraging. However, the rate of photoresponse shutoff in “compensated” rods was significantly slower that in WT with the normal complement of RK, suggesting that more potent phosphorylation-independent mutants are needed. Additional mutations further enhancing arrestin-1 binding to unphosphorylated light-activated rhodopsin without compromising thermal stability were introduced (Vishnivetskiy et al., 2013), and these new mutants need to be tested in vivo. In photoreceptor cells, rhodopsin resides in a specialized “signaling” compartment, the outer segment, whereas in the dark, the bulk of arrestin-1 is localized in the inner segment and cell body, likely due to its binding to microtubules, abundant in these compartments (Gurevich et al., 2011). Upon illumination, arrestin-1 moves to the outer segment due to its binding to light-activated phosphorylated rhodopsin (Nair et al., 2005). Interestingly, both enhanced mutants have higher affinity for microtubules than WT arrestin-1 (Nair et al., 2004) and upon illumination move to the outer segment slower than WT protein (Nair et al., 2005). From a practical standpoint, the most useful unexpected result was the finding that in rods expressing constitutively active rhodopsin, which constitutively binds arrestin, the low affinity of arrestin-1 for the clathrin adaptor AP2 is sufficient to recruit significant amounts of it to the outer segment, where in WT mice AP2 is undetectable (Moaven et al., 2013). It appears that the depletion of AP2 contributes to the death of rod photoreceptors, as the expression of truncated arrestin-1 lacking the AP2 binding site protects these rods (Moaven et al., 2013). These data suggest that an arrestin-1 mutant that combines high binding to unphosphorylated active rhodopsin with a lack of AP2 binding has higher therapeutic potential.
Another result that might be useful for future gene therapy is the discovery that high levels of arrestin-1-3A mutant per se, even in the dark, are harmful for photoreceptors (Song et al., 2013). Interestingly, similar supra-physiological levels of WT arrestin-1 are totally harmless (Song et al., 2011). Since the 3A mutation significantly reduces the ability of arrestin-1 to self-associate (Song et al., 2013), whereas robust oligomerization at biologically relevant concentrations in rods is a common feature of bovine, mouse, and human arrestin-1 (Kim et al., 2011), it was hypothesized that the monomeric nature of the mutant makes it cytotoxic, in contrast to harmless oligomers (Song et al., 2013). The idea that defective self-association per se is harmful needs to be tested experimentally in vivo, because 3A mutation also detaches the C-tail of arrestin-1, thereby exposing its AP2 binding site, and recruitment of AP2 to the abundant arrestin-1 appears to harm rods (Moaven et al., 2013). The third potentially useful finding was that relatively low levels of enhanced arrestin-1, down to 2.5% of the WT level, are sufficient for photoreceptor health and functional performance (Cleghorn et al., 2011; Song et al., 2011). In summary, it appears that therapeutically usable phosphorylation-independent forms of arrestin-1 should either have normal self-association or be expressed at relatively low levels to prevent cytotoxicity. In all cases, it would be beneficial to disable its AP2 binding site.
Importantly, these data show the difficulty of changing one arrestin function without affecting others: mutations that rendered arrestin-1 phosphorylation-independent also increased its affinity for microtubules, impaired its self-association, likely enhanced its binding to AP2 in a rhodopsin-independent manner, and possibly changed some other functions that nobody tested. These unintended changes certainly complicate matters and must be investigated. Still, design and characterization of enhanced arrestin mutants remains a promising direction for therapeutic development because they have the capacity to shut off signaling by unphosphorylated rhodopsin, which WT arrestin-1 cannot do.
3.2.2.2. Clathrin and AP2 binding-deficient arrestins
As expected, non-visual arrestins in which the well-defined binding sites for clathrin, AP2 or both in the arrestin C-tail are disabled still bind receptors, but do not support GPCR internalization (Goodman et al., 1996; Kim and Benovic, 2002; Laporte et al., 1999). These studies clearly demonstrated that direct binding to arrestins of both clathrin and AP2 contributes to GPCR internalization via the coated pits. Recently, clathrin binding-deficient mutants of arrestin-2 and -3 were used to demonstrate that arrestin-dependent clathrin recruitment to microtubules targeting focal adhesions plays an important role in focal adhesion disassembly, which is necessary for normal cell spreading and motility (Cleghorn et al., 2015). No effects of these mutations on other arrestin functions have been detected so far.
3.2.2.3. Receptor preference
Arrestins have an extensive receptor-binding surface that includes large parts of the concave sides of both arrestin domains (Hanson et al., 2006; Hanson and Gurevich, 2006; Vishnivetskiy et al., 2004). However, fairly few residues within this surface appear to determine the receptor specificity of arrestins (Vishnivetskiy et al., 2011). Substitutions of individual “receptor-discriminator” residues significantly changes the relative ability of the very promiscuous arrestin-3 to bind different GPCRs. Double mutations yielded proteins with up to 60-fold preference for certain receptors over others (Gimenez et al., 2014; Gimenez et al., 2012). Most cells express numerous GPCR subtypes. Constructed, more receptor-specific arrestin mutants can be used to determine the contribution of different receptors to particular cellular functions in cultured cells and even in vivo. Gain-of-function (GOF) GPCR mutations cause a variety of human disorders, from retinal degeneration to several forms of cancer (Restagno et al., 1993; Schoneberg et al., 2004; Stoy and Gurevich, 2015). Conceivably, excessive signaling by a GOF mutant can be subdued by an enhanced arrestin with higher than normal ability to block G protein-mediated signaling. The feasibility of this approach was demonstrated in rod photoreceptors in vivo (Song et al., 2009). While non-visual arrestins can be pre-activated by mutations homologous to those that pre-activate visual arrestin-1 (Celver et al., 2002; Gurevich et al., 1997; Kovoor et al., 1999; Pan, Gurevich, and Gurevich, 2003), enhanced non-visual arrestin must be made receptor-specific to target only the mutant receptor and avoid suppressing signaling by other perfectly normal GPCRs co-expressed by the same cell. Thus, the combination of activating mutations and those that increase receptor specificity must be tested in cells. Then potential usefulness of receptor-specific, enhanced non-visual arrestins for gene therapy should be tested in vivo.
However, it appears that additional arrestin residues important for receptor discrimination need to be identified: mutagenesis of the ten residues tested did not significantly reduce the binding to D1 dopamine and Y1 neuropeptide Y receptors (Gimenez et al., 2014; Gimenez et al., 2012), indicating that other elements must be involved. Simultaneous replacement of all receptor-discriminator residues with alanines prevented the binding of arrestin-2 and -3 to most GPCRs, as could be expected (Vishnivetskiy et al., 2011). Unexpectedly, these mutations on the receptor-binding surface (Hanson et al., 2006; Hanson and Gurevich, 2006; Vishnivetskiy et al., 2011) apparently changed the other side of the molecule, where MAP kinases bind (Zhan et al., 2014)(Fig. 2). As a result, the arrestin-3 mutant in which GPCR binding was disabled by these ten alanine substitutions plus replacement of the two phosphate-binding lysines with alanines lost the ability to facilitate JNK3 activation in cells, even though the binding to all kinases in the ASK1-MKK4-JNK3 cascade was preserved (Breitman et al., 2012). Similarly, quite a few mutations on the non-receptor-binding convex side of the two domains significantly affected the ability of arrestin-2 to bind receptors (Coffa et al., 2011). Thus, it appears that we still do not fully understand the structural communications between the two sides of the arrestin molecule.
3.2.2.4,5. Arrestins that do not bind MEK1 or c-Raf1
Modified arrestin-2 was constructed by replacing the key MEK1-binding residues Asp26 and Asp29 (Meng et al., 2009). As could be expected, the mutations prevented arrestin-dependent ERK1/2 activation and precluded arrestin-2 phosphorylation at the C-terminal Ser-412. In agreement with previous data that the phosphorylation of the arrestin-2 C-terminus by ERK1/2 prevents its binding to clathrin (Lin et al., 1997), mutations preventing MEK1 binding facilitated arrestin-dependent internalization of agonist-stimulated β2AR (Meng et al., 2009). The R307A mutation in arrestin-2 prevented c-Raf1 binding and arrestin-dependent receptor-induced ERK1/2 activation (Coffa et al., 2011). As c-Raf1 is an upstream kinase that activates MEK1, it is reasonable to expect that the R307A mutation should also reduce arrestin-2 phosphorylation by ERK1/2, thereby facilitating receptor internalization. This prediction needs to be tested experimentally.
3.2.2.6. Arrestins that do not bind PDE4
Arrestins have been shown to recruit several isoforms of PDE4D to β2AR to facilitate the degradation of cAMP in the vicinity of the receptor, thereby facilitating the desensitization of β2AR, which couples to Gs and activates adenylyl cyclase (Perry et al., 2002). The identification of PDE binding sites in both domains of arrestin-3 enabled the construction of mutants that do not bind PDE and therefore do not recruit it to activated β2AR (Baillie et al., 2007). Expression of WT arrestin-3 in MEFs lacking both non-visual arrestins significantly reduced PKA activity in response to β2AR stimulation. In contrast, the expression of PDE binding-deficient mutants that interact with β2AR normally did not affect PKA activity as much, indicating that PDE recruitment is necessary for rapid desensitization of Gs-coupled receptors (Baillie et al., 2007).
4. Conclusions and future prospects
Collectively, existing data suggest that it is feasible to construct mutants of multi-functional proteins with a single function disabled, although in many cases we need to know much more about the structural effects of particular mutations to achieve this level of selectivity. Virtually every signaling-biased mutant of a multi-functional protein is a useful research tool and many also have clear therapeutic potential (Gurevich and Gurevich, 2012). Wherever feasible, expression of mono-functional elements (in many cases separated domains) of multi-functional proteins is equally instructive in mechanistic studies and can also be used for gene therapy. Functionally defined protein mutants are widely used in cell culture studies, albeit their repertoire remains limited. However, an analogous strategy could also be successful in vivo, when mutants are expressed in living animals, particularly on the knockout background in the rescue paradigm. Such experiments would provide information on the role of individual functions of the protein responsible for the phenotype observed in knockout animals. If knockout is intended to represent a disease state associated with loss-of-function mutations of the protein in question, knowing which function of the multi-functional protein is involved would be valuable in exploring therapeutic options. The combined use of global knockout lines, many of which are already available, and fine manipulation of the protein function via viral transfer of functionally defined mutants into specific cells could serve as a relatively cheap and fast alternative to the generation of conditional and cell-specific knockin and knockout mouse lines. Such approaches would allow for relatively rapid teasing apart of the complex functions of proteins in normal and diseased tissues.
Acknowledgments
Supported in part by NIH grants GM077561, GM109955, and EY011500 (VVG), NS045117 and NS065868 (EVG).
Abbreviations
- GPCR
G protein-coupled receptor
- WT
wild type
- GRK
G protein-coupled receptor kinase
- MEFs
mouse embryonic fibroblasts
- ARRDC
arrestin domain-containing protein
- PD
Parkinson’s disease
- LID
L-DOPA-induced dyskinesia
- PH
pleckstrin homology
- βARKct
the C-terminus of GRK 2 containing the PH domain
- RH
RGS homology
- RK
rhodopsin kinase (systematic name: GRK1)
- KO
knockout
- GOF
gain-of-function
- β2AR
β2-adrenergic receptor
Footnotes
Declaration of Interest
The authors declare that they have no conflict of interest.
References
- 1.Ahmed MR, Berthet A, Bychkov E, Porras G, Li Q, Bioulac BH, Carl YT, Bloch B, Kook S, Aubert I, Dovero S, Doudnikoff E, Gurevich VV, Gurevich EV, Bezard E. Lentiviral overexpression of GRK6 alleviates L-dopa-induced dyskinesia in experimental Parkinson's disease. Sci Transl Med. 2010;2:28ra28. doi: 10.1126/scitranslmed.3000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahmed MR, Bychkov E, Li L, Gurevich VV, Gurevich EV. GRK3 suppresses L-DOPA-induced dyskinesia in the rat model of Parkinson’s disease via its RGS homology domain. Sci Rep. 2015;5:10920. doi: 10.1038/srep10920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ahmed MR, Zhan X, Song X, Kook S, Gurevich VV, Gurevich EV. Ubiquitin ligase parkin promotes Mdm2-arrestin interaction but inhibits arrestin ubiquitination. Biochemistry. 2011;50:3749–3763. doi: 10.1021/bi200175q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akhter SA, Eckhart AD, Rockman HA, Shotwell K, Lefkowitz RJ, Koch WJ. In vivo inhibition of elevated myocardial beta-adrenergic receptor kinase activity in hybrid transgenic mice restores normal beta-adrenergic signaling and function. Circulation. 1999;100:648–653. doi: 10.1161/01.cir.100.6.648. [DOI] [PubMed] [Google Scholar]
- 5.Attramadal H, Arriza JL, Aoki C, Dawson TM, Codina J, Kwatra MM, Snyder SH, Caron MG, Lefkowitz RJ. Beta-arrestin2, a novel member of the arrestin/beta-arrestin gene family. J Biol Chem. 1992;267:17882–17890. [PubMed] [Google Scholar]
- 6.Baillie GS, Adams DR, Bhari N, Houslay TM, Vadrevu S, Meng D, Li X, Dunlop A, Milligan G, Bolger GB, Klussmann E, Houslay MD. Mapping binding sites for the PDE4D5 cAMP-specific phosphodiesterase to the N- and C-domains of beta-arrestin using spot-immobilized peptide arrays. Biochem J. 2007;404:71–80. doi: 10.1042/BJ20070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Benovic JL, DeBlasi A, Stone WC, Caron MG, Lefkowitz RJ. Beta-adrenergic receptor kinase: primary structure delineates a multigene family. Science. 1989;246:235–240. doi: 10.1126/science.2552582. [DOI] [PubMed] [Google Scholar]
- 8.Bhandari D, Trejo J, Benovic JL, Marchese A. Arrestin-2 interacts with the ubiquitin-protein isopeptide ligase atrophin-interacting protein 4 and mediates endosomal sorting of the chemokine receptor CXCR4. J Biol Chem. 2007;282:36971–36979. doi: 10.1074/jbc.M705085200. [DOI] [PubMed] [Google Scholar]
- 9.Bockaert J, Pin JP. Molecular tinkering of G protein-coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–1729. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Breitman M, Kook S, Gimenez LE, Lizama BN, Palazzo MC, Gurevich EV, Gurevich VV. Silent scaffolds: inhibition of c-Jun N-terminal kinase 3 activity in the cell by a dominant-negative arrestin-3 mutant. J Biol Chem. 2012;287:19653–19664. doi: 10.1074/jbc.M112.358192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruchas MR, Macey TA, Lowe JD, Chavkin C. Kappa opioid receptor activation of p38 MAPK is GRK3- and arrestin-dependent in neurons and astrocytes. J Biol Chem. 2006;281:18081–18089. doi: 10.1074/jbc.M513640200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caplen NJ. RNAi as a gene therapy approach. Expert Opin Biol Ther. 2003;3:575–586. doi: 10.1517/14712598.3.4.575. [DOI] [PubMed] [Google Scholar]
- 13.Carman CV, Benovic JL. G-protein-coupled receptors: turn-ons and turn-offs. Curr Opin Neurobiol. 1998;8:335–344. doi: 10.1016/s0959-4388(98)80058-5. [DOI] [PubMed] [Google Scholar]
- 14.Carman CV, Parent JL, Day PW, Pronin AN, Sternweis PM, Wedegaertner PB, Gilman AG, Benovic JL, Kozasa T. Selective regulation of Galpha(q/11) by an RGS domain in the G protein-coupled receptor kinase, GRK2. J Biol Chem. 1999;274:34483–34492. doi: 10.1074/jbc.274.48.34483. [DOI] [PubMed] [Google Scholar]
- 15.Carter JM, Gurevich VV, Prossnitz ER, Engen JR. Conformational differences between arrestin2 and pre-activated mutants as revealed by hydrogen exchange mass spectrometry. J Mol Biol. 2005;351:865–878. doi: 10.1016/j.jmb.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 16.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J Biol Chem. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 17.Cleghorn WM, Branch KM, Kook S, Arnette C, Bulus N, Zent R, Kaverina I, Gurevich EV, Weaver AM, Gurevich VV. Arrestins regulate cell spreading and motility via focal adhesion dynamics. Mol Biol Cell. 2015 doi: 10.1091/mbc.E14-02-0740. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cleghorn WM, Tsakem EL, Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, Gurevich VV. Progressive reduction of its expression in rods reveals two pools of arrestin-1 in the outer segment with different roles in photoresponse recovery. PLoS One. 2011;6:e22797. doi: 10.1371/journal.pone.0022797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coffa S, Breitman M, Hanson SM, Callaway K, Kook S, Dalby KN, Gurevich VV. The Effect of Arrestin Conformation on the Recruitment of c-Raf1, MEK1, and ERK1/2 Activation. PLoS One. 2011;6:e28723. doi: 10.1371/journal.pone.0028723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coffa S, Breitman M, Spiller BW, Gurevich VV. A single mutation in arrestin-2 prevents ERK1/2 activation by reducing c-Raf1 binding. Biochemistry. 2011;50:6951–6958. doi: 10.1021/bi200745k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cookson MR. The role of leucine-rich repeat kinase 2 (LRRK2) in Parkinson's disease. Nat Rev Neurosci. 2010;11:791–797. doi: 10.1038/nrn2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daaka Y, Pitcher JA, Richardson M, Stoffel RH, Robishaw JD, Lefkowitz RJ. Receptor and G betagamma isoform-specific interactions with G protein-coupled receptor kinases. Proc Natl Acad Sci U S A. 1997;94:2180–2185. doi: 10.1073/pnas.94.6.2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Danial NN, Korsmeyer SJ. Cell death: critical control points. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 24.Day PW, Carman CV, Sterne-Marr R, Benovic JL, Wedegaertner PB. Differential interaction of GRK2 with members of the G alpha q family. Biochemistry. 2003;42:9176–9184. doi: 10.1021/bi034442+. [DOI] [PubMed] [Google Scholar]
- 25.Day PW, Wedegaertner PB, Benovic JL. Analysis of G-protein-coupled receptor kinase RGS homology domains. Methods Enzymol. 2004;390:295–310. doi: 10.1016/S0076-6879(04)90019-5. [DOI] [PubMed] [Google Scholar]
- 26.DeFea KA, Vaughn ZD, O'Bryan EM, Nishijima D, Déry O, Bunnett NW. The proliferative and antiapoptotic effects of substance P are facilitated by formation of a beta -arrestin-dependent scaffolding complex. Proc Natl Acad Sci U S A. 2000;97:11086–11091. doi: 10.1073/pnas.190276697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. Beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 28.Farrer M, Chan P, Chen R, Tan L, Lincoln S, Hernandez D, Forno L, Gwinn-Hardy K, Petrucelli L, Hussey J, Singleton A, Tanner C, Hardy J, Langston W. Lewy bodies and parkinsonism in families with parkin mutations. Ann Neurol. 2001;50:293–300. doi: 10.1002/ana.1132. [DOI] [PubMed] [Google Scholar]
- 29.Feil S, Valtcheva NRF. Inducible Cre mice. Methods Mol Biol. 2009;530:343–363. doi: 10.1007/978-1-59745-471-1_18. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs S, Nakazawa M, Maw M, Tamai M, Oguchi Y, Gal A. A homozygous 1-base pair deletion in the arrestin gene is a frequent cause of Oguchi disease in Japanese. Nat Genet. 1995;10:360–362. doi: 10.1038/ng0795-360. [DOI] [PubMed] [Google Scholar]
- 31.Gibbs CS, Zoller MJ. Rational scanning mutagenesis of a protein kinase identifies functional regions involved in catalysis and substrate interactions. J Biol Chem. 1991;266:8923–8931. [PubMed] [Google Scholar]
- 32.Gimenez LE, Babilon S, Wanka L, Beck-Sickinger AG, Gurevich VV. Mutations in arrestin-3 differentially affect binding to neuropeptide Y receptor subtypes. Cell Signal. 2014;26:1523–1531. doi: 10.1016/j.cellsig.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gimenez LE, Kook S, Vishnivetskiy SA, Ahmed MR, Gurevich EV, Gurevich VV. Role of receptor-attached phosphates in binding of visual and non-visual arrestins to G protein-coupled receptors. J Biol Chem. 2012;287:9028–9040. doi: 10.1074/jbc.M111.311803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gimenez LE, Vishnivetskiy SA, Baameur F, Gurevich VV. Manipulation of very few receptor discriminator residues greatly enhances receptor specificity of non-visual arrestins. J Biol Chem. 2012;287:29495–29505. doi: 10.1074/jbc.M112.366674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goodman OB, Jr, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 36.Gray-Keller MP, Detwiler PB, Benovic JL, Gurevich VV. Arrestin with a single amino acid sustitution quenches light-activated rhodopsin in a phosphorylation0independent fasion. Biochemistry. 1997;36:7058–7063. doi: 10.1021/bi963110k. [DOI] [PubMed] [Google Scholar]
- 37.Gurevich EV, Ahmed MR, Carl YT. In vivo gene silencing by virally delivered microRNA. In: Brambilla R, editor. Viral Vector Approaches in Neurobiology and Brain Diseases. New York: Humana Press; 2014. pp. 245–268. [Google Scholar]
- 38.Gurevich EV, Gurevich VV. Arrestins are ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7:236. doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gurevich EV, Gurevich VV. Dopamine receptors and the treatment of Parkinson’s disease. In: Neve K, editor. Dopamine Receptors. New York: Humana Press; 2010. pp. 525–84. [Google Scholar]
- 40.Gurevich EV, Tesmer JJ, Mushegian A, Gurevich VV. G protein-coupled receptor kinases: More than just kinases and not only for GPCRs. Pharmacol Ther. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gurevich VV. The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. J Biol Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]
- 42.Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin: diverse functional roles of positively charged residues within the phosphorylation-recignition region of arrestin. J Biol Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 43.Gurevich VV, Benovic JL. Mechanism of phosphorylation-recognition by visual arrestin and the transition of arrestin into a high affinity binding state. Mol Pharmacol. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- 44.Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 45.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:59–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 46.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G protein-coupled receptors. Pharm Ther. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gurevich VV, Gurevich EV. Synthetic biology with surgical precision: Targeted reengineering of signaling proteins. Cell Signal. 2012;24:1899–1908. doi: 10.1016/j.cellsig.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurevich VV, Gurevich EV. Extensive shape shifting underlies functional versatility of arrestins. Curr Opin Cell Biol. 2014;27:1–9. doi: 10.1016/j.ceb.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gurevich VV, Hanson SM, Song X, Vishnivetskiy SA, Gurevich EV. The functional cycle of visual arrestins in photoreceptor cells. Prog Retin Eye Res. 2011;30:405–430. doi: 10.1016/j.preteyeres.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gurevich VV, Pals-Rylaarsdam R, Benovic JL, Hosey MM, Onorato JJ. Agonist-receptor-arrestin, an alternative ternary complex with high agonist affinity. J Biol Chem. 1997;272:28849–28852. doi: 10.1074/jbc.272.46.28849. [DOI] [PubMed] [Google Scholar]
- 51.Gurevich VV, Richardson RM, Kim CM, Hosey MM, Benovic JL. Binding of wild type and chimeric arrestins to the m2 muscarinic cholinergic receptor. J Biol Chem. 1993;268:16879–16882. [PubMed] [Google Scholar]
- 52.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 53.Han SO, Kommaddi RP, Shenoy SK. Distinct roles for β-arrestin2 and arrestin-domain-containing proteins in β2 adrenergic receptor trafficking. EMBO Rep. 2013;14:164–171. doi: 10.1038/embor.2012.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanson SM, Francis DJ, Vishnivetskiy SA, Klug CS, Gurevich VV. Visual arrestin binding to microtubules involves a distinct conformational change. J Biol Chem. 2006;281:9765–9772. doi: 10.1074/jbc.M510738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. Proc Natl Acad Sci U S A. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J Biol Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 A crystal structure of visual arrestin: a model for arrestin's regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 58.Hüttemann M, Pecina P, Rainbolt M, Sanderson TH, Kagan VE, Samavati L, Doan JW, Lee I. The multiple functions of cytochrome c and their regulation in life and death decisions of the mammalian cell: From respiration to apoptosis. Mitochondrion. 2011;11:369–381. doi: 10.1016/j.mito.2011.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Imamura T, Huang J, Dalle S, Ugi S, Usui I, Luttrell LM, Miller WE, Lefkowitz RJ, Olefsky JM. beta -Arrestin-mediated recruitment of the Src family kinase Yes mediates endothelin-1-stimulated glucose transport. J Biol Chem. 2001;276:43663–43667. doi: 10.1074/jbc.M105364200. [DOI] [PubMed] [Google Scholar]
- 60.Jonchere V, Bennett D. Validating RNAi phenotypes in Drosophila using a synthetic RNAi-resistant transgene. PLoS One. 2013;8:e70489. doi: 10.1371/journal.pone.0070489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Khan NL, Scherfler C, Graham E, Bhatia KP, Quinn N, Lees AJ, Brooks DJ, Wood NW, Piccini P. Dopaminergic dysfunction in unrelated, asymptomatic carriers of a single parkin mutation. Neurology. 2005;64:134–136. doi: 10.1212/01.WNL.0000148725.48740.6D. [DOI] [PubMed] [Google Scholar]
- 62.Kim M, Hanson SM, Vishnivetskiy SA, Song X, Cleghorn WM, Hubbell WL, Gurevich VV. Robust self-association is a common feature of mammalian visual arrestin-1. Biochemistry. 2011;50:2235–2242. doi: 10.1021/bi1018607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim YM, Benovic JL. Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. J Biol Chem. 2002;277:30760–30768. doi: 10.1074/jbc.M204528200. [DOI] [PubMed] [Google Scholar]
- 64.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, Yokochi M, Mizuno Y, Shimizu N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:544–545. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 65.Kohout TA, Lin FS, Perry SJ, Conner DA, Lefkowitz RJ. beta-Arrestin 1 and 2 differentially regulate heptahelical receptor signaling and trafficking. Proc Nat Acad Sci USA. 2001;98:1601–1606. doi: 10.1073/pnas.041608198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kong G, Penn R, Benovic JL. A beta-adrenergic receptor kinase dominant negative mutant attenuates desensitization of the beta 2-adrenergic receptor. J Biol Chem. 1994 May 6;269(18):13084–13087. [PubMed] [Google Scholar]
- 67.Kook S, Zhan X, Kaoud TS, Dalby KN, Gurevich VV, Gurevich EV. Arrestin-3 binds JNK1α1 and JNK2α2 and facilitates the activation of these ubiquitous JNK isoforms in cells via scaffolding. J Biol Chem. 2013;288:37332–37342. doi: 10.1074/jbc.M113.510412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. Targeted construction of phosphorylation-independent b-arrestin mutants with constitutive activity in cells. J Biol Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 69.Krupnick JG, Goodman OB, Jr, Keen JH, Benovic JL. Arrestin/clathrin interaction. Localization of the clathrin binding domain of nonvisual arrestins to the carboxy terminus. J Biol Chem. 1997;272:15011–15016. doi: 10.1074/jbc.272.23.15011. [DOI] [PubMed] [Google Scholar]
- 70.Krupnick JG, Gurevich VV, Benovic JL. Mechanism of quenching of phototransduction. Binding competition between arrestin and transducin for phosphorhodopsin. J Biol Chem. 1997;272:18125–18131. doi: 10.1074/jbc.272.29.18125. [DOI] [PubMed] [Google Scholar]
- 71.Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 72.Laporte SA, Oakley RH, Zhang J, Holt JA, Ferguson sSG, Caron MG, Barak LS. The 2-adrenergic receptor/arrestin complex recruits the clathrin adaptor AP-2 during endocytosis. Proc Nat Acad Sci USA. 1999;96:3712–3717. doi: 10.1073/pnas.96.7.3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. doi: 10.1038/35093537. [DOI] [PubMed] [Google Scholar]
- 74.Lin FT, Krueger KM, Kendall HE, Daake Y, Fredericks ZL, Pitcher JA, Lefkowitz RJ. Clathrin-mediated endocytosis of the beta-adrenergic receptor is regulated by phosphorylation/dephosphorylation of beta-arrestin1. J Biol Chem. 1997;272:31051–31057. doi: 10.1074/jbc.272.49.31051. [DOI] [PubMed] [Google Scholar]
- 75.Lohse MJ, Andexinger S, Pitcher J, Trukawinski S, Codina J, Faure JP, Caron MG, Lefkowitz RJ. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J Biol Chem. 1992;267:8558–8564. [PubMed] [Google Scholar]
- 76.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 77.Lücking CB, Dürr A, Bonifati V, Vaughan J, De Michele G, Gasser T, Harhangi BS, Meco G, Denèfle P, Wood NW, Agid Y, Brice A French Parkinson's Disease Genetics Study Group, European Consortium on Genetic Susceptibility in Parkinson's Disease. Association between Early-Onset Parkinson's Disease and Mutations in the Parkin Gene. N Engl J Med. 2000;342:1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 78.Luttrell LM, Ferguson SS, Daaka Y, Miller WE, Maudsley S, Della Rocca GJ, Lin F, Kawakatsu H, Owada K, Luttrell DK, Caron MG, Lefkowitz RJ. Beta-arrestin-dependent formation of beta2 adrenergic receptor-Src protein kinase complexes. Science. 1999;283:655–661. doi: 10.1126/science.283.5402.655. [DOI] [PubMed] [Google Scholar]
- 79.Luttrell LM, Roudabush FL, Choy EW, Miller WE, Field ME, Pierce KL, Lefkowitz RJ. Activation and targeting of extracellular signal-regulated kinases by beta-arrestin scaffolds. Proc Natl Acad Sci U S A. 2001;98:2449–2454. doi: 10.1073/pnas.041604898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Manteniotis S, Lehmann R, Flegel C, Vogel F, Hofreuter A, Schreiner BS, Altmüller J, Becker C, Schöbel N, Hatt H, Gisselmann G. Comprehensive RNA-Seq Expression Analysis of Sensory Ganglia with a Focus on Ion Channels and GPCRs in Trigeminal Ganglia. PLoS One. 2013;8:e79523. doi: 10.1371/journal.pone.0079523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McDonald PH, Chow CW, Miller WE, Laporte SA, Field ME, Lin FT, Davis RJ, Lefkowitz RJ. Beta-arrestin 2: a receptor-regulated MAPK scaffold for the activation of JNK3. Science. 2000;290:1574–1577. doi: 10.1126/science.290.5496.1574. [DOI] [PubMed] [Google Scholar]
- 82.McDonald PH, Cote NL, Lin FT, Premont RT, Pitcher JA, Lefkowitz RJ. Identification of NSF as a beta-arrestin1-binding protein. Implications for beta2-adrenergic receptor regulation. J Biol Chem. 1999;274:10677–10680. doi: 10.1074/jbc.274.16.10677. [DOI] [PubMed] [Google Scholar]
- 83.Meng D, Lynch MJ, Huston E, Beyermann M, Eichhorst J, Adams DR, Klusmann E, Houslay MD, Baillie GS. MEK1 binds directly to betaarrestin1, influencing both its phosphorylation by ERK and the timing of its isoprenaline-stimulated internalization. J Biol Chem. 2009;284:11425–11435. doi: 10.1074/jbc.M806395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Moaven H, Koike Y, Jao CC, Gurevich VV, Langen R, Chen J. Visual arrestin interaction with clathrin adaptor AP-2 regulates photoreceptor survival in the vertebrate retina. Proc Natl Acad Sci U S A. 2013;110:9463–9468. doi: 10.1073/pnas.1301126110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nabhan JF, Pan H, Lu Q. Arrestin domain-containing protein 3 recruits the NEDD4 E3 ligase to mediate ubiquitination of the beta2-adrenergic receptor. EMBO Rep. 2010;11:605–611. doi: 10.1038/embor.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nair KS, Hanson SM, Kennedy MJ, Hurley JB, Gurevich VV, Slepak VZ. Direct binding of visual arrestin to microtubules determines the differential subcellular localization of its splice variants in rod photoreceptors. J Biol Chem. 2004;279:41240–41248. doi: 10.1074/jbc.M406768200. [DOI] [PubMed] [Google Scholar]
- 87.Nair KS, Hanson SM, Mendez A, Gurevich EV, Kennedy MJ, Shestopalov VI, Vishnivetskiy SA, Chen J, Hurley JB, Gurevich VV, Slepak VZ. Light-dependent redistribution of arrestin in vertebrate rods is an energy-independent process governed by protein-protein interactions. Neuron. 2005;46:555–567. doi: 10.1016/j.neuron.2005.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakajima K, Wess J. Design and functional characterization of a novel, arrestin-biased designer G protein-coupled receptor. Mol Pharmacol. 2012;82:575–582. doi: 10.1124/mol.112.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nelson CD, Perry SJ, Regier DS, Prescott SM, Topham MK, Lefkowitz RJ. Targeting of diacylglycerol degradation to M1 muscarinic receptors by beta-arrestins. Science. 2007;315:663–666. doi: 10.1126/science.1134562. [DOI] [PubMed] [Google Scholar]
- 90.Ohno S, Konno Y, Akita Y, Yano A, Suzuki K. A point mutation at the putative ATP-binding site of protein kinase C alpha abolishes the kinase activity and renders it down-regulation-insensitive. A molecular link between autophosphorylation and down-regulation. J Biol Chem. 1990;265:6296–6300. [PubMed] [Google Scholar]
- 91.Pan L, Gurevich EV, Gurevich VV. The nature of the arrestin x receptor complex determines the ultimate fate of the internalized receptor. J Biol Chem. 2003;278:11623–11632. doi: 10.1074/jbc.M209532200. [DOI] [PubMed] [Google Scholar]
- 92.Perry SJ, Baillie GS, Kohout TA, McPhee I, Magiera MM, Ang KL, Miller WE, McLean AJ, Conti M, Houslay MD, Lefkowitz RJ. Targeting of cyclic AMP degradation to beta 2-adrenergic receptors by beta-arrestins. Science. 2002;298:834–836. doi: 10.1126/science.1074683. [DOI] [PubMed] [Google Scholar]
- 93.Peterson SM, Pack TF, Wilkins AD, Urs NM, Urban DJ, Bass CE, Lichtarge O, Caron MG. Elucidation of G-protein and β-arrestin functional selectivity at the dopamine D2 receptor. Pro Natl Acad Sci USA. 2015;112:7097–7102. doi: 10.1073/pnas.1502742112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pramstaller PP, Scaravilli F, Eskelson C, Pepivani I, Hedrich K, Gonzales-McNeal M, Hilker R, Kramer PL, Klein C. Lewy body Parkinson's disease in a large pedigree with 77 Parkin mutation carriers. Ann Neurol. 2005;58:411–422. doi: 10.1002/ana.20587. [DOI] [PubMed] [Google Scholar]
- 95.Pronin A, Levay K, Velmeshev D, Faghihi M, Shestopalov VI, Slepak VZ. Expression of olfactory signaling genes in the eye. PLoS One. 2014;9:e96435. doi: 10.1371/journal.pone.0096435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Raveh A, Cooper A, Guy-David L, Reuveny E. Nonenzymatic rapid control of GIRK channel function by a G protein-coupled receptor kinase. Cell. 2010;143:750–760. doi: 10.1016/j.cell.2010.10.018. [DOI] [PubMed] [Google Scholar]
- 97.Restagno G, Maghtheh M, Bhattacharya S, Ferrone M, Garnerone S, Samuelly R, Carbonara A. A large deletion at the 3' end of the rhodopsin gene in an Italian family with a diffuse form of autosomal dominant retinitis pigmentosa. Hum Mol Genet. 1993;2:207–208. doi: 10.1093/hmg/2.2.207. [DOI] [PubMed] [Google Scholar]
- 98.Roman G, He J, Davis RL. kurtz, a novel nonvisual arrestin, is an essential neural gene in Drosophila. Genetics. 2000;155:1281–1295. doi: 10.1093/genetics/155.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Salazar NC, Vallejos X, Siryk A, Rengo G, Cannavo A, Liccardo D, De Lucia C, Gao E, Leosco D, Koch WJ, Lymperopoulos A. GRK2 blockade with βARKct is essential for cardiac β2-adrenergic receptor signaling towards increased contractility. Cell Commun Signal. 2013;11:64. doi: 10.1186/1478-811X-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Schoneberg T, Schulz A, Biebermann H, Hermsdorf T, Rompler H, Sangkuhl K. Mutant G-protein-coupled receptors as a cause of human diseases. Pharmacol Ther. 2004;104:173–206. doi: 10.1016/j.pharmthera.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 101.Seo J, Tsakem EL, Breitman M, Gurevich VV. Identification of arrestin-3-specific residues necessary for JNK3 activation. J Biol Chem. 2011;286:27894–27901. doi: 10.1074/jbc.M111.260448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shaw AS, Kornev AP, Hu J, Ahuja LG, Taylor SS. Kinases and pseudokinases: lessons from RAF. Mol Cell Biol. 2014;34:1538–1546. doi: 10.1128/MCB.00057-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shenoy SK, McDonald PH, Kohout TA, Lefkowitz RJ. Regulation of receptor fate by ubiquitination of activated beta 2-adrenergic receptor and beta-arrestin. Science. 2001;294:1307–1313. doi: 10.1126/science.1063866. [DOI] [PubMed] [Google Scholar]
- 104.Shenoy SK, Modi AS, Shukla AK, Xiao K, Berthouze M, Ahn S, Wilkinson KD, Miller WE, Lefkowitz RJ. Beta-arrestin-dependent signaling and trafficking of 7-transmembrane receptors is reciprocally regulated by the deubiquitinase USP33 and the E3 ligase Mdm2. Proc Natl Acad Sci U S A. 2009;106:6650–6655. doi: 10.1073/pnas.0901083106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shenoy SK, Xiao K, Venkataramanan V, Snyder PM, Freedman NJ, Weissman AM. Nedd4 mediates agonist-dependent ubiquitination, lysosomal targeting, and degradation of the beta2-adrenergic receptor. J Biol Chem. 2008;283:22166–22176. doi: 10.1074/jbc.M709668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Song X, Coffa S, Fu H, Gurevich VV. How does arrestin assemble MAPKs into a signaling complex? J Biol Chem. 2009;284:685–695. doi: 10.1074/jbc.M806124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Song X, Seo J, Baameur F, Vishnivetskiy SA, Chen Q, Kook S, Kim M, Brooks EK, Altenbach C, Hong Y, Hanson SM, Palazzo MC, Chen J, Hubbell WL, Gurevich EV, Gurevich VV. Rapid degeneration of rod photoreceptors expressing self-association-deficient arrestin-1 mutant. Cell Signal. 2013;25:2613–2624. doi: 10.1016/j.cellsig.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Song X, Vishnivetskiy SA, Gross OP, Emelianoff K, Mendez A, Chen J, Gurevich EV, Burns ME, Gurevich VV. Enhanced Arrestin Facilitates Recovery and Protects Rod Photoreceptors Deficient in Rhodopsin Phosphorylation. Curr Biol. 2009;19:700–705. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Song X, Vishnivetskiy SA, Seo J, Chen J, Gurevich EV, Gurevich VV. Arrestin-1 expression in rods: balancing functional performance and photoreceptor health. Neuroscience. 2011;174:37–49. doi: 10.1016/j.neuroscience.2010.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sriram SR, Li X, Ko HS, Chung KK, Wong E, Lim KL, Dawson VL, Dawson TM. Familial-associated mutations differentially disrupt the solubility, localization, binding and ubiquitination properties of parkin. Hum Mol Genet. 2005;14:2571–2586. doi: 10.1093/hmg/ddi292. [DOI] [PubMed] [Google Scholar]
- 111.Sterne-Marr R, Dhami GK, Tesmer JJ, Ferguson SS. Characterization of GRK2 RH domain-dependent regulation of GPCR coupling to heterotrimeric G proteins. Methods Enzymol. 2004;390:310–336. doi: 10.1016/S0076-6879(04)90020-1. [DOI] [PubMed] [Google Scholar]
- 112.Sterne-Marr R, Gurevich VV, Goldsmith P, Bodine RC, Sanders C, Donoso LA, Benovic JL. Polypeptide variants of beta-arrestin and arrestin3. J Biol Chem. 1993;268:15640–15648. [PubMed] [Google Scholar]
- 113.Sterne-Marr R, Tesmer JJ, Day PW, Stracquatanio RP, Cilente JA, O'Connor KE, Pronin AN, Benovic JL, Wedegaertner PB. G protein-coupled receptor Kinase 2/G alpha q/11 interaction. A novel surface on a regulator of G protein signaling homology domain for binding G alpha subunits. J Biol Chem. 2003;278:6050–6058. doi: 10.1074/jbc.M208787200. [DOI] [PubMed] [Google Scholar]
- 114.Stoy H, Gurevich VV. How genetic errors in GPCRs affect their function: Possible therapeutic strategies. Genes Dis. 2015;2 doi: 10.1016/j.gendis.2015.02.005. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sun F, Zhou Q, Pang X, Xu Y, Rao Z. Revealing various coupling of electron transfer and proton pumping in mitochondrial respiratory chain. Curr Opin Struct Biol. 2013;23:526–538. doi: 10.1016/j.sbi.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 116.Taylor SS, Shaw A, Hu J, Meharena HS, Kornev A. Pseudokinases from a structural perspective. Biochem Soc Trans. 2013;41:981–986. doi: 10.1042/BST20130120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Violin JD, Crombie AL, Soergel DG, Lark MW. Biased ligands at G-protein-coupled receptors: promise and progress. Trends Pharmacol Sci. 2014;35:308–316. doi: 10.1016/j.tips.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 118.Violin JD, DiPilato LM, Yildirim N, Elston TC, Zhang J, Lefkowitz RJ. beta2-adrenergic receptor signaling and desensitization elucidated by quantitative modeling of real time cAMP dynamics. J Biol Chem. 2008;283:2949–2961. doi: 10.1074/jbc.M707009200. [DOI] [PubMed] [Google Scholar]
- 119.Vishnivetskiy SA, Chen Q, Palazzo MC, Brooks EK, Altenbach C, Iverson TM, Hubbell WL, Gurevich VV. Engineering visual arrestin-1 with special functional characteristics. J Biol Chem. 2013;288:11741–11750. doi: 10.1074/jbc.M112.445437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vishnivetskiy SA, Francis DJ, Van Eps N, Kim M, Hanson SM, Klug CS, Hubbell WL, Gurevich VV. The role of arrestin alpha-helix I in receptor binding. J Mol Biol. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vishnivetskiy SA, Gimenez LE, Francis DJ, Hanson SM, Hubbell WL, Klug CS, Gurevich VV. Few residues within an extensive binding interface drive receptor interaction and determine the specificity of arrestin proteins. J Biol Chem. 2011;286:24288–24299. doi: 10.1074/jbc.M110.213835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface: structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 123.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez M-G, Gurevich VV. An additional phosphate-binding element in arrestin molecule: implications for the mechanism of arrestin activation. J Biol Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 124.Wang C, Tan JM, Ho MW, Zaiden N, Wong SH, Chew CL, Eng PW, Lim TM, Dawson TM, Lim KL. Alterations in the solubility and intracellular localization of parkin by several familial Parkinson's disease-linked point mutations. J Neurochem. 2005;93:422–431. doi: 10.1111/j.1471-4159.2005.03023.x. [DOI] [PubMed] [Google Scholar]
- 125.Wilden U. Duration and amplitude of the light-induced cGMP hydrolysis in vertebrate photoreceptors are regulated by multiple phosphorylation of rhodopsin and by arrestin binding. Biochemistry. 1995;34:1446–1454. doi: 10.1021/bi00004a040. [DOI] [PubMed] [Google Scholar]
- 126.Wilden U, Hall SW, Kühn H. Phosphodiesterase activation by photoexcited rhodopsin is quenched when rhodopsin is phosphorylated and binds the intrinsic 48-kDa protein of rod outer segments. Proc Natl Acad Sci USA. 1986;83:1174–1178. doi: 10.1073/pnas.83.5.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Xiao K, McClatchy DB, Shukla AK, Zhao Y, Chen M, Shenoy SK, Yates JR, Lefkowitz RJ. Functional specialization of beta-arrestin interactions revealed by proteomic analysis. Proc Natl Acad Sci U S A. 2007;104:12011–12016. doi: 10.1073/pnas.0704849104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Xu J, Dodd RL, Makino CL, Simon MI, Baylor DA, Chen J. Prolonged photoresponses in transgenic mouse rods lacking arrestin. Nature. 1997;389:505–509. doi: 10.1038/39068. [DOI] [PubMed] [Google Scholar]
- 129.Yu Y, Ping J, Chen H, Jiao L, Zheng S, Han ZG, Hao P, Huang J. A comparative analysis of liver transcriptome suggests divergent liver function among human, mouse and rat. Genomics. 2010;96:281–289. doi: 10.1016/j.ygeno.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 130.Zhan X, Gimenez LE, Gurevich VV, Spiller BW. Crystal structure of arrestin-3 reveals the basis of the difference in receptor binding between two non-visual arrestins. J Mol Biol. 2011;406:467–478. doi: 10.1016/j.jmb.2010.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zhan X, Kaoud TS, Kook S, Dalby KN, Gurevich VV. JNK3 binding to arrestin-3 differentially affects the recruitment of upstream MAP kinase kinases. J Biol Chem. 2013;288:28535–28547. doi: 10.1074/jbc.M113.508085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhan X, Perez A, Gimenez LE, Vishnivetskiy SA, Gurevich VV. Arrestin-3 binds the MAP kinase JNK3α2 via multiple sites on both domains. Cell Signal. 2014 doi: 10.1016/j.cellsig.2014.01.001. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhuo Y, Vishnivetskiy SA, Zhan X, Gurevich VV, Klug CS. Identification of receptor binding-induced conformational changes in non-visual arrestins. J Biol Chem. 2014;289:20991–21002. doi: 10.1074/jbc.M114.560680. [DOI] [PMC free article] [PubMed] [Google Scholar]