Abstract
The inheritance of information beyond DNA sequence, known as epigenetic inheritance, has been implicated in a multitude of biological processes from control of plant flowering time to cancer in humans. In addition to epigenetic inheritance that occurs in dividing cells of a multicellular organism, it is also increasingly clear that at least some epigenetic information is transmitted via the gametes in a multitude of organisms, including mammals. Here, I review the evidence for epigenetic information carriers in mammalian sperm, and explore the emerging field of intergenerational transfer of environmental information.
A male’s experience and environment (e.g., diet or stress) can influence the cytosine methylation patterns, chromatin structure, and RNA populations of his sperm. These changes may, in turn, affect the phenotypes of his offspring.
In sexual organisms, information transfer from one generation to the next—inheritance—is primarily mediated by the merging of two half complements of the genetic material, DNA. That the vast majority of heritable information is encoded in the DNA sequence is shown by the spectacular success of the enterprise of genetics. Nonetheless, two considerations make it clear that a small amount of information is passed from one generation to the next in the absence of DNA sequence changes. This is known as “epigenetic” inheritance (Jablonka and Lamb 2002). First, genetically identical organisms—human twins, inbred mouse strains, and so forth—can nonetheless show extensive phenotypic variation, and a subset of this variability is heritable. This is known as epivariation, and is extremely well-documented in many model organisms—particularly plants—but has also been reliably documented for at least a few phenotypes in mammals (Daxinger and Whitelaw 2012; Rando 2012). Second, although genome-wide association studies (GWAS) can successfully identify a multitude of genetic contributions to complex traits, at present it is believed that all significant sequence polymorphisms identified in a given GWAS can only explain a small fraction of the overall heritability of the trait in question. Of course, this “missing heritability” could be explained by DNA sequence in many ways—by a multitude of rare polymorphisms, by epistasis (genetic interactions between polymorphisms that do not conform to the linear additivity model used to identify hits and calculate variance explained in GWAS surveys), etc.—but in some cases there is evidence that ancestral environmental conditions contribute to the propensity for certain complex diseases, including diabetes and schizophrenia (Rakyan et al. 2011). In other words, although the contribution of epigenetics to mammalian inheritance is clearly minor relative to genetic inheritance, its relevance to complex heritable diseases is currently unknown and is likely to be underestimated by the scientific community based on the fact that epigenetic inheritance plays no role in the far better-understood case of Mendelian traits. Moreover, the unusual properties of epigenetic information make such information transfer interesting both mechanistically and teleologically (Jablonka and Lamb 1995; Rando and Verstrepen 2007). In this work, I will survey evidence for epigenetic information carriers in mammalian sperm.
THE MOLECULAR CARRIERS OF EPIGENETIC INFORMATION
Epigenetic inheritance is far more robust and widespread in key model organisms such as worms, fission yeast, and plants than it is in mammals (Rando and Verstrepen 2007). As a result, genetic dissection of epigenetic inheritance pathways has largely been performed in these organisms, with efforts in mammalian systems generally lagging for reasons of (1) convenience—the generation time of the mouse is inconveniently long relative to that of worms, and (2) biology—mammals lack key enzymes, such as RNA-dependent RNA polymerase, that make epigenetic inheritance in many model systems far more robust and long-lived. I will therefore introduce each epigenetic information carrier using evidence from model systems and then will turn to the relevant evidence from mammalian studies.
Five of the best-characterized epigenetic information carriers are transcription factors, prions, cytosine methylation, chromatin structure, and RNAs. As there is presently little evidence supporting transcription factors or prion states in intergenerational mammalian inheritance—prion diseases such as kuru appear not to be vertically transmitted, for example—I will focus this review on the remaining three “epigenomes” listed.
CYTOSINE METHYLATION
In addition to the four canonical nucleotides—A, G, C, and T—the DNA of many organisms also carries a substantial fraction of alternative bases. In eukaryotes, the most common and best-studied of these alternative bases is 5-methyl cytosine (5meC) (Jaenisch and Bird 2003). Cytosine methylation is a widespread base modification, occurring in many plants, fungi, and animals, but is absent, or nearly so, in several of the best-studied model organisms such as budding yeast, fission yeast, fruit flies, and nematode worms. In mammals, the majority of cytosine methylation occurs in the symmetric context of a CpG dinucleotide, although many organisms carry substantially more 5meC in asymmetric contexts such as CHH (where H is A, T, or C) and CHG (Reik et al. 2001; Chan et al. 2005; Law and Jacobsen 2010; Cedar and Bergman 2012). Cytosine methylation has garnered the greatest interest in mammals as an epigenetic information carrier, as it is associated with a clear copying mechanism. The “maintenance” methyltransferase, Dnmt1, preferentially acts on the hemimethylated CG dinucleotides found after replication of a symmetrically methylated CG duplex (Bestor 1992). Cytosine methylation is implicated in wide variety of biological processes in mammals, garnering particular attention for its roles in oncogenesis and in allele-specific gene expression, or imprinting.
Two types of heritable cytosine methylation have been described—“programmed” cases such as found in imprinted gene expression, and apparently random “epivariation” as occurs at the Avy locus in mouse. Examples of programmed cytosine methylation being inherited through the gametes involve a number of genomic loci associated with genes that are monoallelically expressed solely from either the maternal or the paternal allele (Bartolomei and Ferguson-Smith 2011). These genomic loci are typically associated with a “differentially methylated region” (DMR), which is only methylated on the genomic copy that was passed down, for example, via the oocyte. Most individuals will therefore carry a single methylated copy of this locus—in this case the maternal allele—along with the unmethylated paternal copy of the same locus. DMRs for imprinted loci can be stably inherited mitotically. An imprint established during female or male germline development can be maintained in children, on the maternal or the paternal allele respectively, into adulthood, but are reliably erased in the child’s germline. Thus, imprinted loci represent well-established cases in which information beyond genomic sequence alone is transmitted from parent to child. This information transfer is generally fairly stereotyped, as imprinting disorders are rare and result most commonly from genetic disorders in which deletion of one copy of an imprinted gene results in disease attributed to the lack of expression of the intact copy of the gene. That said, there is some evidence supporting the idea that epigenetic marks on imprinted genes are modestly susceptible to environmental perturbations. For example, assisted reproductive technologies have been linked to an increase in imprinting disorders (de Waal et al. 2012). Hypomorphic alleles of the maintenance methyltransferase Dnmt1 also show epigenetic defects at a specific subset of imprinted loci (Biniszkiewicz et al. 2002), supporting the idea that cytosine methylation at imprinted DMRs could plausibly be susceptible to dietary or environmental control.
Cytosine methylation also plays a role in “epivariation” in which heritable phenotypic differences are observed between genetically identical organisms. Epivariation is widespread in plants, with examples including variability in maize pigmentation caused by paramutation at the B locus (Arteaga-Vazquez and Chandler 2010), and variation in Arabidopsis flowering phenotypes owing to heritable silencing of the SUPERMAN locus (Jacobsen and Meyerowitz 1997). Genetic studies of epigenetic silencing of these and other loci in plants have implicated a number of epigenetic information carriers including small RNAs, chromatin packaging, and cytosine methylation in heritable silencing. Although epivariation is far more widespread in plants than in mammals, a handful of well-documented cases of mammalian epivariation have been described. The best-studied example of epivariation in mice is found in a mouse mutant, called Agouti variable yellow, or Avy, in which an intracisternal A particle (IAP) retro-element has inserted adjacent to the gene encoding transcription factor Agouti, which plays a role in coat coloration (Morgan et al. 1999). Avy mice have coat colors ranging from brown to yellow, and this coat color variation is somewhat heritable—yellow mothers give birth to more yellow than brown pups, whereas brown mothers give birth to a greater fraction of brown pups. Epivariation in coat color is correlated with the extent of cytosine methylation surrounding the IAP element, with wider domains of methylation being linked to decreased expression of the downstream gene.
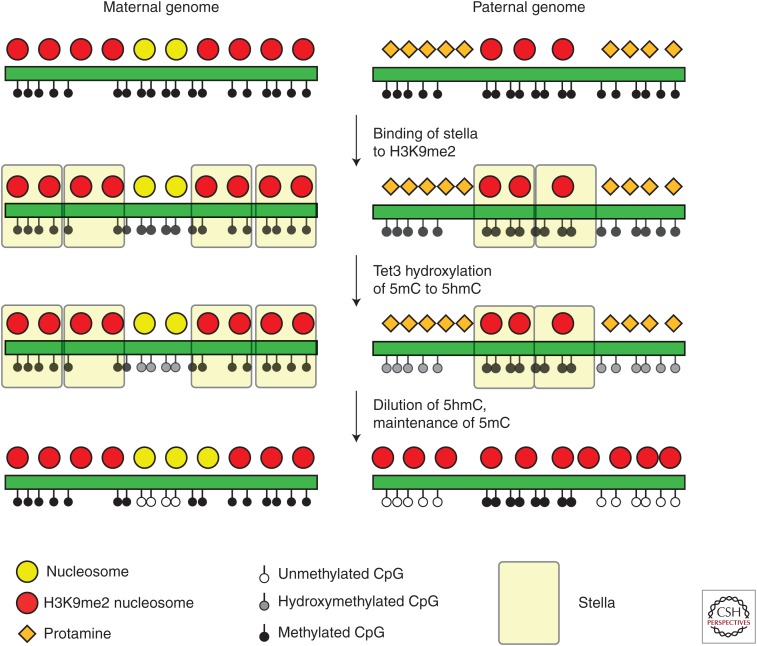
Global patterns of cytosine methylation are massively reconfigured on fertilization in mammals, with nearly global erasure of cytosine methylation on the paternal genome occurring within the first one or two cleavage divisions. This finding raises the question of how any information could be transmitted from father to offspring using cytosine methylation. The mechanistic basis for global demethylation remains incompletely understood, but recent findings implicate active erasure by oxidation of paternal 5meC by the Tet3 demethylase as a major mechanism for this erasure (Inoue and Zhang 2011; Iqbal et al. 2011; Wossidlo et al. 2011). The maternal epigenome is protected from this erasure by a factor variously known as Stella/Dppa3/PGC7, which associates specifically with the maternal, but not the paternal, genome via its association with H3K9me2-marked nucleosomes (Nakamura et al. 2007, 2012). As the paternal genome is primarily packaged in protamines rather than histones (see below), the majority of the paternal genome cannot recruit Stella/Dppa3/PGC7 and thus is susceptible to Tet3-mediated demethylation. A small fraction of the paternal genome does remain associated with histones, including loci at which methylated imprints are contributed paternally (Fig. 1). This suggests that the rare loci that retain histones in sperm may represent “windows” in which the paternal epigenome may be passed on to offspring in mammals.
Figure 1.
Model for epigenetic events occurring immediately after fertilization. Schema shows the maternal genome (left) and paternal genome (right). The maternal genome is maintained in chromatin, whereas the paternal genome is primarily packaged into protamines, with a small fraction of genomic regions remaining associated with histones. After fertilization, binding of Stella to H3K9me2 protects underlying genomic regions from Tet3, which converts methylcytosine to hydroxymethylcytosine. Regions subject to hydroxymethylation are then passively demethylated during replication, resulting in near-global erasure of paternal cytosine methylation patterns. The maternal genome and a small fraction of the paternal genome are protected from this initial demethylation event because of their association with H3K9me2-marked histones.
CHROMATIN PACKAGING
Eukaryotic genomes are packaged into a nucleoprotein complex known as chromatin. The major repeating subunit of chromatin consists of an octamer of histone proteins around which are wrapped 147 bp of DNA (Kornberg and Lorch 1999). Chromatin structure is genetically implicated in mitotic epigenetic inheritance paradigms such as cell state inheritance. Early genetic screens in flies for mutations that interfere with inheritance of active or repressed gene expression states yielded a number of chromatin regulatory factors—the trithorax or polycomb group factors, respectively (Kennison 1995). Despite decades of study of the function and roles of chromatin regulators in cell state inheritance, the question of whether and how chromatin states are propagated during replication remains to be unambiguously answered (Kaufman and Rando 2010).
More complicated still than mitotic inheritance, chromatin state propagation across generations of a multicellular organism requires surviving the dramatic perturbations involved in gametogenesis, fertilization, and early development. In contrast to all other cell types, sperm carry a highly specialized packaging state of the genome to allow the exceptional compaction that is typically observed in the sperm nucleus (Ooi and Henikoff 2007). During mammalian spermatogenesis, histones become hyperacetylated and then evicted (Rousseaux et al. 2005), replaced by so-called transition proteins, which are then in turn replaced by small basic proteins known as protamines (Balhorn 2007). However, a small subset of histone proteins is retained in mammalian sperm. Human sperm carry ∼10%–15% of the histone complement of a somatic cell, whereas in mouse this number is ∼2%. Recent studies on histone retention in human and mouse sperm suggests that there is a bias for promoters of genes expressed early during development to be specifically packaged in histones (Gardiner-Garden et al. 1998; Arpanahi et al. 2009; Hammoud et al. 2009; Brykczynska et al. 2010; Erkek et al. 2013). These findings contrast with several lines of evidence suggesting that histone retention in sperm primarily occurs over repeat elements. Small-scale cloning of DNA released by nuclease digestion of sperm revealed primarily repeat elements such as LINE and SINE sequences (Pittoggi et al. 1999) and pericentric repeats (Govin et al. 2007), whereas immunostaining studies on mature sperm reveal colocalization of histone proteins with the repeat-enriched sperm chromocenter (van der Heijden et al. 2006; Govin et al. 2007). More recently, genome-wide assays for mononucleosome retention in several mammals have reported histone retention over broad gene-poor domains, which are enriched for repeat elements (Carone et al. 2014; Samans et al. 2014), and relative depletion over promoters.
These discrepancies argue that assays for sperm histone retention are sensitive to the precise protocol used for identifying histone-enriched loci. Nonetheless, the contrasting views of histone retention are not incompatible, and are most consistent with the idea that two or more biochemically distinct populations of nucleosomes are present in mature sperm. In this view, the majority of nucleosome retention occurs over long gene-poor regions of the genome, and likely is comprised of nucleosomes carrying the canonical H3.1 molecule marked with H3K9 methylation. These histones represent the majority of histone retention in sperm, consistent with the major histone immunofluorescence signal associated with the DAPI-dense chromocenter of sperm. In contrast, the nucleosomes associated with developmental promoters are biochemically unusual. They are highly resistant to micrococcal nuclease and are only revealed after nuclease overdigestion, and they carry the H3.3 histone variant, marked with H3K4me3 and H3K27me3 (Erkek et al. 2013).
It remains to be seen whether these different classes of nucleosome carry meaningful information into the zygote. In one study, however, changes in histone retention at promoters in mutant animals was seen to correlate with changes in embryonic gene activation at the promoters in question (Ihara et al. 2014). In addition, there is evidence for transgenerational effects of chromatin-related mutations in several model organisms. Most notably, mutations in the H3K4 methylation machinery have transgenerational effects in worms, with mutations in the H3K4 methylase leading to several generations of worms with increased lifespan (Greer et al. 2011) and mutations in the H3K4 demethylase LSD1 resulting in sterility manifesting some 20 generations after introduction of the mutation (Katz et al. 2009). Whether the information transmitted in these systems is carried in the histone proteins or results from altered RNA populations observed in chromatin mutants is unknown.
RNAs
In one of their seminal papers on gene regulation, Jacob and Monod (1961) suggested the possibility that RNA molecules would be ideal candidates for gene regulators. Although the repressor identified for the lac operon proved to be a protein, the past two decades have seen an explosion in roles for RNA in gene regulation. Long intergenic noncoding RNAs (lincRNAs), exemplified by Xist, play diverse roles in gene regulation, from chromosome-wide gene silencing to recruiting chromatin regulators to specific genomic regulatory elements (Rinn and Chang 2012). lincRNAs are often among the transcripts expressed from imprinted domains in mammals, the most famous being the H19 RNA involved in Prader–Willi and Angelman syndromes. More likely to be involved in transgenerational systems are a large variety of smaller RNA species. Here I will focus on RNAs 40 nt and shorter. These include relatively well-studied species such as microRNAs, siRNAs, and the strongly germline-enriched piRNAs (Ghildiyal and Zamore 2009), as well as more mysterious entities such as enhancer-derived RNAs (eRNAs) and tRNA fragments (tRFs) (Peng et al. 2012).
Small RNAs are implicated in a large number of transgenerational epigenetic inheritance paradigms, largely in nonmammalian model organisms. Examples of such models include, among others: (1) RNA interference in Caenorhabditis elegans in which injection of double-stranded RNA results in silencing of target genes for ∼4–5 generations (Fire et al. 1998); (2) paramutation in maize in which an inactive copy of a plant pigmentation gene induces heritable silencing of an active copy in a heterozygote (Arteaga-Vazquez and Chandler 2010); (3) epigenetic repression of FWA and SUPERMAN and many other genes in Arabidopsis (Chan et al. 2004); (4) epigenetic silencing of pericentromeric genes in fission yeast (Grewal 2010); and (5) silencing of transposons in the Drosophila germline (Khurana et al. 2011). Most of these cases occur in organisms whose genome encodes an RNA-dependent RNA polymerase, providing a mechanism by which an initiating signal can be maintained over multiple organismal generations. The detailed mechanisms underlying transgenerational passage of information in these paradigms remain a subject of intensive investigation and are complicated to some extent by extensive cross talk between different “epigenomes.” For example, small RNAs have been shown to play a role in directing deposition of heterochromatic histone modifications in all the organisms detailed above, and in a subset of organisms small RNAs also direct cytosine methylation. Understanding the details of such cross talk pathways will be required for a deep understanding of why some loci are more susceptible to multigenerational epigenetic inheritance than others.
In mammals, sperm carry relatively small amounts of RNA (Ostermeier et al. 2002, 2004, 2005; Krawetz et al. 2011). Until the past decade, it was commonly suggested that sperm lack any meaningful RNA. Moreover, RNA in sperm tends to be extensively degraded. Even ribosomal RNAs are present only as a smear of degradation products, leading to the general idea that the sperm RNA pool simply represents leftover remnants of the RNAs required for the processes of spermatogenesis and spermiogenesis. However, several lines of evidence support the potential for sperm RNAs to play a role in altering early events in the mammalian preimplantation embryo. The first is the abundant evidence for RNAs in a multitude of transgenerational epigenetic inheritance paradigms in other model organisms. Second, Whitelaw and colleagues have described paternal effect mutations that alter penetrance of coat color in the Avy model system described above. Here, heterozygous males can sire offspring with altered coat color even in offspring not inheriting the mutation of interest (Chong et al. 2007; Daxinger and Whitelaw 2012). Interestingly, in some of these mutations, Avy expression is altered even when it was transmitted maternally, which is inconsistent with Avy expression being altered in cis by cytosine methylation or chromatin changes having occurred in the sperm of mutant fathers. Although this trans effect on Avy could be mediated by a variety of indirect mechanisms, RNAs are of course a prime candidate for diffusible factors that can act in trans. Finally, several paradigms have been described in which RNAs from the sperm of males carrying specific mutations, or having been treated with a particular environmental stressor, can be microinjected into zygotes and recapitulate some of the phenotypes observed in the relevant natural mating paradigm. Most famously, Rassoulzadegan et al. (2006) found that mice heterozygous for a β-Gal insertion into the c-Kit locus sired wild-type offspring with altered tail coloration, and this effect could be recapitulated by injecting total RNA from the mutant male’s brain into control zygotes. Although this result has been subject to some controversy (Arnheiter 2007), similar results have been described in several other systems (Wagner et al. 2008; Gapp et al. 2014).
What is the RNA payload of mature mammalian sperm? As noted above, long RNAs show extensive degradation in mature sperm, with mRNAs and rRNAs showing a range of behaviors from rare RNAs being reasonably intact through the majority situation of extensive degradation (Johnson et al. 2011; Sendler et al. 2013). Presumably, more relevant to intergenerational information transfer are small RNA populations found in sperm. Short RNAs (<40 nt) include a variety of species, including relatively well-understood species such as microRNAs and siRNAs, moderately understood species such as piRNAs, and relatively understudied entities such as small fragments derived from rRNAs or tRNAs (Ghildiyal and Zamore 2009; Peng et al. 2012). It is currently unclear whether small RNAs delivered by sperm have much effect on regulation of early development. Sperm carry very little RNA relative to oocytes and the biochemistry of argonautes in somatic cells indicates that only highly abundant microRNAs are likely to exert measureable regulatory effects (Amanai et al. 2006; Suh and Blelloch 2011; Wee et al. 2012). Furthermore, the fate of sperm RNAs on fertilization is unclear as many reports suggest that RNAs delivered by sperm are undetectable in the zygote.
Altogether, sperm RNAs represent a compelling candidate for intergenerational information transfer in mammals, but the enterprise of understanding biological functions for sperm RNAs remains in its infancy.
EVIDENCE FOR TRANSGENERATIONAL TRANSFER OF EPIGENETIC INFORMATION
There is abundant evidence in model organisms that epigenetic information can be transmitted from one generation to the next and, in general, epigenetic information carriers are far more responsive to environmental perturbations than is genomic sequence. Countless studies have shown that perturbing tissue culture cells (by adding signaling molecules, heat stress, etc.) can alter the expression of specific microRNAs or can alter cytosine methylation patterns or histone modification profiles, whereas DNA sequence remains largely or entirely unaffected by these treatments. Taken together, these facts have motivated intense interest in the possibility that information about ancestral environments could be transmitted to offspring via epigenetic marks in the gametes. This concept is often called the “inheritance of acquired characters” or “Lamarckian” inheritance, although it is worth noting that the latter term is somewhat inaccurate. Both Darwin and Lamarck believed in the inheritance of acquired characters, and it is also worth noting that there are far more incorrect aspects of Lamarck’s theory of evolution than the inheritance of acquired characters. The classic test of the inheritance of acquired characters was performed by August Weismann in the late 19th century. He cut the tails off of mice, mated them, and measured tail length in the offspring (Weismann et al. 1904). The absence of any change in tail length in offspring was an influential piece of evidence against the inheritance of acquired characters, although in retrospect many problems with this experiment can be identified.
Modern variants of the Weismann experiment differ in two key ways from the tail amputation protocol. First, conditions used to treat the “ancestral” generation are somewhat more plausible as conditions that might have been experienced repeatedly over the evolutionary history of the organism (Jablonka et al. 1995). Second, instead of one quantitative character (tail length), many current iterations of this experiment involve genome-wide measurements in offspring to capture unanticipated responses in offspring. In mammals, the two main paradigms for paternal environmental exposures are dietary perturbations and stress conditions.
NUTRITIONAL PARADIGMS
A large and increasing number of rodent studies have linked paternal dietary perturbations to metabolic alterations in offspring. Such paradigms include paternal feeding with high-fat diet (Ng et al. 2010; Fullston et al. 2012, 2013) or low-protein diet (Carone et al. 2010; Watkins and Sinclair 2014), intermittent fasting (Anderson et al. 2006), and paternal prediabetes induced by high-fat diet and streptozotocin administration (Wei et al. 2014). In addition to these systems focused on dietary changes after weaning, a large number of studies use in utero nutritional perturbations. Here, pregnant females are subject to high-fat diet or caloric restriction and male offspring of these mothers are then used as the treated paternal generation to sire offspring (Jimenez-Chillaron et al. 2009; Dunn and Bale 2011; Radford et al. 2012). In each of the above studies, males provided with the test diet sired offspring with altered metabolic traits relative to the offspring sired by control males. Most of the above studies document altered glucose control in offspring of treated males, with altered cholesterol and lipid metabolism also being found in several studies. The precise details differ in the different studies. For example, Ng et al. (2010) describe a stronger effect of paternal diet on glucose control specifically in daughters, whereas Carone et al. (2010) reported little difference between paternal low protein diet effects on sons versus daughters.
Paternal dietary effects on offspring metabolism have also been documented in other mammals. Most relevant to human health are human epidemiological studies. It has long been known that maternal undernutrition can influence children’s susceptibility to metabolic disorders (Hales and Barker 2001), as is best-documented for children born following the “Dutch Hunger Winter” of 1944–1945 (Kyle and Pichard 2006; Lumey et al. 2007; Heijmans et al. 2008). In addition to such maternal effects, male line passage of ancestral nutritional status has also been suggested by analysis of the “Overkalix cohort.” Here, Pembrey et al. (2006) found that a man’s risk of diabetes and cardiovascular disease was linked to his paternal grandfather’s access to adequate food. Similarly, a woman’s risk of metabolic disease was traced to her paternal grandmother’s food access. Interestingly, the time of exposure to diet was a key in both cases. The grandson’s risk of metabolic disease was linked to access to an abundance of food in early adolescence (ages 10–13), but to inadequate food availability in early adulthood (ages 18–20). Taken together, the studies in rodents and humans link ancestral food supply to key metabolic outcomes in future generations.
STRESS AND TOXIN PARADIGMS
The other major group of paternal effect paradigms in mammals involves some form of aversive stress. This is not to suggest that poor diet is not stressful. There will likely be a great deal of overlap between outcomes of stress and dietary paternal-effect paradigms when compared in the same study. But it does indicate the separate approaches taken by physiologists and neurobiologists/psychologists to the field of intergenerational inheritance. “Stressful” paradigms can be separated into toxin administration and psychological stress.
Toxin-related paradigms tend to focus on endocrine disruptors, and include one of the earliest paternal effect experiments described in rodents. Administration of endocrine disruptors to pregnant female rats has been reported to alter reproductive success of several generations of male descendants (Anway et al. 2005). Several other environmental toxins, including DDT and the hepatotoxin carbon tetrachloride, have been reported to induce paternal effects. In the case of carbon tetrachloride, exposure of a rat’s father or even grandfather to this hepatotoxin resulted in a suppression of fibrosis induced by hepatotoxin exposure (Zeybel et al. 2012). More commonly, rodent studies focus on behavior-related stress paradigms. These include scenarios in which male mice suffer “defeat stress” after being placed in a cage with a rat, in which male mice suffer chronic variable stress (with random exposure to unfamiliar objects, damp bedding, etc.), and early life stress because of intermittent maternal separation (Dietz et al. 2011; Morgan and Bale 2011; Rodgers et al. 2013; Gapp et al. 2014). Phenotypes observed in offspring include both behavioral alterations (changes in exploratory behavior, etc.) and altered metabolism (altered insulin levels).
SPECIFICITY IN PATERNAL EFFECT PARADIGMS: THE CASE OF ODORANT TRAINING
As is clear from the examples above, a wide variety of fairly distinct environmental stressors can alter some aspect of offspring phenotype in inbred mammalian model systems. In general, the studies described above focus on disparate phenotypes. Dietary perturbations alter metabolism in offspring, stress paradigms alter anxiety and depression in offspring, carbon tetrachloride alters fibrosis in response to toxins, and so forth, thus raising the question of the information content of the sperm epigenome. Do sperm transmit millions of bits of information (calcium was plentiful in the drinking water, the average temperature was 10°C, protein was hard to come by, etc.) or an overall “quality of life” measure? Supporting the “sick sperm” hypothesis is evidence that many of the phenotypes observed in paternal effect experiments can be observed in response to multiple stimuli. For example, both glucose control phenotypes and reproductive success are seen in stress, toxin, and dietary paradigms (Rando 2012). The answer to the question of sperm’s information content has clear mechanistic importance. Cytosine methylation patterns could potentially transmit millions of bits of information to offspring, although the ability of a single day of in vitro embryo culture to induce metabolic phenotypes in offspring is consistent with many inputs leading to alterations in the kinetics of early development, with nonspecific secondary effects occurring downstream from growth rate.
A stunning report from Dias and Ressler (2014) provides the only compelling evidence for the “high bandwidth” hypothesis. In this study, one of two odorants was paired with foot shock. Offspring of the males treated in this way showed increased sensitivity to the odorant administered to the father, but no change in sensitivity to the alternative odorant. This increased susceptibility to the odorant was paired with an increase in the number of the relevant olfactory receptor-positive neurons. Given the specificity of odorant receptors for their ligands, this study suggests the somewhat shocking possibility that fathers could inform offspring of an extraordinarily complex chemical milieu via sperm. Moreover, it seems unlikely that such information transfer would be limited to olfactory receptor loci, which would suggest an essentially arbitrary level of complexity to the information transmitted from father to child. Naturally, understanding the bandwidth available in the sperm epigenome requires understanding the mechanistic basis for paternal effects.
ENVIRONMENTAL EFFECTS ON THE SPERM EPIGENOME
Although demonstrations that paternal environment can influence offspring phenotype are increasingly being replicated and extended to new stimuli and systems, at present we do not have any systems for which a convincing mechanism for information transfer has been definitively established. In fact, few studies have even tested the hypothesis that paternal dietary information is carried in sperm as opposed to seminal fluid (Dietz et al. 2011; Dias and Ressler 2014). That said, a substantial number of studies have identified epigenomic changes in the sperm in response to paternal environmental perturbations.
The majority of studies have reported changes either in cytosine methylation patterns (Chang et al. 2006; Ng et al. 2010; Dias and Ressler 2014; Radford et al. 2014; Wei et al. 2014) or RNA populations in sperm (Carone et al. 2010; Rodgers et al. 2013; Gapp et al. 2014), although chromatin changes have also been reported (Carone et al. 2010; Zeybel et al. 2012). Functional tests of cytosine methylation or chromatin changes in sperm are challenging, but are feasible. Heterozygous deletion of a differentially methylated genomic locus used in a scheme in which heterozygous males are crossed to wild-type females or vice versa, can be used to test whether a given stretch of DNA must be transmitted from the father to allow “reprogramming” of offspring. Alternatively, rapid advances in genomic targeting using ZNFs, TALENs, and CRISPR systems, could plausibly be used in the near future to “edit the epigenome.” That said, reported cytosine methylation changes generally are somewhat subtle changes of ∼10% methylation at a given CpG, and are thus unlikely to explain penetrant paternal effects. Briefly, this is because of the “digital” nature of sperm. As each sperm carries only a single copy of a given CpG and is responsible for generating one embryo, a change from 10% to 20% methylation at some locus should only alter the penetrance of a methylation-dependent phenotype from one in 10 offspring to two in 10 offspring. This concern holds for every single methylation change documented in paternal effect studies to date. That said, distributive methylation changes over a large number of CpGs, coupled with a readout mechanism that integrates total number of methyl marks across a locus, could conceivably allow digital sperm to achieve penetrant analog control of gene expression.
In contrast to methylation and chromatin alterations in sperm, functional tests for differentially regulated RNAs are straightforward as individual RNAs or RNA pools can be microinjected into control zygotes. Several studies have shown that RNA injections into zygotes can alter offspring phenotypes, in some cases recapitulating effects of paternal genetic lesions (Rassoulzadegan et al. 2006; Wagner et al. 2008). Moreover, a recent study of intergenerational effects of early life stress found that microinjection of sperm RNA from stressed males into zygotes could recapitulate some of the phenotypes observed in natural matings (Gapp et al. 2014). Future studies will hopefully identify which RNAs from the sperm pool are responsible for altering behavioral and metabolic phenotypes in offspring.
CONCLUSIONS
It is now clear that sperm carry not only genetic information to the next generation, but also carry some imprint of a male’s prior experiences in the form of epigenetic information carriers. Diet and stress in particular are common paradigms for rodent models of intergenerational transfer of information about paternal conditions. The next decade should be an exciting time for this fledgling field as rigorous interventions are used to definitively identify the relevant information carrier in various dietary and stress paradigms.
Footnotes
Editors: Diana W. Bianchi and Errol R. Norwitz
Additional Perspectives on Molecular Approaches to Reproductive and Newborn Medicine available at www.perspectivesinmedicine.org
REFERENCES
- Amanai M, Brahmajosyula M, Perry AC. 2006. A restricted role for sperm-borne microRNAs in mammalian fertilization. Biol Reprod 75: 877–884. [DOI] [PubMed] [Google Scholar]
- Anderson LM, Riffle L, Wilson R, Travlos GS, Lubomirski MS, Alvord WG. 2006. Preconceptional fasting of fathers alters serum glucose in offspring of mice. Nutrition 22: 327–331. [DOI] [PubMed] [Google Scholar]
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. 2005. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science 308: 1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnheiter H. 2007. Mammalian paramutation: A tail’s tale? Pigment Cell Res 20: 36–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpanahi A, Brinkworth M, Iles D, Krawetz SA, Paradowska A, Platts AE, Saida M, Steger K, Tedder P, Miller D. 2009. Endonuclease-sensitive regions of human spermatozoal chromatin are highly enriched in promoter and CTCF binding sequences. Genome Res 19: 1338–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arteaga-Vazquez MA, Chandler VL. 2010. Paramutation in maize: RNA mediated trans-generational gene silencing. Curr Opin Genet Dev 20: 156–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balhorn R. 2007. The protamine family of sperm nuclear proteins. Genome Biol 8: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei MS, Ferguson-Smith AC. 2011. Mammalian genomic imprinting. Cold Spring Harb Perspect Biol 3: a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bestor TH. 1992. Activation of mammalian DNA methyltransferase by cleavage of a Zn binding regulatory domain. EMBO J 11: 2611–2617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biniszkiewicz D, Gribnau J, Ramsahoye B, Gaudet F, Eggan K, Humpherys D, Mastrangelo MA, Jun Z, Walter J, Jaenisch R. 2002. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol 22: 2124–2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brykczynska U, Hisano M, Erkek S, Ramos L, Oakeley EJ, Roloff TC, Beisel C, Schubeler D, Stadler MB, Peters AH. 2010. Repressive and active histone methylation mark distinct promoters in human and mouse spermatozoa. Nat Struct Mol Biol 17: 679–687. [DOI] [PubMed] [Google Scholar]
- Carone BR, Fauquier L, Habib N, Shea JM, Hart CE, Li R, Bock C, Li C, Gu H, Zamore PD, et al. 2010. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell 143: 1084–1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carone BR, Hung JH, Hainer SJ, Chou MT, Carone DM, Weng Z, Fazzio TG, Rando OJ. 2014. High-resolution mapping of chromatin packaging in mouse embryonic stem cells and sperm. Dev Cell 30: 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedar H, Bergman Y. 2012. Programming of DNA methylation patterns. Annu Rev Biochem 81: 97–117. [DOI] [PubMed] [Google Scholar]
- Chan SW, Zilberman D, Xie Z, Johansen LK, Carrington JC, Jacobsen SE. 2004. RNA silencing genes control de novo DNA methylation. Science 303: 1336. [DOI] [PubMed] [Google Scholar]
- Chan SW, Henderson IR, Jacobsen SE. 2005. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet 6: 351–360. [DOI] [PubMed] [Google Scholar]
- Chang HS, Anway MD, Rekow SS, Skinner MK. 2006. Transgenerational epigenetic imprinting of the male germline by endocrine disruptor exposure during gonadal sex determination. Endocrinology 147: 5524–5541. [DOI] [PubMed] [Google Scholar]
- Chong S, Vickaryous N, Ashe A, Zamudio N, Youngson N, Hemley S, Stopka T, Skoultchi A, Matthews J, Scott HS, et al. 2007. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat Genet 39: 614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxinger L, Whitelaw E. 2012. Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet 13: 153–162. [DOI] [PubMed] [Google Scholar]
- de Waal E, Yamazaki Y, Ingale P, Bartolomei M, Yanagimachi R, McCarrey JR. 2012. Primary epimutations introduced during intracytoplasmic sperm injection (ICSI) are corrected by germline-specific epigenetic reprogramming. Proc Natl Acad Sci 109: 4163–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dias BG, Ressler KJ. 2014. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nat Neurosci 17: 89–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz DM, Laplant Q, Watts EL, Hodes GE, Russo SJ, Feng J, Oosting RS, Vialou V, Nestler EJ. 2011. Paternal transmission of stress-induced pathologies. Biol Psychiatry 70: 408–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn GA, Bale TL. 2011. Maternal high-fat diet effects on third-generation female body size via the paternal lineage. Endocrinology 152: 2228–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schubeler D, van der Vlag J, Stadler MB, Peters AH. 2013. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol 20: 868–875. [DOI] [PubMed] [Google Scholar]
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811. [DOI] [PubMed] [Google Scholar]
- Fullston T, Palmer NO, Owens JA, Mitchell M, Bakos HW, Lane M. 2012. Diet-induced paternal obesity in the absence of diabetes diminishes the reproductive health of two subsequent generations of mice. Hum Reprod 27: 1391–1400. [DOI] [PubMed] [Google Scholar]
- Fullston T, Ohlsson Teague EM, Palmer NO, DeBlasio MJ, Mitchell M, Corbett M, Print CG, Owens JA, Lane M. 2013. Paternal obesity initiates metabolic disturbances in two generations of mice with incomplete penetrance to the F2 generation and alters the transcriptional profile of testis and sperm microRNA content. FASEB J 27: 4226–4243. [DOI] [PubMed] [Google Scholar]
- Gapp K, Jawaid A, Sarkies P, Bohacek J, Pelczar P, Prados J, Farinelli L, Miska E, Mansuy IM. 2014. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nat Neurosci 17: 667–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Ballesteros M, Gordon M, Tam PP. 1998. Histone- and protamine-DNA association: Conservation of different patterns within the β-globin domain in human sperm. Mol Cell Biol 18: 3350–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. 2009. Small silencing RNAs: An expanding universe. Nat Rev Genet 10: 94–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govin J, Escoffier E, Rousseaux S, Kuhn L, Ferro M, Thevenon J, Catena R, Davidson I, Garin J, Khochbin S, et al. 2007. Pericentric heterochromatin reprogramming by new histone variants during mouse spermiogenesis. J Cell Biol 176: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer EL, Maures TJ, Ucar D, Hauswirth AG, Mancini E, Lim JP, Benayoun BA, Shi Y, Brunet A. 2011. Transgenerational epigenetic inheritance of longevity in Caenorhabditis elegans. Nature 479: 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal SI. 2010. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev 20: 134–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales CN, Barker DJ. 2001. The thrifty phenotype hypothesis. Br Med Bull 60: 5–20. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460: 473–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heijmans BT, Tobi EW, Stein AD, Putter H, Blauw GJ, Susser ES, Slagboom PE, Lumey LH. 2008. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc Natl Acad Sci 105: 17046–17049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihara M, Meyer-Ficca ML, Leu NA, Rao S, Li F, Gregory BD, Zalenskaya IA, Schultz RM, Meyer RG. 2014. Paternal poly (adp-ribose) metabolism modulates retention of inheritable sperm histones and early embryonic gene expression. PLoS Genet 10: e1004317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue A, Zhang Y. 2011. Replication-dependent loss of 5-hydroxymethylcytosine in mouse preimplantation embryos. Science 334: 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal K, Jin SG, Pfeifer GP, Szabo PE. 2011. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci 108: 3642–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonka E, Lamb MJ. 1995. Epigenetic inheritance and evolution: The Lamarckian dimension. Oxford University Press, New York. [Google Scholar]
- Jablonka E, Lamb MJ. 2002. The changing concept of epigenetics. Ann NY Acad Sci 981: 82–96. [DOI] [PubMed] [Google Scholar]
- Jablonka E, Oborny B, Molnar I, Kisdi E, Hofbauer J, Czaran T. 1995. The adaptive advantage of phenotypic memory in changing environments. Philos Trans R Soc Lond B 350: 133–141. [DOI] [PubMed] [Google Scholar]
- Jacob F, Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J Mol Biol 3: 318–356. [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Meyerowitz EM. 1997. Hypermethylated SUPERMAN epigenetic alleles in arabidopsis. Science 277: 1100–1103. [DOI] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. 2003. Epigenetic regulation of gene expression: How the genome integrates intrinsic and environmental signals. Nat Genet 33: 245–254. [DOI] [PubMed] [Google Scholar]
- Jimenez-Chillaron JC, Isganaitis E, Charalambous M, Gesta S, Pentinat-Pelegrin T, Faucette RR, Otis JP, Chow A, Diaz R, Ferguson-Smith A, et al. 2009. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes 58: 460–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GD, Sendler E, Lalancette C, Hauser R, Diamond MP, Krawetz SA. 2011. Cleavage of rRNA ensures translational cessation in sperm at fertilization. Mol Hum Reprod 17: 721–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz DJ, Edwards TM, Reinke V, Kelly WG. 2009. A C. elegans LSD1 demethylase contributes to germline immortality by reprogramming epigenetic memory. Cell 137: 308–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman PD, Rando OJ. 2010. Chromatin as a potential carrier of heritable information. Curr Opin Cell Biol 22: 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennison JA. 1995. The Polycomb and trithorax group proteins of Drosophila: Trans-regulators of homeotic gene function. Annu Rev Genet 29: 289–303. [DOI] [PubMed] [Google Scholar]
- Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, Li C, Zamore PD, Weng Z, Theurkauf WE. 2011. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell 147: 1551–1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD, Lorch Y. 1999. Twenty-five years of the nucleosome, fundamental particle of the eukaryote chromosome. Cell 98: 285–294. [DOI] [PubMed] [Google Scholar]
- Krawetz SA, Kruger A, Lalancette C, Tagett R, Anton E, Draghici S, Diamond MP. 2011. A survey of small RNAs in human sperm. Hum Reprod 26: 3401–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle UG, Pichard C. 2006. The Dutch famine of 1944–1945: A pathophysiological model of long-term consequences of wasting disease. Curr Opin Clin Nutr Metab Care 9: 388–394. [DOI] [PubMed] [Google Scholar]
- Law JA, Jacobsen SE. 2010. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11: 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumey L, Stein AD, Kahn HS, van der Pal-de Bruin KM, Blauw G, Zybert PA, Susser ES. 2007. Cohort profile: The Dutch hunger winter families study. Int J Epidemiol 36: 1196–1204. [DOI] [PubMed] [Google Scholar]
- Morgan CP, Bale TL. 2011. Early prenatal stress epigenetically programs dysmasculinization in second-generation offspring via the paternal lineage. J Neurosci 31: 11748–11755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan HD, Sutherland HG, Martin DI, Whitelaw E. 1999. Epigenetic inheritance at the agouti locus in the mouse. Nat Genet 23: 314–318. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, Sekimoto T, Ikawa M, Yoneda Y, Okabe M, et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol 9: 64–71. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Liu YJ, Nakashima H, Umehara H, Inoue K, Matoba S, Tachibana M, Ogura A, Shinkai Y, Nakano T. 2012. PGC7 binds histone H3K9me2 to protect against conversion of 5mC to 5hmC in early embryos. Nature 486: 415–419. [DOI] [PubMed] [Google Scholar]
- Ng SF, Lin RC, Laybutt DR, Barres R, Owens JA, Morris MJ. 2010. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature 467: 963–966. [DOI] [PubMed] [Google Scholar]
- Ooi SL, Henikoff S. 2007. Germline histone dynamics and epigenetics. Curr Opin Cell Biol 19: 257–265. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Dix DJ, Miller D, Khatri P, Krawetz SA. 2002. Spermatozoal RNA profiles of normal fertile men. Lancet 360: 772–777. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Miller D, Huntriss JD, Diamond MP, Krawetz SA. 2004. Reproductive biology: Delivering spermatozoan RNA to the oocyte. Nature 429: 154. [DOI] [PubMed] [Google Scholar]
- Ostermeier GC, Goodrich RJ, Moldenhauer JS, Diamond MP, Krawetz SA. 2005. A suite of novel human spermatozoal RNAs. J Androl 26: 70–74. [PubMed] [Google Scholar]
- Pembrey ME, Bygren LO, Kaati G, Edvinsson S, Northstone K, Sjostrom M, Golding J. 2006. Sex-specific, male-line transgenerational responses in humans. Eur J Hum Genet 14: 159–166. [DOI] [PubMed] [Google Scholar]
- Peng H, Shi J, Zhang Y, Zhang H, Liao S, Li W, Lei L, Han C, Ning L, Cao Y, et al. 2012. A novel class of tRNA-derived small RNAs extremely enriched in mature mouse sperm. Cell Res 22: 1609–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittoggi C, Renzi L, Zaccagnini G, Cimini D, Degrassi F, Giordano R, Magnano AR, Lorenzini R, Lavia P, Spadafora C. 1999. A fraction of mouse sperm chromatin is organized in nucleosomal hypersensitive domains enriched in retroposon DNA. J Cell Sci 112: 3537–3548. [DOI] [PubMed] [Google Scholar]
- Radford EJ, Isganaitis E, Jimenez-Chillaron J, Schroeder J, Molla M, Andrews S, Didier N, Charalambous M, McEwen K, Marazzi G, et al. 2012. An unbiased assessment of the role of imprinted genes in an intergenerational model of developmental programming. PLoS Genet 8: e1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford EJ, Ito M, Shi H, Corish JA, Yamazawa K, Isganaitis E, Seisenberger S, Hore TA, Reik W, Erkek S, et al. 2014. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science 345: 1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakyan VK, Down TA, Balding DJ, Beck S. 2011. Epigenome-wide association studies for common human diseases. Nat Rev Genet 12: 529–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. 2012. Daddy issues: Paternal effects on phenotype. Cell 151: 702–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ, Verstrepen KJ. 2007. Timescales of genetic and epigenetic inheritance. Cell 128: 655–668. [DOI] [PubMed] [Google Scholar]
- Rassoulzadegan M, Grandjean V, Gounon P, Vincent S, Gillot I, Cuzin F. 2006. RNA-mediated non-Mendelian inheritance of an epigenetic change in the mouse. Nature 441: 469–474. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293: 1089–1093. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Chang HY. 2012. Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers AB, Morgan CP, Bronson SL, Revello S, Bale TL. 2013. Paternal stress exposure alters sperm microRNA content and reprograms offspring HPA stress axis regulation. J Neurosci 33: 9003–9012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousseaux S, Caron C, Govin J, Lestrat C, Faure AK, Khochbin S. 2005. Establishment of male-specific epigenetic information. Gene 345: 139–153. [DOI] [PubMed] [Google Scholar]
- Samans B, Yang Y, Krebs S, Sarode GV, Blum H, Reichenbach M, Wolf E, Steger K, Dansranjavin T, Schagdarsurengin U. 2014. Uniformity of nucleosome preservation pattern in mammalian sperm and its connection to repetitive DNA elements. Dev Cell 30: 23–35. [DOI] [PubMed] [Google Scholar]
- Sendler E, Johnson GD, Mao S, Goodrich RJ, Diamond MP, Hauser R, Krawetz SA. 2013. Stability, delivery and functions of human sperm RNAs at fertilization. Nucleic Acids Res 41: 4104–4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh N, Blelloch R. 2011. Small RNAs in early mammalian development: From gametes to gastrulation. Development 138: 1653–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Heijden GW, Derijck AA, Ramos L, Giele M, van der Vlag J, de Boer P. 2006. Transmission of modified nucleosomes from the mouse male germline to the zygote and subsequent remodeling of paternal chromatin. Dev Biol 298: 458–469. [DOI] [PubMed] [Google Scholar]
- Wagner KD, Wagner N, Ghanbarian H, Grandjean V, Gounon P, Cuzin F, Rassoulzadegan M. 2008. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Dev Cell 14: 962–969. [DOI] [PubMed] [Google Scholar]
- Watkins AJ, Sinclair KD. 2014. Paternal low protein diet affects adult offspring cardiovascular and metabolic function in mice. Am J Physiol Heart Circ Physiol 306: H1444–H1452. [DOI] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. 2012. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151: 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yang CR, Wei YP, Zhao ZA, Hou Y, Schatten H, Sun QY. 2014. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc Natl Acad Sci 111: 1873–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weismann A, Thomson JA, Thomson MR. 1904. The evolution theory. Arnold Edward, London. [Google Scholar]
- Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, Arand J, Nakano T, Reik W, Walter J. 2011. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun 2: 241. [DOI] [PubMed] [Google Scholar]
- Zeybel M, Hardy T, Wong YK, Mathers JC, Fox CR, Gackowska A, Oakley F, Burt AD, Wilson CL, Anstee QM, et al. 2012. Multigenerational epigenetic adaptation of the hepatic wound-healing response. Nat Med 18: 1369–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]