Abstract
Genome sequences have become the new phenotype for microbial evolutionists. The patterns of diversity revealed in the first 100 bacterial genomes fostered development of a comprehensive framework that can explain their contents, organization, and evolution.
The elucidation of the complete sequences of bacterial genomes, beginning in 1995, revealed the general principles governing their structure, organization, and evolution.
The study of microbes has been at the forefront of research for some time but has changed considerably over the past century. What were originally witnessed as agents of disease became heralded as models for all life forms with the advent of molecular biology. And this, in turn, led to two of the most notable accomplishments in microbial evolution: an understanding of both their variation and their relationships at the deepest and shallowest taxonomic depths. These two areas expanded—motivated largely by polymerase chain reaction (PCR)—into complementary fields: population genetics, which analyzes the source and appointment of genetic variation within bacterial species, and phylogenetics, which arranges organisms, even those noncultivable, into a molecular tree of life.
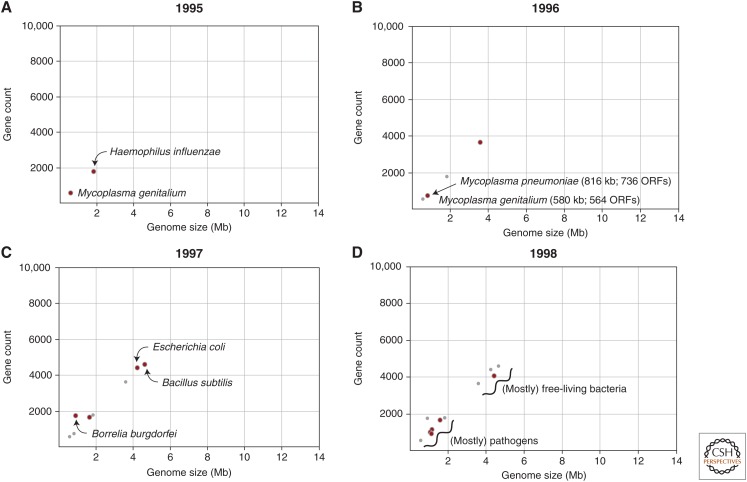
It is undeniable that a revolution in the study of bacteria has been fostered by genomics (i.e., the sequencing and analysis of entire genomes). Original interest in sequencing bacterial genomes was probably more technical than biological—bacteria had small, gene-dense, single-chromosome genomes, which made assembly more tractable, and their compact size was viewed as the logical next step after elucidation of the several mitochondrial and viral genomes over the previous decade. The era of genomics, particularly bacterial genomics, is commonly viewed to have begun in 1995 with the publication of the genome sequence of Haemophilus influenzae (Fleishmann et al. 1995) followed by the Mycoplasma genitalium genome sequence 3 months later (Fig. 1A) (Fraser et al. 1995).
Figure 1.
Genome size and gene count in bacterial genomes sequenced from 1995 to 1998. Red dots indicate genomes that were published in the designated year and smaller gray dots represent genomes published in all prior years. Panels A–D show results for the year indicated. ORF, Open reading frame.
The field of bacterial genomics was actually well underway before the appearance of the first genome sequences. Before 1995, it was already known (1) that bacterial chromosomes are, with few exceptions, circular, possessing a single replication origin; (2) that most bacterial genomes comprise a single chromosome but that smaller extrachromosomal elements in the form of plasmids and phage are common; (3) that genome sizes range from 500 to 10,000 kb; (4) that their chromosomes are tightly packed with genes that average ∼1 kb in length, that their genes contain no introns, are assorted onto both strands, and are arranged in operons; (5) that genomic base composition varies widely among bacterial species (from 25% to 75% G+C), but that base composition is relatively homogeneous over the entire chromosome within a given species; (6) that gene order is conserved among related species; and (7) that the rates and patterns of mutations can vary within a gene, and according to chromosomal location and transcriptional status of a gene.
So, what was learned from the first sequenced bacterial genome? Their multipage foldout figure rendered the size, location, orientation, and putative function of each of the 1727 genes in the 1.8-Mb genome, the culmination of spending a reported 13 months and one million dollars (although it is difficult to see how this amount could cover the expense of reagents and the 40 coinvestigators for a year). Naturally, the genome contained and corroborated many of the features listed above, but there were also two inimitable benefits. First and foremost was the credence it gave to the whole-genome shotgun (WGS) approach to sequencing. This methodology eliminated the need for genetic maps or manipulation, and could be applied to any organism yielding a sufficient amount of starting material. Second was that the complete genome sequence not only defines the complete gene inventory but also reveals those genes that are not present. Knowledge of the entire genome sequence allows no room to hypothesize activities specified by unknown genes. And, finally, my personal delight was that this genome set the standard for genome quality. A published bacterial genome needed to be a closed circle, with every gap closed and nucleotide confirmed.
Even with only two sequenced bacterial genomes, there was a consistent relationship between genome size and total gene number, and the two genomes that appeared in 1996 closely followed this trend (Fig. 1B). But more importantly, the slate of four genomes now included Mycoplasma pneumoniae, and direct comparisons with the already-sequenced M. genitalium genome indicated that members of the same bacterial genus can differ by more than 30% in genome size and contents (Himmelreich et al. 1996).
The genome size difference between these congeners was attributed to the horizontal acquisition of numerous genes by M. pneumoniae, as opposed to the loss of genes by M. genitalium. Typically, conclusions about gene gain and gene loss are based on the presence or absence of genes in a common ancestor of the focal species, but no such genome was available. But by exhuming those long-established principles of bacterial genome organization (hint, point 5 above), it is possible to infer the ancestry of a sequence without any comparisons whatsoever. Because base composition varies widely among bacterial species but is homogeneous within a species, genes possessing atypical features (such as anomalously high or low base compositions) are most plausibly gained from outside sources. This makes it possible to scan bacterial genomes to assess the extent of laterally acquired genes, and it was subsequently shown that most bacterial genomes were subject to high amounts of gene transfer and that many strain- or species-specific phenotypes were conferred by unique genomic segments of atypical base composition (Ochman et al. 2000).
In 1997, the genomes of the well-studied model systems, Escherichia coli and Bacillus subtilis, were published (Blattner et al. 1997; Kunst et al. 1997), and at more than 4 Mb in length, they constituted the largest bacterial genomes yet sequenced (with the B. subtilis publication surpassing the 100-author mark) (Fig. 1C). Also that year, were the genomes of Helicobacter pylori (Tomb et al. 1997) and Borrelia burgdorferi, whose relatively high gene number reflects the inclusion of genes encoded on its 11 extrachromosomal elements (Fraser et al. 1997). By the time there were a dozen bacterial genome sequences (Fig. 1D), it was apparent that there was an association between genome size and bacterial lifestyle. Those species with smaller genomes tended to be pathogens, whereas those with larger genomes were generally free-living species. There were two notable exceptions: the hot-spring bacterium, Aquifex aeolicus, had a genome size of only 1.6 Mb and the human pathogen (Deckert et al. 1998) and Mycobacterium tuberculosis, a genome size of more than 4.4 Mb (Cole et al. 1998).
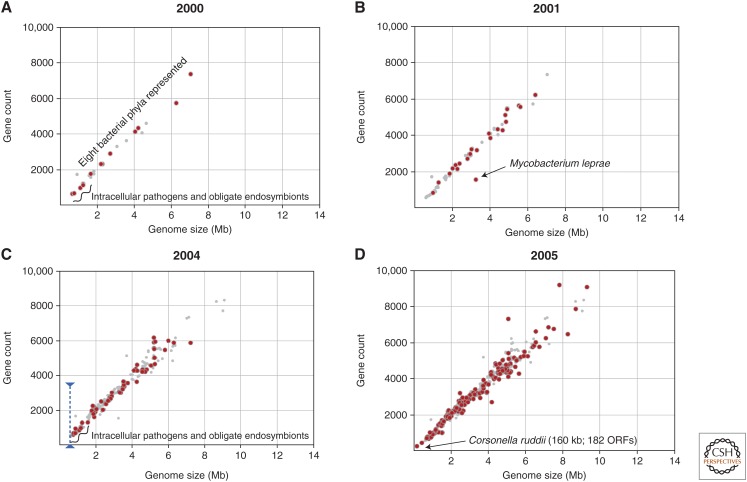
Moving forward a few years, we see that by 2000, after the resolution of 30 bacterial genomes, there remained a remarkable relationship between genome size and gene number across eight phyla and a >10-fold range in genome size (Fig. 2A). But this tenet of bacterial genome contents was undermined by elucidation of the Mycobacterium leprae genome (Fig. 2B) (Cole et al. 2001). M. leprae was replete with pseudogenes such that its 3.3-Mb genome contained only 1600 functional genes, the majority of which are also present in M. tuberculosis.
Figure 2.
Genome size and gene count in bacterial genomes sequenced from 2001 to 2005. Red dots indicate genomes that were published in the designated year and smaller gray dots represent genomes published in all prior years. Panels A–D show results for the year indicated. ORF, Open reading frame.
Discovering that M. leprae harbored large numbers of pseudogenes raised questions about why other bacterial genomes lacked similarly large numbers of inactivated and nonfunctional regions. Organisms are continually sieged by mutations and pseudogenes are continually being generated. So, why is there the same monotonic relationship between size and gene number across genomes large and small?
First, it is necessary to understand that M. leprae pseudogenes were discovered using a comparative approach—by gene-by-gene alignments with orthologs in closely related M. tuberculosis. The increasing availability of sequenced genomes offered several such opportunities for comparisons, and it became clear that many—possibly all—genomes contained pseudogenes, identified as truncated versions of genes in related genomes. Most interesting, however, was that the genomes with the largest numbers of pseudogenes were, like M. leprae, relatively recent pathogens of humans: for example, Shigella flexneri, an agent of dysentery that is descended from E. coli, and Yersinia pestis, which causes plague, each possessing more than 300 pseudogenes when compared with their closest sequenced relatives (Lerat and Ochman 2004, 2005).
But the reason that pseudogenes do not accumulate in genomes is that the mutational process in bacteria is biased toward deletions of all sizes, such that superfluous sequences are eliminated from the genomes (Mira et al. 2001). This deletion bias is apparent at all scales—from continuously cultured populations that manifest deletions spanning ∼5% of their genome (Nilsson et al. 2005) to the broad phylogenetic level in which lineages of bacteria with small genomes derive from large-genomed ancestors over evolutionary time scales (Ochman 2005).
Synthesizing information about the size and contents of sequenced genomes has led to a comprehensive understanding of the principles that underlie the progression toward compact genomes. Large-genomed, free-living bacteria move into a host or any environment that serves as a ready source of previously scarce or unavailable nutrients. The nutrient-rich environment renders many genes superfluous, which are inactivated by mutations causing pseudogenes to accumulate. Finally, the pervasive deletional bias rids genomes of pseudogenes. Note that the primary force countering gene erosion and elimination is natural selection, with the result that bacterial genomes, both large and small, contain mostly functional genes (Ochman and Moran 2001).
Which brings us to the topic of tiny genomes and the minimal gene set required for life. In the decades before full genome sequencing, the minimal set of genes that would support a cellular life form was reasoned to number between 500 and 600. This figure was arrived at by recovering genome sizes, using microscopic and DNA renaturation procedures, in the 400-MDa (≈600-kb) range for the small-celled Mycoplasma and estimating gene lengths as averaging 1200 bp, based on the mass of a typical protein.
Despite the vagaries of these calculations, the numbers were remarkably correct. And, when the complete genome sequence of M. genitalium was elucidated in 1995 (Fraser et al. 1995), its 580-kb genome encoded a mere 506 genes (470 proteins and 36 structural RNAs). Other experimental and computational approaches in estimating the minimal gene sets seemed to point to rather similar numbers. For example, based on the proportion of essential genes detected in an early mutagenesis screen of several dozen loci in B. subtilis, a minimal genome size was calculated to be 318 kb to 562 kb (or about 300 to 500 genes) (Itaya 1995). And by comparing the genes common to the first two fully sequenced bacterial genomes (H. influenzae and M. genitalium; Fig. 1A), the absolute minimal gene set sufficient for cellular life was estimated to number 256 genes (Mushegian and Koonin 1996), which matched well with some of the lower estimates obtained through large-scale gene disruptions in various bacterial species (see Table 2 in Zhang et al. 2010 for a compilation of these studies).
With nearly 200 genomes sequenced, it was generally agreed that the minimal size of a bacterial genome was ∼500 kb (or 500 genes) because numerous genomes from divergent taxa converged and hovered around that size (Fig. 2C). Clearly, there are many routes to minimal genomes, but it seemed that it took about 500 genes to thrive. But as the poet A.R. Ammons observed—“In nature there are few sharp lines”—and in 2005 came the report of Carsonella ruddii, whose 159,622-bp genome encoded only 182 genes (Fig. 2D) (Nakabachi et al. 2006). Not only did this genome surpass the lowest estimates of the number of genes necessary to sustain life, but its genome was extreme in its base composition (16.5% G+C) and in its lack of several genes that are essential to replication and repair in E. coli. Once the 500-gene barrier was broken, there have been numerous other reports of tiny bacterial genomes—all of which are endosymbionts of insects—with the current record held by Tremblaya princeps, which has only 121 genes and encodes no recognizable tRNA synthetases (McCutcheon and Von Dohlen 2011), although it clearly needs to translate its genes into proteins.
Integrating what we currently know about the process of bacterial genome evolution, we can conclude that most bacterial genomes are already at their minimal size. Genome-wide patterns of sequence conservation indicate that the vast majority of protein-coding genes in bacterial genomes are under functional constraints. The maintenance of an intact coding region, given the constant onslaught of mutations, is evidence that a sequence has been required over the evolutionary history of the lineage, because genes that serve no function incur debilitating mutations and are eventually eliminated by deletions. The genomes of recent pathogens, which often contain substantial numbers of nonfunctional genes, can be viewed as a transitional stage in the progression of events that result in the typically high coding densities observed in other bacterial genomes. Therefore, most bacterial genomes, regardless of their size or gene number, are already at a minimal size for the environment in which they evolved—there needs to be changes either in the challenges faced by bacteria or in the overall efficacy of selection to alter genomes in any appreciable way.
The goal of this essay is to show that the contents of bacterial genomes can be understood in a context above that includes—in most genome publications—that there is something beyond the compendia of genomic properties and gene inventories. Very little about the evolution of bacterial genomes can be gleaned from a single publication produced during the glory days of bacterial genome sequencing, but collectively we have been able to render the entire landscape.
Footnotes
Editor: Howard Ochman
Additional Perspectives on Microbial Evolution available at www.cshperspectives.org
REFERENCES
- Blattner FR, Plunkett G III, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277: 1453–1462. [DOI] [PubMed] [Google Scholar]
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE III, et al. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537–544. [DOI] [PubMed] [Google Scholar]
- Cole ST, Eiglmeier K, Parkhill J, James KD, Thomson NR, Wheeler PR, Honoré N, Garnier T, Churcher C, Harris D, et al. 2001. Massive gene decay in the leprosy bacillus. Nature 409: 1007–1011. [DOI] [PubMed] [Google Scholar]
- Deckert G, Warren PV, Gaasterland T, Young WG, Lenox AL, Graham DE, Overbeek R, Snead MA, Keller M, Aujay M, et al. 1998. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature 392: 353–358. [DOI] [PubMed] [Google Scholar]
- Fleishmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM, et al. 1995. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269: 496–512. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O, Adams MD, Clayton RA, Fleischmann RD, Bult CJ, Kerlavage AR, Sutton G, Kelley JM, et al. 1995. The minimal gene complement of Mycoplasma genitalium. Science 270: 397–403. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Casjens S, Huang WM, Sutton GG, Clayton R, Lathigra R, White O, Ketchum KA, Dodson R, Hickey EK, et al. 1997. Genomic sequence of a Lyme disease spirochaete, Borrelia burgdorferi. Nature 390: 580–586. [DOI] [PubMed] [Google Scholar]
- Himmelreich R, Hilbert H, Plagens H, Prikl E, Li BC, Herrmann R. 1996. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res 24: 4420–4449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itaya M. 1995. An estimation of minimal genome size required for life. FEBS Lett 362: 257–260. [DOI] [PubMed] [Google Scholar]
- Kunst F, Ogasawara N, Moszer I, Albertini AM, Alloni G, Azevedo V, Bertero MG, Bessières P, Bolotin A, Borchert S, et al. 1997. The complete genome sequence of the Gram-positive bacterium Bacillus subtilis. Nature 390: 249–256. [DOI] [PubMed] [Google Scholar]
- Lerat E, Ochman H. 2004. Ψ−Φ: Exploring the outer limits of bacterial pseudogenes. Genome Res 14: 2273–2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerat E, Ochman H. 2005. Recognizing pseudogenes in bacterial genomes. Nucleic Acids Res 33: 3125–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon JM, Von Dohlen CD. 2011. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol 21: 1366–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet 10: 589–596. [DOI] [PubMed] [Google Scholar]
- Mushegian AR, Koonin EV. 1996. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci 93: 10268–10273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabachi A, Yamashita A, Toh H, Ishikawa H, Dunbar HE, Moran NA, Hattori M. 2006. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science 314: 267. [DOI] [PubMed] [Google Scholar]
- Nilsson A, Koskiniemi S, Eriksson S, Kugelberg E, Hinton JCD, Andersson DI. 2005. Bacterial genome size reduction by experimental evolution. Proc Natl Acad Sci 102: 12112–12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H. 2005. Genomes on the shrink. Proc Natl Acad Sci 102: 11959–11960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Moran NA. 2001. Genes lost and genes found: The molecular evolution of bacterial pathogenesis and symbiosis. Science 292: 1096–1098. [DOI] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. 2000. Lateral gene transfer and the nature of bacterial innovation. Nature 405: 299–305. [DOI] [PubMed] [Google Scholar]
- Tomb J, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388: 539–547. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Chang SH, Wang J. 2010. How to make a minimal genome for synthetic minimal cell. Protein Cell 1: 427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]