Abstract
From 2003 to 2013, Indonesia had the highest number of avian influenza A cases in humans, with 192 cases and 160 fatalities. Avian influenza is caused by influenza virus type A, such as subtype H5N1. This virus has two glycoproteins: hemagglutinin and neuraminidase, which will become the primary target to be neutralized by vaccine. Vaccine is the most effective immunologic intervention. In this study, we use the epitope-based vaccine design from hemagglutinin and neuraminidase of H5N1 Indonesian strain virus by using immunoinformatics approach in order to predict the binding of B-cell and T-cell epitopes (class I and class II human leukocyte antigen [HLA]). BCPREDS was used to predict the B-cell epitope. Propred, Propred I, netMHCpan, and netMHCIIpan were used to predict the T-cell epitope. Two B-cell epitopes of hemagglutinin candidates and one B-cell epitope of neuraminidase candidates were obtained to bind T-cell CD4+ (class II HLA), and also five T-cell epitope hemagglutinin and four T-cell epitope neuraminidase were obtained to bind T-cell CD8+ (class I HLA). The visualization of epitopes was done using MOE 2008.10. It shows that the binding affinity of epitope–HLA was based on minimum binding free energy (ΔGbinding). Based on this result, visualization, and dynamic simulation, four hemagglutinin epitopes (MEKIVLLLA, CPYLGSPSF, KCQTPMGAI, and IGTSTLNQR) and two neuraminidase epitopes (NPNQKIITI and CYPDAGEIT) were computed as having the best binding affinity from HLA ligand. The results mentioned above are from in silico experiments and need to be validated using wet experiment.
Keywords: H5N1, hemagglutinin, neuraminidase, epitope, vaccine, B-cell, T-cell
Introduction
Avian influenza, also known as bird flu or avian flu, has become a global health issue in several countries and has spread from Asia to Africa, Middle East, and Europe continent, with an estimated 622 cases that occurred within 2003–2013 worldwide.1–3 According to the World Health Organization, in 2012, Indonesia had the highest mortality of avian influenza disease out of 15 countries, since it was first found in Banten in 2005, with 160 deaths out of 192 cases. It has spread out to 11 provinces, including West Java, Bali, North Sumatera, and South Sulawesi.4,5 While this disease has been a serious threat across the country, there are still no effective vaccines or antiviral drugs yet for combating this highly pathogenic disease.6,7 The development of new, efficient vaccines or drugs is urgently needed in order to decrease the mortality rate of avian influenza.
Avian influenza is caused by several subtypes of influenza type A viruses, such as H5N1, H5N2, and H7N9, which belong to the Orthomyxoviridae family.8–10 The influenza A virus genome has eight ssRNA segments that encode 10 proteins with two major surface glycoproteins: hemagglutinin and neuraminidase.11 Hemagglutinin is the essential antigen that neutralizes human antibody and binds to the host cell receptor, and it mediates the sialic acid binding receptor’s surface cell that initiates the virus infection on the host cell.12,13 Neuraminidase is a glycoprotein that is known for its role to catalyze the cleavage of the sialic acid bond from d-galactose or d-galactosamine residues by cutting off the α-keto acid bond between neuraminic acid and glycosyl residue of glycoprotein, glycolipid, or colominic acid.14 The recent utilization of serological analysis has categorized influenza A viruses into 18 hemagglutinin subtypes (H1–H18) and 9 neuraminidase subtypes (N1–N9), which allow them to form 162 different kinds of influenza subtypes.15–19 Because of their vital role in the pathogenicity of the virus, hemagglutinin and neuraminidase have become primary targets in the vaccine design and development fields for neutralizing the avian influenza virus.20
Usage of vaccine is one of the most effective immunological interventions in controlling the infection of H5N1 virus. To date, several H5N1 vaccine design and development efforts have been conducted, yet the effective, affordable vaccine has to be found.21,22 Most immunization has not provided sufficient protection from epitope antigens.23,24 An ideal vaccine should have humoral immune and cellular responses that can be used to trigger the B-cells and T-cells selectively. As in the inactivated vaccine, the vaccine provides only the humoral immune response and causes little or no cellular immune response.23 The latest influenza vaccine has been developed by using the epitope-based approach. The epitope is an amino acid that binds to the antibodies, and it is located on the surface of the antigen. The specific epitope identification of pathogens has significantly improved the epitope-based vaccine design and development.25
The epitope itself is related to human leukocyte antigen (HLA). Every individual has his or her own specific set of class I and class II HLA.26 The epitope that has high binding affinity to several HLA alleles tends to be a potential candidate for epitope-based vaccine design.27 The epitope-based vaccine has already been designed and tested in the scientific community.28,29 Hence, the ideal vaccine design should be able to cope with the challenge of antigenic drift.30 The purpose of this study is to design a new epitope-based H5N1 vaccine that can bind to the HLA by using in silico approach. Therefore, the epitope-based vaccines of hemagglutinin and neuraminidase from H5N1 of Indonesian strain virus were designed to elicit the immune response of class I and class II HLA.
Research Methodology
The utilized pipeline for this research was modified and extended from the existing ones.31–33 The established pipelines were also taken into account.34,35 Moreover, our research is based on immunoinformatics approach that was already validated in the scientific community.36–39 Thus, immunoinformatics approach has been proven to be useful for examining the response of immune system toward pathogens.37,40–43 All of the experiment methods were done by using a personal computer with 2.1 GHz Intel Pentium Dual Core, RAM 1 GB, and Windows XP Professional Edition. The experiment was also conducted with the Internet support and several kinds of online and offline softwares.
Sequence of hemagglutinin and neuraminidase H5N1 virus
The sequences of hemagglutinin and neuraminidase of H5N1 Indonesian strain were retrieved from Influenza Virus Resource of National Center Biotechnology Information website (http://www.ncbi.nlm.nih.gov/genomes/FLU/FLU.html/).44
Prediction B-cell and T-cell epitope hemagglutinin and neuraminidase H5N1 virus
Following Velkov et al, the mapping of our epitopes has taken into account their tendency of antigenic drifting. The prediction of proteosomal cleavage H5N1 virus was analyzed by using PAPROC-I tools (http://www.paproc.de/), while the transport antigen presentation (TAP) binding was predicted by using TAPPRED server (http://www.imtech.res.in/raghava/tappred/).45,46 Propred and netMHCpan servers were used to identify the bond region on the class I HLA of the antigen.47,48
Visualization and molecular dynamics using MOE 2008.10
The selected epitopes were further analyzed their conformation and stability by performing epitope visualization and molecular dynamics simulation, respectively. All of these steps were completed by using MOE 2008.10 software.49 The epitope visualization itself was used to determine class I and class II HLA of the selected epitopes. The evaluation of the residues was conducted by counting the free energy minimization for the interactions between HLA and epitope. The 10 ns molecular dynamics simulation was executed in order to observe the interaction and stability between epitope and HLA.50
Confirmation of the origin of hemagglutinin epitopes
Following Velkov et al, the origin of Indonesian hemagglutinin epitopes was confirmed by using BLASTp tools (http://blast.ncbi.nlm.nih.gov/).
Results and Discussion
Hemagglutinin and neuraminidase H5N1 sequence
In this study, the sequences of Indonesian H5N1 virus, including both hemagglutinin and neuraminidase, were collected from National Center Biotechnology Information (GenBank Nos BAL61222.1 for hemagglutinin and BAL61230.1 for neuraminidase). The result of Basic Local Alignment Search Tool indicated that all of the Indonesian H5N1 virus sequences showed a high similarity, from 97% to 100%. Group A viruses have the highest percentage of homology with chicken virus isolated in Indonesia in 2010.51
B-cell epitope prediction
An antibody would bind to a specific epitope on the surface of the virus. When a virus is destroyed by macrophage, it would facilitate epitope binding.52 In this study, the epitope prediction of H5N1 hemagglutinin and neuraminidase of the B-cells was done through BCPREDS 1.0 at the following website address: http://ailab.cs.iastate.edu/bcpreds/. The peptide sequence is the required input, and the output will provide information about the position of the peptide sequences along with scores based on its accessibility. The analysis was performed every nine amino acids (nonamer) on a series of peptide sequences with a specificity of 80%. Accessibility is very important because if the score is increasing, the epitope peptide will be having a great accessibility to be recognized by B-cells or bind to the antibody. Table 1 is the predicted epitope of H5N1 hemagglutinin that has accessibility scores against B-cell epitope prediction from the server BCPREDS. It predicts 15 hemagglutinin epitope candidates. They will be expressed in the CD4+ T-cells and bind to class II HLA on the IGTSTLNQR and MVSLVKSDQ.
Table 1.
Results of hemagglutinin epitope prediction against B-cells.
| POSITION | EPITOPE | VALUE |
|---|---|---|
| 98 | KANPNNDLC | 0.998 |
| 137 | SWSDHEASS | 0.997 |
| 181 | NNTNQEDLL | 0.996 |
| 338 | QRESRRKKR | 0.985 |
| 216 | IGTSTLNQR | 0.984 |
| 288 | GNCNTKCQT | 0.979 |
| 510 | EEARLKREE | 0.973 |
| 10 | MVSLVKSDQ | 0.964 |
| 557 | CSNGSLQCR | 0.935 |
| 25 | ANNSTEQVD | 0.931 |
| 447 | LMENERTLD | 0.918 |
| 371 | HHSNEQGSG | 0.909 |
| 361 | QGMVDGWYG | 0.893 |
| 427 | NKKMEDGFL | 0.892 |
| 266 | PEYAYKIVK | 0.856 |
Table 2 shows H5N1 neuraminidase epitope that has an accessibility score against B-cells. A total of 17 candidates of neuraminidase epitope derived from the B-cell epitope prediction. Epitopes will be expressed with the introduction of the CD4+ T-cells and bind to class II HLA on the YNGIITDTI.
Table 2.
Results of neuraminidase epitope prediction against B-cells.
| POSITION | EPITOPE | VALUE |
|---|---|---|
| 342 | TKSTNSRSG | 0.996 |
| 174 | GISGPDNEA | 0.995 |
| 142 | PVGEAPSPY | 0.995 |
| 303 | GDNPRPNDG | 0.989 |
| 84 | NNIRIGSKG | 0.977 |
| 188 | YNGIITDTI | 0.975 |
| 257 | EESCSCYPDA | 0.972 |
| 242 | KVVKSVELD | 0.970 |
| 414 | STIWTSGSS | 0.963 |
| 3 | PNQKIITIG | 0.951 |
| 355 | WDPNGWTGT | 0.943 |
| 317 | PMSPNGAYG | 0.941 |
| 223 | TDGPSNGQA | 0.928 |
| 42 | NQHQAESIS | 0.895 |
| 66 | AGNSSLCPI | 0.883 |
| 273 | RDNWHGSNR | 0.868 |
| 434 | SWSWPDGAE | 0.841 |
Humoral immune systems have an important role in relieving the infection of H5N1 avian influenza virus. During the process of infection, antibodies are produced for all the major glycoproteins (hemagglutinin and neuraminidase) on the surface of the H5N1 virus. Specific antibodies to hemagglutinin are very important for virus neutralization at the mucosal surface by blocking the entry of the virus into the cell. Specific antibodies for neuraminidase are effective in reducing the release of virus from infected cells. Antibodies bind to a particular epitope on the surface of the virus and act as opsonins that facilitate the binding and virus destruction by macrophages.
B-cell epitope candidates of hemagglutinin H5N1 were recognized by the receptor of CD4+ T-cell and bound with class II HLA, which were IGTSTLNQR and MVS-LVKSDQ. Furthermore, YNGIITDTI was an epitope candidate of neuraminidase for B-cell.
T-cell epitope-bound class I HLA prediction
Proteasome is a key factor in the degradation of cytosolic proteins in which the C-terminal ligand of class I HLA is selected by proteome.45 In defining the epitope of hemagglutinin and neuraminidase H5N1 that can binds to class I HLA, the protein sequences were selected by using PaProC server to see the sequences that can be interrupted by proteome. In general, the effective epitope antigen should have a sequence that do not split the proteome and can be transported by TAP.27 In this study, we use TAPpred server for predicting the sequences of the effective epitope antigen. Data analysis results of the epitope of hemagglutinin and neuraminidase with TAP are shown in Tables 3 and 4, respectively.
Table 3.
The analyzed results of TAP (transport antigen presentation) on H5N1 virus hemagglutinin by using TAPpred server.
| PEPTIDE RANK | INITIAL POSITION | SEQUENCES | SCORE | AFFINITY PREDICTION |
|---|---|---|---|---|
| 68 | 2 | EKIVLLLAM | 4.819 | Intermediate |
| 167 | 1 | MEKIVLLLA | 3.416 | Intermediate |
| 194 | 293 | KCQTPMGAI | 3.090 | Intermediate |
| 250 | 389 | KAVDGVTNK | 2.525 | Low or undetectable |
| 297 | 151 | CPYLGSPSF | 2.027 | Low or undetectable |
Table 4.
The analyzed results of TAP (transport antigen presentation) on H5N1 virus neuraminidase by using TAPpred server.
| PEPTIDE RANK | INITIAL POSITION | SEQUENCES | SCORE | AFFINITY PREDICTION |
|---|---|---|---|---|
| 140 | 398 | IRPCFWVEL | 2.929 | Low or undetectable |
| 196 | 2 | NPNQKIITI | 2.102 | Low or undetectable |
| 220 | 399 | RPCFWVELI | 1.818 | Low or undetectable |
| 406 | 261 | CYPDAGEIT | −1.441 | Low or undetectable |
The immune response is induced by T-cell epitope, which has an important role in vaccine designing. T-cell epitopes are presented on the surface of an antigen-presenting cell, which are bound to class I and II HLA.53 After selecting a sequence of hemagglutinin and neuraminidase, we proceeded with the determination of epitope potential vaccine candidates. In this study, the determination of hemagglutinin and neuraminidase H5N1 epitopes on class I HLA has been done and analyzed by using two servers’ prediction. The first prediction is net-MHCpan server version 2.4 with 4% threshold that can be accessed at the website address of http://www.cbs.dtu.dk/services/NetMHCpan/. This server predicts the potential epitope binding to class I HLA of supertypes A and B, so it can be recognized by the receptor CD8+ T-cells. This method consists of 326 HLA, divided into 43 different alleles at the HLA-A and HLA-B.54 In addition, class I HLA-bound epitopes were also predicted using the ProPred 1 server on the threshold of 4%, which can be accessed online at the website address of http://www.imtech.res.in/raghava/propred1/. ProPred 1 server is derived from TEPITOPE, namely a matrix program position. The server was developed for 47 class I HLA with an algorithm based on the linear coefficient matrix.48
Table 5 shows that there are five hemagglutinin epitope candidates bound by CD8+ T-cell, such as MEKIVLLA, EKIVLLAM, CPYLGSPSF, KCQTPMGAI, and KAVDGVTNK. Otherwise, there are four neuraminidase epitope candidates bound to class I HLA: NPNQKIITI, CYPDAGEIT, IRPCFWVEL, and RPCFWVELI (Table 6).
Table 5.
The prediction results of hemagglutinin epitopes toward class I HLA.
| POSITION | EPITOPE | ALLELE | PREDICTION | VALUE |
|---|---|---|---|---|
| 1 | MEKIVLLLA | {HLA-I,B61(4)} | Propred I | 3.41 |
| 2 | EKIVLLLAM | {HLA-I,B14(4)} {HLA-I,B*3902(2)} {HLA-I,B*3501} |
Propred I, netMHCpan Propred I, netMHCpan netMHCpan |
4.81;WB 4.81;WB WB |
| 151 | CPYLGSPSF | {HLA-I,B*0702} {HLA-I,B*1402} {HLA-I,B*3501} {HLA-I,B*3902} |
netMHCpan netMHCpan Propred I, netMHCpan netMHCpan |
SB SB 2.02;SB SB |
| 293 | KCQTPMGAI | {HLA-I,B*0702(4)} | Propred I | 3.08 |
| 389 | KAVDGVTNK | {HLA-I,A*1101(3)} | Propred I, netMHCpan | 2.52;WB |
Abbreviations: SB, strong binder; WB, weak binder.
Table 6.
The prediction result of neuraminidase epitopes toward class I HLA.
| POSITION | EPITOPE | ALLELE | PREDICTION | VALUE |
|---|---|---|---|---|
| 2 | NPNQKIITI | {HLA-I,B*0702} | Propred I, netMHCpan | 2.10;WB |
| {HLA-I,B*0801} | Propred I, netMHCpan | 2.10;WB | ||
| {HLA-I,B*5101} | Propred I, netMHCpan | 2.10;SB | ||
| {HLA-I,B*5102} | Propred I, netMHCpan | 2.10;WB | ||
| {HLA-I,B*5103} | Propred I, netMHCpan | 2.10;SB | ||
|
| ||||
| 261 | CYPDAGEIT | {HLA-I,A24(4)} | Propred I | −1.44 |
|
| ||||
| 398 | IRPCFWVEL | {HLA-I,B14(2)} | Propred I | 2.92 |
| {HLA-I,B*2705(2)} | Propred I | 2.92 | ||
| {HLA-I,B*3901(2)} | Propred I | 2.92 | ||
| {HLA-I,B*2702(3)} | Propred I, netMHCpan | 2.92;WB | ||
|
| ||||
| 399 | RPCFWVELI | {HLA-I,B*5101(4)} | Propred I, netMHCpan | 1.81;WB |
| {HLA-I,B*5102(4)} | Propred I, netMHCpan | 1.81;WB | ||
| {HLA-I,B*5103} | netMHCpan | WB | ||
| {HLA-I,B*0702(2)} | Propred I | 1.81 | ||
Abbreviations: SB, strong binder; WB, weak binder.
T-cell epitope-bound class II HLA prediction
Any peptide molecule that can bind to class II HLA will be presented on CD4+ T-cells, which is very important in the regulation of the B-cell responses and CD8+ T-cells.26 If the response of CD4+ does not exist, then the quality of the cytotoxic response to the antigen administration gradually declined and failed to respond effectively.55 The determination of hemagglutinin and neuraminidase epitopes on class II HLA has been done by using two prediction servers. The first one is netMHCIIpan version 2.1 with 4% threshold that can be accessed at the website address of http://www.cbs.dtu.dk/services/NetMHCIIpan/. This server predicts epitope binding to class II HLA for two supertypes of DRB (antigen D Related, Beta chain), so it can be recognized by receptors CD4+ T-cells. The selection of these alleles are most often found in Asian populations, especially in the parts of Malay distribution.56 NetMHCIIpan method was used to evaluate the epitope binding to class II HLA with a measuring accuracy of prediction on the area under the receiver operating characteristic curve (AROC), which is the curve for evaluating the accuracy of diagnostic markers. AROC values that are close or equal to one indicate that the marker has a high diagnostic accuracy.57 netMHCpan method has been identified as the best predictive value with AROC >0.9. Epitope-binding affinity to HLA-DR alleles was analyzed by netMHCIIpan. Identification of potential epitope is classified as a strong binder, weak binder, and no binder to choose the alleles of class II HLA. Selection is based on the binding affinity threshold of [X] ≤ 50 nM, 50 nM < [X] ≤ 500 nM, and [X] > 500 nM.58
The subsequent prediction was done using the ProPred server with 4% threshold that can be accessed online at the website address of http://www.imtech.res.in/raghava/propred/. The server was developed for 51 HLA-DR alleles by extracting the database matrix.48 Prediction of peptides that bind to class II HLA by using the ProPred server in the development of vaccines for Leptospira interrogans serovar Lai was already in place.59 By using ProPred and netMHCpan II, there are two hemagglutinin epitope candidates bound by CD4+ T-cell (class II HLA), MVSLVKSDQ and IGTSTLNQR, and also one neuraminidase epitope candidate, YNGIITDTI (Tables 7 and 8).
Table 7.
The prediction results of hemagglutinin epitopes toward class II HLA
| POSITION | EPITOPE | ALLELE | PREDICTION | VALUE |
|---|---|---|---|---|
| 10 | MVSLVKSDQ | {HLA-II,DRB1*0301} | Propred | 3.47 |
| {HLA-II,DRB1*0405} | Propred | 1.70 | ||
| {HLA-II,DRB1*1202} | netMHCIIpan | WB | ||
| {HLA-II,DRB1*1302} | Propred | 1.90 | ||
|
| ||||
| 216 | IGTSTLNQR | {HLA-II,DRB1*0301} | Propred | 1.81 |
Abbreviation: WB, weak binder.
Table 8.
The prediction results of neuraminidase epitopes toward class II HLA.
| POSITION | EPITOPE | ALLELE | PREDICTION | VALUE |
|---|---|---|---|---|
| 188 | YNGIITDTI | {HLA-II,DRB1*0101} | netMHCIIpan | WB |
| {HLA-II,DRB1*0402} | netMHCIIpan | WB | ||
| {HLA-II,DRB1*0403} | netMHCIIpan | WB | ||
| {HLA-II,DRB1*0405} | Propred, netMHCIIpan | 2.70;WB | ||
| {HLA-II,DRB1*0406} | netMHCIIpan | WB | ||
| {HLA-II,DRB1*0701} | Propred, netMHCIIpan | 4.12;WB | ||
| {HLA-II,DRB1*0803} | NetMHCIIpan | WB | ||
| {HLA-II,DRB1*1001} | netMHCIIpan | WB | ||
| {HLA-II,DRB1*1302} | Propred | 0.30 | ||
| {HLA-II,DRB1*1502} | Propred | 1.56 |
Abbreviation: WB, weak binder.
Visualization and molecular dynamics of hemagglu-tinin and neuraminidase epitope using MOE 2008.10
The visualization was useful to predict the binding free energy from class I and class II HLA–epitope of hemagglutinin and neuraminidase. Position and affinity binding from class I and class II HLA–epitope of hemagglutinin and neuraminidase are shown in Tables 9 and 10, respectively.
Table 9.
The position and affinity of hemagglutinin epitope H5N1-bound class I and class II HLA.
| POSITION | EPITOPE | ALLELE | HYDROGEN BONDING | ΔGBINDING (kcal/mol) |
|---|---|---|---|---|
| 1 | MEKIVLLLA | HLA A61 | Glu 76, Arg 97, Tyr 136, and Lys 146 | −40.0148 |
| 2 | EKIVLLLAM | HLA B*3501 | Arg 97, Arg 151, Gln 155, and Leu 156 | −22.4538 |
| 10 | MVSLVKSDQ | HLA DRB1*0301 | Glu 9, Glu 11, Ser 11, Tyr 30, Phe 31, Trp 61, Asp 66, Asn 69, and Arg 76 | −11.7756 |
| 151 | CPYLGSPSF | HLA B*0702 | Tyr 7, Tyr 99, Glu 152, Gln 155, and Tyr 171 | −28.0471 |
| 216 | IGTSTLNQR | HLA DRB1*0301 | Glu 11, Glu 55, Asn 62, Asp 66, Lys 71, Arg 74, and Asn 82 | −56.9580 |
| 293 | KCQTPMGAI | HLA B*0702 | Tyr 7, Glu 152, Tyr 159, and Tyr 171 | −33.6109 |
| 389 | KAVDGVTNK | HLA A*1101 | Glu 63, Asp 77, Thr 80, Arg 114, Asp 116, Lys 146, Gln 155, Arg 163 and Trp 167 | −48.9974 |
Table 10.
The position and affinity of neuraminidase epitope H5N1-bound class I and class II HLA.
| POSITION | EPITOPE | ALLELE | HYDROGEN BONDING | ΔGBINDING (kcal/mol) |
|---|---|---|---|---|
| 2 | NPNQKIITI | HLA B*0702 | Asp 114, Arg 156, Tyr 159, Glu 163, Val 165, Arg 169, Arg 170 | −23.8779 |
| 188 | YNGIITDTI | HLA DRB1-0101 | Glu 28, Arg 71, Arg 72, Val 75, Thr 77, Cys 79, dan Asn 82 | −17.8049 |
| 261 | CYPDAGEIT | HLA A*24 | Lys 12, Asp 74, Tyr 85, Tyr 123, Asp 137, Met 138, dan Ala 139 | −33.0200 |
| 398 | IRPCFWVEL | HLA B*2705 | Asp 77, Tyr 84, His 93, Thr 94, Gln 96, Asp 119, dan Thr 143 | −33.6376 |
| 399 | RPCFWVELI | HLA B*0702 | Tyr 7, Tyr 159, Tyr 9, Tyr 99, Arg 62, Glu 152, dan Gln 155 | −24.8928 |
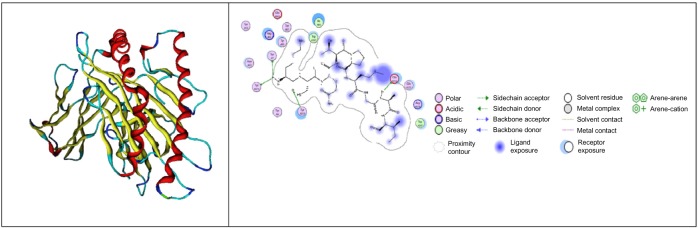
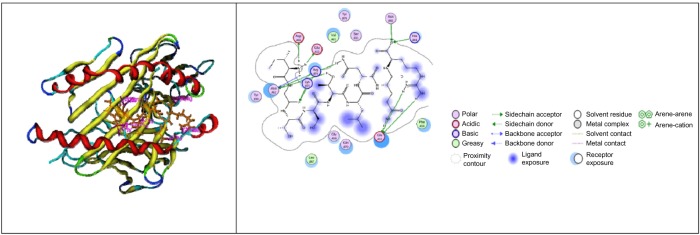
For hemagglutinin epitope-bound class I HLA, the weak binder is EKIVLLLAM, which has ΔG of −22.4538 kcal/mol, and KCQTPMGAI is one of the strong binder candidates, which has ΔG of −33.6109 kcal/mol (Fig. 1, the description of legend could be observed in the Supplementary material here: http://staff.ui.ac.id/system/files/users/aditya.parikesit/material/supplementary_material_feimmy.pdf). The interaction between class II HLA and hemagglutinin epitope has the weak binder candidate (MVSLVKSDQ) with ΔG of −11.7756 kcal/mol and the strong binder candidate (IGTSTLNQR) with ΔG of −56.9580 kcal/mol (Fig. 2).
Figure 1.
The visualization of KCQTPMGAI with HLA B*0702 PDB: 3VCL (MOE 2008.10).62
Figure 2.
The visualization of IGTSTLNQR with HLA DRB1*0301 PDB: 1A6A (MOE 2008.10).63
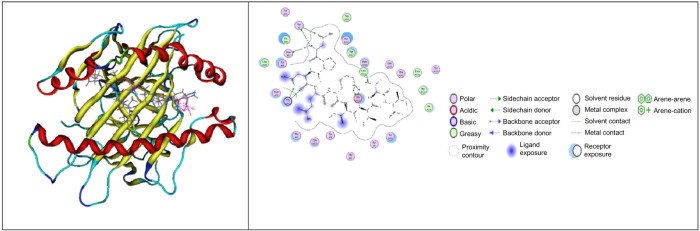
For neuraminidase, there are two epitope candidates, which have strong interactions, such as CYPDAGEIT and NPNQKIITI. The strongest interaction between epitope and class I HLA is NPNQKIITI (ΔG −43.5280 kcal/mol) (Fig. 3). The KCQTPMGAI epitope was visualized because of its role as strong binder candidate for class I HLA, while NPNQKIITI was visualized as having the strongest interaction with class I HLA.
Figure 3.
The visualization of NPNQKIITI with HLA B*5101 PDB: 1E27 (MOE 2008.10).64
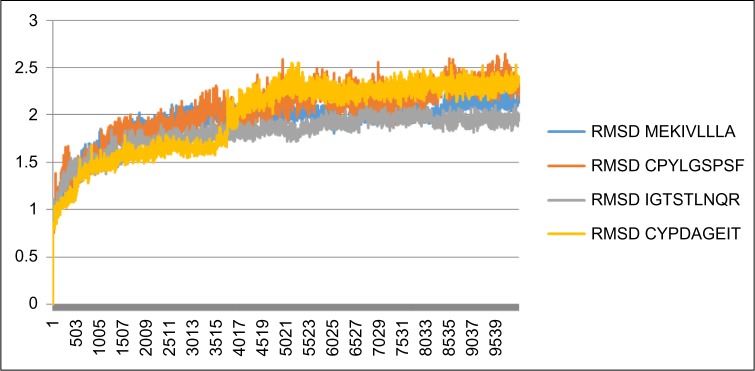
Figure 4 shows epitope–HLA interaction in the initialization process of 0–100 ps that fluctuates and stabilizes in more than 100 ps. It tends to be in a linear form graphically. The variation of the curve form is determined for three epitopes of hemagglutinin (MEKIVLLLA, CPYLGSPSF, and IGTSTLNQR) and one epitope of neuraminidase (CYPDAGEIT) with high fluctuation. It is indicated that epitope–HLA structure conformity changes during the time of forming. The curve becomes linear on more than 100 ps, confirming that the epitope–HLA structure conformity has not changed significantly. The IGTSTLNQR epitope was included both in molecular dynamics and in visualization due its tendency as having the most spontaneous ΔGbinding of all epitopes and acting as both T-cell and B-cell epitopes. The CYPDAGEIT epitope was included in the molecular dynamics simulation because of its role as the strongest binding neuraminidase-based epitope. Concerning the molecular dynamics of hemagglutinin epitopes, out from the data range, MEKIVLLA was taken as the mean value, CPYLGSPSF was the minimum value, and IGTSTLNQR was the maximum value.
Figure 4.
The molecular dynamics simulation of MEKIVLLLA, CPYLGSPSF, IGTSTLNQR, and CYPDAGEIT epitopes by constructing the plot of RMSD (in nm) vs t (in ps).
In order to confirm the homology of our epitopes with the marketed vaccine, CLUSTAL was applied to seek homology with hemagglutinin and neuraminidase of H5N1 NIBRG-14 strain.60 Based on the CLUSTAL results, the hemagglutinin epitopes of MEKIVLLLA, CPYLGSPSF, and IGTSTLNQR were confirmed to be highly conserved with hemagglutinin of H5N1 NIBRG-14 strain that commonly used in the wet experimentation of vaccine development. Thus, the neuraminidase epitope of CYPDAGEIT was confirmed to be highly conserved with neuraminidase of H5N1 NIBRG-14 strain also.
Confirmation of the origin of hemagglutinin epitopes
Following Velkov et al and Basic Local Alignment Search Tool computation, the epitope of CPYLGSPSF was derived from hemagglutinin of influenza A virus (A/Indonesia/TLL014/2006(H5N1)), the epitope of KCQTPMGAI was derived from borne transmissible avian influenza H5 hemagglutinin mutant from the influenza A virus (A/Indonesia/5/2005(H5N1)), and the epitope of IGTSTLNQR was derived from hemagglutinin influenza A virus (A/Indonesia/TLL014/2006(H5N1)). Thus, the other hemagglutinin epitopes were not detected as part of H5N1 of Indonesian strain as mentioned earlier.61).
Conclusion
There were four epitopes of hemagglutinin and two epitopes of neuraminidase, based on visualization and dynamics molecular simulation that have high binding affinity to several human HLA. The epitopes were MEKIVLLLA (ΔGbinding −40.0148 kcal/mol), CPYLGSPSF (ΔGbinding −28.0471 kcal/mol), KCQTPMGAI (ΔGbinding−33.6109 kcal/mol), IGTSTLNQR (ΔGbinding −56.9580 kcal/mol), NPNQKIITI (ΔGbinding−23.8779 kcal/mol), and CYPDAGEIT (ΔGbinding−33.0200 kcal/mol). Most of the hemagglutinin epitopes were derived from H5N1 of Indonesian strain. Both hemagglutinin and neuraminidase epitopes were confirmed to have a high degree of homology with the H5N1 NIBRG-14 strain that is commonly used in vaccine development.
The evaluation of epitopes based on dynamics molecular simulation on normal human body temperature confirms that the epitope–HLA structure conformity has not changed significantly. Therefore, it is concluded that the epitopes can be improved as H5N1 vaccine candidate. We also suggest testing this potential epitopes through in vitro and in vivo studies to validate the result of this study.
Acknowledgments
The authors thank Abdul Hakim from Statistics Study Program, Faculty of Mathematics and Science, University of Indonesia, and Mochammad Arfin Fardiansyah Nasution from Bioinformatics Research Group, Faculty of Mathematics and Science, University of Indonesia, for the excellent proofreading of this article.
Footnotes
ACADEMIC EDITOR: Thomas Dandekar, Associate Editor
PEER REVIEW: Three peer reviewers contributed to the peer review report. Reviewers’ reports totaled 1,656 words, excluding any confidential comments to the academic editor.
FUNDING: The authors thank Hibah PUPT BOPTN Ditjen Dikti No: 0528/UN2.R12/HKP.05.00/2015 for supporting this research. The authors confirm that the funder had no influence over the study design, content of the article, or selection of this journal.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Supervised this research: USFT, AAP. Worked on the technical details: FRPS. Suggested the improvement of our pipelines: DK. All authors reviewed and approved the final article.
REFERENCES
- 1.WHO . Situation Updates – Avian Influenza. 2012. p. 6. [Google Scholar]
- 2.Morgan A. Avian influenza: an agricultural perspective. J Infect Dis. 2006;194(Suppl_2):S139–46. doi: 10.1086/507561. [DOI] [PubMed] [Google Scholar]
- 3.Kilpatrick AM, Chmura AA, Gibbons DW, Fleischer RC, Marra PP, Daszak P. Predicting the global spread of H5 N1 avian influenza. Proc Natl Acad Sci U S A. 2006;103(51):19368–73. doi: 10.1073/pnas.0609227103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Depkes RI. Flu burung. Jakarta Litbang Depkes RI; Jakarta: 2005. [Google Scholar]
- 5.Depkes RI. Pedoman Pengambilan dan Pengiriman Spesimen yang Berhubungan Dengan Flu Burung. Jakarta Puslitbang Biomedis dan Farm Litbangkes Depkes RI; Jakarta: 2006. [Google Scholar]
- 6.Ferguson NM, Cummings DAT, Cauchemez S, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437(7056):209–14. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 7.Stöhr K, Esveld M. Will vaccines be available for the next influenza pandemic? Science. 2004;306(5705):2195–6. doi: 10.1126/science.1108165. [DOI] [PubMed] [Google Scholar]
- 8.Yuen K, Chan P, Peiris M, et al. Clinical features and rapid viral diagnosis of human disease associated with avian influenza A H5N1 virus. Lancet. 1998;351(9101):467–71. doi: 10.1016/S0140-6736(98)01182-9. [DOI] [PubMed] [Google Scholar]
- 9.García M, Crawford JM, Latimer JW, Rivera-Cruz E, Perdue ML. Heterogeneity in the haemagglutinin gene and emergence of the highly pathogenic phenotype among recent H5N2 avian influenza viruses from Mexico. J Gen Virol. 1996;77(7):1493–504. doi: 10.1099/0022-1317-77-1493. [DOI] [PubMed] [Google Scholar]
- 10.Gao R, Cao B, Hu Y, et al. Human infection with a novel avian-origin influenza A (H7 N9) virus. N Engl J Med. 2013;368(20):1888–97. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- 11.Jia D. Influenza Virus H5N1 Non-Structural Protein 1 Alters Interferon- α/β Signaling. 2009. [Google Scholar]
- 12.Mulyanto CC, Saleh R. Prediction of a neutralizing epitope of a H5N1 virus hemagglutinin complexed with an antibody variable fragment using molecular dynamics simulation. J Biophys Chem. 2011;2(3):258–67. doi: 10.4236/jbpc.2011.23031. [DOI] [Google Scholar]
- 13.Russell RJ, Haire LF, Stevens DJ, et al. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443(7107):45–9. doi: 10.1038/nature05114. [DOI] [PubMed] [Google Scholar]
- 14.Ilyushina NA, Bovin NV, Webster RG. Decreased neuraminidase activity is important for the adaptation of H5N1 influenza virus to human airway epithelium. J Virol. 2012;86(9):4724–33. doi: 10.1128/JVI.06774-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moemen A, Mohamed SEAS. Molecular characterization of highly pathogenic avian influenza viruses circulating in commercial broiler chickens in some localities in Sohag Province. Assiut Vet Med J. 2012;58(133):1–13. [Google Scholar]
- 16.Gamblin SJ, Skehel JJ. Influenza hemagglutinin and neuraminidase membrane glycoproteins. J Biol Chem. 2010;285(37):28403–9. doi: 10.1074/jbc.R110.129809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He F, Leyrer S, Kwang J. Strategies towards universal pandemic influenza vaccines. Expert Rev Vaccines. 2016;15(2):215–25. doi: 10.1586/14760584.2016.1115352. [DOI] [PubMed] [Google Scholar]
- 18.Rabadan R, Levine AJ, Krasnitz M. Non-random reassortment in human influenza A viruses. Influenza Other Respi Viruses. 2008;2(1):9–22. doi: 10.1111/j.1750-2659.2007.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith DJ, Lapedes AS, de Jong JC, et al. Mapping the antigenic and genetic evolution of influenza virus. Science. 2004;305(5682):371–6. doi: 10.1126/science.1097211. [DOI] [PubMed] [Google Scholar]
- 20.Horimoto T, Kawaoka Y. Strategies for developing vaccines against H5N1 influenza A viruses. Trends Mol Med. 2006;12(11):506–14. doi: 10.1016/j.molmed.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 21.Lazer D, Kennedy R, King G, Vespignani A. The parable of Google flu: traps in big data analysis. Science. 2014;343(6167):1203–5. doi: 10.1126/science.1248506. [DOI] [PubMed] [Google Scholar]
- 22.Oyarzun P, Kobe B. Recombinant and epitope-based vaccines on the road to the market and implications for vaccine design and production. Hum Vaccin Immunother. 2015 Oct;:00–00. doi: 10.1080/21645515.2015.1094595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ben-Yedidia T, Arnon R. Epitope-based vaccine against influenza. Expert Rev Vaccines. 2007;6(6):939–48. doi: 10.1586/14760584.6.6.939. [DOI] [PubMed] [Google Scholar]
- 24.Subbarao K, Joseph T. Scientific barriers to developing vaccines against avian influenza viruses. Nat Rev Immunol. 2007;7(4):267–78. doi: 10.1038/nri2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Somvanshi P, Seth PK. Prediction of T cell epitopes for the utility of vaccine development from structural proteins of dengue virus variants using in silico methods. Indian J Biotechnol. 2009;8(2):193–8. [Google Scholar]
- 26.Gustiananda M. Immunoinformatics analysis of H5N1 proteome for designing an epitope-derived vaccine and predicting the prevalence of pre-existing cellular-mediated immunity toward bird flu virus in Indonesian population. Immunome Res. 2011;7(3):1–11. [Google Scholar]
- 27.Rai J, Lok KI, Mok CY, et al. Immunoinformatic evaluation of multiple epitope ensembles as vaccine candidates: E coli 536. Bioinformation. 2012;8(6):272–5. doi: 10.6026/97320630008272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodman AG, Heinen PP, Guerra S, et al. A human multi-epitope recombinant vaccinia virus as a universal T cell vaccine candidate against influenza virus. PLoS One. 2011;6(10):e25938. doi: 10.1371/journal.pone.0025938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Correia BE, Bates JT, Loomis RJ, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507(7491):201–6. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carrat F, Flahault A. Influenza vaccine: the challenge of antigenic drift. Vaccine. 2007;25:6852–62. doi: 10.1016/j.vaccine.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 31.Tambunan USF, Parikesit AA. In silico analysis of envelope Dengue Virus-2 and envelope Dengue Virus-3 protein as the backbone of Dengue Virus tetravalent vaccine by using homology modeling method. Online J Biol Sci 9.1. 2009:6–16. [Google Scholar]
- 32.Tambunan USF, Sugiono D, Tochary TA, Parikesit AA. Computational study of post translation modification in chimeric virus like particles vaccine of human papilloma virus with virion capsid L1. Makara Seri Sains. 2007;11(2):56–64. [Google Scholar]
- 33.Tambunan USF, Hikmawan O, Tockary TA. In silico mutation study of haemagglutinin and neuraminidase on Banten province strain influenza A H5N1 virus. Trends in Bioinformatics. 2008;1(1):18–24. [Google Scholar]
- 34.Tan PT, Khan AM, August JT. Highly conserved influenza A sequences as T cell epitopes-based vaccine targets to address the viral variability. Hum Vaccin. 2011;7(4):402–9. doi: 10.4161/hv.7.4.13845. [DOI] [PubMed] [Google Scholar]
- 35.Khan AM, Miotto O, Nascimento EJM, et al. Conservation and variability of dengue virus proteins: implications for vaccine design. PLoS Negl Trop Dis. 2008;2(8):e272. doi: 10.1371/journal.pntd.0000272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moise L, De Groot AS. Putting immunoinformatics to the test. Nat Biotechnol. 2006;24(7):791–2. doi: 10.1038/nbt0706-791. [DOI] [PubMed] [Google Scholar]
- 37.Tomar N, De RK. Immunoinformatics: an integrated scenario. Immunology. 2010;131(2):153–68. doi: 10.1111/j.1365-2567.2010.03330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.De Groot AS, Ardito M, Moise L, et al. Immunogenic consensus sequence T helper epitopes for a pan-burkholderia biodefense vaccine. Immunome Res. 2011;7(2):e7. doi: 10.4172/1745-7580.1000043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagen JB. The origins of bioinformatics. Nat Rev Genet. 2000;1(3):231–6. doi: 10.1038/35042090. [DOI] [PubMed] [Google Scholar]
- 40.Flower DR. Immunoinformatics: Predicting Immunogenicity in Silico. Humana; 2007. [DOI] [PubMed] [Google Scholar]
- 41.Korber B, LaBute M, Yusim K. Immunoinformatics comes of age. PLoS Comput Biol. 2006;2(6):e71. doi: 10.1371/journal.pcbi.0020071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brusic V, Petrovsky N. Immunoinformatics and its relevance to understanding human immune disease. Expert Rev Clin Immunol. 2005;1(1):145–57. doi: 10.1586/1744666X.1.1.145. [DOI] [PubMed] [Google Scholar]
- 43.Pappalardo F, Brusic V, Castiglione F, Schönbach C. Computational and bioinformatics techniques for immunology. Biomed Res Int. 2014;2014:1–2. doi: 10.1155/2014/263189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bao Y, Bolotov P, Dernovoy D, et al. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82(2):596–601. doi: 10.1128/JVI.02005-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nussbaum AK, Kuttler C, Hadeler K-P, Rammensee H-G, Schild H. PAProC: a prediction algorithm for proteasomal cleavages available on the WWW. Immunogenetics. 2001;53(2):87–94. doi: 10.1007/s002510100300. [DOI] [PubMed] [Google Scholar]
- 46.Bhasin M, Lata S, Raghava GPS. TAPPred prediction of TAP-binding peptides in antigens. Methods Mol Biol. 2007;409:381–6. doi: 10.1007/978-1-60327-118-9_28. [DOI] [PubMed] [Google Scholar]
- 47.Karosiene E, Rasmussen M, Blicher T, Lund O, Buus S, Nielsen M. NetMHCIIpan-3.0, a common pan-specific MHC class II prediction method including all three human MHC class II isotypes, HLA-DR, HLA-DP and HLA-DQ. Immunogenetics. 2013;65:711–24. doi: 10.1007/s00251-13-0720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17(12):1236–7. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 49.Vilar S, Cozza G, Moro S. Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Curr Top Med Chem. 2008;8(18):1555–72. doi: 10.2174/156802608786786624. [DOI] [PubMed] [Google Scholar]
- 50.Bakri R, Parikesit AA, Satriyanto CP, Kerami D, Tambunan USF. Utilization of boron compounds for the modification of suberoyl anilide hydroxamic acid as inhibitor of histone deacetylase class II Homo sapiens. Adv Bioinformatics. 2014;2014:10. doi: 10.1155/2014/104823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nidom CA, Yamada S, Nidom RV, et al. Genetic characterization of H5N1 influenza viruses isolated from chickens in Indonesia in 2010. Virus Genes. 2012;44(3):459–65. doi: 10.1007/s11262-012-0722-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkata R. Immunogenicity of Hemagglutinin T Cell Epitopes in Influenza. 2011 [Google Scholar]
- 53.Patronov A, Doytchinova I. T-cell epitope vaccine design by immunoinformatics. Open Biol. 2013;3(1):120139. doi: 10.1098/rsob.120139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen M, Lundegaard C, Blicher T, et al. NetMHCpan, a method for quantitative predictions of peptide binding to any HLA-A and -B locus protein of known sequence. PLoS One. 2007;2(8):e796. doi: 10.1371/journal.pone.0000796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heiny AT, Miotto O, Srinivasan KN, et al. Evolutionarily conserved protein sequences of influenza a viruses, avian and human, as vaccine targets. PLoS One. 2007;2(11):e1190. doi: 10.1371/journal.pone.0001190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mack SJ, Bugawan TL, Moonsamy PV, et al. Evolution of Pacific/Asian populations inferred from HLA class II allele frequency distributions. Tissue Antigens. 2000;55(5):383–400. doi: 10.1034/j.1399-0039.2000.550501.x. [DOI] [PubMed] [Google Scholar]
- 57.Faraggi D, Reiser B. Estimation of the area under the ROC curve. Stat Med. 2002;21(20):3093–106. doi: 10.1002/sim.1228. [DOI] [PubMed] [Google Scholar]
- 58.Duvvuri VR, Marchand-Austin A, Eshaghi A, Patel SN, Low DE, Gubbay JB. Potential T cell epitopes within swine-origin triple reassortant influenza A (H3 N2) variant virus which emerged in 2011: an immunoinformatics study. Vaccine. 2012;30(42):6054–63. doi: 10.1016/j.vaccine.2012.07.054. [DOI] [PubMed] [Google Scholar]
- 59.Rakesh S, Pradhan D, Umamaheswari A. In Silico approach for future development of subunit vaccines against Leptospira interrogans serovar lai. Int J Bioinforma Res. 2009;1(2):85–92. doi: 10.9735/0975-3087.1.2.85-92. [DOI] [Google Scholar]
- 60.Leroux-Roels I, Borkowski A, Vanwolleghem T, et al. Antigen sparing and cross-reactive immunity with an adjuvanted rH5N1 prototype pandemic influenza vaccine: a randomised controlled trial. Lancet. 2007;370(9587):580–9. doi: 10.1016/S0140-6736(07)61297-5. [DOI] [PubMed] [Google Scholar]
- 61.Velkov T, Ong C, Baker MA, et al. The antigenic architecture of the hemagglutinin of influenza H5 N1 viruses. Mol Immunol. 2013;56(4):705–19. doi: 10.1016/j.molimm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 62.Brennan RM, Petersen J, Neller MA, et al. The impact of a large and frequent deletion in the human TCR β locus on antiviral immunity. J Immunol. 2012;188(6):2742–8. doi: 10.4049/jimmunol.1102675. [DOI] [PubMed] [Google Scholar]
- 63.Ghosh P, Amaya M, Mellins E, Wiley DC. The structure of an intermediate in class II MHC maturation: CLIP bound to HLA-DR3. Nature. 1995;378(6556):457–62. doi: 10.1038/378457a0. [DOI] [PubMed] [Google Scholar]
- 64.Maenaka K, Maenaka T, Tomiyama H, Takiguchi M, Stuart DI, Jones EY. Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes. J Immunol. 2000;165(6):3260–7. doi: 10.4049/jimmunol.165.6.3260. [DOI] [PubMed] [Google Scholar]