Abstract
Rapamycin is a Food and Drug Administration (FDA)-approved immunosuppressant and anticancer agent discovered in the soil of Easter Island in the early 1970s. Rapamycin is a potent and selective inhibitor of the mechanistic target of rapamycin (mTOR) protein kinase, which acts as a central integrator of nutrient signaling pathways. During the last decade, genetic and pharmaceutical inhibition of mTOR pathway signaling has been found to promote longevity in yeast, worms, flies, and mice. In this article, we will discuss the molecular biology underlying the effects of rapamycin and its physiological effects, evidence for rapamycin as an antiaging compound, mechanisms by which rapamycin may extend life span, and the potential limitations of rapamycin as an antiaging molecule. Finally, we will discuss possible strategies that may allow us to inhibit mTOR signaling safely while minimizing side effects, and reap the health, social, and economic benefits from slowing the aging process.
Rapamycin influences several physiological processes (via mTOR inhibition) to extend life span. But because it has serious side effects in humans (e.g., increased risk of infections and diabetes), alternatives must be explored.
The mechanistic target of rapamycin (mTOR) is a phosphatidylinositol-3-kinase (PI3K)-like serine/threonine protein kinase that is conserved in eukaryotes including yeast, worms, flies, and mammals. mTOR was discovered as a result of the search for the target of rapamycin, a polyketide produced by Streptomyces hygroscopicus, which originally attracted attention because of its ability to inhibit the growth of Candida albicans and other fungi (Vezina et al. 1975). It was soon determined that rapamycin also acts against mammalian cells, with effects on both cell size and proliferation, leading to its development as an immunosuppressant (Seto 2012). Its immunosuppressive effects led to very cautious exploration of the potential of rapamycin as an anticancer agent, but several rapamycin derivatives, including everolimus and temsirolimus, as well as rapamycin itself (sirolimus) are FDA-approved both as immunosuppressants and anticancer agents. Rapamycin has attracted significant interest with the finding in 2009 that rapamycin treatment can robustly extend the life span of mice (Harrison et al. 2009). In this article, we discuss the molecular biology of mTOR, research into the mechanism by which mTOR inhibition promotes life span, the side effects of rapamycin, and possible ways in which rapamycin or alternative strategies to inhibit mTOR signaling may enable us to extend human life span and health span.
MOLECULAR BIOLOGY OF RAPAMYCIN
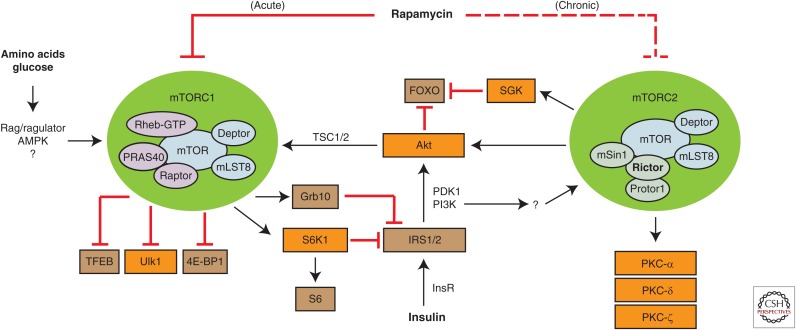
The mTOR protein kinase is found in two evolutionarily conserved protein complexes with distinct functions, substrates, and sensitivity to rapamycin (Fig. 1). mTOR complex 1 (mTORC1) consists of the mTOR protein kinase, RAPTOR, and mLST8, along with the regulatory proteins PRAS40 and DEPTOR. mTORC1 plays a key role in the regulation of translation and cell growth through substrates that include S6 kinase 1 (S6K1) and the eukaryotic initiation factor eIF4E-binding protein 1 (4E-BP1) (reviewed in Caron et al. 2015). Other mTORC1 substrates include unc-51-like autophagy-activating kinase 1 (ULK-1), a key regulator of autophagy, transcription factor EB (TFEB), a regulator of lysosome biogenesis, and Grb-10, an insulin-receptor binding protein (Hsu et al. 2011; Kim et al. 2011; Settembre et al. 2012). The activity of mTORC1 toward many substrates is acutely sensitive to rapamycin, but mTORC1 also possess rapamycin-resistant activity toward certain substrates (Thoreen et al. 2012; Kang et al. 2013).
Figure 1.
The mechanistic target of rapamycin (mTOR) signaling pathway. Rapamycin is an acute inhibitor of mTOR complex 1 (mTORC1), which phosphorylates substrates including S6 kinase 1 (S6K1), eIF4E-binding protein 1 (4E-BP1), transcription factor EB (TFEB), unc-51-like autophagy-activating kinase 1 (Ulk1), and growth factor receptor-bound protein 10 (GRB-10). Rapamycin dosed chronically also inhibits mTOR complex 2 (mTORC2), which regulates the phosphorylation of Akt, serum/glucocorticoid regulated kinase (SGK), and members of the protein kinase C (PKC) family. mTORC2 is primarily responsive to insulin/insulin-like growth factor 1 (IGF-1) signaling, whereas mTORC1 is sensitive to insulin as well as amino acids and glucose. AMPK, Adenosine monophosphate-stimulated kinase.
The activity of mTORC1 is dependent on its localization to the lysosome by the Rag/ragulator complex, where it can interact with its activator Rheb, but these proteins are not, strictly speaking, components of mTORC1 itself (Sancak et al. 2010; Bar-Peled and Sabatini 2014). The regulation of mTORC1 activity is extremely complex, but in brief, the Rag/ragulator complex recruits mTORC1 to the lysosome when amino acids and glucose are plentiful, whereas the tuberous sclerosis complex 1/2 (TSC1/2) complex, which negatively regulates mTORC1 signaling, departs from the lysosome in response to insulin (Bar-Peled and Sabatini 2014; Menon et al. 2014). The regulation of the Rag/ragulator complex has been a subject of intensive investigation, resulting in the identification of the additional upstream regulators of mTORC1 signaling, including the GATOR1/2 complex and Sestrin1–Sestrin3 (Bar-Peled et al. 2013; Chantranupong et al. 2014).
mTOR complex 2 (mTORC2) consists of mTOR, rapamycin-insensitive companion of mammalian target of rapamycin (RICTOR), mLST8, PROTOR1/2, and mSin1, as well as the regulatory protein DEPTOR. In contrast to mTORC1, mTORC2 is relatively resistant to the effects of rapamycin both in vitro and in vivo, but can be disrupted by prolonged treatment (Sarbassov et al. 2006; Lamming et al. 2012). mTOR complex 2 (mTORC2) regulates a diverse set of substrates downstream from the insulin/insulin-like growth factor 1 (IGF-1) receptor, the best characterized of which include AKT on residues T450, S473, and S477/T479, serum/glucocorticoid-regulated kinase (SGK) S422, and protein kinase C α (PKC-α) (Guertin et al. 2006; Garcia-Martinez and Alessi 2008; Ikenoue et al. 2008; Liu et al. 2014a). More recently, mTORC2 has been shown to regulate control of other PKC family members, including PKC-δ and PKC-ζ, and mTORC2 also regulates the stability of insulin receptor substrate 1 (IRS1) via phosphorylation of the ubiquitin ligase subunit Fbw8 (Gan et al. 2012; Kim et al. 2012; Li and Gao 2014). It is apparent from this diverse set of substrates that mTORC2 is a key effector of the insulin signaling pathway.
Although the pathway that mediates activation of mTORC2 by the insulin/IGF-1 receptor is not fully understood, at least some mTORC2 localizes to the mitochondrial-associated endoplasmic reticulum membrane, and the activity of mTORC2 may be dependent on its association with ribosomal subunits (Zinzalla et al. 2011; Betz et al. 2013). A variety of other proteins, including TSC1/2, P-rex1, Rac1, sestrin3, and XPLN, have also been implicated in the regulation of mTORC2 (Hernandez-Negrete et al. 2007; Huang et al. 2008; Saci et al. 2011; Khanna et al. 2013; Tao et al. 2014). However, a cohesive model integrating all of these additional proteins is still lacking.
RAPAMYCIN TREATMENT EXTENDS LIFE SPAN
The role of the mTOR-signaling pathway in longevity was first discovered in 2003 in Caenorhabditis elegans; mutation of worm mTOR or RNAi against mTOR more than doubled the life span (Vellai et al. 2003). A similar effect was found in Drosophila, in which expression of dominant negative mTOR or S6K similarly increased life span (Kapahi et al. 2004). The interest surrounding the mTOR-signaling pathway increased still further when inhibition of mTOR signaling in yeast was found to increase both chronological and replicative life span (Kaeberlein et al. 2005; Powers et al. 2006). Importantly, Kaeberlein and colleagues found that calorie restriction (CR), an intervention that extends life span in yeast as well as mammals, was unable to extend the life span of long-lived tor1Δ yeast.
A CR diet has been the gold standard for life span interventions since its discovery in the 1930s, and extends the life span of yeast, worms, flies, mice, dogs, and nonhuman primates (reviewed in Lamming and Anderson 2014). The mechanism behind the effects of a CR diet on life span have been elusive and highly debated, and the possibility that CR might function by inhibiting mTOR pathway signaling spurred significant efforts into understanding how the mTOR-signaling pathway regulates life span. It also suggested the possibility that rapamycin, as an inhibitor of mTOR, could function as a small molecule CR mimetic and extend life span. Indeed, rapamycin was soon shown to extend life span in yeast (Powers et al. 2006; Lamming et al. 2007).
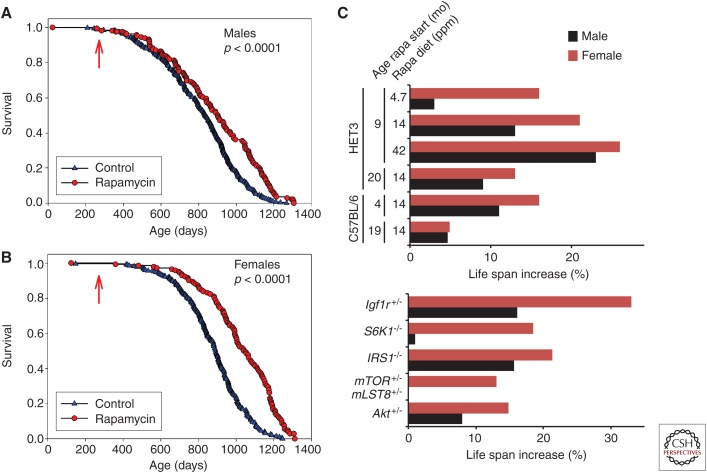
Although interest in testing rapamycin in flies and worms was intense, and has now been shown to extend life span (Bjedov et al. 2010; Robida-Stubbs et al. 2012), investigation of the effects of rapamycin on life span jumped directly to mice with the aid of the National Institute of Aging (NIA) Interventions Testing Program (ITP). The ITP was able to solve the technical challenges of delivering rapamycin to mice in chow by microencapsulating it in an enteric polymer to protect rapamycin from the acidic environment of the stomach. In 2009, the ITP published the first of several manuscripts on the effects of rapamycin on mice, showing that rapamycin could extend the life span of genetically heterogeneous HET3 mice when treatment began at 20 mo of age (Harrison et al. 2009). Subsequent studies by the ITP determined that rapamycin had a similar effect on life span when delivered starting at 9 mo of age (Fig. 2A,B), and that the response to rapamycin was dose dependent (Miller et al. 2011b, 2014).
Figure 2.
Sexually dimorphic impact of rapamycin and genetic interventions in the insulin/insulin-like growth factor 1 (IGF-1)/mechanistic target of rapamycin (mTOR)-signaling pathway. (A,B) Treatment with 14 ppm rapamycin begun at 9 mo of age extends the life span of genetically heterogeneous HET3 (A) males and (B) females. (C) Rapamycin and genetic interventions in the insulin/IGF-1/mTOR-signaling pathway that promote life span have a stronger effect on median female life span than on male life span (data from Holzenberger et al. 2003; Harrison et al. 2009; Selman et al. 2009, 2011; Lamming et al. 2012; Nojima et al. 2013; Fok et al. 2014b; Miller et al. 2014; Zhang et al. 2014). Mean life span is shown for HET3 mice initiated on 14 ppm rapamycin at 20 mo of age as median is not available. (Panels A and B from Miller et al. 2011a; reprinted by permission of Oxford University Press.)
In addition to HET3 mice, rapamycin has now been shown to extend the life span of both male and female C57BL/6J mice (Fok et al. 2014b; Zhang et al. 2014), male C57BL/6J Rj mice (Neff et al. 2013), female 129/Sv mice (Anisimov et al. 2011), and female FVB/N HER-2/neu mice (Popovich et al. 2014). Fascinatingly, all studies that have compared the effect of rapamycin on both males and females have found that rapamycin promotes longevity in females more effectively than in males (Fig. 2C). We will discuss a possible reason for this effect below, but it is interesting to note that many genetic interventions in the insulin/IGF-1/mTOR signaling pathway also show greater benefits in females than males (Fig. 2C). This includes mice null for either Irs1 or S6K1 (Selman et al. 2009, 2011) and mice heterozygous for both mTOR and mLST8 (Lamming et al. 2012).
HOW DOES RAPAMYCIN EXTEND LIFE SPAN, AND WHAT CAN IT TEACH US?
Because of the involvement of mTOR in so many key physiological processes, rapamycin has many different biological effects in pathways that are important in health and longevity. Interestingly, although rapamycin initially attracted attention as a CR mimetic, a microarray and metabolome study found that rapamycin and CR have surprisingly divergent effects on gene expression and metabolites in the liver (Fok et al. 2014a). As we have previously detailed (Lamming et al. 2013), the proposed mechanisms by which rapamycin extends life span include suppression of cancer, inhibition of translation, maintenance of protein quality, and effects on stem cells. We provide a brief overview of these areas with the latest research on these areas below.
Cancer
Cancer is an important cause of mortality in both mice and humans. Overall, rapamycin and derivatives such as everolimus and temsirolimus have been only modestly effective in humans (Miller et al. 2011b), although targeted use of rapamycin against cancers with hyperactivating mutations in the mTOR protein kinase shows significant promise (Grabiner et al. 2014; Wagle et al. 2014). Approximately 70% of HET3 mice die from lymphoma, hemangiosarcoma, and lung carcinoma, the frequency of which is not significantly shifted by rapamycin (Miller et al. 2011b), suggesting that rapamycin does not prevent cancer. Rapamycin significantly reduces the proportion of 16-mo-old mice with cancer or precancerous lesions, suggesting that rapamycin does delay cancer in mice, and it has been argued that the effect of rapamycin may be limited to delaying cancer, not aging (Neff et al. 2013). However, it is clear that rapamycin delays many forms of age-dependent changes and preserves health span (Wilkinson et al. 2012). Although the anticancer effect of rapamycin may be important, it likely does not account for the full effects of rapamycin on the aging process.
Protein Translation
mTORC1 is an important regulator of protein translation through two distinct mechanisms: the regulation of ribosomal biogenesis via S6K1, and the regulation of mRNA translation mediated by the 4E-BPs. The importance of translation in regulating longevity in model organisms is undisputed, as experiments in Saccharomyces cerevisiae, C. elegans, and Drosophila melanogaster clearly show (Kapahi et al. 2004; Hansen et al. 2007; Syntichaki et al. 2007; Steffen et al. 2008; Zid et al. 2009). In these experiments, deletion or RNAi-mediated knockdown of specific ribosomal proteins or translation initiation factors extend life span. In yeast, the life span extension resulting from reduced expression of large ribosomal subunits is dependent upon the increased translation of the transcriptional activator GCN4, providing a mechanistic explanation for how the efficiency of translation initiation can regulate life span (Steffen et al. 2008). However, this has not been shown in higher organisms. Indeed, recent findings in C. elegans show that mTOR pathway inhibition can further promote longevity in long-lived C. elegans with RNAi-depressed translation initiation factors (Hansen et al. 2007; Syntichaki et al. 2007). Moreover, a 50% decrease in protein translation is not sufficient to extend C. elegans life span (Hansen et al. 2007).
In mice, it is unclear whether decreased protein translation is sufficient to extend life span. Although deletion of S6K1 significantly extends life span (Selman et al. 2009), initial studies found that loss of S6K1 does not impair protein translation in skeletal muscle (Mieulet et al. 2007). Although yeast lacking Rpl22a have extended life span, loss of Rpl22 in mice has essentially no effect on translation because of compensatory expression of a paralog, Rpl22l1 (O’Leary et al. 2013). A more recent study found that rapamycin does decrease skeletal muscle protein synthesis, but the amount of the change is very small, and rapamycin does not decrease protein synthesis in heart (Drake et al. 2013). A study comparing the effect of rapamycin and S6K1 deletion in multiple tissues of mice found that although a single dose of rapamycin did decrease translation in multiple tissues, chronic treatment with rapamycin for 4 wk did not (Garelick et al. 2013). Moreover, mice lacking S6K1 have normal translational activity and respond normally to rapamycin, suggesting that the benefits of chronic rapamycin on life span are not a result of decreased translation (Garelick et al. 2013).
Protein Quality
Maintaining protein quality is an important challenge during aging. One of the most interesting effects of rapamycin that was recently discovered is that rapamycin treatment of aged mice rejuvenates the aging heart proteome. Despite increased protein half-life, the hearts of rapamycin treated mice had a decreased abundance of damaged proteins (Dai et al. 2014). Such a change could result from increased clearance of damaged proteins.
One of the ways in which damaged proteins are cleared is autophagy, a process in which proteins, especially damaged ones, are broken down to their constituent amino acids. mTOR normally suppresses autophagy by phosphorylating S757 of Ulk1, a kinase required for initiation of autophagy (Kim et al. 2011). In C. elegans, autophagy is required for either CR or mTOR inhibition to extend life span (Hansen et al. 2008). Autophagy is up-regulated during CR in mice, and may mediate the beneficial effects of CR on many organ systems, including the liver (Cuervo et al. 2005; Zhang and Cuervo 2008; Kume et al. 2010; Han et al. 2012). The regulation of autophagy is likely a critical part of how rapamycin promotes life span, and it may also impact cancer, as stimulation of autophagy may be an important mechanism of tumor suppression (White et al. 2010).
A second way in which damaged proteins are cleared is proteasome activity, which has been shown to be important in yeast life span (Kruegel et al. 2011). Enhanced proteasome activity has also been found in the exceptionally long-lived naked mole rat (Rodriguez et al. 2012). It was recently found that rapamycin boosts proteasome activity in the brains of female mice treated with rapamycin (Rodriguez et al. 2014), suggesting that regulation of proteasome activity may be important for life span.
Stem Cells and Cell Senescence
Loss of stem-cell proliferative capacity, either because of a decrease in stem cell number or decreased potency, may explain many of the phenotypes of aging. Some of the first work on mTOR signaling in stem cells was performed with hematopoietic stem cells (HSCs), which show age-related declines in self-renewal and function. The function of HSCs declines during aging, and Chen et al. (2009) determined that mTOR signaling was elevated in HSCs from aged mice, and that treatment with rapamycin restored self-renewal of aged HSCs. Similarly, rapamycin treatment or CR increases the self-renewal of aged intestinal stem cells (Yilmaz et al. 2012). More recent experiments conducted in vitro have found that rapamycin can preserve mesenchymal stem-cell self-renewal and prevent epithelial stem cell senescence (Iglesias-Bartolome et al. 2012; Gharibi et al. 2014). In both cases, this appears to be largely a result of decreased damage from reactive oxygen species rather than more general protection from aging. Stem cells remain an important research area for the biology of aging, and hopefully more in vivo data will determine whether rapamycin can protect or even rejuvenate other populations of stem cells. In vivo data suggests that rapamycin may increase transcription of oxidative stress response genes in C. elegans and mouse liver (Robida-Stubbs et al. 2012).
WILL THE SIDE EFFECTS OF RAPAMYCIN LIMIT ITS USE AS A HUMAN ANTIAGING THERAPEUTIC?
Although rapamycin shows many beneficial effects in mice, in humans, rapamycin and rapamycin derivatives are used primarily as immunosuppressants following organ transplantation, and in the treatment of several specific types of cancer, including renal cell carcinoma, pancreatic neuroendocrine tumors, and HER2-negative breast cancer (Pusceddu et al. 2014). Serious side effects in humans include an increased incidence of viral and fungal infections including pneumonia, chronic edema, painful oral aphthous ulceration, and hair loss (Mahe et al. 2005; McCormack et al. 2011). Metabolic effects of long-term rapamycin treatment have also been observed, including decreased insulin sensitivity, glucose intolerance, and an increased risk of new-onset diabetes (Johnston et al. 2008). Finally, rapamycin treatment of mice consistently benefits females more than males, suggesting the possibility that rapamycin treatment of humans may show a similar sexually dimorphic effect on health span and life span.
Short-term treatment with rapamycin is acceptable in the context of cancer treatment and organ transplantation, and might be acceptable for short-term treatment of specific age-related pathologies. For instance, 10 wk of rapamycin treatment reverses age-related cardiac hypertrophy and diastolic dysfunction in aged mice while rejuvenating the heart at the level of the proteome (Dai et al. 2014). However, short-term rapamycin treatment is likely to be insufficient in the case of many age-related diseases, including Alzheimer’s disease. Although prophylactic dosing with rapamycin in mouse models of Alzheimer’s disease significantly reduces amyloid-β, plaques, tangles, and cognitive defects, dosing older mice has no beneficial effects (Spilman et al. 2010; Majumder et al. 2012). The risks of long-term prophylactic treatment with rapamycin are therefore likely to be unacceptable.
One area in which the side effects of rapamycin treatment may be less important is in the treatment of diseases of rapid aging such as Hutchinson–Gilford progeria syndrome (HGPS). HGPS is a rare, fatal genetic disorder resulting from a mutation in LMNA, with no known treatment or cure. The cause of death in most cases of HGPS is progressive arterial occlusive disease, with death from heart attack or stroke occurring at an average age of 13 years (Varga et al. 2006). Treatment of human HGPS fibroblasts and mice lacking Lmna with rapamycin reverses HGPS phenotypes at the cellular level and promotes life span and health at the organismal level (Cao et al. 2011; Ramos et al. 2012). Although long-term treatment with rapamycin poses risks, the fatal nature of HGPS suggests that clinical trials of rapamycin in HGPS patients should be considered.
The majority of the data on the side effects of rapamycin have come from mice and from relatively sick humans, not from relatively healthy humans, and healthy humans might experience fewer serious side effects. The potential benefits of rapamycin are so large that trials in other mammals, which may be better models for humans, are getting underway. Rapamycin pharmacology studies in a nonhuman primate, the marmoset, show that rapamycin can be dosed to socially housed marmosets for more than a year without causing anemia, fibrotic lung changes, or mouth ulcers (Tardif et al. 2014). The effects on metabolism and immunity in marmosets, however, are as yet unknown. Also, these marmosets have been maintained in a relatively pathogen-free environment, not the relatively pathogen-rich environment in which humans live. To address some of these issues, a new study at the University of Washington will test the effect of rapamycin on aging phenotypes in pet dogs (Check Hayden 2014). Although these experiments have the potential to significantly advance our understanding of the real-world effects of rapamycin, they must be pursued cautiously, as negative outcomes, such as the development of diabetes in pet dogs, could taint the public perception of rapamycin as a pro-longevity intervention.
INTERMITTENT RAPAMYCIN TREATMENT: A WAY TO SIDESTEP SIDE EFFECTS
How can we use the exciting potential of rapamycin to reap the longevity dividend? Importantly, recent discoveries suggest that at least some of the negative side effects of rapamycin may be separable from its deleterious side effects. In particular, it was recently discovered that long-term treatment with rapamycin disrupts not only mTORC1, but also disrupts mTORC2 in vivo in multiple tissues, including the liver, white adipose tissue, and skeletal muscle (Lamming et al. 2012). Research from many laboratories has identified many positive roles for mTORC2 in health and longevity, and negative consequences from its disruption.
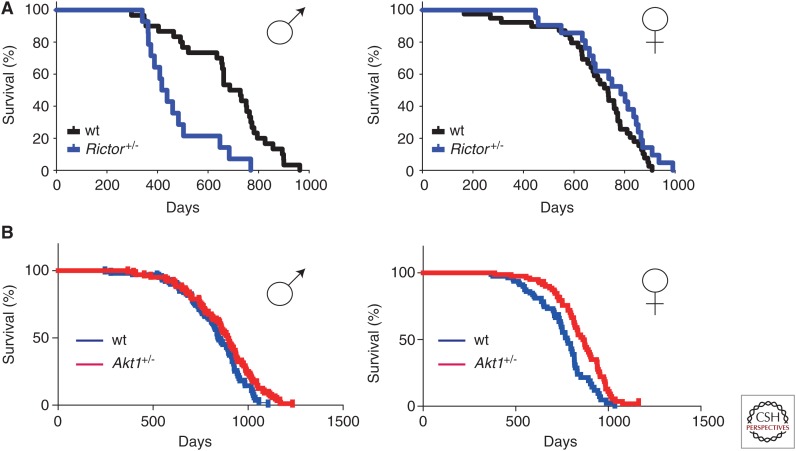
Specifically, hepatic mTORC2 is important for the regulation of gluconeogenesis, and disruption of hepatic mTORC2 by rapamycin causes hepatic insulin resistance and decreased glucose tolerance (Lamming et al. 2012, 2014a). mTORC2 is also important in the proper functioning and proliferation of β cells (Zahr et al. 2008; Yang et al. 2012). The critical role of mTORC2 in the immune system has only been recently uncovered, with mTORC2 playing a role in the function, proliferation, and differentiation of T cells, B cells, and macrophages (Haxhinasto et al. 2008; Maier et al. 2012; Powell et al. 2012; Byles et al. 2013). A significant decrease in T-cell number and the expansion of regulatory T cells (Tregs) are the likely cause of many of the effects of rapamycin on the immune system (Powell et al. 2012), and mTORC2 activity normal suppresses the differentiation of Tregs (Haxhinasto et al. 2008). Finally, mTORC2 is extremely important in male longevity, and genetic depletion of Rictor, a key component of mTORC2, severely shortens male, but not female, life span (Lamming et al. 2014b).
It is possible that this male-specific effect of mTORC2 inhibition on life span explains the sexually dimorphic impact of rapamycin and genetic inhibition of insulin/IGF-1/mTOR signaling pathway on life span, but the mechanistic basis for this sexually dimorphic effect remains unknown. Many of the physiological effects of hepatic Rictor deletion are mediated by reduced Akt activity; however, mice heterozygous for Rictor, although having decreased male longevity (Fig. 3A), have normal Akt activity (Lamming et al. 2014b). A recent article examining the life span of mice heterozygous for Akt1 found that these mice had a significant increase in life span (Fig. 3B) (Nojima et al. 2013). It is, therefore, likely that one or more additional mTORC2 substrates mediate the decreased male life span resulting from Rictor depletion. Although the role of the mTORC2 substrate SGK in mammalian life span is not known, recent findings in C. elegans suggest that SGK may play an important role in determining life span (Mizunuma et al. 2014).
Figure 3.
Haploinsufficiency of Rictor, but not Akt, significantly decreases male life span. (A) Kaplan–Meier plots showing life spans of male and female mice heterozygous for Rictor. (B) Kaplan–Meier survival plots showing life spans of mice heterozygous for Akt. (Panel A from Lamming et al. 2014b; reprinted, with permission, from the author. Panel B from Nojima et al. 2013; adapted, with permission, from the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium.)
Since many of the negative side effects of rapamycin are mediated by inhibition of mTORC2, drugs that specifically inhibit mTORC1 without inhibiting mTORC2 could allow us to realize the full power of rapamycin (Lamming et al. 2013). We have reported that the rapamycin analogs everolimus and temsirolimus have a decreased impact on glucose tolerance in male C57BL/6J mice, suggesting that these analogs may have a reduced impact on mTORC2, but this remains to be proven (Arriola Apelo et al. 2016). Unfortunately, although the scientific literature suggests several compounds specifically inhibit mTORC1, we have found that some of these results may be specific to particular cell lines or time points. Regrettably, the efforts of the pharmaceutical industry have been focused on the development of mTOR kinase inhibitors, such as Torin 1, PP242, KU63794, and WYE354 for the treatment of cancer (Liu et al. 2012). These inhibitors are extremely effective at inhibiting both mTOR complexes, and are therefore likely to have increased side effects as compared with rapamycin.
A more promising strategy is the possibility that intermittent rapamycin treatment might be sufficient to extend life span, while minimizing the time period that an individual might be immunosuppressed or at risk of diabetes. Recent findings that the effects of rapamycin on (at least) glucose tolerance and mTORC1 signaling can be washed out within a few weeks suggest this may be a feasible approach (Yang et al. 2012; Liu et al. 2014b). A fairly intensive dosing schedule (2 wk on, 2 wk off) extends the life span of inbred female 129/Sv mice (Anisimov et al. 2011), but this dosing schedule still leads to mice spending >50% of their lives exposed to rapamycin and subject to glucose intolerance, in addition to any other metabolic and immunological impacts. We recently identified an intermittent rapamycin treatment regimen with decreased metabolic and immunological effects (Arriola Apelo et al. 2016), but it remains to be determined whether this regimen can extend life span and health span.
SUSTAINABLE DIETARY INTERVENTIONS TO INHIBIT mTORC1
An alternative approach that has not been fully explored is the possibility of inhibiting mTORC1 by altering the diet. mTORC1, but not mTORC2, is specifically sensitive to glucose and amino acid levels among other stimuli (Bar-Peled and Sabatini 2014). Interventions that focus on either amino-acid sensing or on the availability of glucose and amino acids may act to inhibit mTORC1 signaling. A CR diet has been suggested to function in part by lowering fasting blood glucose levels, which is one of the most well-documented, reproducible, and widely conserved response to a CR diet in mammals (Lamming and Anderson 2014). Treatment with acarbose, a compound that acts to slow glucose uptake from food, has been shown to extend life span (Harrison et al. 2014), and it will be interesting to learn the effect of acarbose on mTORC1 activity.
mTORC1 activity in cultured cells is extremely sensitive to leucine (Long et al. 2005), and in rodents, the branched-chain amino acids—leucine, isoleucine, and valine—promote mTORC1 activity in the liver, skeletal muscle, adipose tissue, and the pancreas (Blomstrand et al. 2006; Sans et al. 2006; Li et al. 2011; Xiao et al. 2011). Consumption of leucine also significantly affects mTORC1 activity in humans (Moberg et al. 2014). A short-term protein-free diet leads to a significant decrease in mTORC1 signaling (Harputlugil et al. 2014), and we recently found that a low protein diet can inhibit mTORC1 signaling in both the tumors and somatic tissues of a mouse xenograft model (Lamming et al. 2015).
Recent studies have clearly shown that a low-protein diet significantly extends rodent life span and is associated with reduced cancer and mortality in humans (Levine et al. 2014; Solon-Biet et al. 2014), although it is not clear whether this effect is mediated by mTOR signaling. Low-protein diets may be an attractive and more sustainable alternative to CR in humans (Fontana and Partridge 2015). A CR diet is extremely difficult for humans in the developed world to maintain, surrounded by the sights and smells of abundant food. In contrast, vegan diets may be maintainable, and it has been suggested that such plant-based diets may be particularly low in methionine (McCarty et al. 2009), which when restricted significantly extends the life span of rodents (Anthony et al. 2013). Diets restricted in specific amino acids are often used in the treatment of inborn errors of metabolism, suggesting that diets with reduced dietary protein or specific amino acids may be a sustainable intervention for a large population.
CONCLUSION
There has been significant excitement over the discovery that rapamycin can prolong rodent life span and may be a potent antiaging drug. Although rapamycin is very promising, its side-effect profile may limit its clinical use for the treatment of diseases of aging in humans. Although it is still unclear how mTOR inhibition extends life span, it appears that many of the side effects are mediated by the “off-target” inhibition of mTORC2, whose beneficial effects are primarily mediated by inhibition of mTORC1. Alternative dosing strategies for rapamycin that limit its effects on mTORC2, or the development of compounds that specifically inhibit mTORC1, may allow us to fully realize the health, social, and economic benefits of slowed aging. While we await these developments, consumption of a low-protein diet may promote health, perhaps in part by inhibiting mTORC1 signaling.
ACKNOWLEDGMENTS
D.W.L. is supported in part by a K99/R00 Pathway to Independence Award from the National Institutes of Health/National Institute of Aging (NIH/NIA) (AG041765). This work was supported using facilities and resources from the William S. Middleton Memorial Veterans Hospital. This work does not represent the views of the Department of Veterans Affairs or the United States Government.
Footnotes
Editors: S. Jay Olshansky, George M. Martin, and James L. Kirkland
Additional Perspectives on Aging available at www.perspectivesinmedicine.org
REFERENCES
- Anisimov VN, Zabezhinski MA, Popovich IG, Piskunova TS, Semenchenko AV, Tyndyk ML, Yurova MN, Rosenfeld SV, Blagosklonny MV. 2011. Rapamycin increases lifespan and inhibits spontaneous tumorigenesis in inbred female mice. Cell Cycle 10: 4230–4236. [DOI] [PubMed] [Google Scholar]
- Anthony TG, Morrison CD, Gettys TW. 2013. Remodeling of lipid metabolism by dietary restriction of essential amino acids. Diabetes 62: 2635–2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriola Apelo SI, Neuman JC, Baar EL, Syed FA, Cummings NE, Brar HK, Pumper CP, Kimple ME, Lamming DW. 2016. Alternative rapamycin treatment regimens mitigate the impact of rapamycin on glucose homeostasis and the immune system. Aging Cell 15: 28–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM. 2014. Regulation of mTORC1 by amino acids. Trends Cell Biol 24: 400–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Peled L, Chantranupong L, Cherniack AD, Chen WW, Ottina KA, Grabiner BC, Spear ED, Carter SL, Meyerson M, Sabatini DM. 2013. A tumor suppressor complex with GAP activity for the Rag GTPases that signal amino acid sufficiency to mTORC1. Science 340: 1100–1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz C, Stracka D, Prescianotto-Baschong C, Frieden M, Demaurex N, Hall MN. 2013. Feature article: mTOR complex 2-Akt signaling at mitochondria-associated endoplasmic reticulum membranes (MAM) regulates mitochondrial physiology. Proc Natl Acad Sci 110: 12526–12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjedov I, Toivonen JM, Kerr F, Slack C, Jacobson J, Foley A, Partridge L. 2010. Mechanisms of life span extension by rapamycin in the fruit fly Drosophila melanogaster. Cell Metab 11: 35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomstrand E, Eliasson J, Karlsson HK, Kohnke R. 2006. Branched-chain amino acids activate key enzymes in protein synthesis after physical exercise. J Nutr 136: 269S–273S. [DOI] [PubMed] [Google Scholar]
- Byles V, Covarrubias AJ, Ben-Sahra I, Lamming DW, Sabatini DM, Manning BD, Horng T. 2013. The TSC-mTOR pathway regulates macrophage polarization. Nat Commun 4: 2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao K, Graziotto JJ, Blair CD, Mazzulli JR, Erdos MR, Krainc D, Collins FS. 2011. Rapamycin reverses cellular phenotypes and enhances mutant protein clearance in Hutchinson–Gilford progeria syndrome cells. Sci Transl Med 3: 89ra58. [DOI] [PubMed] [Google Scholar]
- Caron A, Richard D, Laplante M. 2015. The roles of mTOR complexes in lipid metabolism. Annu Rev Nutr 35: 321–348. [DOI] [PubMed] [Google Scholar]
- Chantranupong L, Wolfson RL, Orozco JM, Saxton RA, Scaria SM, Bar-Peled L, Spooner E, Isasa M, Gygi SP, Sabatini DM. 2014. The sestrins interact with GATOR2 to negatively regulate the amino-acid-sensing pathway upstream of mTORC1. Cell Rep 9: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Check Hayden E. 2014. Pet dogs set to test anti-ageing drug. Nature 514: 546. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu Y, Liu Y, Zheng P. 2009. mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci Signal 2: ra75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo AM, Bergamini E, Brunk UT, Droge W, Ffrench M, Terman A. 2005. Autophagy and aging: The importance of maintaining “clean” cells. Autophagy 1: 131–140. [DOI] [PubMed] [Google Scholar]
- Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, Chen T, Gu H, Djukovic D, Raftery D, et al. 2014. Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart. Aging Cell 13: 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake JC, Peelor FF III, Biela LM, Watkins MK, Miller RA, Hamilton KL, Miller BF. 2013. Assessment of mitochondrial biogenesis and mTORC1 signaling during chronic rapamycin feeding in male and female mice. J Gerontol A Biol Sci Med Sci 68: 1493–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok WC, Bokov A, Gelfond J, Yu Z, Zhang Y, Doderer M, Chen Y, Javors M, Wood WH III, Zhang Y, et al. 2014a. Combined treatment of rapamycin and dietary restriction has a larger effect on the transcriptome and metabolome of liver. Aging Cell 13: 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fok WC, Chen Y, Bokov A, Zhang Y, Salmon AB, Diaz V, Javors M, Wood WH III, Zhang Y, Becker KG, et al. 2014b. Mice fed rapamycin have an increase in lifespan associated with major changes in the liver transcriptome. PLoS ONE 9: e83988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, Partridge L. 2015. Promoting health and longevity through diet: From model organisms to humans. Cell 161: 106–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan X, Wang J, Wang C, Sommer E, Kozasa T, Srinivasula S, Alessi D, Offermanns S, Simon MI, Wu D. 2012. PRR5L degradation promotes mTORC2-mediated PKC-δ phosphorylation and cell migration downstream of Gα12. Nat Cell Biol 14: 686–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Martinez JM, Alessi DR. 2008. mTOR complex 2 (mTORC2) controls hydrophobic motif phosphorylation and activation of serum- and glucocorticoid-induced protein kinase 1 (SGK1). Biochem J 416: 375–385. [DOI] [PubMed] [Google Scholar]
- Garelick MG, Mackay VL, Yanagida A, Academia EC, Schreiber KH, Ladiges WC, Kennedy BK. 2013. Chronic rapamycin treatment or lack of S6K1 does not reduce ribosome activity in vivo. Cell Cycle 12: 2493–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharibi B, Farzadi S, Ghuman M, Hughes FJ. 2014. Inhibition of Akt/mTOR attenuates age-related changes in mesenchymal stem cells. Stem Cells 32: 2256–2266. [DOI] [PubMed] [Google Scholar]
- Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, Jordan A, Beck AH, Sabatini DM. 2014. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov 4: 554–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. 2006. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKC-α, but not S6K1. Dev Cell 11: 859–871. [DOI] [PubMed] [Google Scholar]
- Han X, Turdi S, Hu N, Guo R, Zhang Y, Ren J. 2012. Influence of long-term caloric restriction on myocardial and cardiomyocyte contractile function and autophagy in mice. J Nutr Biochem 23: 1592–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. 2007. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6: 95–110. [DOI] [PubMed] [Google Scholar]
- Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. 2008. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS Genet 4: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harputlugil E, Hine C, Vargas D, Robertson L, Manning BD, Mitchell JR. 2014. The TSC complex is required for the benefits of dietary protein restriction on stress resistance in vivo. Cell Rep 8: 1160–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. 2009. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature 460: 392–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Allison DB, Ames BN, Astle CM, Atamna H, Fernandez E, Flurkey K, Javors MA, Nadon NL, et al. 2014. Acarbose, 17-α-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell 13: 273–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haxhinasto S, Mathis D, Benoist C. 2008. The AKT–mTOR axis regulates de novo differentiation of CD4+Foxp3+ cells. J Exp Med 205: 565–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Negrete I, Carretero-Ortega J, Rosenfeldt H, Hernandez-Garcia R, Calderon-Salinas JV, Reyes-Cruz G, Gutkind JS, Vazquez-Prado J. 2007. P-Rex1 links mammalian target of rapamycin signaling to Rac activation and cell migration. J Biol Chem 282: 23708–23715. [DOI] [PubMed] [Google Scholar]
- Holzenberger M, Dupont J, Ducos B, Leneuve P, Geloen A, Even PC, Cervera P, Le Bouc Y. 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421: 182–187. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. 2011. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 332: 1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Dibble CC, Matsuzaki M, Manning BD. 2008. The TSC1-TSC2 complex is required for proper activation of mTOR complex 2. Mol Cell Biol 28: 4104–4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglesias-Bartolome R, Patel V, Cotrim A, Leelahavanichkul K, Molinolo AA, Mitchell JB, Gutkind JS. 2012. mTOR inhibition prevents epithelial stem cell senescence and protects from radiation-induced mucositis. Cell Stem Cell 11: 401–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenoue T, Inoki K, Yang Q, Zhou X, Guan KL. 2008. Essential function of TORC2 in PKC and Akt turn motif phosphorylation, maturation and signalling. Embo J 27: 1919–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston O, Rose CL, Webster AC, Gill JS. 2008. Sirolimus is associated with new-onset diabetes in kidney transplant recipients. J Am Soc Nephrol 19: 1411–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Powers RW III, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. 2005. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310: 1193–1196. [DOI] [PubMed] [Google Scholar]
- Kang SA, Pacold ME, Cervantes CL, Lim D, Lou HJ, Ottina K, Gray NS, Turk BE, Yaffe MB, Sabatini DM. 2013. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science 341: 1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. 2004. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol 14: 885–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna N, Fang Y, Yoon MS, Chen J. 2013. XPLN is an endogenous inhibitor of mTORC2. Proc Natl Acad Sci 110: 15979–15984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kundu M, Viollet B, Guan KL. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol 13: 132–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SJ, DeStefano MA, Oh WJ, Wu CC, Vega-Cotto NM, Finlan M, Liu D, Su B, Jacinto E. 2012. mTOR complex 2 regulates proper turnover of insulin receptor substrate-1 via the ubiquitin ligase subunit Fbw8. Mol Cell 48: 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruegel U, Robison B, Dange T, Kahlert G, Delaney JR, Kotireddy S, Tsuchiya M, Tsuchiyama S, Murakami CJ, Schleit J, et al. 2011. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet 7: e1002253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume S, Uzu T, Horiike K, Chin-Kanasaki M, Isshiki K, Araki S, Sugimoto T, Haneda M, Kashiwagi A, Koya D. 2010. Calorie restriction enhances cell adaptation to hypoxia through Sirt1-dependent mitochondrial autophagy in mouse aged kidney. J Clin Invest 120: 1043–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Anderson RM. 2014. Metabolic effects of caloric restriction. eLS. Wiley, Chichester, NY. [Google Scholar]
- Lamming DW, Medvedik O, Kim KD, Sinclair DA. 2007. Calorie restriction and TOR signaling converge on sirtuin-mediated lifespan extension in Saccharomyces cerevisiae. Age 29: 118–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, et al. 2012. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335: 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Sabatini DM, Baur JA. 2013. Rapalogs and mTOR inhibitors as anti-aging therapeutics. J Clin Invest 123: 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Demirkan G, Boylan JM, Mihaylova MM, Peng T, Ferreira J, Neretti N, Salomon A, Sabatini DM, Gruppuso PA. 2014a. Hepatic signaling by the mechanistic target of rapamycin complex 2 (mTORC2). FASEB J 28: 300–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Mihaylova MM, Katajisto P, Baar EL, Yilmaz OH, Hutchins A, Gultekin Y, Gaither R, Sabatini DM. 2014b. Depletion of rictor, an essential protein component of mTORC2, decreases male lifespan. Aging Cell 13: 911–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamming DW, Cummings NE, Rastelli AL, Gao F, Cava E, Bertozzi B, Spelta F, Pili R, Fontana L. 2015. Restriction of dietary protein decreases mTORC1 in tumors and somatic tissues of a tumor-bearing mouse xenograft model. Oncotarget 6: 31233–31240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine ME, Suarez JA, Brandhorst S, Balasubramanian P, Cheng CW, Madia F, Fontana L, Mirisola MG, Guevara-Aguirre J, Wan J, et al. 2014. Low protein intake is associated with a major reduction in IGF-1, cancer, and overall mortality in the 65 and younger but not older population. Cell Metab 19: 407–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Gao T. 2014. mTORC2 phosphorylates protein kinase C to regulate its stability and activity. EMBO Rep 15: 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Yin Y, Tan B, Kong X, Wu G. 2011. Leucine nutrition in animals and humans: mTOR signaling and beyond. Amino Acids 41: 1185–1193. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kirubakaran S, Hur W, Niepel M, Westover K, Thoreen CC, Wang J, Ni J, Patricelli MP, Vogel K, et al. 2012. Kinome-wide selectivity profiling of ATP-competitive mammalian target of rapamycin (mTOR) inhibitors and characterization of their binding kinetics. J Biol Chem 287: 9742–9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Wang Z, Wei W. 2014a. Phosphorylation of Akt at the C-terminal tail triggers Akt activation. Cell Cycle 13: 2162–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Diaz V, Fernandez E, Strong R, Ye L, Baur JA, Lamming DW, Richardson A, Salmon AB. 2014b. Rapamycin-induced metabolic defects are reversible in both lean and obese mice. Aging (Albany NY) 6: 742–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X, Ortiz-Vega S, Lin Y, Avruch J. 2005. Rheb binding to mammalian target of rapamycin (mTOR) is regulated by amino acid sufficiency. J Biol Chem 280: 23433–23436. [DOI] [PubMed] [Google Scholar]
- Mahe E, Morelon E, Lechaton S, Sang KH, Mansouri R, Ducasse MF, Mamzer-Bruneel MF, de Prost Y, Kreis H, Bodemer C. 2005. Cutaneous adverse events in renal transplant recipients receiving sirolimus-based therapy. Transplantation 79: 476–482. [DOI] [PubMed] [Google Scholar]
- Maier E, Duschl A, Horejs-Hoeck J. 2012. STAT6-dependent and -independent mechanisms in Th2 polarization. Eur J Immunol 42: 2827–2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder S, Caccamo A, Medina DX, Benavides AD, Javors MA, Kraig E, Strong R, Richardson A, Oddo S. 2012. Lifelong rapamycin administration ameliorates age-dependent cognitive deficits by reducing IL-1β and enhancing NMDA signaling. Aging Cell 11: 326–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty MF, Barroso-Aranda J, Contreras F. 2009. The low-methionine content of vegan diets may make methionine restriction feasible as a life extension strategy. Med Hypotheses 72: 125–128. [DOI] [PubMed] [Google Scholar]
- McCormack FX, Inoue Y, Moss J, Singer LG, Strange C, Nakata K, Barker AF, Chapman JT, Brantly ML, Stocks JM, et al. 2011. Efficacy and safety of sirolimus in lymphangioleiomyomatosis. N Engl J Med 364: 1595–1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menon S, Dibble CC, Talbott G, Hoxhaj G, Valvezan AJ, Takahashi H, Cantley LC, Manning BD. 2014. Spatial control of the TSC complex integrates insulin and nutrient regulation of mTORC1 at the lysosome. Cell 156: 771–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mieulet V, Roceri M, Espeillac C, Sotiropoulos A, Ohanna M, Oorschot V, Klumperman J, Sandri M, Pende M. 2007. S6 kinase inactivation impairs growth and translational target phosphorylation in muscle cells maintaining proper regulation of protein turnover. Am J Physiol Cell Physiol 293: C712–C722. [DOI] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Rodriguez Boyd A, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, et al. 2011a. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci 66A: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Baur JA, Boyd AR, de Cabo R, Fernandez E, Flurkey K, Javors MA, Nelson JF, et al. 2011b. Rapamycin, but not resveratrol or simvastatin, extends life span of genetically heterogeneous mice. J Gerontol A Biol Sci Med Sci 66: 191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RA, Harrison DE, Astle CM, Fernandez E, Flurkey K, Han M, Javors MA, Li X, Nadon NL, Nelson JF, et al. 2014. Rapamycin-mediated lifespan increase in mice is dose and sex dependent and metabolically distinct from dietary restriction. Aging Cell 13: 468–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma M, Neumann-Haefelin E, Moroz N, Li Y, Blackwell TK. 2014. mTORC2-SGK-1 acts in two environmentally responsive pathways with opposing effects on longevity. Aging Cell 13: 869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moberg M, Apro W, Ohlsson I, Ponten M, Villanueva A, Ekblom B, Blomstrand E. 2014. Absence of leucine in an essential amino acid supplement reduces activation of mTORC1 signalling following resistance exercise in young females. Appl Physiol Nutr Metab 39: 183–194. [DOI] [PubMed] [Google Scholar]
- Neff F, Flores-Dominguez D, Ryan DP, Horsch M, Schroder S, Adler T, Afonso LC, Aguilar-Pimentel JA, Becker L, Garrett L, et al. 2013. Rapamycin extends murine lifespan but has limited effects on aging. J Clin Invest 123: 3272–3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima A, Yamashita M, Yoshida Y, Shimizu I, Ichimiya H, Kamimura N, Kobayashi Y, Ohta S, Ishii N, Minamino T. 2013. Haploinsufficiency of akt1 prolongs the lifespan of mice. PLoS ONE 8: e69178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary MN, Schreiber KH, Zhang Y, Duc AC, Rao S, Hale JS, Academia EC, Shah SR, Morton JF, Holstein CA, et al. 2013. The ribosomal protein Rpl22 controls ribosome composition by directly repressing expression of its own paralog, Rpl22l1. PLoS Genet 9: e1003708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovich IG, Anisimov VN, Zabezhinski MA, Semenchenko AV, Tyndyk ML, Yurova MN, Blagosklonny MV. 2014. Lifespan extension and cancer prevention in HER-2/neu transgenic mice treated with low intermittent doses of rapamycin. Cancer Biol Therapy 15: 586–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JD, Pollizzi KN, Heikamp EB, Horton MR. 2012. Regulation of immune responses by mTOR. Annu Rev Immunol 30: 39–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers RW III, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. 2006. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev 20: 174–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pusceddu S, Tessari A, Testa I, Procopio G. 2014. Everolimus in advanced solid tumors: When to start, early or late? Tumori 100: 2e–3e. [DOI] [PubMed] [Google Scholar]
- Ramos FJ, Chen SC, Garelick MG, Dai DF, Liao CY, Schreiber KH, MacKay VL, An EH, Strong R, Ladiges WC, et al. 2012. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci Transl Med 4: 144ra103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. 2012. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell Metab 15: 713–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KA, Edrey YH, Osmulski P, Gaczynska M, Buffenstein R. 2012. Altered composition of liver proteasome assemblies contributes to enhanced proteasome activity in the exceptionally long-lived naked mole-rat. PLoS ONE 7: e35890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez KA, Dodds SG, Strong R, Galvan V, Sharp ZD, Buffenstein R. 2014. Divergent tissue and sex effects of rapamycin on the proteasome-chaperone network of old mice. Front Mol Neurosci 7: 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saci A, Cantley LCt, Carpenter CL. 2011. Rac1 regulates the activity of mTORC1 and mTORC2 and controls cellular size. Mol Cell 42: 50–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. 2010. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans MD, Tashiro M, Vogel NL, Kimball SR, D’Alecy LG, Williams JA. 2006. Leucine activates pancreatic translational machinery in rats and mice through mTOR independently of CCK and insulin. J Nutr 136: 1792–1799. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell 22: 159–168. [DOI] [PubMed] [Google Scholar]
- Selman C, Tullet JM, Wieser D, Irvine E, Lingard SJ, Choudhury AI, Claret M, Al-Qassab H, Carmignac D, Ramadani F, et al. 2009. Ribosomal protein S6 kinase 1 signaling regulates mammalian life span. Science 326: 140–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman C, Partridge L, Withers DJ. 2011. Replication of extended lifespan phenotype in mice with deletion of insulin receptor substrate 1. PLoS ONE 6: e16144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto B. 2012. Rapamycin and mTOR: A serendipitous discovery and implications for breast cancer. Clin Trans Med 1: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Zoncu R, Medina DL, Vetrini F, Erdin S, Erdin S, Huynh T, Ferron M, Karsenty G, Vellard MC, et al. 2012. A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J 31: 1095–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solon-Biet SM, McMahon AC, Ballard JW, Ruohonen K, Wu LE, Cogger VC, Warren A, Huang X, Pichaud N, Melvin RG, et al. 2014. The ratio of macronutrients, not caloric intake, dictates cardiometabolic health, aging, and longevity in ad libitum-fed mice. Cell Metab 19: 418–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spilman P, Podlutskaya N, Hart MJ, Debnath J, Gorostiza O, Bredesen D, Richardson A, Strong R, Galvan V. 2010. Inhibition of mTOR by rapamycin abolishes cognitive deficits and reduces amyloid-beta levels in a mouse model of Alzheimer’s disease. PLoS ONE 5: e9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, Tchao BN, et al. 2008. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 133: 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syntichaki P, Troulinaki K, Tavernarakis N. 2007. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature 445: 922–926. [DOI] [PubMed] [Google Scholar]
- Tao R, Xiong X, Liangpunsakul S, Dong XC. 2014. Sestrin 3 protein enhances hepatic insulin sensitivity by direct activation of the mTORC2-Akt signaling. Diabetes 64: 1211–1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardif S, Ross C, Bergman P, Fernandez E, Javors M, Salmon A, Spross J, Strong R, Richardson A. 2014. Testing efficacy of administration of the antiaging drug rapamycin in a nonhuman primate, the common marmoset. J Gerontol A Biol Sci Med Sci 70: 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485: 109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga R, Eriksson M, Erdos MR, Olive M, Harten I, Kolodgie F, Capell BC, Cheng J, Faddah D, Perkins S, et al. 2006. Progressive vascular smooth muscle cell defects in a mouse model of Hutchinson–Gilford progeria syndrome. Proc Natl Acad Sci 103: 3250–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. 2003. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature 426: 620. [DOI] [PubMed] [Google Scholar]
- Vezina C, Kudelski A, Sehgal SN. 1975. Rapamycin (AY-22,989), a new antifungal antibiotic. I: Taxonomy of the producing streptomycete and isolation of the active principle. J Antibiot (Tokyo) 28: 721–726. [DOI] [PubMed] [Google Scholar]
- Wagle N, Grabiner BC, Van Allen EM, Hodis E, Jacobus S, Supko JG, Stewart M, Choueiri TK, Gandhi L, Cleary JM, et al. 2014. Activating mTOR mutations in a patient with an extraordinary response on a phase I trial of everolimus and pazopanib. Cancer discovery 4: 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E, Karp C, Strohecker AM, Guo Y, Mathew R. 2010. Role of autophagy in suppression of inflammation and cancer. Curr Opin Cell Biol 22: 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JE, Burmeister L, Brooks SV, Chan CC, Friedline S, Harrison DE, Hejtmancik JF, Nadon N, Strong R, Wood LK, et al. 2012. Rapamycin slows aging in mice. Aging Cell 11: 675–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F, Huang Z, Li H, Yu J, Wang C, Chen S, Meng Q, Cheng Y, Gao X, Li J, et al. 2011. Leucine deprivation increases hepatic insulin sensitivity via GCN2/mTOR/S6K1 and AMPK pathways. Diabetes 60: 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SB, Lee HY, Young DM, Tien AC, Rowson-Baldwin A, Shu YY, Jan YN, Jan LY. 2012. Rapamycin induces glucose intolerance in mice by reducing islet mass, insulin content, and insulin sensitivity. J Mol Med (Berl) 90: 575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz OH, Katajisto P, Lamming DW, Gultekin Y, Bauer-Rowe KE, Sengupta S, Birsoy K, Dursun A, Yilmaz VO, Selig M, et al. 2012. mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahr E, Molano RD, Pileggi A, Ichii H, San Jose S, Bocca N, An W, Gonzalez-Quintana J, Fraker C, Ricordi C, et al. 2008. Rapamycin impairs β-cell proliferation in vivo. Transplant Proc 40: 436–437. [DOI] [PubMed] [Google Scholar]
- Zhang C, Cuervo AM. 2008. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat Med 14: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bokov A, Gelfond J, Soto V, Ikeno Y, Hubbard G, Diaz V, Sloane L, Maslin K, Treaster S, et al. 2014. Rapamycin extends life and health in C57BL/6 mice. J Gerontol A Biol Sci Med Sci 69: 119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 2009. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell 139: 149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinzalla V, Stracka D, Oppliger W, Hall MN. 2011. Activation of mTORC2 by association with the ribosome. Cell 144: 757–768. [DOI] [PubMed] [Google Scholar]