Abstract
The bacterial community composition and structure of water from an established teleost fish system was examined before, during and after a major water change to explore the impact of such a water-change disturbance on the stability of the aquarium water microbiome. The diversity and evenness of the bacterial community significantly increased following the 90% water replacement. While the change in bacterial community structure was significant, it was slight, and was also weakly correlated with changes in physicochemical parameters. Interestingly there was a significant shift in the correlative network relationships between operational taxonomic units from before to after the water replacement. We suggest this shift in network structure is due to the turnover of many taxa during the course of water replacement. These observations will inform future studies into manipulation of the microbiome by changing system environmental parameter values to optimize resident animal health.
Keywords: aquatic microbes, aquarium life support, veterinary, dysbiosis, fish health
INTRODUCTION
The microbial communities associated with animals and their environs have been significantly associated with numerous disease states, as well as physiological and neurological conditions; and a rapidly growing body of literature highlights causative relationships between changes in the microbiome and host health (Balter, 2012). Hygienic practices, antibiotic therapy, and other antimicrobial activities have been linked to multiple health problems, which have become prevalent in Western societies; including asthma, allergies, and other inappropriate immune responses, even several behavioral disorders (Okada et al., 2010). The water in aquatic systems housing animals supports a massive array of microbes (Kramer et al., 2013). A single milliliter of surface seawater may contain a million bacterial cells and 10 million virus particles (Breitbart, 2012). Preliminary evidence strongly suggests the aquatic microbiome influences the immune responsiveness of resident white whales in managed systems (Spoon and Romano, 2012; Van Bonn unpublished data). We believe the microbiome of aquarium systems housing other aquatic mammals and fish also influences health and thus welfare. (Van Bonn, 2015).
To investigate the impact of aquarium management practices on animal health, we have established the Aquarium Microbiome Project (AMP; www.aquariummi-crobiomeproject.org) This program of research, centered at Shedd Aquarium in Chicago, is focused on characterizing the microbial communities in aquarium systems and animals. The primary aim of this activity is to examine associations or correlations with environmental parameters under the control of husbandry and veterinary staff. Such associations can help inform decision-making protocols to control aquarium environments so as to optimize the microbiome for the health of the resident animals. In the current study we examine the microbiome of an established 1600-gallon artificial salt-water ‘estuary’ exhibit prior to, during and immediately following a ninety-percent water change.
MATERIALS AND METHODS
Exhibit Life Support
We studied a 1600-gallon artificial salt-water open top system located within the Shedd Aquarium’s oceanarium space. Salinity of the water was maintained at 33 ± 3 ppt by additions of commercial artificial sea salt mix (Instant Ocean®, Instant Ocean United Pet Group, Blacksburg, VA 24060). Routine surveillance included twice-weekly measures of water pH, temperature, salinity, ammonia, nitrite and nitrate. Mean values for these parameters for the six months immediately prior to the study period were 8.16, 54.5° F, 33.8 ppt, 0.0, 0.009, and 12.3 ppm respectively. System water circulates through a life support loop composed of a mixed media biofilter tower, a 40-watt UV unit, and sand filters before return to the exhibit. The entire volume of water turns over 2.8 times per hour. Resident animals in the exhibit include 1 Sunflower Sea Star (Pycnopodia helianthoides), 1 Kelp Greenling (Hexagrammos decagrammus), 3 C-O Sole (Pleuronichthys coenosus), 3 Red Urchin (Mesocentrotus franciscanus), 1 Staghorn Sculpin (Leptocottus armatus), 3 Painted Greenling (Oxylebius pictus), 3 China Rockfish (Sebastes nebulosus), 10 Striped Surf Perch (Embiotoca lateralis), 1 Copper Rockfish (Sebastes caurinus), 1 Rosy Rockfish (Sebastes rosaceus), 1 Vermillion Rockfish (Sebastes miniatus), 1 Blue Rockfish (Sebastes mystinus), 1 Tiger Rockfish (Sebastes nigrocinctus), 1 Brown Rockfish (Sebastes auriculatus), 1 Clifornia Spiny Lobster (Panulirus interruptus), and 2 Red Irish Lord (Hemilepidotus hemilepidotus). Exhibit animals are fed 40 gm of large krill, 15 gm white shrimp, 15 gm chopped clam, 35 gm silversides, 75 gm capelin, and 75 gm herring twice a week.
The aquarist responsible for this exhibit planned a ninety-percent water change to evaluate any potential influence on some unusual behavior observed in several of the Rockfish. These fish had previously been examined by the veterinary staff and no etiology for the behavior was determined. To rule out an association with some unmeasured water parameter value the 90% change was elected. It was also recognized as an opportunity to explore the potential for a water change of this magnitude to influence the aquatic microbiome of the exhibit. Typically water changes in this system are less than 25% and conducted twice per month.
On day 1 of the study period an automated submersible data logger (Hydrolab DataSonde, Stevens Water Monitoring Systems, Inc. Portland, OR 97220) was deployed in the water column of the system and programed to measure water temperature, water pH, total dissolved gas pressure (TDG), delta-P, oxidation-reduction potential (ORP) and salinity at one hour intervals. The device was also programmed to measure dissolved oxygen and calculate oxygen saturation of the system. These parameters values are not shown due to a device malfunction part-way through data collection and these data were thus excluded from analysis. The data logger remained in the system until day 8 of the study. A ninety-percent water change of the exhibit was conducted on day 4 of the study.
Characterizing the Microbial Community Assemblage of the Water
Beginning four days prior to the planned ninety-percent water change (study day 1), triplicate water samples were acquired daily. Triplicate 250 mL water samples were aseptically filtered through a 0.2 µm Sterivex cartridge (Millipore Corporation, Billerica, MA 01821) on-site at the exhibit using 50 mL sterile plastic syringes to manually pump water. Sterivex filters were sealed with parafilm, labeled and then stored in a −80°C freezer. On the water change date, samples were similarly collected and processed at one-hour intervals beginning immediately following dropping the system water level to 10% and continuing for 8 hours as water was replaced back up to 100% of the initial volume. The source of replacement water was municipal water that was initially passed through a granulated activated charcoal contact chamber and then to which Instant Ocean® was added in a mixing basin, until a salinity of 34 ppt was reached. The water was then sent to a temporary storage basin to continue mixing and to bring the water to the desired temperature of 78°Fahrenheit.
Finally, triplicate samples were collected and processed once per day for 7 more days following this change. This resulted in four daily time points prior to exchange, 8 hourly time points during exchange, and 7 daily time points after the exchange.
Genomic DNA was extracted from Sterivex filters using the PowerWater Sterivex DNA Isolation Kit (MO BIO) following the manufacturer’s suggested protocol. Genomic DNA was amplified using the Earth Microbiome Project (EMP); www.earthmicrobiome.org; (Gilbert et al., 2014) barcoded primer set adapted for MiSeq by adding nine extra bases in the adapter region of the forward amplification primer that support paired-end sequencing (Caporaso et al., 2012). The V4 region of the 16S rRNA gene (515F-806R) was amplified with region-specific primers that includedthe Illumina flowcell adapter sequences. The reverse amplification primer also contained a twelve base barcode sequence that supports pooling of up to 2,167 different samples in each lane. Each 25 µL PCR reaction contained 12 µL of MO BIO PCR Water (Certified DNA-free), 10 µL of 5 Prime HotMasterMix (1×), 1 µL of Forward Primer (5 µM concentration, 200 pM final), 1 µL Golay Barcode Tagged Reverse Primer (5 µM concentration, 200 pM final), and 1 µL of genomic DNA. The PCR conditions are as follows: 94°C for 3 min to denature the DNA, with 35 cycles at 94°C for 45 sec, 50°C for 60 sec and 72°C for 90 sec, with a final extension of 10 minutes at 72°C to ensure complete amplification. Following PCR, amplicons were quantified using PicoGreen (Invitrogen) and a plate reader. Once quantified, different volumes of each of the products were pooled into a single tube so that each amplicon was represented equally. This pool was then cleaned using the UltraClean® PCR Clean-Up Kit (MO BIO) and quantified using Qubit (Invitrogen). After quantification, the molarity of the pool was determined and diluted to 2 nM, denatured, and then diluted to a final concentration of 2 pM with a 30% PhiX spike for loading on the Illumina MiSeq sequencer.
Data Processing and Statistical Analysis
Alpha and beta diversity were generated using QIIME v1.8.0 and R statistical software v3.1.2 (Caporaso et al., 2010). To avoid sampling biases, the OTUs containing fewer than 130 sequences (0.01%) were discarded followed by rarifying the OTU table to an even depth of 12,300 sequences per sample. Within each sample type, alpha diversity and evenness were calculated utilizing Shannon, Simpson, Chao1 and observed species indices. Beta-diversity was analyzed using weighted UniFrac distances. An analysis of varience (ANOVA) and analysis of similarity (ANOSIM) assessed significant variation in taxonomic abundance and composition utilizing UniFrac distances. Matrices were randomized, 999 permutations, to assess correlation across categorical variables.
The changes in water chemical parameters before, during and after the water change were assessed for statistical significance (P < 0.01) using multivariate repeated measure tests. Chemical parameters influencing microbial community function and differentiation were analyzed utilizing a canonical correspondence analysis (CCA) from R’s vegan package (R Core Team, 2013). BEST analysis was used to select the optimal n-parameter model of environmental parameters that could account for the largest percentage of microbial community variance (i.e., the model with the maximum rho-value). This optimal parameter set was then used to constrain a canonical correspondence analysis (CCA) of microbial community structure. Tracing a line from the tip of a vector to an ordination axis (perpendicular) indicates the relative contribution of a particular environmental parameter to the constrained variance represented by that axis.
Only abundant taxa were retained for network analyses (at least 0.1% relative abundance across the entire data set). OTU matrices containing abundant taxa were normalized to an even sequence depth of 2,100 sequences per sample. The OTU table was then split into ‘before’, ‘during’, and ‘after’ time series. The resulting matrices were then processed using SparCC, to calculate all pairwise linear correlations between OTUs across each time series (Friedman & Alm, 2012). Matrices were then randomized 1000 times, and were used to generate correlation tables. The randomized correlation matrices were then used to calculate bootstrapped p-values for each pairwise correlation (Friedman & Alm, 2012). Correlation matrices were filtered to only include significant correlations (P < 0.05) with |rho-values| ≥0.75. These filtered correlation matrices were then used to construct OTU networks, where each OTU represents a node and each correlation an edge. Network statistics were then calculated using the NetworkX package in python, and plotting was done using Matplotlib and Cytoscape (Hunter, 2007; Hagberg et al., 2008; Lopes et al., 2010).
RESULTS
Mean hourly physiochemical parameter values for the system captured by the data logger, prior to, during and after the water change are presented as Table 1. During the water change process, water temperature, pH, and delta-P parameters experienced statistically significant increase from the prior values, in addition the ORP experienced a statistically significant decrease. There were no statistical significant changes from the prior levels during the water change phase in the remaining physiochemical measures including TDG and Salinity parameters. However, when comparing between data taken before and after the water change, there were statistically significant differences for all the six water structure measures.
TABLE 1.
Mean hourly system water values of test system calculated from measures taken by data logger during study period prior to, during and post 90% water change
| Mean hourly system water values | ||||||
|---|---|---|---|---|---|---|
| Temperature (F) | pH | TDG (mmHg) | Delta-P (mmHg) | ORP | Salinity (ppt) | |
| Prior | 54.93 | 8.49 | 747.54 | 5.34 | 337.80 | 37.91 |
| During | 55.24 | 8.62 | 747.79 | 9.38 | 316.42 | 37.71 |
| Post | 54.62 | 8.67 | 753.33 | 5.00 | 313.38 | 37.39 |
To explore how the microbial community changed during the manipulation, a total of 1,139,278 sequences were generated from 44 samples. These data were processed at a rarefaction depth of 12,900 sequences, which comprised 1,197 OTUs (Operational Taxonomic Units; 97% identity).
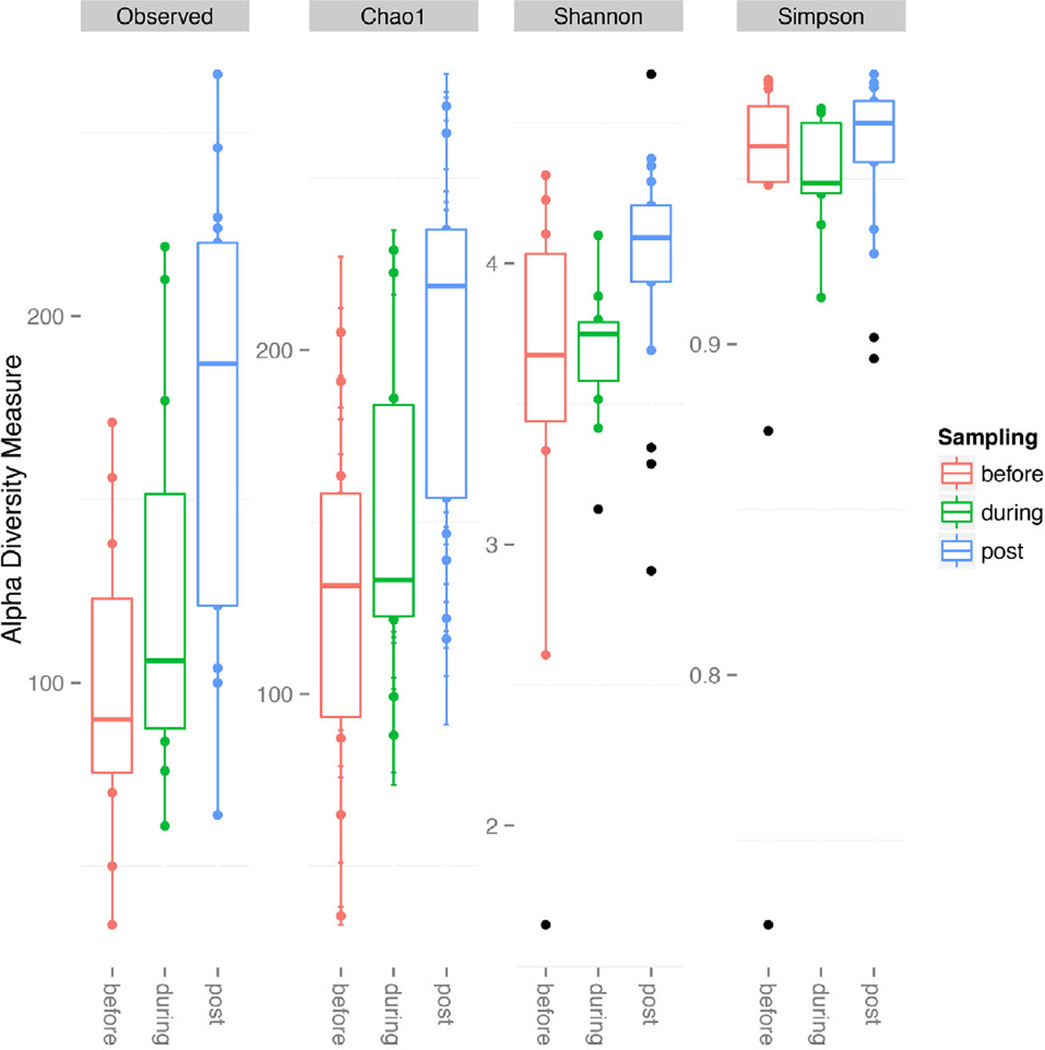
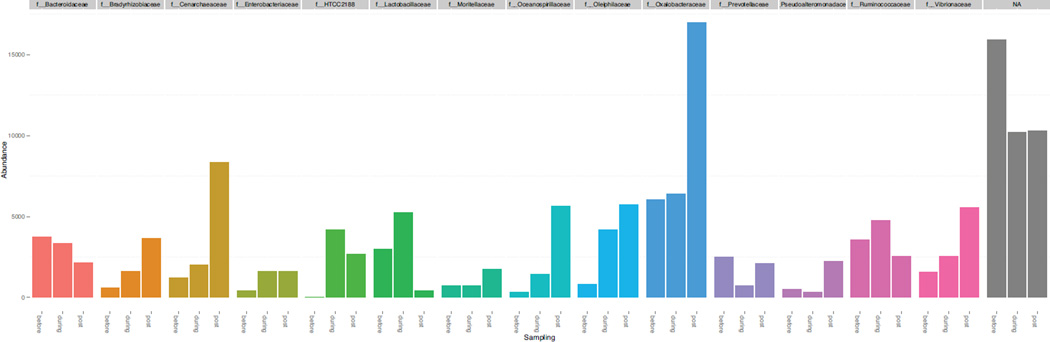
Bacterial community richness and evenness increased in response to the 90% water replacement (Fig. 1), with a significantly greater species richness and evenness in samples taken after the water change. The bacterial community structure was significantly different when comparing between water samples taken before and after the water change. However, this difference was weak, whether measured as unweighted (ANOSIM, P < 0.01, R = 0.33) or weighted UniFrac distance (ANOSIM, P < 0.01, R = 0.25). Family level taxonomic groups did demonstrate a significant shift in relative abundance. Bacteroidaceae (7.1%) and Lactobacillaceae (2.6%) were significantly reduced due to the water change, while Cenarchaeaceae (5.6%), Oceanospirillaceae (3.1%), Vibrionaceae (5.3%), and Bradyrhizobiaceae (2.2%) all showed significant increases in relative abundance (P < 0.01; Fig. 2).
Fig. 1.
Grouping samples into pre, during and post water replacement, observed species and chao1 indices characterized sample richness. Shannon and Simpson indices were generated to observe alpha diversity and evenness. Samples taken post water replacement displayed higher diversity and richness than pre and during water replacement samples.
Fig. 2.
Microbial community composition pre, during and post water replacement were characterized at the Family taxonomic level. Cenarchaeaceae, Oceanospirillaceae, Bradyrhizobiaceae and Vibrionaceae showed a significant increase post water replacement.
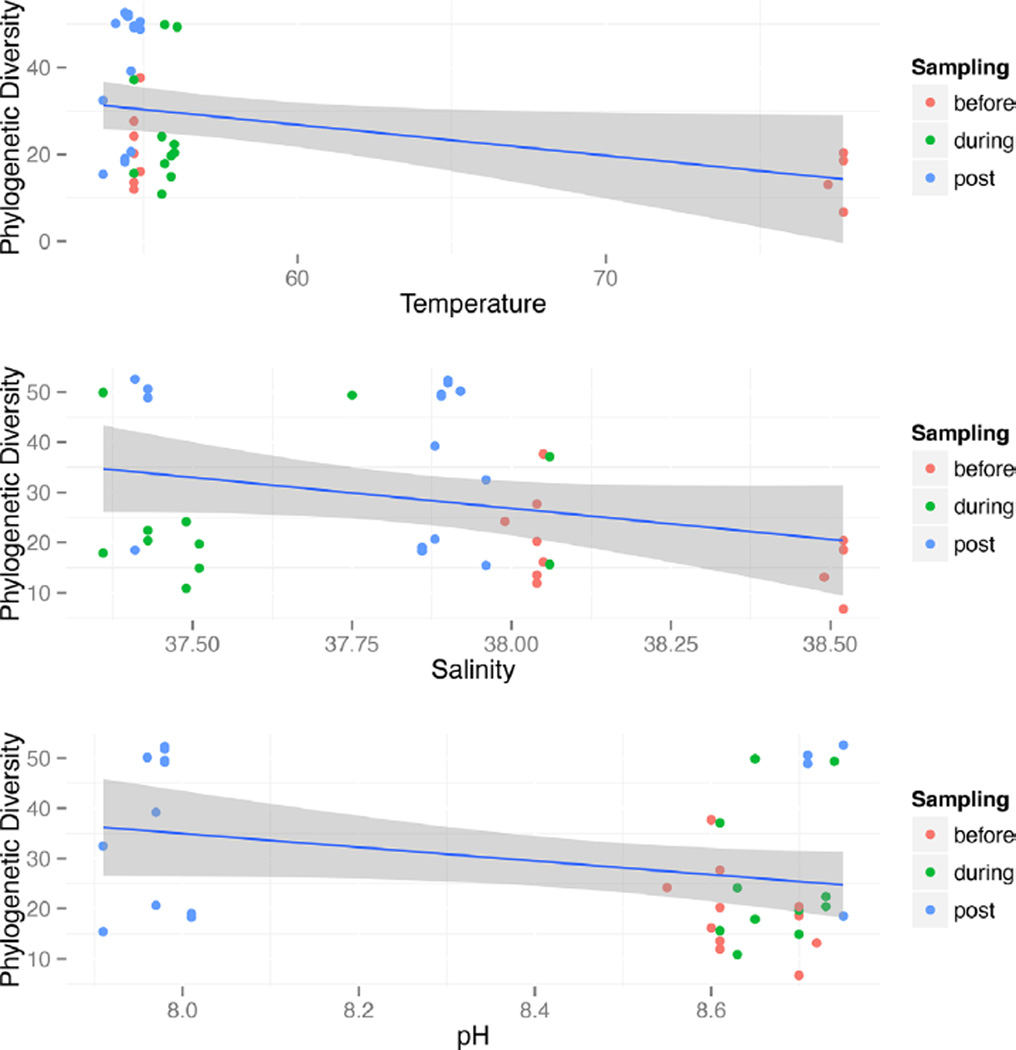
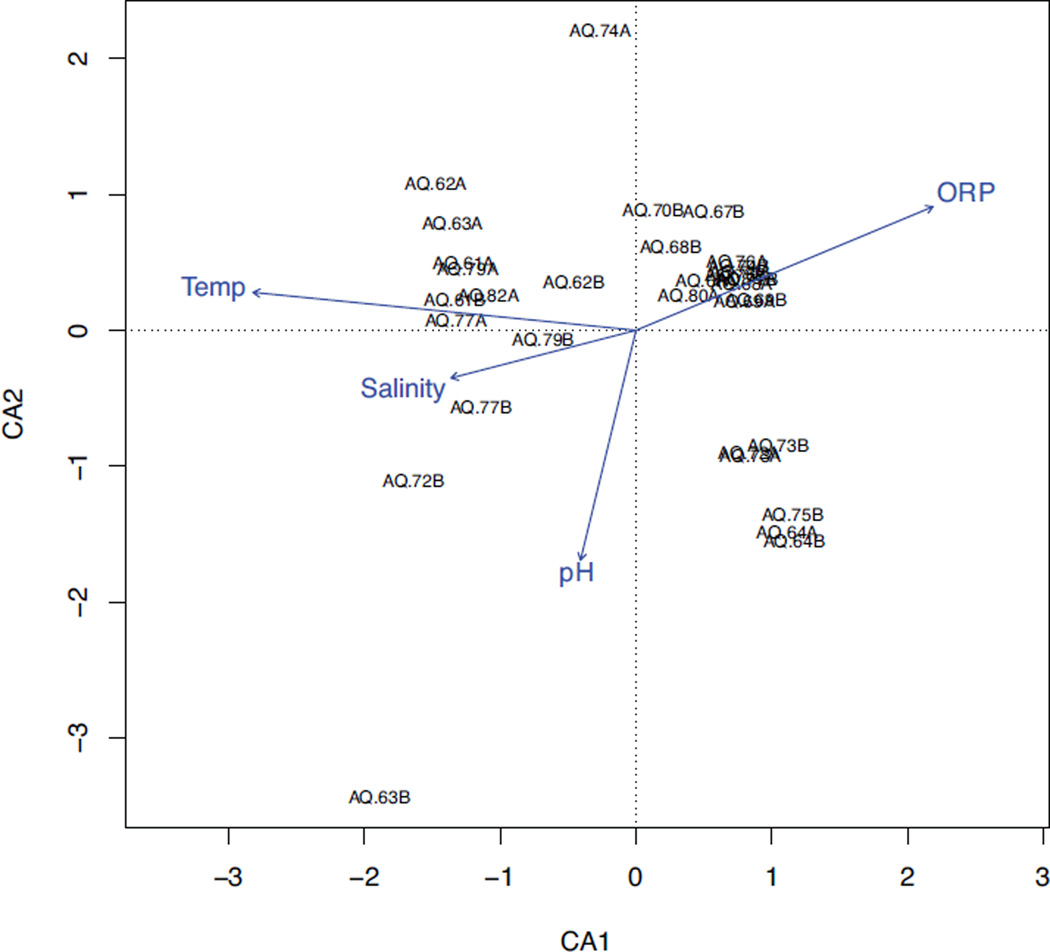
Temperature showed the strongest correlation (BEST, rho = 0.259, P < 0.01) with unweighted UniFrac distances (Fig. 3), compared to oxidative reduction potential, salinity and pH. A slight increase in the correlation statistic was observed when combining temperature with salinity (rho = 0.264), with no increased correlation when adding pH (rho = 0.252). A canonical correspondence analysis on these four chemical parameters supports rho correlation statistics in differentiating community structure (Fig. 4).
Fig. 3.
The diversity of the community (phylogenetic diversity) was plotted against each chemical parameter (oxidative reduction potential [ORP], salinity, and pH) that influenced community differentiation using Spearman’s rho value. Samples were colored by the water replacement sampling (pre, during and post) to understand whether the community diversity was impacted using a linear regression model. Salinity and pH introduced diversity into the community as time elapsed, but oxidative reduction potential (ORP) decreased diversity.
Fig. 4.
A canonical correspondence analysis (CCA) showed correlation of variable chemical parameters (barometric pressure, oxidative reduction potential [ORP], salinity, temperature and pH) on samples pre, during and post water replacement. A CCA constrains the ordination to better predict influence. The arrow points to the environmental parameter that differentiates the community, and the length of the arrow signifies magnitude of the influence.
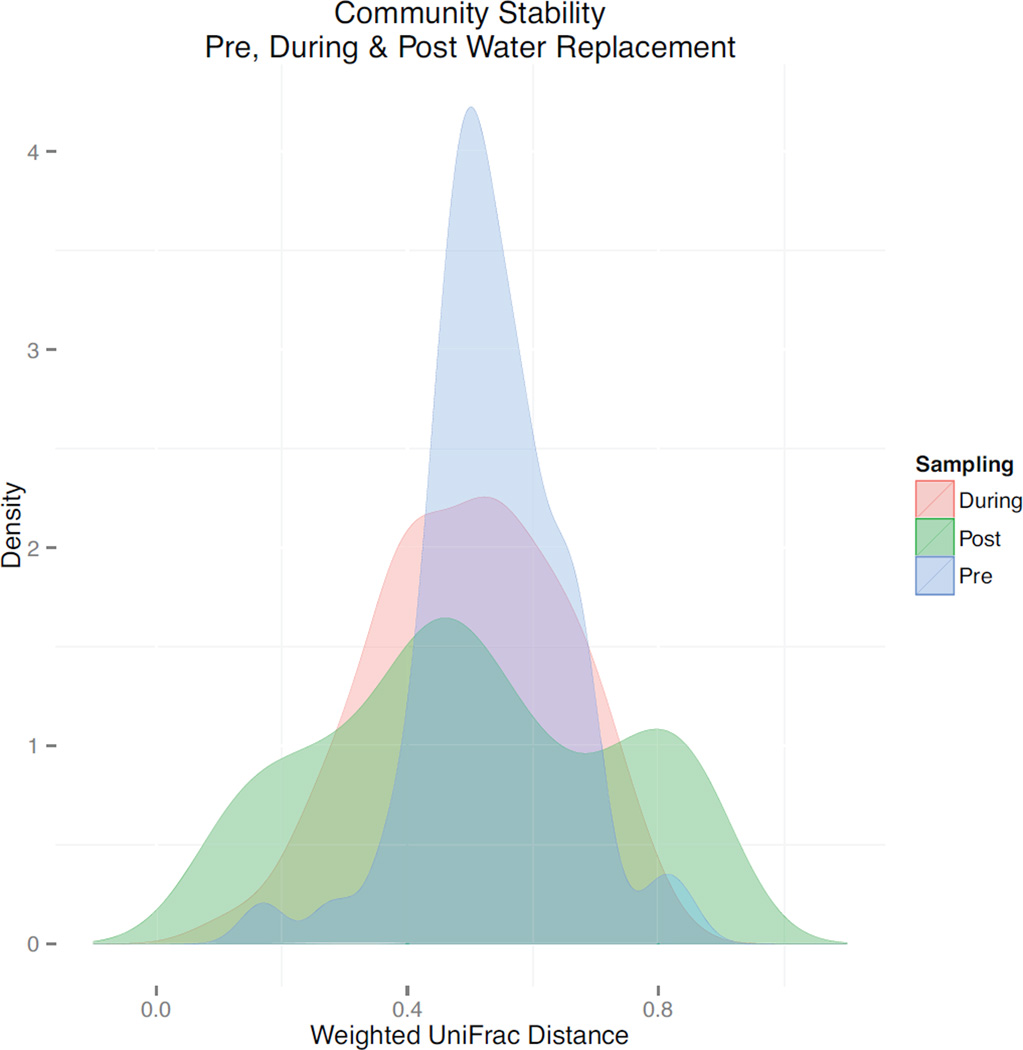
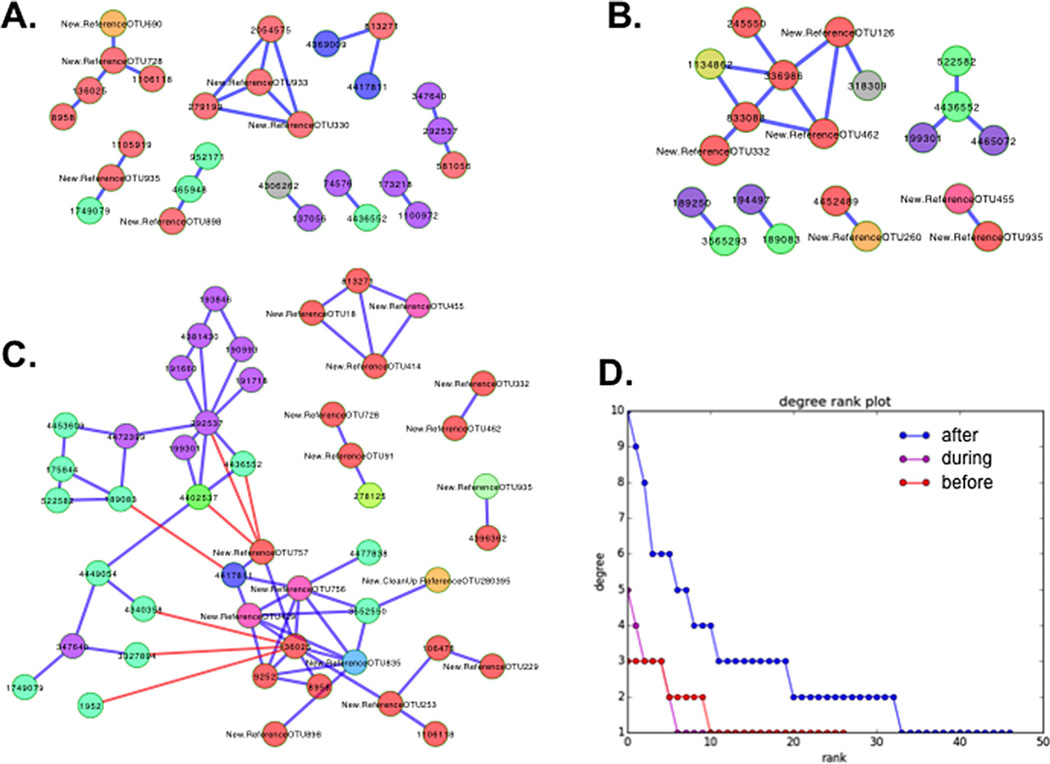
Samples taken prior to, during and post water replacement displayed the least to most variance in bacterial community structure, respectively (Fig. 5). Correlations between OTUs over time within each time period (before, during and after water replacement) demonstrated that the network of correlative interactions is relatively unstructured before the replacement. After the replacement there is a significant increase in correlative network structure (Fig. 6A–C). A large connected network of Proteobacteria is negatively correlated with a large connected network of Firmicutes and Bacteroidetes after the water replacement (Fig. 6C). The number of connections for both high and low degree nodes, increased from before through during to after the replacement (Fig. 6D), highlighting the significant increase in correlative relationships between OTUs.
Fig. 5.
Density plots comparing the distribution of pairwise weighted UniFrac distances calculated between pre, during and post water replacement. Values of 0 would suggest a state that is completely stable, while 1 suggests a state that is in constant flux. If more sample comparisons are closer to zero then the community is not changing dramatically. Samples taken prior to the water replacement showed the least variation in community stability/volatility, where variation increased successively during and post water replacement. The x-axis represents the relative stability/volatility state of the community. The y-axis indicates the abundance of the community within its relative stability/volatility state.
Fig. 6.
The number of putative interactions between bacterial taxa significantly increased post water change. But this may be due to the statistical frequency of potential interactions as the communities were also much more variable post-water change. OTU networks were generated for pre, during and post water replacements using SparCC by calculating OTU-OTU pairwise interactions, which were randomized 1000 times to generate filtering OTUs with P-values <0.05 and rho correlations ≥0.75. Nodes represent OTUs, and the edges represent the interaction between them with blue and red signifying a positive and negative correlation, respectively.
DICSUSSION
The removal and replacement of system water is a standard method for refreshing aquaria in both professional and amateur systems. The husbandry benefits of conducting water changes are numerous and indisputable. However, as resource management and water conservation becomes increasingly important there is a need to improve understanding of the impacts of water change. Although we observed statistically significant changes in water temperature, pH, delta-P and ORP of the system during the water change, these changes would not be considered clinically significant by animal health or husbandry managers. This is as expected because these values are controlled for to maintain them within known tolerance ranges for the species housed within the exhibit. Our interest in this study was to investigate the influence of water change on the exhibit’s microbiome.
Here, we demonstrate the impact of water replacement on the diversity, structure and stability of the water-associated bacterial community. Bacterial community diversity and evenness was significantly increased following the water replacement, which was indicative of a significant reduction in the relative abundance of dominant taxa that made up the stable microbial assemblage prior to exchange. The environmental factors that most significantly correlated with changes in microbial community structure were temperature and salinity, suggesting that the metabolism of these microorganisms is influence by the slight changes in environmental conditions brought on by water replacement. However, the correlation was weak, and environmental factors only accounted for a minority of the community variance detected. Water replacement resulted in a significant increase in microbial community variance. Increased stochasticity in microbial community structure is expected following a disturbance, due to a disruption of ecological interactions (Shade et al., 2012). Greater flux in the microbial community composition increases the likelihood of identifying correlative relationships between co-varying taxonomic units. Therefore, the correlation network structure we observed is likely due to a successional process that is driven by adding freshwater (mixed with salt) to the older aquarium water. As the new water community mixes with the remnants of the old water community, the system is forced into disequilibrium. The primary mechanical disturbance did significantly affect microbial community diversity and structure. However, the impact in this study on the microbial community structure was very slight and likely had a minimal influence on metabolic, functional and virulence potential. While these aspects were not the aim of the current study, it is interesting to note that the limited change observed in the bacterial community is likely to have not resulted in any significant health status outcomes for the resident fauna and flora of this system.
CONCLUSIONS
Aquarium systems are intensively monitored with standardized and high confidence values for multiple physiochemical parameters. These systems provide unique test platforms for controlled artificial environments and may lead to discoveries about managing the microbiomes of these types of environments and our increasingly managed global environment. The importance of this knowledge is growing as the body of evidence supporting the influence of microbiomes on optimal animal health continues to grow. This study demonstrates the exceptional power of microbial community characterization using contemporary DNA extraction, amplification, and sequencing methods followed by data processing and statistical analyses routinely employed by molecular microbial ecologists. It also informs future planned studies into manipulation of environmental parameters to identify means of optimizing microbial assemblages that promote resident animal health. Aquarium scientists and managers now also have a means of evaluating the impacts of life support system changes aimed at resource conservation (for example reduced water changes, reduced turn-over rates, changes from one disinfection technology to another, and so on) on the microbiome of the system and resulting ‘downstream’ consequences.
Acknowledgments
We are grateful to Mr. James Clark, aquarist responsible for study system, Ms. Caryn Svienty, Ms. Julie Nagler and Ms. Jennifer Bozych for water sample collection and processing. We thank Ms. Lei Zhao for assistance with statistical analyses. Sean Gibbons was supported by an EPA STAR Graduate Fellowship.
Footnotes
The authors confirm no conflict of interest involved in this work product.
REFERENCES
- Balter M. Taking Stock of the Human Microbiome and Disease. Science. 2012;336:1246–1247. doi: 10.1126/science.336.6086.1246. [DOI] [PubMed] [Google Scholar]
- Breitbart M. Marine viruses: truth or dare. Annu Rev Mar Sci. 2012;4:425–448. doi: 10.1146/annurev-marine-120709-142805. [DOI] [PubMed] [Google Scholar]
- Caporaso J, Gregory, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Alm EJ. Inferring correlation networks from genomic survey data. PLoS Comput Biol. 2012;8:9, e1002687. doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, Jansson JK, Knight R. The Earth Microbiome project: successes and aspirations. BMC Biol. 2014;12:69. doi: 10.1186/s12915-014-0069-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagberg A, Schult D, Swart P. Exploring network structure, dynamics, and function using networkx; Proceedings of the 7th Python in Science Conference; 2008. pp. 11–15. [Google Scholar]
- Hunter JD. Matplotlib: a 2D graphics environment. Comput Sci Eng. 2007;9:90. [Google Scholar]
- Kramer A, Bekeschus S, Bröker BM, et al. Maintaining health by balancing microbial exposure and prevention of infection: the hygiene hypothesis versus the hypothesis of early immune challenge. J Hosp Infect. 2013;83S1:S29–S34. doi: 10.1016/S0195-6701(13)60007-9. [DOI] [PubMed] [Google Scholar]
- Lopes CT, Franz M, Kazi F, et al. Cytoscape Web: an interactive web based browser. Bioinformatics. 2010;26:18. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: An update. Clin Exp Immunol. 2010;160:1–9. doi: 10.1111/j.1365-2249.2010.04139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. URL http://www.R-project.org/ [Google Scholar]
- Shade A, Peter H, Allison S, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:417. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoon TR, Romano TA. Neuroimmunological response of beluga whales (Delphinapterus leucas) to translocation and a novel social environment. Brain Behav Immun. 2012;26:122–131. doi: 10.1016/j.bbi.2011.08.003. [DOI] [PubMed] [Google Scholar]
- Van Bonn WG. Pinnipedia. In: Miller RE, Fowler ME, editors. Zoo and Wild Animal Medicine. Vol. 8. St. Louis: Elsevier Saunders; 2015. pp. P436–P450. [Google Scholar]