ABSTRACT
Collagen VI myopathies are genetic disorders caused by mutations in collagen 6 A1, A2 and A3 genes, ranging from the severe Ullrich congenital muscular dystrophy to the milder Bethlem myopathy, which is recapitulated by collagen-VI-null (Col6a1−/−) mice. Abnormalities in mitochondria and autophagic pathway have been proposed as pathogenic causes of collagen VI myopathies, but the link between collagen VI defects and these metabolic circuits remains unknown. To unravel the expression profiling perturbation in muscles with collagen VI myopathies, we performed a deep RNA profiling in both Col6a1−/− mice and patients with collagen VI pathology. The interactome map identified common pathways suggesting a previously undetected connection between circadian genes and collagen VI pathology. Intriguingly, Bmal1−/− (also known as Arntl) mice, a well-characterized model displaying arrhythmic circadian rhythms, showed profound deregulation of the collagen VI pathway and of autophagy-related genes. The involvement of circadian rhythms in collagen VI myopathies is new and links autophagy and mitochondrial abnormalities. It also opens new avenues for therapies of hereditary myopathies to modulate the molecular clock or potential gene–environment interactions that might modify muscle damage pathogenesis.
KEY WORDS: RNAseq, Circadian rhythms, Interactome pathway, Myopathies
Summary: A new association between hereditary myopathy and CLOCK circuit genes opens novel avenues of research on circadian genes and their role in skeletal muscle hereditary pathologies.
INTRODUCTION
Collagen VI (ColVI) is a major component of the extracellular matrix (ECM) crucial for muscle cell adhesion, stability and regeneration. ColVI forms a microfibrillar network involved in maintaining tissue integrity through a structural link between different components of connective-tissue basement membranes and cells (Allamand et al., 2011). ColVI is a heterotrimeric molecule composed of three α-chains encoded by distinct genes (COL6A1, COL6A2 and COL6A3). The three α-chains first assemble in equimolar association into a triple helical monomer, and successively form dimers and tetramers stabilized by disulfide bonds. The secreted tetramers give rise to a network of microfilaments that bridge the sarcolemma with the extracellular connective tissue (Bonaldo et al., 1990). Mutations occurring in the three ColVI genes cause two major disease phenotypes: a mild form, Bethlem myopathy, and a severe form, Ullrich congenital muscular dystrophy (UCMD) (Gualandi et al., 2012). Three additional ColVI genes have recently been described, COL6A4, COL6A5 and COL6A6, and code for the α4(VI), α5(VI) and α6(VI) protein chains, respectively. However, only α6(VI) is highly expressed in skeletal muscle, although not associated with basement membranes (Tagliavini et al., 2014a) and none of these additional ColVI genes are known as disease genes in humans.
ColVI myopathies are well-defined conditions, characterized by a broad spectrum of clinical features, ranging from prenatal onset with decreased fetal movements; hypotonia or torticollis, early-childhood onset, characterized by delayed motor milestones, muscle weakness and contractures; and adult onset (4th–6th decade), characterized by proximal weakness and Achilles tendon or long finger flexor contractures (De Visser et al., 2004). Owing to the progressive nature of the condition, 75% of patients over age 50 years need assistance for outdoor mobility (Jöbsis et al., 1999).
The severe phenotype with early onset, UCMD, is characterized by severe muscle weakness, striking hyperlaxity of distal joints, and proximal joint contractures. In contrast to Bethlem myopathy, UCMD is marked by an early loss of ambulation and a rapid disease progression leading to early death caused by respiratory failure (Maraldi et al., 2009). In addition, two other conditions have been associated with ColVI myopathy: autosomal-dominant limb-girdle muscular dystrophy (LGMD), and autosomal-recessive myosclerosis myopathy (Merlini et al., 2008; Scacheri et al., 2002). Recently, the COL12A1 gene has been shown to be mutated in patients with a Bethlem-myopathy-like phenotype (Hicks et al., 2014).
Despite this broad clinical spectrum, both muscle function and respiratory performance evaluation can address the diagnosis. Nevertheless, UCMD and Bethlem myopathy can be inherited accordingly to both dominant and recessive models, and generally, neither the type of mutation (missense, nonsense, splicing insertion or deletion), nor the effect of the mutation on the protein structure or function allows precise discrimination between two phenotypes (Allamand et al., 2011; Maraldi et al., 2009; Bönnemann, 2011). This suggests that the genetic background of patients might influence the expression of the mutated genes, therefore impacting on the resulting phenotype.
The Col6a1−/− mouse is a well studied animal model for ColVI myopathies. These mice carry a targeted inactivation of the α1(VI) chain and exhibit an early onset myopathic phenotype that strongly resembles Bethlem myopathy (Bonaldo et al., 1998). Muscle fibers of Col6a1−/− mice show loss of contractile strength and a number of structural defects in the hind limb muscle and especially in the diaphragm. Although the pathology associated with collagen VI myopathy is nonspecific and variable, it has been widely documented (Grumati et al., 2010; Urciuolo et al., 2013). Both muscles from mice and patients have been found to show mitochondrial dysfunction accompanied by ultrastructural alterations, increased spontaneous apoptosis and abnormal propensity of the permeability transition pore (PTP) to open (Irwin et al., 2003; Bernardi and Bonaldo, 2008; Tagliavini et al., 2014b).
Col6a1−/− mice and UCMD or Bethlem myopathy patients also show defective autophagy (Grumati et al., 2011). This suggests a mechanism of disease pathogenesis that might be in part due to defective regulation of autophagy, which leads to accumulation of abnormal mitochondria and sarcoplasmic reticulum. Moreover, in Col6a1−/− mice, changes in metabolic proteins resulting in decreased glycolysis and affecting the tricarboxylic acid (TCA) cycle fluxes lead to a different fate of α-ketoglutarate triggering lipotoxicity. The metabolic changes are associated with changes of proteins involved in mechanotransduction at the myotendineous junction, costameric or sarcomeric level, and in energy metabolism (De Palma et al., 2013). Recently, a strong depletion of the α6(VI) chain in skeletal muscle has been reported as potential biomarker of ColVI myopathies (Tagliavini et al., 2014a). In spite of these recent achievements, a full understanding of ColVI disease pathogenic mechanisms remains unknown.
Gene expression profiling and RNA sequencing are powerful tools to explore the transcriptional alterations in disease tissues (Sánchez-Pla et al., 2012). It brought great advances in cancer research and disclosed deregulated transcripts that are now being considered as prognostic or therapeutic biomarkers of neoplastic diseases (Sevov et al., 2012; Campo, 2013). Considering the availability of novel therapeutic possibilities for rare muscle disorders (Aartsma-Rus and Muntoni, 2013), finding prognostic biomarkers for disease severity or drug response will be of great benefit for patient care and treatment (Ferlini et al., 2013; Scotton et al., 2014). In this work, we explored RNA profiling of Col6a1−/− mice and Bethlem myopathy and UCMD patients to identify transcriptional patterns correlating with disease state. Such patterns can identify specific pathway alterations and pathophysiological biomarkers. This unbiased approach revealed marked changes in the expression of genes involved in the circadian rhythm pathway. We further supported these findings by protein studies performed both in mice and in patients. In addition, RNA and protein studies in the Bmal1−/− (also known as Arntl) mice, a known animal model of circadian rhythm disruption, showed profound deregulation of Col6a6 as well as of the autophagy-related genes such as p62/SQSTM1 and AMPK. Although it has been previously reported that disruption of circadian rhythms are linked to skeletal muscle remodeling, function, performance and aging, the involvement of the molecular clock genes in hereditary myopathy as ColVI disease is a new finding. These findings highlight new potential target genes involved in muscle damage possibly addressing new therapies.
RESULTS
Overall gene expression profile in Col6a1−/− muscles reveals changes in genes associated with muscle function
Gene expression array profiles of wild-type (WT) and Col6a1−/− mice were compared in three muscle types: diaphragm, tibialis anterior and gastrocnemius. To determine genes with statistically significant differential expression (P<0.05 in limma package) we analyzed each muscle type (either separately and in pools) and determined average expression change over all muscle types. The degree of transcriptional deregulation was different among Col6a1−/− muscles, with 11,170 differentially expressed genes in tibialis anterior, 6790 in diaphragm and 3759 genes in gastrocnemius. To further support the finding of different tissue susceptibility to ColVI deficiency, we performed sample clustering of expression profiles from each muscle type for Col6a1−/− and in WT mice. The resulting dendrogram is shown in Fig. S1A. We found that the expression profiles of Col6a1−/− and WT gastrocnemius were clustered together and therefore similar. Indeed this supports the existing knowledge about muscle involvement in the Col6a1−/− mouse model, where gastrocnemius is very mildly affected, whereas tibialis and even more diaphragm are severely affected (De Palma et al., 2013). In contrast, diaphragm and tibialis anterior Col6a1−/− muscles clustered together and were clearly separate from their WT counterparts, suggesting that ColVI deficiency severely and similarly affects these muscle types (Fig. S1).
Expression of genes associated with muscle development and function was altered in Col6a1−/− mice (Table S1). The identified changes involved several different muscle biology circuits, including structure, metabolism, transcription and regulation. There was variability between tested muscle types. A total of 60% (26 out of 44) of muscle-specific genes changed their expression in an opposite direction in tibialis anterior versus diaphragm, 23% in tibialis anterior versus gastrocnemius (7 out of 30) and 67% in gastrocnemius versus diaphragm (12 of 18). Tibialis anterior showed differential expression of myosin, troponin and calsequestrin isoforms (predominantly cardiac and slow), and upregulation of Pax3. In diaphragm and tibialis anterior, the differentially expressed genes included almost all members of the dystroglycan complex (dystrophin, dystrobrevin, dystroglycan 1, syntrophin, α-, β-, γ-sarcoglycans) and several transcription factors important for muscle-related processes, such as MyoD1, MyoG, MYF6, NFATc2 and Pax7. Embryonic isoforms of myosin were upregulated in the diaphragm. Pax7, a marker of satellite cell number, which can be regarded as a sign of muscle regeneration, was also upregulated. This muscle type variability in the transcription profile is not surprising given that these three muscles have different metabolic properties and different fiber type composition (Bonaldo et al., 1998).
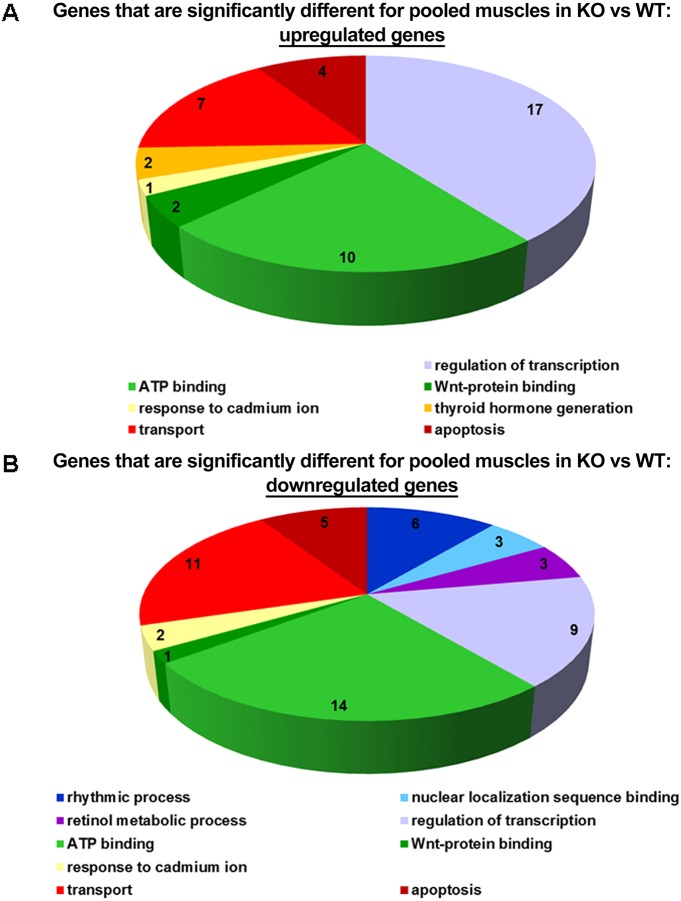
Gene ontology groups and key expression regulators analyses show deregulation of the circadian rhythmic process in Col6a1−/− mice
Among the differentially expressed genes between Col6a1−/− and WT mice, 479 changed expression in the same direction in all tissues with P<0.05. We used Gene Set Enrichment analysis (GSEA) to identify gene ontology groups enriched with such genes. The top ten gene ontology groups are listed in Fig. S2. Identified gene ontology groups were broad categories and, therefore, do not specifically address any definite biological effects occurring in Col6a1−/− mice. We also performed gene ontology analysis of 334 genes differentially expressed in the ‘pooled’ data from all three muscle groups compared with ‘pooled’ WT muscles. This GSEA identified the gene ontology groups shown in Fig. 1. One of the top ten gene ontology groups was ‘rhythmic process’, which contains genes regulating circadian rhythms, as well as known genes that are regulated downstream of the molecular clock, such as DBP, and all were downregulated in Col6a1−/− muscles.
Fig. 1.
Gene ontology groups that were differentially represented in samples from pooled tissues from three muscle tissues. Muscle types examined were the diaphragm, gastrocnemius and tibialis anterior, of four Col6a1−/− and WT mice. A representation of the gene ontology groups for the 334 genes differentially expressed in the pooled data from all three muscle groups (diaphragm, gastrocnemius and tibialis anterior) of Col6a1−/− mice compared with that in pooled wild-type muscles (A shows upregulated genes, and B shows downregulated genes).
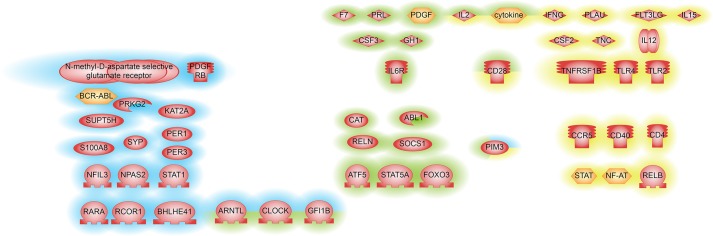
To identify key regulators that change their activity in Col6a1−/− muscles, we used sub-network enrichment analysis (SNEA) in Pathway Studio. Our first SNEA was performed using the list of genes differentially expressed in one direction in all muscle types as an input. Regulators identified by this approach are shown in Fig. 2. Genes changing their expression in one direction in all muscle types are primarily regulated by inflammatory cytokines and their receptors. In good agreement with the gene ontology analysis, SNEA of the ‘pooled’ dataset found BMAL1 (ARNTL), CLOCK, PER1 and PER3 regulators that belong to cellular circadian rhythm pathway (Fig. 2; Table S2). We then explored the intersection between the two SNEA analyses and found consistent results (Fig. 2; Table S2). Indeed, we found that CLOCK, BMAL1, GFI1B, IL2, CD28, PIM3 and inflammatory cytokines were differentially expressed genes that were also identified as being key regulators in all three comparisons.
Fig. 2.
Regulators identified by SNEA upstream genes that are differentially expressed between Col6a1−/− and wild-type mice muscle tissues. Genes with P<0.05 are shown. Blue highlighting represents SNEA-derived regulators identified from differentially expressed genes in samples pooled from three muscle types – diaphragm, gastrocnemius and tibialis anterior – from four Col6a1−/− in respect to WT mice. Yellow highlighting indicates SNEA-derived regulators identified from differentially expressed genes that have the same direction of change in all muscle types. Green highlighting represents SNEA-derived regulators with the same direction of change using both pooled and individual muscle approaches.
To validate the 46 key regulators selected by SNEA, we studied the differential expression of these transcripts using TaqMan low-density arrays (TLDA). By comparing the microarray experiment data to those of the TLDA cards, we observed the same trend of fold change in 37 out of 46 genes (80%) of the diaphragm and tibialis anterior, whereas in the gastrocnemius we found 32 out of 46 genes (70%) with these features (Fig. S3A,B).
Both in the diaphragm and tibialis anterior and, to a lesser extent in gastrocnemius, the TLDA and microarray data for regulators of the rhythmic process (BMAL1, ATF5, CLOCK, DBP, EGR1, FKBP5, PER1, PER2 and PER3) and transcription factors important for muscle-related processes (MyoD1, MyoG, MYF6, PAX7) were in good agreement. This confirmed the deregulation of these two pathways in the Col6a1−/− mice (Fig. S3B).
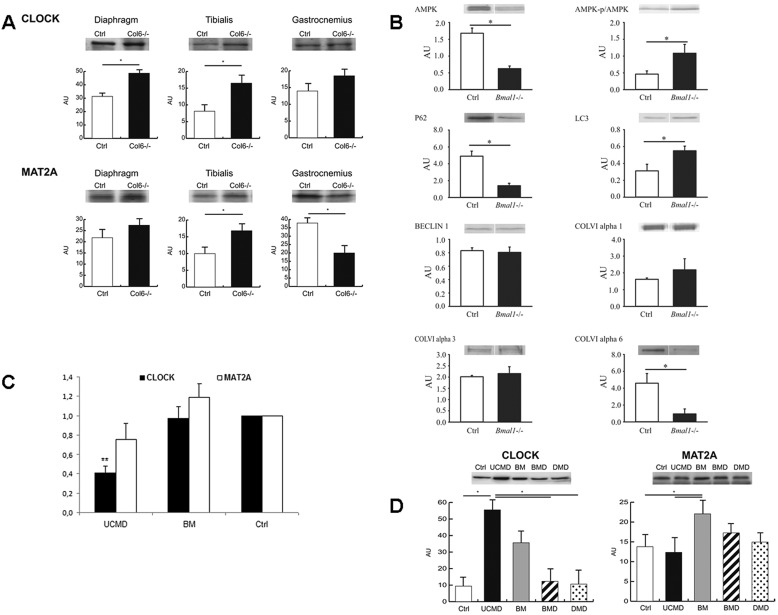
Western blotting revealed that CLOCK levels were significantly (Student's t-test, n=3, P<0.05) increased in diaphragm and tibialis anterior muscles of Col6a1−/− mice compared with their respective WT controls, whereas they remained unchanged in gastrocnemius. MAT2A protein levels did not change significantly in the diaphragm, whereas in tibialis anterior and gastrocnemius muscles they were, respectively, increased and decreased (Fig. 3A).
Fig. 3.
Transcription of circadian genes and expression analysis on mice and human muscles. (A) Western blotting of CLOCK and MAT2A in three different muscle tissues (diaphragm, gastrocnemius and tibialis anterior) of five 6-month-old male Col6a1−/− and wild-type (Ctrl) mice revealed a significant upregulation (*P<0.05, t-test) of CLOCK protein levels in the diaphragm and tibialis anterior, whereas the MAT2a protein level was significantly altered in tibialis anterior and gastrocnemius, which showed an increase and decrease, respectively. (B) Immunoblotting to evaluate the protein levels of p62, α3(VI), α6(VI), α1(VI), Beclin 1, LC3, phosphorylated AMPK (p/AMPK), AMPK and α-tubulin in the gastrocnemius muscle of four male 30–35-week-old wild-type (Ctrl) and Bmal1−/− mice. The analysis identified a downregulation of α6(VI), and upregulation of α3(VI). Components of autophagic pathway were also affected, expressed as an increase in LC3 (LC3A) and decrease in p62. (C) Real-time analysis of CLOCK gene (black bars) and MAT2A gene (white bars) transcription in the cohort of 12 Bethlem myopathy and 12 UCMD patients. The analysis identified a significant deregulation of the CLOCK gene compared to controls (Ctrl) in the UCMD patients only (**P<0.0001, t-test). All the experiments were performed in triplicate [CLOCK analysis, UCMD: 0.4137±0.2146; Bethlem myopathy (BM): 0.9709±0.5846; MAT2A analysis, UCMD: 0.7599±0.4012; Bethlem myopathy (BM): 1.1918±0.5074; mean±s.d.]. (D) Western blotting of CLOCK and MAT2A in four Bethlem myopathy, eight UCMD patients, and 30 patients affected by other diseases (BMD and DMD), as well as five healthy controls (Ctrl), showed a significant increase (*P<0.05, ANOVA) in CLOCK protein levels in UCMD patients only; MAT2A was slightly but significantly increased in Bethlem myopathy patients.
Abnormal collagen VI expression in Bmal1−/− mice, a model for circadian rhythm deregulation
There are multiple genetic mouse models in which molecular clock factors have been knocked out, but of these models only the Bmal1−/− mice exhibit complete loss of behavioral rhythm under normal housing conditions (Bunger et al., 2000). We adopted this last model in order to have a homogeneous phenotype, with complete failure of circadian control. Western blot analysis of gastrocnemius muscles of Bmal1−/− mice revealed significant changes in the levels of ColVI chains, with decreased α6(VI) and increased α3(VI) levels. Interestingly, downregulation of COL6A6 was also recently observed by us in UCMD and Bethlem myopathy muscles (Tagliavini et al., 2014a). In addition to ColVI, components of the autophagy pathway were also affected in the muscle of Bmal1−/− mice with increased LC3A (also known as MAP1LC3A) and decreased p62 (also known as SQSTM1) levels, whereas Beclin 1 levels were unchanged. We also found an increase in the activation status of AMPK, as defined by a higher phosphorylated AMPK to total AMPK ratio; however, this was driven in part by a significant decrease in the levels of total AMPK (Fig. 3B).
Circadian genes show deregulation in muscle from UCMD and Bethlem myopathy patients
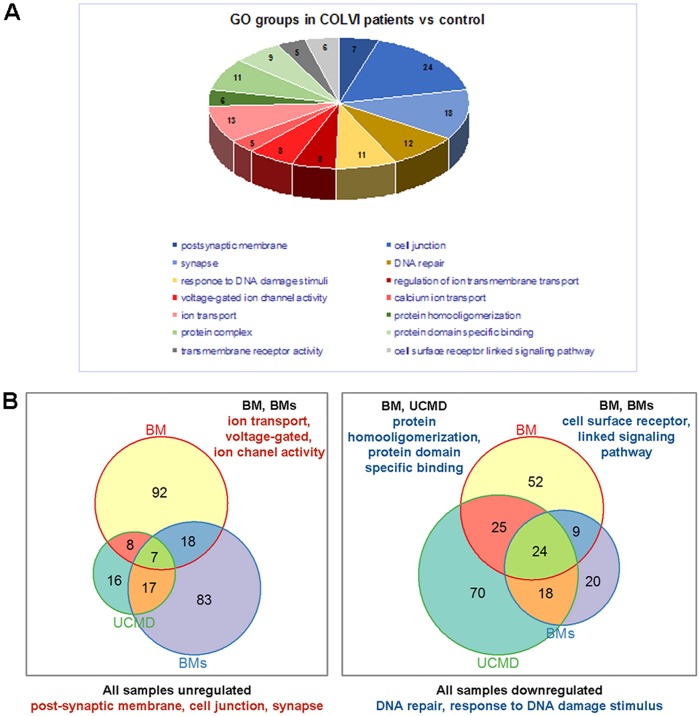
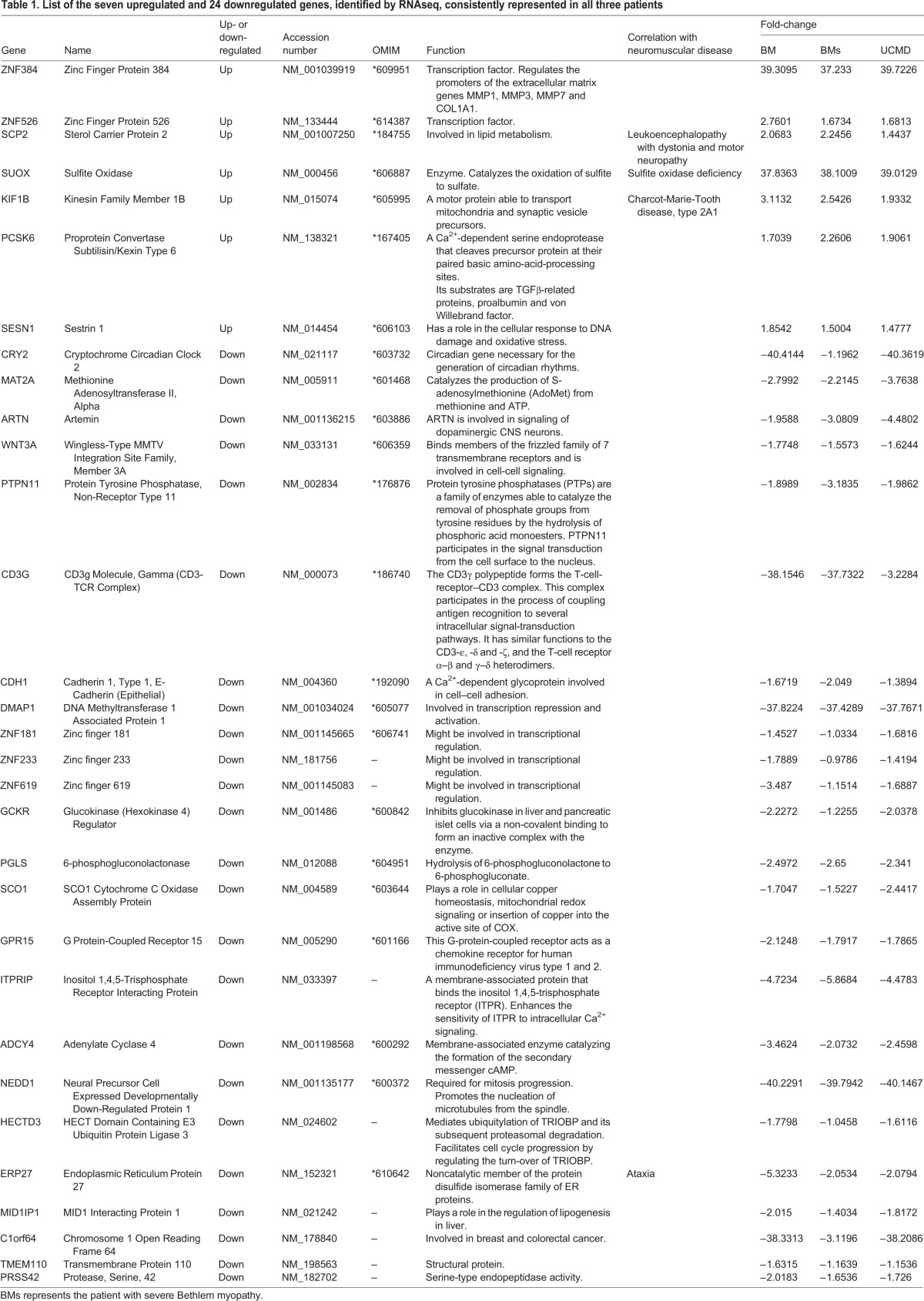
Gene expression profiles in three patients (one with UCMD and two with Bethlem myopathy) were compared to healthy controls by RNAseq, and we identified several altered transcripts, which we grouped into gene ontology categories (Fig. 4A). Genes that were deregulated in muscles from UCMD and Bethlem myopathy patients are reported in Table 1. In all three patients, we identified seven consistently upregulated genes belonging to transcription factor, metabolism process and cellular stress response categories. The 24 consistently downregulated genes were involved in signaling, metabolism, immunity and cell adhesion, axon guidance and circadian rhythm, including DNA repair gene ontology categories (Fig. 4B). Consistent with a link to circadian rhythms, we found that eight of the genes listed in Table 1 are known to be circadian in mouse skeletal muscle and/or changed in expression in the Bmal1−/− mouse.
Fig. 4.
Gene ontology groups that were differentially represented in ColVI patients as determined by RNAseq output. (A) Representation of the differentially expressed genes in the indicated gene ontology (GO) groups in the ColVI patients (UCMD, severe Bethlem myopathy and Bethlem myopathy) compared to control. (B) Venn diagrams showing the number of genes, identified by RNAseq analysis, constantly deregulated in the ColVI patients. The patients shared an upregulation of genes involved in postsynaptic membrane, cell junction and synapse pathways, and a downregulation of genes belonging to response to DNA damage stimuli and DNA repair pathways.
Table 1.
List of the seven upregulated and 24 downregulated genes, identified by RNAseq, consistently represented in all three patients
In order to find gene ontology groups deregulated in different samples, we performed GSEA analysis (P-value 0.05 cut-off) for each patient separately. Bi-clustering was performed by using the Jaccard-based distance between gene ontology groups (based on the number of common genes in groups). This method allowed finding of common deregulated ‘patterns’ in different patients, even though the GSEA method shows different gene ontology groups. We found genes coding proteins belonging to postsynaptic membrane, cell junction and synapse, regulation of ion transmembrane transport, protein homo-oligomerization or protein-specific binding, cell-surface-receptor-linked signaling pathways and DNA repair or response to DNA damage, including circadian rhythm genes (Fig. 4A,B).
Considering the evidence from both mice and human studies for the deregulation of circadian genes in muscles with ColVI defects, we performed an real-time PCR expression analysis of CLOCK and MAT2A genes in another 11 UCMD and 12 Bethlem myopathy patients. We found that the CLOCK gene was significantly downregulated (P<0.0001) in all UCMD muscles. The MAT2A transcript showed a milder downregulation, although not statistically significant (P=0.0753), but only in UCMD patients (Fig. 3C). Bethlem myopathy patients did not show any significant deregulation in the analyzed circadian genes. Western blotting in four UCMD and three Bethlem myopathy patient muscles showed significantly increased CLOCK protein levels (P<0.05, ANOVA plus Tukey, n=3) in UCMD patients only, whereas MAT2A protein levels were increased in Bethlem myopathy patients only (Fig. 3D).
DISCUSSION
ColVI myopathies have been both clinically and genetically defined, and alterations in the apoptosis and autophagy circuits have been reported. Nevertheless, the link between mutations of COL6A1–COL6A3 genes and the muscle dysfunction seen in ColVI myopathy still remains elusive. We therefore adopted RNA profiling to study muscle from both Col6a1−/− mice and Bethlem myopathy or UCMD patient biopsies, searching for unique transcriptional patterns. The idea was to detect specific signatures that might enhance our understanding of the pathogenesis of ColVI myopathies. In order to strengthen our findings, we also evaluated protein expression of the transcripts that we found to be consistently deregulated in both mouse and patient muscle tissue.
Col6a1−/− and Bmal1−/− mice shows connection between ColVI myopathy and circadian genes
To our knowledge, this is the first attempt to study Col6a1−/− mice by RNA profiling. Proteomic studies were performed by Irwin et al. (2003) in Col6a1−/− mice, revealing the diaphragm as the most severely affected muscle, having a deeply subverted ultrastructure and evident loss of contractile strength, accompanied by changes in proteins involved in mechanotransduction. Our pooled data identified 334 differentially expressed genes in Col6a1−/− versus WT, showing consistent deregulation of the genes involved in metabolic processes and regulation of transcription, known to be affected in myopathies, as well as in those of the circadian CLOCK gene. This is an entirely new observation.
We show major transcriptional changes in the diaphragm and tibialis anterior, accordingly to the proteomic findings (De Palma et al., 2013). All muscles shared similar alterations in genes involved in the inflammatory response, cell proliferation, transcriptional regulation and metabolism, but differed substantially in the direction of expression change in a number of genes related to muscle functions, including transcription, metabolism, structure and regulation – a not entirely unexpected finding considering that these three muscles have different morphogenetic origins and functional requirements (Eberhard and Jockusch, 2004; Merrell and Kardon, 2013).
The CLOCK gene deregulation, we also confirmed at the protein level in Col6a1−/− mice, suggests a previously unreported relationship between circadian rhythms, the molecular clock and myopathy. The Col6a1−/− model shows both muscle and osteotendinous junction damage (Izu et al., 2011, 2012), reflecting defective costamerogenesis and altered mechanotransduction (De Palma et al., 2013). Specific activation of both myocardin-related transcription factors (MRTFs) and ternary complex factors (TCFs), whose targets are regulators of cytoskeleton dynamics and mechanosensing, have potential to reset the molecular clock through regulation of PER1, PER2, BMAL1 and other CLOCK genes (Esnault et al., 2014). This first demonstration of a link between the ECM and the profound alterations we detected in the circadian rhythm or molecular clock pathway (BMAL1, ATF5, CLOCK, DBP, EGR1, FKBP5, PER1, PER2 and PER3 genes) supports the known data demonstrating that physical activity can regulate circadian gene expression in skeletal muscle (Wolff and Esser, 2012). In addition, providing a further bridge between protein and RNA data, intersection between the individual muscle groups and the pooled transcription data, to identify key regulators, showed that inflammatory cytokines and their receptors, known to be regulated by PER1–PER3 genes, are significantly underexpressed. The connection between ColVI and circadian genes is also strongly supported by our protein findings in Bmal1−/− mice, which exhibit significant changes in the levels of ColVI chains as well as alterations of the autophagy pathway (Fig. 3B). Given that our expression analyses were only performed at one time point, our results can only allow us to conclude that molecular clock pathway is disrupted. Further studies are needed to determine whether the clock factors are phase shifted, damped or arrhythmic.
RNA profiling of ColVI patients shows that there is a profound deregulation in ECM-mediated structural muscle remodeling and circadian genes
In the three ColVI patients (Bethlem myopathy, severe Bethlem myopathy and UCMD) studied, the highly efficient RNAseq analysis revealed many changes in transcription profile, as expected for this progressive, chronic and severe myopathy. The major changes observed, shared by all patients, were underexpression or overexpression in proteins related to membrane, cell junction or synapse, ion transmembrane transport, protein homo-oligomerization or protein-specific binding, the cell-surface-receptor-linked signaling pathway and circadian gene pathways (Fig. 4B). The overexpressed genes we detected (ZNF384, ZNF526, SCP2, SUOX, KIF1B, PCSK6 and SESN1) are all involved in the control of gene expression and metabolic pathways. As reported in UCMD muscles (Paco et al., 2013), increased expression of these transcripts is associated with muscle regeneration defects and inflammatory signatures. Our RNAseq data confirm that this myopathy is associated with structural alterations coupled with either signaling or regeneration failure (Bönnemann, 2011). The 24 downregulated genes we identified mainly involved transcripts associated with cell–cell communication or adhesion, and signaling. Deregulation of transcription, homeostasis and the cell response was also evident. This RNAseq data combines with that obtained by Paco et al. (2013) using array technology to indicate that ColVI deficiency drives both structural remodeling of muscle tissue, mediated by the ECM, adhesion, cell signaling and metabolism genes, and a massive immune response. The central role of metabolism in muscle remodeling has also been confirmed by De Palma et al. (2014), who described changes in the unfolded protein response coupled with metabolic dysregulation, autophagic impairment and mechanotransduction signaling alterations.
Most intriguingly, and in line with the mouse data, our RNAseq analysis also revealed that circadian-related CRY2 and MAT2A genes were profoundly deregulated. CRY2 is a well-established negative regulator of the molecular clock mechanism that underlies circadian rhythms, and MAT2A has a link with circadian rhythms through regulation of the enzyme required for melatonin synthesis, and mediates photoneural-circadian regulation of the pineal gland (Kim et al., 2005). Using array technology, Paco et al. (2013) also identified significant deregulation of the CLOCK pathway gene EGR1, which also supports circadian rhythm involvement in the pathogenesis of ColVI in both mice and patients. This finding prompted us to profile both MAT2A and CLOCK in larger patient cohorts, and the CLOCK gene was significantly downregulated (P<0.0001) in all UCMD patients, the more severe form of ColVI myopathy. All these data support that circadian genes, and CLOCK in particular, have a role in ColVI myopathy pathogenesis.
Circadian genes and hereditary myopathies – a new perspective
Circadian rhythms are approximate 24-h oscillations in physiology and behavior driven by the molecular clock, a transcription–translation feedback mechanism. A link between circadian genes and skeletal muscle has been identified in both animal models and in humans (Schroder and Esser, 2013). In particular, expression profiling has identified genes expressed in a circadian manner in the gastrocnemius, soleus and tibialis anterior muscles of mice (McCarthy et al., 2007; Dyar et al., 2013; Hodge et al., 2015). Analysis of human muscle samples taken 12 h apart have also shown changes in clock gene expression, consistent with the findings in mice (Zambon et al., 2003). Indeed, Zhang et al. (2012) found that the molecular clock factors BMAL1 and CLOCK transcriptionally regulate expression of MyoD1 in adult muscle, thereby suggesting a link between the daily clock cycle and periods of muscle repair and maintenance. We add to this growing body of evidence by demonstrating, by RNA and protein analysis, a new link between ColVI myopathy and deregulation of CLOCK circadian gene expression in both mouse and human tissues.
In mice, the protein data fully support the transcription results. Altered expression of CLOCK seems to be a marker of ColVI disease severity, being upregulated in the most profoundly affected muscles (tibialis anterior and diaphragm). In this way, CLOCK involvement appears to provide the missing link between autophagic pathway alterations and the physiopathogenesis of ColVI myopathies (Fig. 5). Supporting this link, many mitochondrial genes show a circadian oscillation in expression and are significantly downregulated in the muscle of the CLOCK mutant mice (McCarthy et al., 2007); in addition, the circadian animal models (both ClockΔ19 and Bmal1−/−) have a dramatic reduction (40% less) in the mitochondrial volume. In these mutant mice, the remaining mitochondria display a pathological morphology characterized by swelling and disruption of cristae, associated with an increased uncoupling of respiration (Andrews et al., 2010). These findings suggest that circadian genes can modulate the respiratory defects. Unsurprisingly, therefore, protein analysis in the Bmal1−/− mice identified a reduced expression of the α6(VI) chain. Interestingly, a severe depletion of the α6(VI) chain has recently been identified both in skeletal muscle and muscle cell cultures of ColVI patients, and this protein change has been proposed as a diagnostic marker of ColVI diseases (Tagliavini et al., 2014a). Indeed, timing plays a crucial role in muscle remodeling. For instance, Dadgar et al. (2014) demonstrated that asynchrony drives fibrosis and regeneration failure in the mdx model, finding a link between profound deregulation of global muscle transcription and the muscle damage seen in various dystrophies, including dystrophinopathies and several LGMD types. In addition, BMAL1 has recently been found to be involved in myogenic response to muscle damage, orchestrating the regeneration processes in adult skeletal muscle (Chatterjee et al., 2015).
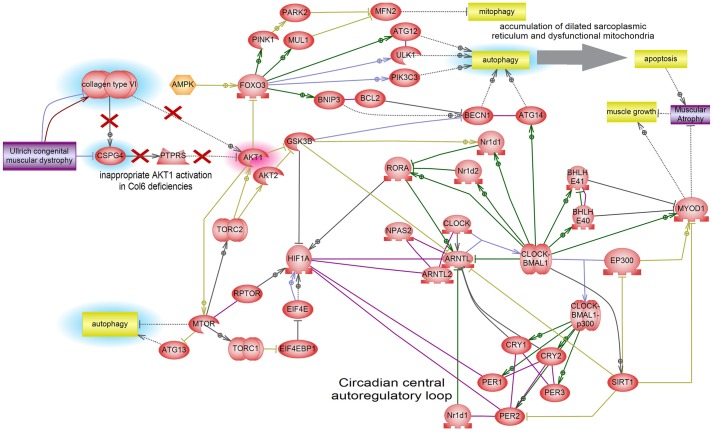
Fig. 5.
Interactome pathway linking ColVI and circadian rhythms. This model is based on the literature data analyzed by Pathway Studio, and shows the connections between autophagy and circadian pathways. In this model the proposed pathophysiological mechanism of UCMD is that mTOR kinase activation caused by ColVI deficiency yields not only downregulation of autophagy but also activation of the HIF1A transcription factor, which in turn represses the expression of autophagy components through FoxO3 inhibition. In addition, HIF1A physically interacts with several major components of the circadian rhythm pathway, bringing about a reduction of MyoD1 levels. AKT1 directly inhibits GSK3-β by phosphorylation. GSK3-β is a central negative regulator of circadian rhythm that phosphorylates its components. GSK3-β repression by AKT1 further derepresses HIF1A, which is under GSK3-β negative control.
Although these studies suggest that the circadian circuit plays a crucial role in muscle function and regeneration, the mechanism mediating these effects in skeletal-muscle-wasting diseases remains to be elucidated. In summary, our results demonstrate, for the first time, a deregulation of circadian genes in skeletal muscle of ColVI myopathy that might lead to characterizing the involvement of circadian clock circuit in many other muscle-wasting diseases, opening new therapeutical perspectives.
Putative models linking collagen VI myopathies with circadian rhythm deregulation
The new data suggest a pathogenic connection between circadian rhythm gene deregulation and collagen-VI-related myopathies. It appears that CLOCK gene deregulation could provide a link between autophagy, mitochondrial abnormalities and ColVI diseases. In light of the asynchrony recently described as the mechanism driving regeneration failure (Dadgar et al., 2014), this relationship could be expressed as one of the two following scenarios. First, that circadian rhythm deregulation is a downstream effect of ColVI deficiency and contributes to muscle damage pathogenesis (Fig. 6A). In this hypothesis, the crucial player interconnecting ColVI deficiency with circadian rhythm could be the inappropriate persistence of AKT1 activation. Owing to the reduced ColVI expression, this would lead not only to the downregulation of autophagy (Grumati and Bonaldo, 2012), but also to the activation of the HIF1A transcription factor and the consequent downregulation of GSK3-β kinase. Indeed, muscle regeneration is a finely regulated process which drives the transcription of myogenic factors through the orchestration of cell contact, growth factors and various hormones. Murine models lacking or constitutively expressing AKT genes show severe muscle deficiency or extensive hypertrophy, respectively (Wilson and Rotwein, 2007; Peng et al., 2003; Lai et al., 2004), implicating AKT genes (AKT1 and AKT2) in both the development and regeneration of muscle. Serra et al. (2007) hypothesize that this might arise through convergence of the AKT pathway and the p38 MAPK circuit at the chromatin level controlling the assembly of myogenic transcriptome and driving the regeneration process.
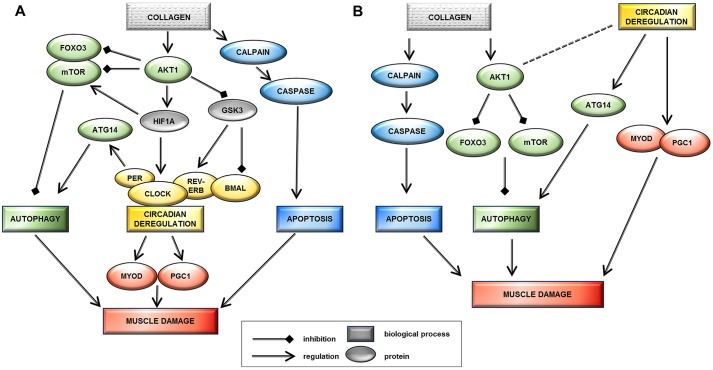
Fig. 6.
Proposed models that potentially link collagen VI myopathy and circadian genes. Model A suggests that circadian rhythm deregulation is a downstream effect of ColVI deficiency and contributes to muscle damage pathogenesis. The inappropriate persistence of AKT1 activation could follow reduced ColVI expression, leading not only to the downregulation of autophagy (also related to mTOR overexpression) but also to the activation of HIF1A transcription factor and the downregulation of the GSK3-β kinase. HIF1A physically interacts with several central components of the circadian rhythm pathway: BMAL1 protein (Hogenesch et al., 1998), PER1 (Chilov et al., 2001), PER2 (Koyanagi et al., 2003) and ROR-α. HIF1A activation has been shown to increase levels of PER1 and CLOCK proteins (Chilov et al., 2001). Increased levels of PER1, a negative regulator of circadian rhythm, might lead to decreased transcriptional activity of the BMAL1–CLOCK heterodimer, thereby slowing down circadian oscillations. GSK3-β also plays a role in the post-translational regulation of molecular clock components, targeting BMAL1 for degradation at the end of the circadian cycle and stabilizing Rev-ErbA, a negative regulator of BMAL1 transcription (Pritchett et al., 2012). Model B suggests that circadian rhythm deregulation in collagen VI myopathies is an independent event acting as a secondary modifier of disease severity. The collagen VI α6 chain is downregulated in the mouse circadian model, suggesting that circadian genes can influence the primary ColVI defect, as supported by the direct control circadian genes have on MyoD1 (muscle development) and ATG14 (autophagy circuit) proteins. Downregulation of CLOCK induces ATG14 persistence and autophagy activation, whereas MyoD1 overexpression alters regeneration and induces muscle damage.
The second scenario is that circadian rhythm deregulation in ColVI myopathies is an independent event acting as a secondary modifier of disease severity (Fig. 6B). Our observation that the collagen VI α6 chain is downregulated in a mouse circadian model could suggest that circadian genes are genetic modifiers able to influence the primary ColVI defect. It is well recognized that the molecular clock in skeletal muscle can be modified by environmental factors including time of feeding or time of physical activity (Wolff and Esser, 2012; Hatori and Panda, 2015). Merging this concept with the known direct control that circadian genes have on MyoD1 (muscle development), PGC1 (mitochondria) and ATG14 (autophagy circuit) proteins provides the model through which circadian rhythm disruption could function independently to modify disease severity. It might therefore be enlightening to explore circadian genes in the many ColVI patients whose causative mutations are still unknown (recently mutations in COL12A1 gene have been linked to the UCMD and Bethlem myopathy phenotype) (Hicks et al., 2014).
In conclusion, our demonstration of circadian gene involvement in the pathogenesis of ColVI myopathy in both an animal model and human patients opens new avenues for both pathogenesis and treatment of hereditary muscle diseases. Timing and synchronization might have different effects on gene expression, depending on the specific gene mutation (COL6 or DMD). Different key regulators, such as AKT1 for COL6 myopathies and TGFB1 for dystrophin mutations (Dadgar et al., 2014), might be implicated, as these two muscle diseases have different pathogenic signatures, represented by defective autophagy and massive muscle necrosis, respectively. Finally, given that CLOCK is downregulated only in the severe UCMD phenotype, it should be further explored as a severity biomarker in patients with myopathies characterized by a severe muscle remodeling, with a view to applications in both disease stratification in patients and monitoring in clinical trials.
MATERIALS AND METHODS
Analysis of Col6a1−/− mice
RNA extraction
For the focus of this work, we used standardized timing of muscle tissue collection from the Col6a1−/− mice and the Bmal1−/− mice. Mice in all colonies were maintained in normal 12-h-light–12-h-dark schedules with lights on at 7 am (Zeitgeber time – ZT0). Muscle tissues were collected between 10 am and 12 pm (ZT3–5) and frozen immediately in liquid N2. By following these strict time of collection procedures this allows comparisons across tissues and between animals. Both WT and Col6a1−/− mice belong to C57BL/6 strain regarding the fiber type composition of different muscles as previously described (Augusto et al., 2004).
Total RNA was isolated from three muscles (diaphragm, gastrocnemius and tibialis) of male mice. Mice were wild type (four) and knock out (four), at an age of 6 months. TRIzol extraction (Invitrogen), as specified by the manufacturer, was used. RNA from each muscle was quantified with Nanodrop (Thermo Scientific) spectrophotometer. Its integrity was evaluated with a 2100 Bioanalyzer (Agilent) prior to pooling equal amounts of RNA from muscles of the same kind.
Analysis of whole genome expression microarray data
We used the whole mouse genome gene expression kit including four arrays of 44,000 probes (Agilent Technologies, G4122F) to measure the transcriptional profile of muscle tissue samples of four mice. Sample labeling and hybridization were performed in triplicate according to protocols provided by Agilent (One-Color Microarray-Based Gene Expression Analysis version 5.0.1). The hybridized array was processed by Agilent scanner and Feature Extraction software (v9.5). The uniformness of array hybridization was verified through Agilent Quality Controls (Spike-in), consisting of a mixture of ten in-vitro-synthesized, polyadenylated transcripts derived from the Adenovirus E1A gene, premixed at concentrations spanning six logs and differing by one-log or half-log increments.
Microarray data analysis
Agilent's Feature Extraction Software (v9.5) was used for array image processing. Intensity data with local background adjustment by spatial detrending were log-transformed and quantile-normalized. Synonymous probes from the same gene were averaged. The software selected 28,052 probes for further analysis. To perform multi-way comparisons we utilized the limma package (Smyth et al., 2005). This package fits a separate linear model for each gene and uses moderated t-statistics to estimate differences between arrays. Empirical Bayes analysis is used to improve statistical power in small sample sizes. P-values were corrected for multiple testing using the Benjamini and Hochberg correction (Benjamini and Hochberg, 1995).
To ascribe biological changes in knockout versus WT muscles we used Gene Ontology term enrichment analysis. Gene ontology terms enriched in the differentially expressed genes were identified using Fisher exact test. We also performed Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) using Pathway Studio 9 from Elsevier.
Potential transcription regulators of the observed expression changes were identified using sub-network enrichment analysis (Sivachenko et al., 2007) with default parameters implemented in Pathway Studio. This algorithm identifies sub-networks consisting of a single expression regulator (for example, transcription factor) and its downstream genes showing concordant changes in expression between two conditions. Information about gene–gene regulations is extracted from the literature using natural language processing and stored in the Pathway Studio database. At the time of analysis, the database contained over 250,000 expression relationships between proteins. Relationships were extracted by natural language processing of more than 22 million PubMed abstracts and 880,000 full-text articles.
Microarray data analysis validation by fluidic cards
A total of 46 genes were selected for fluidic card analysis based on both differential expression between wild-type and knockout mice and literature analysis in Pathway Studio to find genes associated with the circadian rhythm pathway, muscle regeneration, autophagy and apoptosis. Selected genes were used to set up a custom TaqMan low density array (TLDA or fluidic card) according to manufacturer specifications in order to validate results obtained using the microarray platform. The major intrinsic protein of lens fiber (MIP) gene was selected as negative control for its absence of expression in the microarray data. Single fluidic card can analyze 48 genes (47 test and the 18S control housekeeping gene) simultaneously in eight samples and was run five times for each sample, it also included major autophagic effector protein Beclin 1 (Grumati et al., 2010).
150 ng for each RNA pool was retrotranscribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems), according to the manufacturer's instructions and loaded in a single port of the TLDA run on ABI 7900HT System (Applied Biosystems) for 2 min at 50°C, 10 min at 95°C, followed by 40 cycles for 15 s at 97°C and 1 min at 60°C. Data analysis was performed following the ΔΔCT Method (Applied Biosystems User Bulletin #2) and using the 18S gene as calibrator and the WT samples as controls with respect to the knockout samples. Calculated mean 2−ΔΔCT values were log transformed and compared with normalized fold change microarray data.
Western blotting and RNA analysis in Bmal1−/− mice
Immunoblotting was used to evaluate the protein levels of LC3A (rabbit polyclonal, Thermo Scientific, PA1-16930; 1:1000), BECLIN1 (D40C5 rabbit mAb, Cell Signaling Technology, 3495; 1:1000), p62/SQSTM1 (Guinea pig polyclonal, Progen GP62-C; 1:1000), AMPK (rabbit polyclonal, Cell Signaling Technology, 2532; 1:1000), phosphorylated AMPK (rabbit polyclonal, Cell Signaling Technology, 2531; 1:1000), COL6A1 (COL6A1 H-200, Santa Cruz Biotechnology, sc-20649, 1:1000), COL6A3 and COL6A6 [rabbit polyclonal antisera against mouse a3(VI) and a6(VI) were kindly supplied by R. Wagener, Cologne, Germany]. Gastrocnemius muscles from 30–35-week-old male wild-type and Bmal1−/− (Andrews et al., 2010; Bunger et al., 2000) mice (both n=4) were homogenized using extraction buffer (2% SDS, 10% glycerol, 2% 2-mercaptoethanol, 50 mM Tris-HCl, pH 8.8). The homogenized samples were immediately incubated at 80°C for 5 min and protein concentration was measured using a Bio-Rad DC protein assay kit. The protein samples (50 µg/lane) were then subjected to 7.5% (for ColVI) or 10% SDS-PAGE. After electrophoresis, proteins were transferred to PVDF membrane (Millipore). The membrane was blocked in Odyssey blocking buffer (LI-COR) for 1 h and then incubated with primary antibody in 50% Odyssey blocking buffer (LI-COR) diluted with 1× PBS containing 0.1% Tween 20 for 1 h. The membrane was then washed and incubated with horseradish peroxidase (HRP)-conjugated secondary Ab (for all collagen VI proteins and p62) or Alexa-Fluor-680-conjugated secondary antibody diluted in in 50% Odyssey blocking buffer (LI-COR). The protein signal was detected either by incubation with ECL substrate and developing signals by exposing the membrane to CL-Xposure film (for HRP-conjugated secondary antibody), or by using the Odyssey infrared imager (LI-COR, for Alexa Fluor 680 antibody). The protein bands were analyzed by using ImageJ. The plots in the figures are average of four samples for both wild type and Bmal1−/−. For the Bmal1−/− studies, the animal procedures were conducted in accordance with institutional guidelines for the care and use of laboratory animals as approved by the AAALAC accredited, University of Kentucky Institutional Animal Care and Use Committees with the protocol number: 00728M2004.
Analysis of Bethlem myopathy and UCMD patients
RNA extraction
The three ColVI patients have the following mutations: UCMD patient, heterozygous c.819_833del p.Pro274_Gly278del in COL6A1 gene; severe Bethlem myopathy patient: homozygous c.1393C>T p.Arg465* in COL6A3 gene; Bethlem myopathy patient, heterozygous c.428+1G>A p. Tyr122_Gly143del in COL6A1 gene. We obtained muscle biopsies from ColVI patients after informed consent and approval of the Ethics Committee of the University of Ferrara (no. 02/2009, 26 February 2009). The research involving human subjects was performed according to the Helsinki Declaration.
Muscle biopsy procedures were taken at a time according to the local clinic rules, ranging from 8–10 am (central Europe time) or 10–12 am (Greenwich time), depending on the sites. Fresh biopsies were frozen following routine procedures. Because of the similarity in time of collection, comparison of molecular clock components across patient samples can be made. The muscle fiber composition in these three muscles shows a predominance of fiber type I (Fig. S4). Generally, hereditary myopathies, including the collagen VI myopathies, are characterized by the fiber type disproportion with type I fiber hypotrophy and predominance (Schessl et al., 2008).
Total RNA was isolated from skeletal muscle biopsies of three patients with different phenotypes: UCMD, dominant and recessive Bethlem myopathy and from one healthy control biopsy using the RNeasy-kit (Qiagen, Chatsworth, CA) according to the manufacturer's instructions. We verified high-quality of isolated RNA and RNA integrity using an Agilent Technologies 2100 Bioanalyzer. The isolated RNA had an RNA Integrity Number (RIN) value of greater than 8.
RNAseq
1 μg of total RNA sample was used for poly(A)+ mRNA selection using streptavidin-coated magnetic beads, followed by thermal mRNA fragmentation. Fragmented mRNA was retro-transcribed using reverse transcriptase (Super-Script II) and random primers. The cDNA was converted into double-stranded cDNA, its ends were repaired using Klenow fragment, T4 polynucleotide kinase and T4 polymerase, and then cDNA was ligated to Illumina paired end adaptors. Size selection was performed using a 2% agarose gel, generating cDNA libraries ranging in size from 200 to 250 bp. Finally, the libraries were enriched using 15 cycles of PCR (30 s at 98°C; 15 cycles of 10 s at 98°C, 30 s at 65°C, and 30 s at 72°C; then 5 min at 72°C) and purified by the QIAquick PCR purification kit (Qiagen). The enriched libraries were diluted with elution buffer to a final concentration of 10 nM. Each library was run at a concentration of 7 pM on one Genome Analyzer (GAIIe) lane using paired-end 100-bp sequencing.
We performed mapping of reads to human genome using TopHat software. TopHat, the splice junction mapper for RNASeq reads, was run with default parameters. Supplied annotations from hg19 Refseq refGene were downloaded from the UCSC Genome Browser. The mapping results from TopHat were then passed to Cuffdiff, a tool which finds significant changes in transcript expression. In Cuffdiff quartile normalization was turned on, bias correction was set on, the minimum alignment count was set to 10, and all other parameters were set to default. Both TopHat and Cuffdiff were run on the free pubic Galaxy server (http://galaxyproject.org/). To identify consistently deregulated genes, differential probes were identified by the OK test in Cufflinks (Trapnell et al., 2010). Log ratios were calculated as the difference between patient and control.
Quantification of CLOCK and MAT2A transcripts by real-time PCR
To validate the deregulated expression of CLOCK and MAT2A genes, we enlarged our patient cohort. We analyzed 12 UCMD and nine Bethlem myopathy, three patients with intermediate phenotypes and a pool made of muscle controls. All ColVI patients have been clinically characterized with regard to ‘circadian involvement’, date and time of biopsy, if the patients were fasting or taking drugs, if the patients were ambulatory and the presence of ventilation at the time of biopsy (Table S3). Total RNA was isolated from muscle sections using the RNeasy Kit (QIAGEN, Chatsworth, CA) and reverse transcribed by using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). In order to quantify the steady state level of CLOCK and MAT2A transcripts, commercially available TaqMan expression assays (Applied Biosystems) were used (CLOCK: Hs00231857_m1 NM_001267843 exon boundary 19–20; MAT2A: Hs01553622_g1 NM_005911 exon boundary 5–6) as for β-actin as housekeeping reference gene (ACTB Endogenous Control). Real-time PCR was performed in triplicate on the Applied Biosystems Prism 7900HT system, using 10 ng of cDNA and default parameters. Evaluation of transcripts levels was performed by the comparative CT method (ΔΔCT method). Statistical analysis was performed with a Student's t-test.
Western blotting analysis in Col6a1−/− mice and ColVI patients
For immunoblotting analysis, frozen muscle tissues from both patients and mice were ground in a dry-ice-cooled mortar and stored at −80°C. An aliquot of each frozen muscle was suspended in lysis buffer (urea 7 M, thiourea 2 M, CHAPS 4%, Tris-HCl 30 mM, and PMSF 1 mM) and solubilized by sonication on ice. Samples were pooled and protein concentrations were determined using PlusOne 2D-Quant kit (GE Healthcare, Milan, Italy). Total protein extracts (50 µg) from pooled Col6a1−/− (5 mice) and WT (5 mice) animal models, and from pooled UCMD (4 patients), Bethlem myopathy (8 patients), 30 unrelated patients with Duchenne (DMD) or Becker (BMD) muscular dystrophies, and healthy control (5 subjects) muscles, were loaded in triplicate and resolved on 14–8% gradient polyacrylamide gels. Blots were incubated with rabbit polyclonal primary antibodies as follows: anti-MAT2A (Novus Biologicals, NB110-94158; 1:1000), anti-CLOCK (Santa Cruz Biotechnology, sc-25361; 1:500). After washing, the membranes were incubated with anti-rabbit-IgG (GE Healthcare) secondary antibody conjugated to horseradish peroxidase (1:5000). The signals were visualized by chemiluminescence using the ECL Prime (GE Healthcare) detection kit and the Image Quant LAS 4000 (GE Healthcare) analysis system. Statistical analysis was performed applying the ANOVA and Tukey tests (n=3, P<0.05).
Manipulations of Col6a1−/− animals, required for the experiments described, conform to the rules of the Animal Experimental Commission of the ‘Complesso Interdipartimentale A. Vallisneri, University of Padua (Italy)’, where the work has been carried out. These regulations represent the application of the D.L. 27/1/92 n. 116 of the Italian Government, which meets Directive 86/609/EEC of the EU Council concerning the protection of animals used for experiments and scientific purposes. The presented procedures have been approved by the Animal Experimental Commission with the certificate no. 22675.
Acknowledgements
Special thanks are due to Mikhail Pyatnitskiy from Ariadne-Dx for his data analysis. We are grateful to Raimund Wagener (University of Cologne, Germany) for a3(VI) and a6(VI) for kindly providing collagen antibodies. The Eurobiobank is also acknowledged.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
A. Ferlini designed the rationale of the research and supervised the work; C. Scotton, M. Bovolenta, M. S. Falzarano, E. Martoni, C. Passarelli, A. Armaroli, H. Osman and R. Selvatici performed the experimental procedures related to expression profile analysis and data validation; T. Castrignanò and G. Pesole performed the bioinformatics analysis of RNAseq. E. Schwartz, E. Kotelnikova, A. Yuryev and M. Lebowitz performed the interactome analysis and statistical analysis; C. Scotton, E. Schwartz, K. Esser and A. Ferlini performed the data interpretation; C. Rodolico, S. Messina, E. Pegoraro, A. D'Amico, E. Bertini, F. Gualandi, M. Neri, , P. Boffi, M. A. Maioli, H. Lochmüller, V. Straub, K. Bushby, P. Sabatelli, L. Merlini, R. Foley, S. Cirak, I. Zaharieva and F. Muntoni performed the clinical assessment and provided patient samples; P. Braghetta, P. Bonaldo and P. Bernardi provided and characterized the Col6a1−/− mice; D. Capitanio and C. Gelfi performed the proteomic analysis on Col6a1−/− mice and patients. X. Zhang, B. Hodge and K. A. Esser performed the proteomic analysis on Bmal1−/− mice; E. Schwartz, P. Bonaldo, P. Bernardi, F. Muntoni, H. Lochmüller and K. A. Esser critically revised the paper; C. Scotton and A. Ferlini wrote the paper. The authors M. Bovolenta and E. Schwartz contributed equally to this work.
Funding
This study was funded by the BIO-NMD European Union Seventh Framework Programme [project 241665 to A.F., E.S., F.M., H.L., C.G. and P. Bonaldo); the Fondazione Telethon [grant numbers GGP08017 to P. Bernardi, P. Bonaldo., C.G. and A.F.; GUP11007 to L.M. and P.S.]; the RD-Connect EU project 305444 [to H.L.]; and the National Institutes of Health AR066082 to K.A.E.]. The financial support of the Muscular Dystrophy Campaign Centre Grant and of the MRC Neuromuscular Centre at UCL; of the Biomedical Research Centre at Great Ormond Street Hospital and of the Great Ormond Street Hospital Children's Charity (in support of F.M.) are also gratefully acknowledged. Deposited in PMC for release after 12 months.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.175927/-/DC1
References
- Aartsma-Rus A. and Muntoni F. (2013). 194th ENMC international workshop. 3rd ENMC workshop on exon skipping: towards clinical application of antisense-mediated exon skipping for Duchenne muscular dystrophy 8-10 December 2012, Naarden, The Netherlands. Neuromuscul. Disord. 23, 934-944. 10.1016/j.nmd.2013.06.369 [DOI] [PubMed] [Google Scholar]
- Allamand V., Briñas L., Richard P., Stojkovic T., Quijano-Roy S. and Bonne G. (2011). ColVI myopathies: where do we stand, where do we go? Skelet. Muscle. 23, 1-30. 10.1186/2044-5040-1-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews J. L., Zhang X., McCarthy J. J., McDearmon E. L., Hornberger T. A., Russell B., Campbell K. S., Arbogast S., Reid M. B., Walker J. R. et al. (2010). CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proc. Natl. Acad. Sci. USA 107, 19090-19095. 10.1073/pnas.1014523107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Augusto V., Padovani C. R. and Campos G. E. (2004). Skeletal muscle fiber types in C57BL6J mice. Braz. J. Morphol. Sci. 21, 89-94. [Google Scholar]
- Benjamini Y. and Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57, 289-300. [Google Scholar]
- Bernardi P. and Bonaldo P. (2008). Dysfunction of mitochondria and sarcoplasmic reticulum in the pathogenesis of collagen VI muscular dystrophies. Ann. N. Y. Acad. Sci. 1147, 303-311. 10.1196/annals.1427.009 [DOI] [PubMed] [Google Scholar]
- Bonaldo P., Russo V., Bucciotti F., Doliana R. and Colombatti A. (1990). Structural and functional features of the alpha 3 chain indicate a bridging role for chicken collagen VI in connective tissues. Biochemistry 29, 1245-1254. 10.1021/bi00457a021 [DOI] [PubMed] [Google Scholar]
- Bonaldo P., Braghetta P., Zanetti M., Piccolo S., Volpin D. and Bressan G. M. (1998). Collagen VI deficiency induces early onset myopathy in the mouse: an animal model for Bethlem myopathy. Hum. Mol. Genet. 7, 2135-2140. 10.1093/hmg/7.13.2135 [DOI] [PubMed] [Google Scholar]
- Bönnemann C. G. (2011). The collagen VI-related myopathies Ullrich congenital muscular dystrophy and Bethlem myopathy. Handb. Clin. Neurol. 101, 81-96. 10.1016/B978-0-08-045031-5.00005-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunger M. K., Wilsbacher L. D., Moran S. M., Clendenin C., Radcliffe L. A., Hogenesch J. B., Simon M. C., Takahashi J. S. and Bradfieldet C. A. (2000). Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 103, 1009-1017. 10.1016/S0092-8674(00)00205-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campo E. (2013). Whole genome profiling and other high throughput technologies in lymphoid neoplasms—current contributions and future hopes. Mod. Pathol. 26, S97-S110. 10.1038/modpathol.2012.179 [DOI] [PubMed] [Google Scholar]
- Chatterjee S., Yin H., Nam D., Li Y. and Ma K. (2015). Brain and muscle Arnt-like 1 promotes skeletal muscle regeneration through satellite cell expansion. Exp. Cell Res. 331, 200-210. 10.1016/j.yexcr.2014.08.041 [DOI] [PubMed] [Google Scholar]
- Chilov D., Hofer T., Bauer C., Wenger R. H. and Gassmann M. (2001). Hypoxia affects expression of circadian genes PER1 and CLOCK in mouse brain. FASEB J. 15, 2613-2622. 10.1096/fj.01-0092com [DOI] [PubMed] [Google Scholar]
- Dadgar S., Wang Z., Johnston H., Kesari A., Nagaraju K., Chen Y.-W., Hill D. A., Partridge T. A., Giri M., Freishtat R. J. et al. (2014). Asynchronous remodeling is a driver of failed regeneration in Duchenne muscular dystrophy. J. Cell Biol. 207, 139-158. 10.1083/jcb.201402079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma S., Leone R., Grumati P., Vasso M., Polishchuk R., Capitanio D., Braghetta P., Bernardi P., Bonaldo P. and Gelfi C. (2013). Changes in muscle cell metabolism and mechanotransduction are associated with myopathic phenotype in a mouse model of collagen VI deficiency. PLoS ONE 8, e56716 10.1371/journal.pone.0056716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Palma S., Capitanio D., Vasso M., Braghetta P., Scotton C., Bonaldo P., Lochmüller H., Muntoni F., Ferlini A. and Gelfi C. (2014). Muscle proteomics reveals novel insights into the pathophysiological mechanisms of collagen VI myopathies. J. Proteome Res. 13, 5022-5030. 10.1021/pr500675e [DOI] [PubMed] [Google Scholar]
- De Visser M., van der Kooi A. J. and Jobsis G. J. (2004). Bethlem myopathy. In Myology (ed. Franzini-Amstrong AGEaC), New York: McGraw-Hill pp. 1135-1146. [Google Scholar]
- Dyar K. A., Ciciliot S., Wright L. E., Biensø R. S., Tagliazucchi G. M., Patel V. R., Forcato M., Paz M. I. P., Gudiksen A., Solagna F. et al. (2013). Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Mol. Metab. 3, 29-41. 10.1016/j.molmet.2013.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhard D. and Jockusch H. (2004). Intermingling versus clonal coherence during skeletal muscle development: mosaicism in eGFP/nLacZ-labeled mouse chimeras. Dev. Dyn. 230, 69-78. 10.1002/dvdy.20039 [DOI] [PubMed] [Google Scholar]
- Esnault C., Stewart A., Gualdrini F., East P., Horswell S., Matthews N. and Treisman R. (2014). Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 28, 943-958. 10.1101/gad.239327.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlini A., Scotton C. and Novelli G. (2013). Biomarkers in rare diseases. Public Health Genomics 16, 313-321. 10.1159/000355938 [DOI] [PubMed] [Google Scholar]
- Grumati P. and Bonaldo P. (2012). Autophagy in skeletal muscle homeostasis and in muscular dystrophies. Cells 1, 325-345. 10.3390/cells1030325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grumati P., Coletto L., Sabatelli P., Cescon M., Angelin A., Bertaggia E., Blaauw B., Urciuolo A., Tiepolo T., Merlini L. et al. (2010). Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat. Med. 16, 1313-1320. 10.1038/nm.2247 [DOI] [PubMed] [Google Scholar]
- Grumati P., Coletto L., Sandri M. and Bonaldo P. (2011). Autophagy induction rescues muscular dystrophy. Autophagy 7, 426-428. 10.4161/auto.7.4.14392 [DOI] [PubMed] [Google Scholar]
- Gualandi F., Manzati E., Sabatelli P., Passarelli C., Bovolenta M., Pellegrini C., Perrone D., Squarzoni S., Pegoraro E., Bonaldo P. et al. (2012). Antisense-induced messenger depletion corrects a COL6A2 dominant mutation in Ullrich myopathy. Hum. Gene Ther. 23, 1313-1318. 10.1089/hum.2012.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M. and Panda S. (2015). Response of peripheral rhythms to the timing of food intake. Methods Enzymol. 552, 145-161. 10.1016/bs.mie.2014.10.027 [DOI] [PubMed] [Google Scholar]
- Hicks D., Farsani G. T., Laval S., Collins J., Sarkozy A., Martoni E., Shah A., Zou Y., Koch M., Bönnemann C. G. et al. (2014). Mutations in the collagen XII gene define a new form of extracellular matrix-related myopathy. Hum. Mol. Genet. 23, 2353-2363. 10.1093/hmg/ddt637 [DOI] [PubMed] [Google Scholar]
- Hodge B. A., Wen Y., Riley L. A., Zhang X., England J. H., Harfmann B. D., Schroder E. A. and Esser K. A. (2015). The endogenous molecular clock orchestrates the temporal separation of substrate metabolism in skeletal muscle. Skelet. Muscle 5, 17 10.1186/s13395-015-0039-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch J. B., Gu Y.-Z., Jain S. and Bradfield C. A. (1998). The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc. Natl. Acad. Sci. USA 95, 5474-5479. 10.1073/pnas.95.10.5474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin W. A., Bergamin N., Sabatelli P., Reggiani C., Megighian A., Merlini L., Braghetta P., Columbaro M., Volpin D., Bressan G. M. et al. (2003). Mitochondrial dysfunction and apoptosis in myopathic mice with collagen VI deficiency. Nat. Genet. 35, 367-371. 10.1038/ng1270 [DOI] [PubMed] [Google Scholar]
- Izu Y., Ansorge H. L., Zhang G., Soslowsky L. J., Bonaldo P., Chu M.-L. and Birk D. E. (2011). Dysfunctional tendon collagen fibrillogenesis in collagen VI null mice. Matrix Biol. 30, 53-61. 10.1016/j.matbio.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izu Y., Ezura Y., Mizoguchi F., Kawamata A., Nakamoto T., Nakashima K., Hayata T., Hemmi H., Bonaldo P. and Noda M. (2012). Type VI collagen deficiency induces osteopenia with distortion of osteoblastic cell morphology. Tissue Cell 44, 1-6. 10.1016/j.tice.2011.08.002 [DOI] [PubMed] [Google Scholar]
- Jöbsis G. J., Boers J. M., Barth P. G. and de Visser M. (1999). Bethlem myopathy: a slowly progressive congenital muscular dystrophy with contractures. Brain 122, 649-655. 10.1093/brain/122.4.649 [DOI] [PubMed] [Google Scholar]
- Kim J.-S., Coon S. L., Blackshaw S., Cepko C. L., Møller M., Mukda S., Zhao W.-Q., Charlton C. G. and Klein D. C. (2005). Methionine adenosyltransferase: adrenergic-cAMP mechanism regulates a daily rhythm in pineal expression. J. Biol. Chem. 280, 677-684. 10.1074/jbc.M408438200 [DOI] [PubMed] [Google Scholar]
- Koyanagi S., Kuramoto Y., Nakagawa H., Aramaki H., Ohdo S., Soeda S. and Shimeno H. (2003). A molecular mechanism regulating circadian expression of vascular endothelial growth factor in tumor cells. Cancer Res. 63, 7277-7283. [PubMed] [Google Scholar]
- Lai K.-M. V., Gonzalez M., Poueymirou W. T., Kline W. O., Na E., Zlotchenko E., Stitt T. N., Economides A. N., Yancopoulos G. D. and Glass D. J. (2004). Conditional activation of akt in adult skeletal muscle induces rapid hypertrophy. Mol. Cell. Biol. 24, 9295-9304. 10.1128/MCB.24.21.9295-9304.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraldi N. M., Sabatelli P., Columbaro M., Zamparelli A., Manzoli F. A., Bernardi P., Bonaldo P. and Merlini L. (2009). Collagen VI myopathies: from the animal model to the clinical trial. Adv. Enzyme Regul. 49, 197-211. 10.1016/j.advenzreg.2008.12.009 [DOI] [PubMed] [Google Scholar]
- McCarthy J. J., Andrews J. L., McDearmon E. L., Campbell K. S., Barber B. K., Miller B. H., Walker J. R., Hogenesch J. B., Takahashi J. S. and Esser K. A. (2007). Identification of the circadian transcriptome in adult mouse skeletal muscle. Physiol. Genomics 31, 86-95. 10.1152/physiolgenomics.00066.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L., Martoni E., Grumati P., Sabatelli P., Squarzoni S., Urciuolo A., Ferlini A., Gualandi F. and Bonaldo P. (2008). Autosomal recessive myosclerosis myopathy is a collagen VI disorder. Neurology 71, 1245-1253. 10.1212/01.wnl.0000327611.01687.5e [DOI] [PubMed] [Google Scholar]
- Merrell A. J. and Kardon G. (2013). Development of the diaphragm -- a skeletal muscle essential for mammalian respiration. FEBS J. 280, 4026-4035. 10.1111/febs.12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paco S., Kalko S. G., Jou C., Rodríguez M. A., Corbera J., Muntoni F., Feng L., Rivas E., Torner F., Gualandi F. et al. (2013). Gene expression profiling identifies molecular pathways associated with collagen VI deficiency and provides novel therapeutic targets. PLoS ONE 8, e77430 10.1371/journal.pone.0077430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X.-D., Xu P.-Z., Chen M.-L., Hahn-Windgassen A., Skeen J., Jacobs J., Sundararajan D., Chen W. S., Crawford S. E., Coleman K. G. et al. (2003). Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 17, 1352-1365. 10.1101/gad.1089403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchett D., Wulff K., Oliver P. L., Bannerman D. M., Davies K. E., Harrison P. J., Peirson S. N. and Foster R. G. (2012). Evaluating the links between schizophrenia and sleep and circadian rhythm disruption. J. Neural Transm. 119, 1061-1075. 10.1007/s00702-012-0817-8 [DOI] [PubMed] [Google Scholar]
- Sánchez-Pla A., Reverter F., Ruíz de Villa M. C. and Comabella M. (2012). Transcriptomics: mRNA and alternative splicing. J. Neuroimmunol. 248, 23-31. 10.1016/j.jneuroim.2012.04.008 [DOI] [PubMed] [Google Scholar]
- Scacheri P. C., Gillanders E. M., Subramony S. H., Vedanarayanan V., Crowe C. A., Thakore N., Bingler M. and Hoffman E. P. (2002). Novel mutations in collagen VI genes: expansion of the Bethlem myopathy phenotype. Neurology 58, 593-602. 10.1212/WNL.58.4.593 [DOI] [PubMed] [Google Scholar]
- Schessl J., Goemans N. M., Magold A. I., Zou Y., Hu Y., Kirschner J., Sciot R. and Bönnemann C. G. (2008). Predominant fiber atrophy and fiber type disproportion in early ullrich disease. Muscle Nerve 38, 1184-1191. 10.1002/mus.21088 [DOI] [PubMed] [Google Scholar]
- Schroder E. A. and Esser K. A. (2013). Circadian rhythms, skeletal muscle molecular clocks, and exercise. Exerc. Sport. Sci. Rev. 41, 224-229. 10.1097/JES.0b013e3182a58a70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotton C., Passarelli C., Neri M. and Ferlini A. (2014). Biomarkers in rare neuromuscular diseases. Exp. Cell Res. 325, 44-49. 10.1016/j.yexcr.2013.12.020 [DOI] [PubMed] [Google Scholar]
- Serra C., Palacios D., Mozzetta C., Forcales S. V., Morantte I., Ripani M., Jones D. R., Du K., Jhala U. S., Simone C. et al. (2007). Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell. 28, 200-213. 10.1016/j.molcel.2007.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevov M., Rosenquist R. and Mansouri L. (2012). RNA-based markers as prognostic factors in chronic lymphocytic leukemia. Expert. Rev. Hematol. 5, 69-79. 10.1586/ehm.11.80 [DOI] [PubMed] [Google Scholar]
- Sivachenko A. Y., Yuryev A., Daraselia N. and Mazo I. (2007). Molecular networks in microarray analysis. J. Bioinform. Comput. Biol. 5, 429-456. 10.1142/S0219720007002795 [DOI] [PubMed] [Google Scholar]
- Smyth G. K., Michaud J. and Scott H. S. (2005). Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics 21, 2067-2075. 10.1093/bioinformatics/bti270 [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S. et al. (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545-15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliavini F., Pellegrini C., Sardone F., Squarzoni S., Paulsson M., Wagener R., Gualandi F., Trabanelli C., Ferlini A., Merlini L. et al. (2014a). Defective collagen VI α6 chain expression in the skeletal muscle of patients with collagen VI-related myopathies. Biochim. Biophys. Acta 1842, 1604-1612. 10.1016/j.bbadis.2014.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliavini F., Sardone F., Squarzoni S., Maraldi N. M., Merlini L., Faldini C. and Sabatelli P. (2014b). Ultrastructural changes in muscle cells of patients with collagen VI-related myopathies. Muscles Ligaments Tendons J. 3, 281-286. [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B. A., Pertea G., Mortazavi A., Kwan G., van Baren M. J., Salzberg S. L. Wold B. J. and Pachter L. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511-515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urciuolo A., Quarta M., Morbidoni V., Gattazzo F., Molon S., Grumati P., Montemurro F., Tedesco F. S., Blaauw B., Cossu G. et al. (2013). Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun. 4, 1964 10.1038/ncomms2964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E. M. and Rotwein P. (2007). Selective control of skeletal muscle differentiation by Akt1. J. Biol. Chem. 282, 5106-5110. 10.1074/jbc.C600315200 [DOI] [PubMed] [Google Scholar]
- Wolff G. and Esser K. A. (2012). Scheduled exercise phase shifts the circadian clock in skeletal muscle. Med. Sci. Sports Exerc. 44, 1663-1670. 10.1249/MSS.0b013e318255cf4c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambon A. C., McDearmon E. L., Salomonis N., Vranizan K. M., Johansen K. L., Adey D., Takahashi J. S., Schambelan M. and Conklin B. R. (2003). Time- and exercise-dependent gene regulation in human skeletal muscle. Genome Biol. 4, R61 10.1186/gb-2003-4-10-r61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Patel S. P., McCarthy J. J., Rabchevsky A. G., Goldhamer D. J. and Esser K. A. (2012). A non-canonical E-box within the MyoD core enhancer is necessary for circadian expression in skeletal muscle. Nucleic Acids Res. 40, 3419-3430. 10.1093/nar/gkr1297 [DOI] [PMC free article] [PubMed] [Google Scholar]