1 Introduction
Structure-activity relationships (SAR) are key to many aspects of drug discovery, ranging from primary screening to lead optimization. Working with SAR starts from identifying whether an SAR actually exists in a collection of molecules and their associated activities to trying to elucidate the details of one or more SARs and subsequently using that information to make structural modifications to optimize some property or activity. Fundamentally, an understanding of the SAR for a set of molecules, allows one to rationally explore chemical space, which, in the absence of “sign posts” is essentially infinite [1]. Invariably, the development of a chemical series involves optimizing multiple physicochemical and biological properties simultaneously [2, 3, 4]. For example, most lead optimization projects will try and improve potency, reduce toxicity and ensure sufficient bioavailability, amongst other properties. While the intuition and experience of a medicinal chemist is vital to these efforts, the data generated by modern high throughput experimental techniques can overwhelm the capabilities of a single chemist. For example, in a primary HTS it is possible that one is faced with hundred of chemical series. How does one rapidly identify the most promising series amongst them? In these scenarios, in silico methods allow rapid and efficient characterization of SARs. These methods allow one to build a variety of models to capture and encode one or more SARs, which can then be used to predict activities for new molecules. Coupled with in silico modifications of structures, one can easily prioritize large screening decks or even generate new compounds de novo and ascertain whether they belong to the SAR being studied or not. It should be kept in mind that computational methods do not replace medicinal chemistry domain knowledge. Rather, they can provide a guide to the experienced user by integrating and summarizing large amounts of pre-existing data to suggest useful structural modifications.
While it is certainly true that computational methods can help in identifying, explaining and predicting structure-activity relationships (SAR), it is also true that naïve usage (or even misuse) of these techniques can lead to misleading results. Fundamentally, SAR models are just that: models. That is, a reduced or simplified representation of reality, replete with assumptions and limitations. These methods cover the spectrum in terms of complexity and utility. The goal of this chapter is to highlight the different type of SAR modeling methods, and specifically, how they support the task of exploring chemical space to elucidate and optimize structure-activity relationships in a drug discovery setting. In addition to considering modeling algorithms, I will also briefly discuss the use of databases as a source of SAR data and how they can be used to inform and enhance the exploration of SAR trends.
The remainder of the text is structured as follows. Section 2 provides an overview of common modeling techniques that are used to encode SAR relationships. Section 3 discusses recent work in the area of structure-activity landscapes and how they can be used as an alternative view of SAR data. Section 4 discusses the role of SAR databases. Section 5 discusses some approaches to exploring SAR data that do not involve explicit model development and finally Section 6 provides a summary of this topic.
2 Capturing SAR
Over the last 60 years there have been a multitude of ways to capture structure-activity relationships. We can broadly divide them into two groups - those based on statistical or data mining methods (e.g., regression models) and those based on physical approaches (e.g., pharmacophore models). For a comprehensive review of QSAR methodologies the reader is referred to previous reviews of the field [5, 6, 7]. It is important to realize that the choice of modeling technique can influence the to what extent and in how much detail an SAR can be explored. For example, statistical QSAR approaches bsed on 2D descriptors that ignore stereochemistry can be miss key elements of an SAR that depend on chirality [8]. 3D approaches on the other hand are generally more explicitly informative, in the sense that one can directly understand the nature of ligand - receptor interactions that underlie an observed SAR. Some 3D approaches are more explicit than others - docking versus CoMFA for example. However, it generally remains true that 3D approaches are preferable when a crystal structure is available and when a few chemical series are being explored.
Much of traditional QSAR is based on statistical models that link chemical structure (characterized by numerical descriptors) to biological activities. In some cases, the model makes distributional assumptions (linear regression), whereas others are “model-free” [9]. In either case, one develops a model based on a training set of molecules. The model can then be used to predict the activity for new molecules. While much of QSAR has focused on various forms of linear regression (ranging from ordinary least squares to more robust methods such as PLS or ridge regression), there is no reason to assume, a priori, the structure-activity relationship is linear. Indeed, for nost biological systems it is unreasonable to expect linear relationships simply because of the multiple, complex processes occurring in vivo. Thus, modern non-linear methods such as neural networks and support vector machines have seen extensive use and tend to exhibit high accuracy.
However, building a predictive model is just the first step. For certain scenarios, such as virtual screening, one can apply the model and simply obtain numerical predictions of activity. However, the focus of this article is to consider the use of such predictive models for exploring SAR. Key to such exploration is the ability to interpret the model and understand how exactly it correlates activity to specific structural features [10, 11]. For interpretive purposes, a model should be understandable - both in terms of the descriptors used and the underlying model itself. The predictive ability of the model is not primary (though of course, for statistical models, they should be statistically significant). Examples include linear regression and random forests. In this type of usage, one is interested in what the model can tell us about the effects of specific structural features on the observed activity. It is thus vital that this information can be teased out from the model. Obviously, for more physical methods such as pharmacophore modeling and docking, the interpretability is much more explicit. Clearly, SAR exploration benefits from models that can be dissected. Examples of such interpretive usages have been reported, both for simple models [12] as well as traditional black box models [13]. However, it is certainly true that purely predictive models can also be useful, especially when used to identify more or less active molecules from a large collection. In such a scenario, users could employ a predictive model to provide an initial ranking, allowing them to focus on a small subset using more interpretive methods.
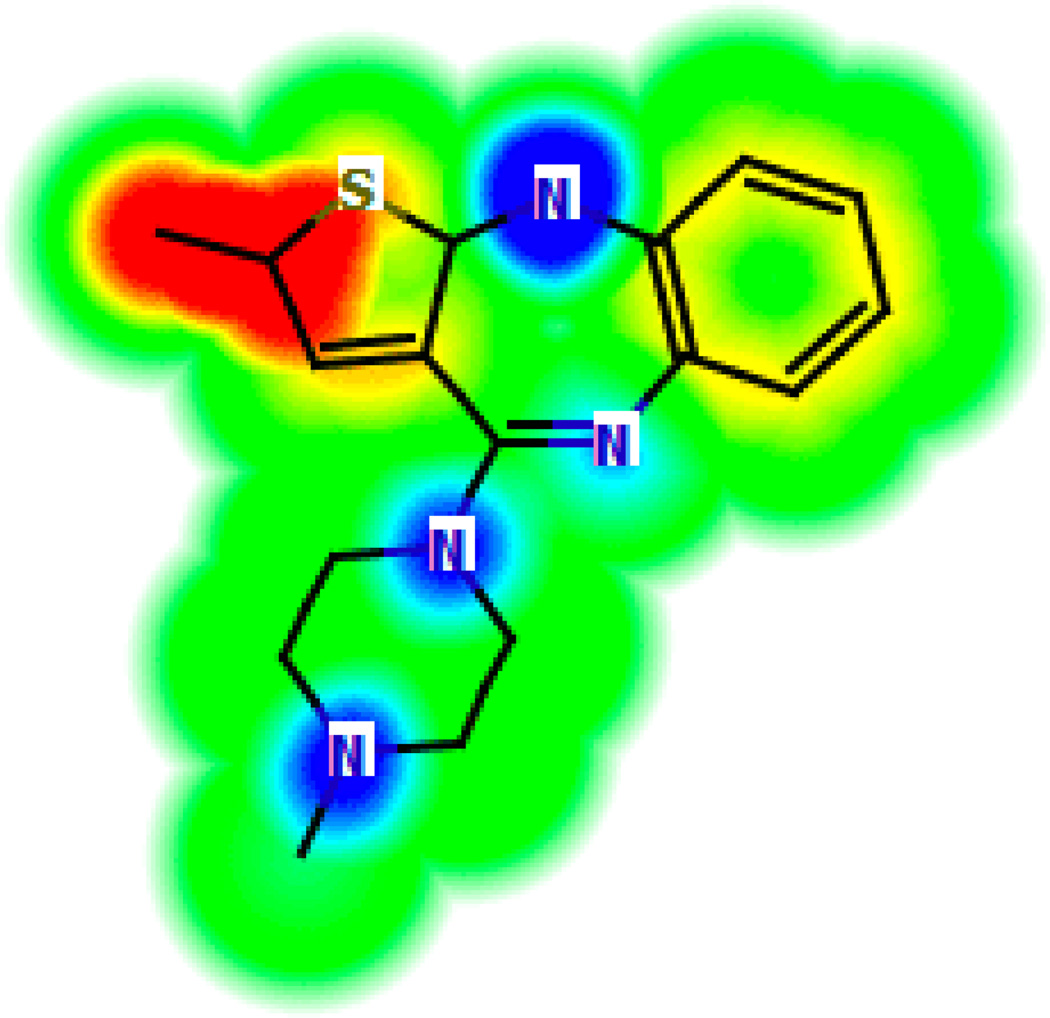
Closely related to analytical interpretations of QSAR models is the ability to visualize the SAR trends encoded in a model. The “glowing molecule” representation developed by Segall et al [14] is an example of direct visualization of a predictive model in terms of the actual chemical structure. Figure 1 shows an example of such a representation, where the color coding corresponds to the influence of that substructural feature on the predicted property. This type of visualization allows the user to directly understand how structural modifications at specific points will affect the property or activity being optimized.
Figure 1.
An example of a glowing molecule representation, developed by Optibrium. The color coding corresponds to the influence of the structural feature on the predicted property (red for a negative influence, blue for a positive influence). Image modified, with permission, from http://www.optibrium.com/community/faq/glowing-molecule.
While capturing SAR trends in a predictive model, used to subsequently predict properties for new molecules, is useful, one can also consider the inverse approach. That is, identify structures that match a given activity or activity profile. Most formulations of this approach aim to derive a set of descriptor values rather than the structure directly. The challenge is in identifying valid structures from a set of descripor values. A number of workers have addressed the inverse QSAR problem. Visco et al employed the signature molecular descriptors [15] to perform an inverse QSAR analysis of 121 HIV protease inhibitors and Churchwell et al [16] employed these same descriptors to explore QSARs of peptides inhibiting ICAM-1. More recently, Wong et al [17] developed a novel descriptor to address inverse QSAR and coupled this to kernel method to allow explicit mapping between points in the high dimensional kernel space (i.e., candidate structures) back to the original descriptor space and thence to a set of candidate molecules.
2.1 Is the model reliable?
While any statistical method or machine learning algorithm can be used to learn from SAR data and then predict activities for new molecules, such predictions are not always reliable. More specifically, these types of approaches to QSAR modeling assume that new molecules to be predicted will have some structural features in common with the training set that they are based on. If the new molecule is sufficiently different, one will obtain an unreliable (or even meaningless) prediction. Thus it is vital to denote the “domain of applicability” (DA) of a model, thus letting the user know when the predictions of the model can be relied upon. In scenarios where a model is built at one time point and then used to make predictions over a period of time, defining the DA can be very useful in determining at what point a model should be rebuilt, due to sufficient divergence of the new molecules to be predicted from original model [18].
A variety of methods have been developed to define domains of applicability [19, 20, 21, 22, 23]. The simplest approach is to determine how similar a new molecule is to the training set for the model. Sheridan et al [24] employed this technique focusing on two approaches - similarity of the molecule to be predicted to the nearest neighbor in the training set and the number of nearest neighbors in the training (decided by a user-defined similarity cutoff). Their results indicated that either of these approaches lead to a robust measure of reliability of predictions. Xu et al [25] also considered similarity to the training set, using a novel distance metric terms the “dimension related distance”, allowing them to measure the similarity of a molecule to the entire training set. Other distance metrics have also been employed including Mahalanobis and Manhattan distances. For models based on linear regression, various diagnostics such as the Cooks distance [26] and leverage [27] have been employed [28, 29, 30]
A number of approaches based on descriptor values have also been proposed. The simplest approach is to determine the range of descriptor values in the training set and the values for the new molecule lie outside the range, the model will have to extrapolate, and hence the prediction will be unreliable. While conceptually simple, this approach is easily mislead by non-uniformly distributed descriptor values. Alternatives have performed PCA and used the ranges of the resulting principal components as the space within which reliable predictions can be obtained [31].
3 Exploring SAR Landscapes
QSAR model predictions are a useful guide for lead optimization [32], but alternative views of SAR data can be useful. Over the last few years, the landscape paradigm of SAR data has gained focus and allows us to explore a number of aspects of structure-activity relationships. This work stems from the work of Lajiness [33] that viewed chemical structure and bioactivity, simultaneously, in a 3D view, with the structure represented in the X-Y plane and the activity along the Z-axis. The immediate consequence of this is that a SAR dataset can be viewed as a landscape of varying “topography”. Smooth regions correspond to molecules that are similar in structure and activity whereas jagged (i.e., discontinuous) regions correspond to structures that are similar but exhibit very different activities (so called activity cliffs). In fact, it has suggested that the latter regions of the landscape represent the most interesting parts of an SAR, as they provide the possibility of making small structural changes to significantly change activities. At the same time, these discontinuities can be problematic as they can lead to poor performance of many QSAR modeling methods (primarily those based on machine learning or statistical models) [34]. As a result, a variety of methods have been developed to characterize and mine SAR landscapes.
Structure-Activity Similarity (SAS) maps were first described by Shanmugasundaram and Maggiora [35], which is a pairwise plot of the structure similarity against the activity similarity. The resultant plot can be divided into four quadrants allowing one to identify molecules characteristic of one of four possible behaviors: smooth regions of the SAR space, (rough) activity cliffs, non-descript (i.e., low structural similarity and low activity similarity) and scaffold hops (low structural similarity but high activity similarity). Recently, there have been a number of extensions of SAS maps that take into account multiple descriptor representations (2D & 3D) [36, 37]. In addition to SAS maps, other pairwise metrics to characterize and visualize SAR landscapes have been developed such as SALI [38] and SARI [39].
Visualization of landscapes via network diagrams has also led to novel developments in the exploration of SAR data. Examples include the SALI networks described by Guha et al [38] and network similarity graphs (NSG’s) described by Wawer et al [40]. Both network representations use compounds as nodes and draw edges between them based on some metric that characterizes the pair of nodes in the context of the landscape (SARI for NSG’s and SALI for SALI networks). The networks can be then analyzed to identify specific SAR trends. For example, Wawer et al [41] described an approach to identifying “SAR pathways” - paths in an NSG that connected regions of low and high SAR discontinuity. Such SAR pathways represent a set of compounds, which when order appropriately exhibit a continuous series of SAR changes. While network based analyses of landscapes have seen much activiy an alternative visualization approach described by Seebeck et al [42] abstracted the idea of the SALI metric and extended it to include the receptor. As a result of this, they were able o highlight specific regions within protein binding sites that are most likely to lead to activity cliffs.
The concept of activity cliffs and the landscape paradigm have also been applied to R-groups, where an “R-cliff” occurs when a pair of compounds differ in a single R-group. This is clearly a specialization of the activity cliff concept, placing this type of analysis in the context of analog series derived via R-group decompositions [43, 44].
4 Canned SAR
Over the last few years a number of large chemical structure databases have become available. Importantly, a number of these databases also provide extensive information of compound activities in addition to compound structures. Notable examples include PubChem, ChEMBL and GVK GOSTAR. The first two are freely available resources, whereas GOSTAR is a commercial offering. While these databases provide structure-activity information, they differ in the nature of the data that is provided. For example, PubChem provides a compound and substance database, where records are individual structures as well as a bioassay database, which contains assay results for various compound sets, deposited by the Molecular Libraries Initiative [45]. The two databases are linked, allowing one to easily identify the assays a compound has been tested in or conversely, the structures tested in an assay. Assay datasets range in size from two or three compounds to more than 300,000 compounds and assay types range from primary screens to secondary and confirmatory screens, both as single point and dose response formats. It is important to note that PubChem data is not curated and thus the assay data can be noisy (which is a given for primary screening data). ChEMBL and GOSTAR are both curated SAR databases, where structures and their activities have been manually extracted form the literature and stored in a standardized form. In addition a variety of annotations have also been added such as assay target, species and so on. In essence, these databases have “canned” structure-activity relationships, making them readily available for analysis.
One obvious application of these database is to use them as sources of training data when developing predictive models. For example, Novotarskyi et al [46] employed PubChem Bioassay data to develop models to predict CYP450 1A2 inhibition and Shen et al [47] employed the database to develop a support vector machine model to predict hERG liabilities. Though there are many other examples of QSAR modeling studies using PubChem as the source of data, the bulk focus on specific targets. In contrast, Chen et al [48] built a series of models using multiple PubChem assays, that could be used to provide a prediction of an “activity profile”. They employed random forest models, aiming for pure predictive ability, over any explanatory power.
A relatively unique database is the GDB-13 database [49] which is an exhaustive enumeration of small molecule structures containing up to 13 heavy atoms (restricted to C, H, N, O, S, P and Cl). While the database does not contain activity information associated with the structures, it can be used a source of structures for virtual screening purposes [50]. In this sense, it is similar in nature to databases such as ZINC [51] - the key differentiator is that the latter are all commercially available, whereas the former are in effect, completely virtual. In general, this class of databases is useful primarily for virtual screening type methods, where the goal is to identify candidates for more in depth study, rather than explicitly understanding SAR trends.
5 Alternatives to QSAR?
While QSAR approaches (in all its forms) are by far the most common ways to capture and explore SAR trends, a number of other approaches are possible. While not quantitative, they can. be useful in the form of “idea generators”.
5.1 Characterizing SAR in series
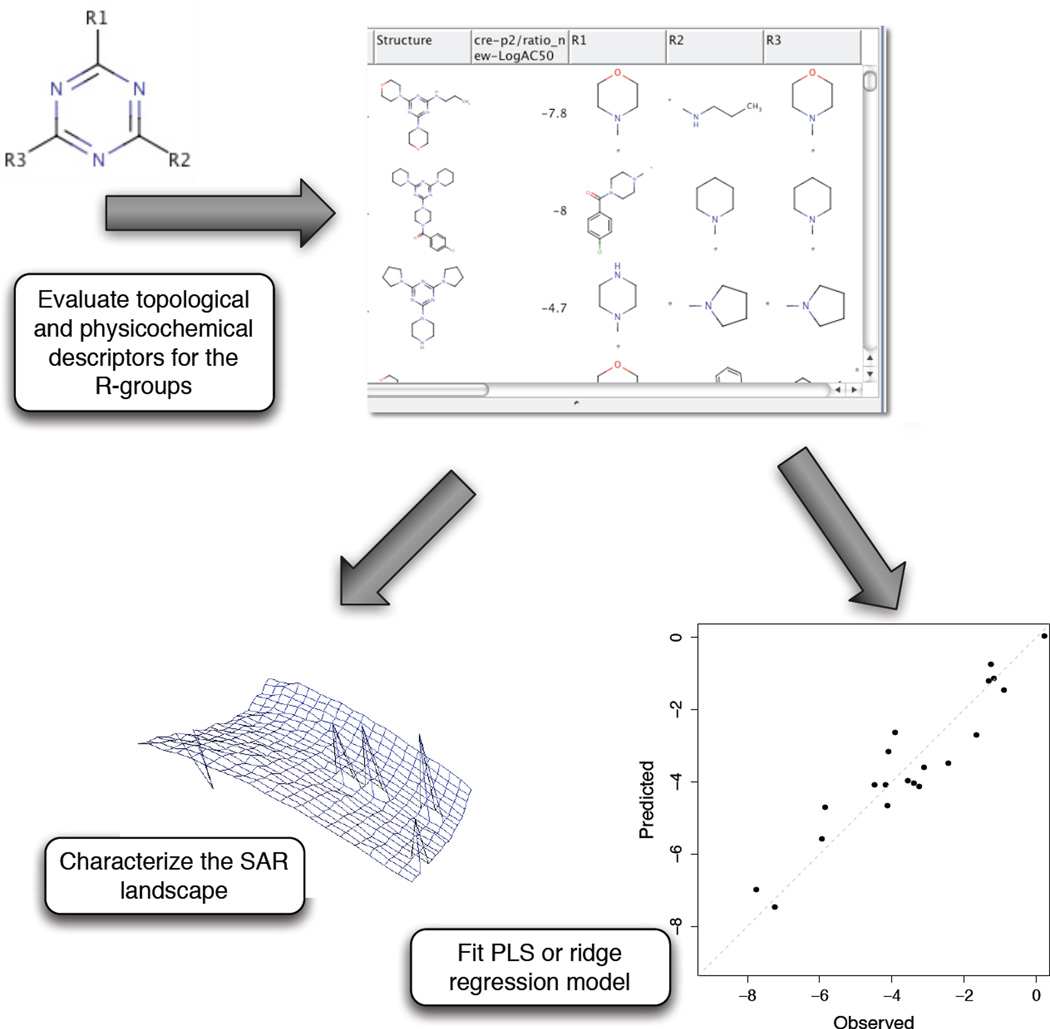
One approach is to consider fragments as the basis for SAR exploration. This is not without precedent as substructure based models have been developed that are useful for both prediction and interpretation [52]. One approach to using fragments for exploring SAR is to develop “R-group QSAR” models, whose goal is to determine whether an SAR exists or not, and if so, how do different R-groups affect it. Given a set of molecules, we perform an R-group decomposition, generating a series of scaffolds and associated substituents. Given a scaffold, we can create an R-group matrix, with observations (i.e., molecules containing the scaffold) in the rows and the R-groups, R1, R2, …, Rn, in the columns. Element (i, j) of the matrix is set to 1 if the i’th molecule contains the j’th substituent. Given this R-group incidence matrix, along with the observed activities for the molecules, one can develop a predictive model. Given that most such R-group matrices will be small, some form of linear regression is likely most suitable. Figure 2 shows a schematic diagram summarizing this technique. However, it is obvious that this approach has a number of limitations. Firstly, the number of observations for many scaffolds will be very small (< 10) and hence a reliable model cann’t be generated. Second, most R-group matrices will be sparse since all substitution points will not be uniformly filled. Also, a binary incidence matrix may not be the most appropriate input for a linear regression model. One way to address this is to replace each of n columns with n × d columns, where d represents a d-dimensional real-valued descriptor vector derived from the structure of the corresponding R-group. For a scaffold with three or more substitution points, this can lead to a R-group matrix with more columns than rows. Even when this is not the case, such a matrix will usually have a large number of features (depending on how many descriptors are employed). In spite of these problems, for those scaffolds where there is a sufficient number of observations and relatively few substitution points a linear regression model can be developed. In such cases, the predictive accuracy of the model is not the main focus. Rather, we are interested in whether the model is statistically significant; the hypothesis being that such a model represents an actual relationship between the substitutions and the observed activities. As a result, this could allow us to provide a ranking to individual series. Obviously, for a diverse set of structures this approach is not very useful.
Figure 2.
A schematic diagram of R-group QSAR workflow that can be used to rank scaffolds (i.e., chemical series) in terms of whether they exhibit an SAR or not.
5.2 Exploring SAR via fragments
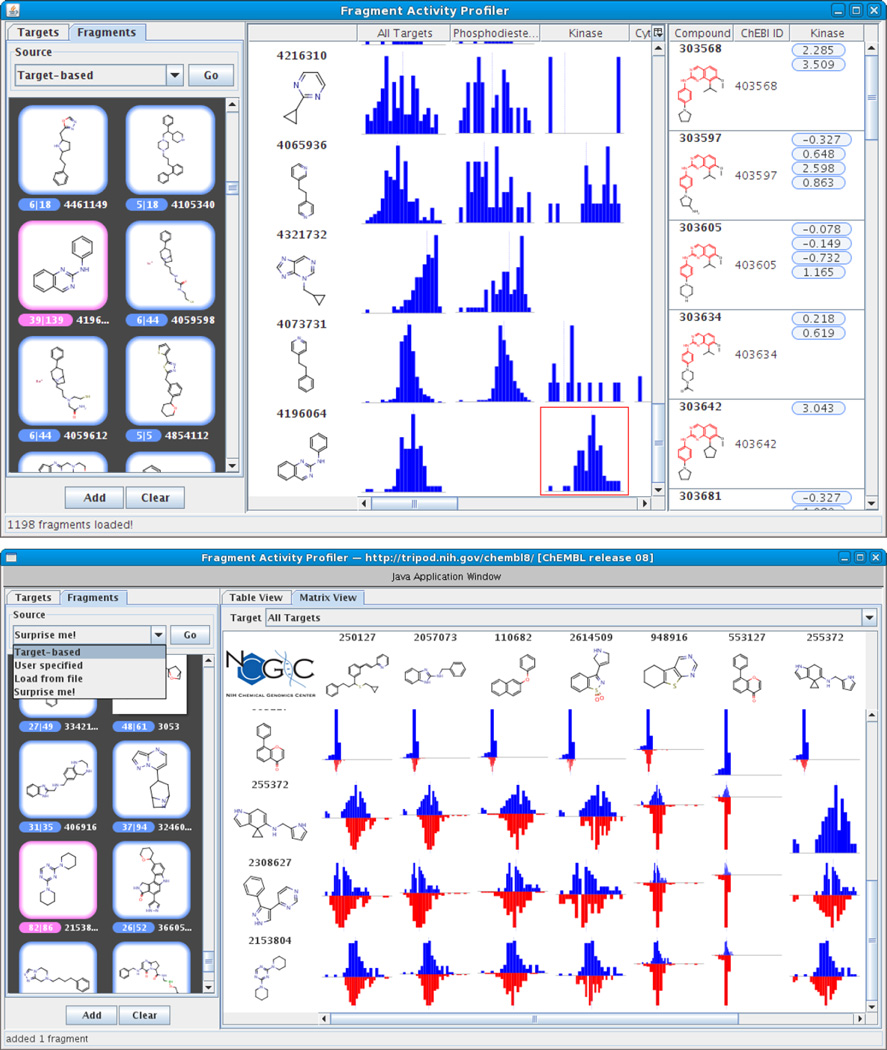
Another approach to using fragments to explore SAR is by making use of SAR databases such as ChEMBL. The underlying motivation is that certain fragments may be associated with high (or low) activities or may show activity preferentially towards a specific target or target family. By precomputing the fragments for a database like ChEMBL and storing the fragment-compound associations, we can take an external set of compounds, fragment them and explore the activity data associated with them in ChEMBL. While a relatively simple task, it does require a number of steps. We recently developed a software tool, the Fragment Activity Profiler (http://tripod.nih.gov/?p=206) that allows one to explore structure collections from the point of view of scaffolds, integrated with activity and target information (Fig. 3). Given a compound collection, with observed activity data, one can generate fragments and identify activity profiles associated with those fragments from from ChEMBL, drilling down to the individual ChEMBL compounds associated with the fragment of interest. Alternatively, one can characterize scaffolds in terms of their activities against different targets, and thus summarize selectivity of compound series. This is especially easy with ChEMBL since structures (and therefore the derived scaffolds) are associated with targets via the assays they are tested in. Other approaches have also been discussed in the literature, such as the Scaffold Explorer described by Agrafiotis et al [53] and the Scaffold Hunter application [54].
Figure 3.
Screenshots of the Fragment Activity Profiler. The top figure highlights the single scaffold view that displays activity profiles for a given scaffold versus individual targets (at a specified level of the ChEMBL target hierarchy. The bottom figure shows the pairwise view, allowing one to compare activity profiles of two scaffolds simultaneously.
6 Summary
It is clear that computational methods play a key role in the process of drug discovery. While there has been some discussion on the actual value added by these models, it is still evident that with informed usage of these techniques together with appropriate testing and control methods [55], these methods have much to offer [56]. Indeed, with the deluge of data being generated by modern high throughput experimental techniques and the evolution of large chemical structure databases, it is all the more important to address how computational techniques can enable practising scientists in exploring the wealth of structure activity data.
Traditional QSAR approaches based on machine learning or statistical methods have been used extensively. However, many are black boxes and can make understanding the actual SAR encoded by the model difficult. This can hinder the use of such models when deciding how to modify a structure to improve an activity or reduce liabilities. While visualizations in some cases can make this job easier, more physical models such as 3D-QSAR methods (CoMFA, CoMSIA) and pharmacophore methods can provide a much more direct view into structural features contributing to the SAR. Given that the much of SAR exploration is done during the lead optimization stage of a drug development project, it makes sense to consider related SAR information that has been reported in the literature or otherwise made public. Databases such as PubChem and ChEMBL allow one to easily access large amounts of SAR data points (both curated and uncurated) and compare those data with our own data. Such comparisons can be performed in a variety of ways ranging from developing predictive models to browsing scaffolds and viewing activity profiles of compounds containing those scaffolds.
In summary, there is an extensive array of computational methods that allows one to explore SAR trends at varying levels of detail. While all methods are not necessarily accurate in a numerical sense, in silico models can serve as idea generators, augmenting the decision making ability of the medicinal chemist.
References
- 1.Bohacek RS, McMartin C, Guida WC. The art and practice of structure based drug design: A molecular modeling perspective. Med. Res. Rev. 1996;16(1):3–50. doi: 10.1002/(SICI)1098-1128(199601)16:1<3::AID-MED1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Nicolotti O, Gillet VJ, Fleming PJ, Green DVS. Multiobjective optimization in quantitative structure-activity relationships: Deriving accurate and interpretable qsars. J. Med. Chem. 2002;23:5069–5080. doi: 10.1021/jm020919o. [DOI] [PubMed] [Google Scholar]
- 3.Cruz-Monteagudo Maykel, Borges Fernanda, Natália M, Cordeiro DS. Desirability-based multiobjective optimization for global QSAR studies: Application to the design of novel NSAIDs with improved analgesic, antiinflammatory, and ulcerogenic profiles. J. Comp. Chem. 2008 Nov;29(14):2445–2459. doi: 10.1002/jcc.20994. [DOI] [PubMed] [Google Scholar]
- 4.Nicolotti Orazio, Giangreco Ilenia, Miscioscia Teresa Fabiola, Carotti Angelo. Improving quantitative structure-activity relationships through multiobjective optimization. J. Chem. Inf. Model. 2009 Oct;49(10):2290–2302. doi: 10.1021/ci9002409. [DOI] [PubMed] [Google Scholar]
- 5.Dudek Arkadiusz Z, Arodz Tomasz, Gálvez Jorge. Computational methods in developing quantitative structure-activity relationships (QSAR): A review. Comb. Chem. High. Throughput Screen. 2006 Mar;9(3):213–228. doi: 10.2174/138620706776055539. [DOI] [PubMed] [Google Scholar]
- 6.Winkler David A. The role of quantitative structure–activity relationships (QSAR) in biomolecular discovery. Brief. Bioinform. 2002 Mar;3(1):73–86. doi: 10.1093/bib/3.1.73. [DOI] [PubMed] [Google Scholar]
- 7.Zvinavashe E, Albertinka JM, Rietjens IMCM. Promises and pitfalls of quantitative structureactivity relationship approaches for predicting metabolism and toxicity. Chem. Res. Toxicol. 2008;21(12):2229–2236. doi: 10.1021/tx800252e. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee Ashutosh, Schepmann Dirk, Köhler Jens, Würthwein Ernst-Ulrich, Wünsch Bernhard. Synthesis and SAR studies of chiral non-racemic dexoxadrol analogues as uncompetitive NMDA receptor antagonists. Bioorg. Med. Chem. 2010 Nov;18(22):7855–7867. doi: 10.1016/j.bmc.2010.09.047. [DOI] [PubMed] [Google Scholar]
- 9.Breiman L. Statistical modeling: Two cultures. Stat. Sci. 2001;16:199–231. [Google Scholar]
- 10.Guha R. On the interpretation and interpretability of qsar models. J. Comp. Aid. Molec. Des. 2008;22(12):857–871. doi: 10.1007/s10822-008-9240-5. [DOI] [PubMed] [Google Scholar]
- 11.Stanton DT. On the physical interpretation of QSAR models. J. Chem. Inf. Comput. Sci. 2003;43(5):1423–1433. doi: 10.1021/ci0340658. [DOI] [PubMed] [Google Scholar]
- 12.Guha R, Jurs PC. The development of QSAR models to predict and interpret the biological activity of artemisinin analogues. J. Chem. Inf. Comput. Sci. 2004;44:1440–1449. doi: 10.1021/ci0499469. [DOI] [PubMed] [Google Scholar]
- 13.Guha R, Stanton DT, Jurs PC. Interpreting computational neural network QSAR models: A detailed interpretation of the weights and biases. J. Chem. Inf. Model. 2005;45:1109–1121. doi: 10.1021/ci050110v. [DOI] [PubMed] [Google Scholar]
- 14.Segall Matthew, Champness Edmund, Obrezanova Olga, Leeding Chris. Beyond profiling: using ADMET models to guide decisions. Chem. Biodivers. 2009 Nov;6(11):2144–2151. doi: 10.1002/cbdv.200900148. [DOI] [PubMed] [Google Scholar]
- 15.Faulon JL, Visco DP, Pophale RS. The signature molecular descriptor. 1. using extended valence sequences in qsar and qspr studies. Journal of Chemical Information and Computer Sciences. 2003;43:707–720. doi: 10.1021/ci020345w. [DOI] [PubMed] [Google Scholar]
- 16.Churchwell Carla J, Rintoul Mark D, Martin Shawn, Visco Donald P, Jr, Kotu Archana, Larson Richard S, Sillerud Laurel O, Brown David C, Faulon Jean-Loup. The signature molecular descriptor. 3. inverse-quantitative structure-activity relationship of ICAM-1 inhibitory peptides. J. Mol. Graph. Model. 2004;22(4):263–273. doi: 10.1016/j.jmgm.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 17.Wong William Wl, Burkowski Forbes J. A constructive approach for discovering new drug leads: Using a kernel methodology for the inverse-QSAR problem. J. Cheminform. 2009;1:4. doi: 10.1186/1758-2946-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weaver Shane, Gleeson M Paul. The importance of the domain of applicability in QSAR modeling. J. Mol. Graph. Model. 2008;26(8):1315–1326. doi: 10.1016/j.jmgm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 19.Schultz Terry W, Hewitt Mark, Netzeva Tatiana I, Cronin Mark TD. Assessing applicability domains of toxicological QSARs: Definition, confidence in predicted values, and the role of mechanisms of action. QSAR. Comb. Sci. 2007;26:238–254. [Google Scholar]
- 20.Roberts DW, Patlewicz G, Kern PS, Gerberick F, Kimber I, Dearman RJ, Ryan CA, Basketter DA, Aptula AO. Mechanistic applicability domain classification of a local lymph node assay dataset for skin sensitization. Chem. Res. Tox. 2007;20(7):1019–1030. doi: 10.1021/tx700024w. [DOI] [PubMed] [Google Scholar]
- 21.Stanforth RW, Kolossov E, Mirkin B. A measure of domain of applicability for qsar modelling based on intelligent k-means clustering. QSAR. Comb. Sci. 2007;26(7):837–844. [Google Scholar]
- 22.Tetko IV, Bruneau P, Mewes HW, Rohrer DC, Poda GI. Can we estimate the accuracy of ADME-tox predictions? Drug Discov. Today. 2006;11(15–16):700–707. doi: 10.1016/j.drudis.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 23.Jaworska J, Nikolova-Jeliazkova Nina, Aldenberg Tom. QSAR applicability domain estimation by projection of the training set in descriptor space: A review. Altern. Lab. Anim. 2005;33:445–459. doi: 10.1177/026119290503300508. [DOI] [PubMed] [Google Scholar]
- 24.Sheridan RP, Feuston BP, Maiorov VN, Kearsley SK. Similarity to molecules in the training set is a good discriminator for prediction accuracy in qsar. J. Chem. Inf. Comput. Sci. 2004;44(6):1912–1928. doi: 10.1021/ci049782w. [DOI] [PubMed] [Google Scholar]
- 25.Xu YJ, Gao H. Dimension related distance and its application in QSAR/QSPR model error estimation. QSAR. Comb. Sci. 2003;22:422–429. [Google Scholar]
- 26.Cook RD. Detecting influential observations in linear regression. Technometrics. 1977;19:15–18. [Google Scholar]
- 27.Chatterjee S, Hadi AS. Influential observations, high leverage points, and outliers in linear regression. Stat. Sci. 1986;1(3):379–416. [Google Scholar]
- 28.Tropsha A, Gramatica P, Gombar VK. The importance of being earnest: Validation is the absolute essential for successful application and interpretation of QSPR models. QSAR. Comb. Sci. 2003;22:69–77. [Google Scholar]
- 29.Eriksson Lennart, Jaworska Joanna, Worth Andrew P, Cronin Mark TD, Mc-Dowell Robert M, Gramatica Paola. Methods for reliability and uncertainty assessment and for applicability evaluations of classification- and regression-based qsars. Environ. Health Perspect. 2003 Aug;111(10):1361–1375. doi: 10.1289/ehp.5758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Papa Ester, Villa Fulvio, Gramatica Paola. Statistically validated qsars, based on theoretical descriptors, for modeling aquatic toxicity of organic chemicals in pimephales promelas (fathead minnow) J. Chem. Inf. Model. 2005;45(5):1256–1266. doi: 10.1021/ci050212l. [DOI] [PubMed] [Google Scholar]
- 31.Nikolova-Jeliazkova Nina, Jaworska J. An approach to determining AD for QSAR group contribution models: An analysis of SRC KOWWIN. Altern. Lab. Anim. 2005;33:461–470. doi: 10.1177/026119290503300510. [DOI] [PubMed] [Google Scholar]
- 32.Brown Nathan, Lewis Richard A. Exploiting QSAR methods in lead optimization. Curr. Opin. Drug Discov. Devel. 2006;9(4):419–424. [PubMed] [Google Scholar]
- 33.Lajiness MS. QSAR: Rational Approaches to the Design of Bioactive Compounds. Netherlands: Escom, Leiden; 1991. pp. 201–204. chapter Evaluation of the Performance of Dissimilarity Selection Methodology. [Google Scholar]
- 34.Maggiora Gerald M. On outliers and activity cliffs–why QSAR often disappoints. J. Chem. Inf. Model. 2006;46(4):1535–1535. doi: 10.1021/ci060117s. [DOI] [PubMed] [Google Scholar]
- 35.Shanmugasundaram V, Maggiora GM. CINF-032. 222nd ACS National Meeting, Chicago, IL, United States. Washington, DC: American Chemical Society; 2001. Characterizing property and activity landscapes using an information-theoretic approach. [Google Scholar]
- 36.Medina-Franco Jose L, Martínez-Mayorga Karina, Bender Andreas, Marín Ray M, Giulianotti Marc A, Pinilla Clemencia, Houghten Richard A. Characterization of activity landscapes using 2D and 3D similarity methods: Consensus activity cliffs. J. Chem. Inf. Model. 2009 Feb;49(2):477–491. doi: 10.1021/ci800379q. [DOI] [PubMed] [Google Scholar]
- 37.Yongye Austin B, Byler Kendall, Santos Radleigh, Martínez-Mayorga Karina, Maggiora Gerald M, Medina-Franco José L. Consensus models of activity landscapes with multiple chemical, conformer, and property representations. J. Chem. Inf. Model. 2011 Jun;51(6):1259–1270. doi: 10.1021/ci200081k. [DOI] [PubMed] [Google Scholar]
- 38.Guha R, Van Drie JH. The structure-activity landscape index: Identifying and quantifying activity-cliffs. J. Chem. Inf. Model. 2008;48(3):646–658. doi: 10.1021/ci7004093. [DOI] [PubMed] [Google Scholar]
- 39.Peltason L, Bajorath J. Sar index: Quantifying the nature of structure-activity relationships. J. Med. Chem. 2007;50(23):5571–5578. doi: 10.1021/jm0705713. [DOI] [PubMed] [Google Scholar]
- 40.Wawer Mathias, Peltason Lisa, Weskamp Nils, Teckentrup Andreas, Bajorath Jürgen. Structure-activity relationship anatomy by network-like similarity graphs and local structure-activity relationship indices. J. Med. Chem. 2008;51(19):6075–6084. doi: 10.1021/jm800867g. [DOI] [PubMed] [Google Scholar]
- 41.Wawer M, Peltason L, Bajorath J. Elucidation of structure-activity relationship pathways in biological screening data. J. Chem. Inf. Model. 2009;52(4):1075–1080. doi: 10.1021/jm8014102. [DOI] [PubMed] [Google Scholar]
- 42.Seebeck Birte, Wagener Markus, Rarey Matthias. From activity cliffs to target-specific scoring models and pharmacophore hypotheses. Chem Med Chem. 2011 Jul; doi: 10.1002/cmdc.201100179. in press. [DOI] [PubMed] [Google Scholar]
- 43.Agrafiotis Dimitris K, Wiener John JM, Skalkin Andrew, Kolpak Jeremy. Single r-group polymorphisms (SRPs) and r-cliffs: An intuitive framework for analyzing and visualizing activity cliffs in a single analog series. J. Chem. Inf. Model. 2011 May;51(5):1122–1131. doi: 10.1021/ci200054u. [DOI] [PubMed] [Google Scholar]
- 44.Sisay Mihiret T, Peltason Lisa, Bajorath Juergen. Structural interpretation of activity cliffs revealed by systematic analysis of structure-activity relationships in analog series. J. Chem. Inf. Model. 2009 Oct;49(10):2179–2189. doi: 10.1021/ci900243a. [DOI] [PubMed] [Google Scholar]
- 45.Austin CP, Brady LS, Insel TR, Collins FS. NIH molecular libraries initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 46.Novotarskyi Sergii, Sushko Iurii, Körner Robert, Pandey Anil Kumar, Tetko Igor V. A comparison of different QSAR approaches to modeling CYP450 1A2 inhibition. J. Chem. Inf. Model. 2011 Jun;51(6):1271–1280. doi: 10.1021/ci200091h. [DOI] [PubMed] [Google Scholar]
- 47.Shen Meng-Yu, Su Bo-Han, Esposito Emilio Xavier, Hopfinger Anton J, Tseng Yufeng J. A comprehensive support vector machine binary hERG classification model based on extensive but biased end point hERG data sets. Chem. Res. Toxicol. 2011 Jun;24(6):934–949. doi: 10.1021/tx200099j. [DOI] [PubMed] [Google Scholar]
- 48.Chen Bin, Wild David J. PubChem BioAssays as a data source for predictive models. J. Mol. Graph. Model. 2010 Jan;28(5):420–426. doi: 10.1016/j.jmgm.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 49.Blum Lorenz C, Reymond Jean-Louis. 970 million druglike small molecules for virtual screening in the chemical universe database GDB-13. J. Am. Chem. Soc. 2009 Jul;131(25):8732–8733. doi: 10.1021/ja902302h. [DOI] [PubMed] [Google Scholar]
- 50.Blum Lorenz C, van Deursen Ruud, Bertrand Sonia, Mayer Milena, Bürgi Justus J, Bertrand Daniel, Reymond Jean-Louis. Discovery of α7-nicotinic receptor ligands by virtual screening of the chemical universe database GDB-13. J. Chem. Inf. Model. 2011 Dec;51(12):3105–3112. doi: 10.1021/ci200410u. [DOI] [PubMed] [Google Scholar]
- 51.Irwin John J, Shoichet Brian K. ZINC - a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 2005;45(1):177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.von Korff M, Sander T. Toxicity-indicating structural patterns. J. Chem. Inf. Model. 2006;46:536–544. doi: 10.1021/ci050358k. [DOI] [PubMed] [Google Scholar]
- 53.Agrafiotis Dimitris K, Wiener John JM. Scaffold explorer: An interactive tool for organizing and mining structure-activity data spanning multiple chemotypes. J. Med. Chem. 2010 Jul;53(13):5002–5011. doi: 10.1021/jm1004495. [DOI] [PubMed] [Google Scholar]
- 54.Wetzel Stefan, Klein Karsten, Renner Steffen, Rauh Daniel, Oprea Tudor I, Mutzel Petra, Waldmann Herbert. Interactive exploration of chemical space with Scaffold Hunter. Nat. Chem. Biol. 2009 Aug;5(8):581–583. doi: 10.1038/nchembio.187. [DOI] [PubMed] [Google Scholar]
- 55.Jain Ajay N, Cleves Ann E. Does your model weigh the same as a Duck? J. Comp. Aid. Molec. Des. 2011 Dec; doi: 10.1007/s10822-011-9530-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cramer Richard D. The inevitable QSAR renaissance. J. Comp. Aid. Molec. Des. 2011 Nov; doi: 10.1007/s10822-011-9495-0. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]