Abstract
The transforming growth factor-β (TGF-β) is the prototype of the TGF-β family of growth and differentiation factors, which is encoded by 33 genes in mammals and comprises homo- and heterodimers. This review introduces the reader to the TGF-β family with its complexity of names and biological activities. It also introduces TGF-β as the best-studied factor among the TGF-β family proteins, with its diversity of roles in the control of cell proliferation and differentiation, wound healing and immune system, and its key roles in pathology, for example, skeletal diseases, fibrosis, and cancer.
Members of the TGF-β family are encoded by 33 genes in mammals and show diverse roles in health and disease. Their effects are contextual; a cell may show differing or opposing responses to them, depending on the situation.
Although initially thought to stimulate cell proliferation, just like many growth factors, it became rapidly accepted that transforming growth factor β (TGF-β) is a bifunctional regulator that either inhibits or stimulates cell proliferation. TGF-β was originally isolated as a cytokine that, together with epidermal growth factor (EGF), induces cellular transformation and anchorage-independent growth of selected fibroblast cell lines (Roberts et al. 1981), yet did not require the presence of EGF to induce phenotypic transformation of other fibroblast cell lines (Shipley et al. 1984). In contrast, TGF-β was also identified as a growth inhibitor secreted from confluent BSC-1 cells, epithelial cells of African green monkey kidney (Tucker et al. 1984). The growth inhibitory activity of TGF-β has been well documented in most cell types, and has been best characterized in epithelial cells. The bifunctional and context-dependent nature of TGF-β activities was further confirmed in a large variety of cell systems and biological responses. For example, TGF-β can inhibit EGF-dependent proliferation of cells in monolayer culture, whereas TGF-β and EGF synergistically enhance anchorage-independent growth of the same cells in soft agar medium (Roberts et al. 1985). Now, it is widely accepted that TGF-β regulates a variety of key events in normal development and physiology, and perturbation of TGF-β signaling has been implicated in the pathogenesis of diseases such as connective tissue disorders, fibrosis, and cancer.
The identification of TGF-β family members and their signaling components has enabled the characterization of the complex biology of the TGF-β family members. Molecular cloning of TGF-β family members and their signaling mediators started in 1985 with the reported characterization of complementary DNA (cDNA) coding for human TGF-β1 (Derynck et al. 1985). Subsequently, various approaches, based on biochemical purification, developmental genetics, and/or targeted cDNA cloning, led to the identification of polypeptides structurally similar to TGF-β1, which together comprise the members of the TGF-β family. Now that the human and mouse genome sequence projects are completed, it is apparent that mammalian genomes encode 33 TGF-β-related polypeptides. Table 1 shows the 33 known human TGF-β family polypeptides, which include three TGF-β isoforms, activins, nodal, bone morphogenetic proteins (BMPs), and growth and differentiation factors (GDFs). Although mostly studied as homodimers, various heterodimeric combinations of these have also been identified and characterized as biologically active proteins.
Table 1.
Names and genes for the TGF-β family proteins
| Protein name | Official gene symbol (human) | Protein name synonyms |
|---|---|---|
| TGF-β1 | TGFB1 | CIF-A (cartilage-inducing factor-A), differentiation-inhibiting factor |
| TGF-β2 | TGFB2 | G-TsF (glioblastoma-derived, T-cell suppressor factor), BSC-1 GI (BSC-1 cell-growth inhibitor), polyergin, CIF-B (cartilage-inducing factor-B) |
| TGF-β3 | TGFB3 | |
| Inhibin α | INHA | Inhibin A and Ba |
| Inhibin βA | INHBA | Inhibin A and activin A or AB,a,b FRP (follicle-stimulating hormone-releasing protein), EDF (erythroid differentiation factor), XTC-MIF (Xenopus XTC cell mesoderm-inducing factor) |
| Inhibin βB | INHBB | Inhibin B and activin B or AB,a,b XTC-MIF |
| Inhibin βC | INHBC | Activin Cc |
| Inhibin βE | INHBE | Activin Ec |
| Nodal | NODAL | BMP-16 (bone morphogenetic protein-16) |
| Myostatin | MSTN | GDF-8 (growth and differentiation factor-8) |
| BMP-2 | BMP2 | |
| BMP-3 | BMP3 | Osteogenin |
| BMP-4 | BMP4 | BMP-2B |
| BMP-5 | BMP5 | |
| BMP-6 | BMP6 | Vgr1 (Vg1-related protein) |
| BMP-7 | BMP7 | OP-1 (osteogenic protein-1) |
| BMP-8A | BMP8A | OP-2 |
| BMP-8B | BMP8B | OP-3 |
| BMP-9 | GDF2 | GDF-2 |
| BMP-10 | BMP10 | |
| GDF-1 | GDF1 | |
| GDF-3 | GDF3 | Vgr2 |
| GDF-5 | GDF5 | CDMP-1 (cartilage-derived morphogenetic protein-1), BMP-14 |
| GDF-6 | GDF6 | CDMP-2, BMP-13 |
| GDF-7 | GDF7 | CDMP-3, BMP-12 |
| GDF-9 | GDF9 | |
| GDF-9B | BMP15 | BMP-15 |
| GDF-10 | GDF10 | BMP-3b |
| GDF-11 | GDF11 | BMP-11 |
| GDF-15 | GDF15 | Placental TGF-β, placental BMP (PLAB), PDF (prostate-derived factor), NAG-1 (nonsteroidal anti-inflammatory drug-activated gene-1), MIC-1 (macrophage inhibitory cytokine-1) |
| MIS (Müllerian-inhibiting substance) | AMH | AMH (anti-Müllerian hormone) |
| Lefty A | LEFTY2 | EBAF (endometrial bleeding-associated factor), TGF-β4, Stra3 |
| Lefty B | LEFTY1 |
aInhibin A or B is a heterodimer of inhibin α and inhibin βA or βB, respectively.
bActivin A or B is a homodimer of inhibin βA or βB, respectively. Activin AB is a heterodimer of inhibin βA and βB.
cActivin C or D is a homodimer of inhibin βC or βD, respectively.
In contrast to the large number of TGF-β ligands, fewer receptors and downstream intracellular effectors, termed Smad proteins, mediate transduction of intracellular signaling. In 1991, a cDNA clone for an activin receptor, currently known as ActRII, was isolated (Mathews and Vale 1991). The kinase domain of this receptor closely resembled the Caenorhabditis elegans daf-1, a transmembrane serine/threonine-specific protein kinase, which at that time was seen as an orphan receptor, and provided the first indication that TGF-β family members signal through transmembrane serine/threonine kinases. In the following years, seven type I receptors and five type II receptors were identified in mammals and were shown to form heteromeric type I/type II receptor complexes. Genetic analysis in Drosophila and C. elegans led to a breakthrough in how signals are transduced from the receptors to the nucleus. In Drosophila, mothers against dpp (Mad) was identified as a gene encoding a component that acts epistatically downstream from Decapentaplegic (Dpp; Drosophila BMP-2/-4 ligand) (Raftery et al. 1995; Sekelsky et al. 1995). In C. elegans, daf-1 and daf-4 turned out to also encode serine/threonine transmembrane kinase receptors for TGF-β family members. Screening for mutants with similar phenotypes with daf-4 revealed three genes, sma-2, sma-3, and sma-4, that were structurally similar to Mad of Drosophila (Savage et al. 1996). In frog, mouse, and human, genes structurally similar to Mad and sma were subsequently identified, and the designation “Smad” (Sma and Mad) was adopted. Ligand binding to specific tetrameric type II/type I receptor complexes stabilizes and activates their signaling capacities, and the receptors then transduce the signals by phosphorylating carboxy-terminal serine residues of receptor-regulated (R-) Smads. In most cell types, TGF-βs and activins induce phosphorylation of Smad2 and Smad3 (activin/TGF-β-specific R-Smads), and BMPs induce phosphorylation of Smad1, Smad5, and Smad8 (BMP-specific R-Smads). The activated R-Smads form hetero-oligomeric complexes with a common-partner (co-) Smad, that is, Smad4 in vertebrate cells (Lagna et al. 1996; Zhang et al. 1996; Kawabata et al. 1998). The complexes translocate into the nucleus where they regulate the expression of target genes, such as those encoding inhibitory (I-) Smads, namely, Smad6 and Smad7 in vertebrates, which can inhibit R-Smad activation by the receptors. Finally, TGF-β family proteins were also shown to induce PI3K-Akt signaling and to activate the common mitogen-associated protein (MAP) kinase pathways that are activated by receptor tyrosine kinases, albeit, generally, to a lower extent. Now that essential players in the signaling pathways have been identified, one of the major questions to be addressed in this field is to reveal the precise molecular mechanisms that define the context-dependent dual roles of TGF-β family members.
In this review, we will introduce the TGF-β family members, which in mammals are encoded by 33 genes. We will cluster them into several subgroups based on the structural or sequence similarities of the encoded polypeptides. We further focus on the three TGF-β isoforms, TGF-β1, -β2, and -β3, as the best-studied factors among the TGF-β family proteins. In addition to the original “transforming” potential, we introduce their diverse roles in the control of cell proliferation, differentiation, wound healing, and the immune system, and TGF-β’s key roles in the context of pathological processes in vivo, for example, connective tissue disorders, fibrosis, and cancer.
TGF-β FAMILY LIGANDS IN MAMMALS
The name TGF-β derives from the transforming activity of the cytokine, which induces anchorage-independent growth when administered to cells together with EGF (Roberts et al. 1981) or without the need to add EGF (Shipley et al. 1984), depending on the cell system. At first, two distinct transforming growth factors, that is, TGF-α and -β, were identified and isolated. TGF-α is related to EGF and binds to the EGF receptor, whereas TGF-β is structurally distinct from TGF-α. The most striking characteristic, which distinguished the two cytokines at that time, was that TGF-β is a 25-kd disulfide-linked dimer that is reduced to a 12.5-kd band on gels following treatment with reducing agents, for example, β-mercaptoethanol (Roberts et al. 1983), whereas TGF-α was a monomeric protein of smaller size (Roberts et al. 1980).
All TGF-β family members are encoded by much larger precursor polypeptides whose sequences have been deduced through cDNA cloning. The precursor polypeptides are composed of three segments: an amino-terminal signal peptide that is removed during translocation of the protein into the lumen of the rough endoplasmic reticulum, a large precursor segment or prosegment, and the carboxy-terminal TGF-β family monomer polypeptide for the active and fully mature TGF-β family protein (112 amino acid residues in the case of TGF-β). The prosegments vary in length from 150 to 450 residues and are remarkably not conserved in sequence among TGF-β family members, even among the three TGF-β isoforms (Derynck et al. 1988). Indeed, the sequence similarities among TGF-β-related proteins pertain exclusively to the sequences of the mature polypeptides that follow the prosegments. In the precursor, the prosegment is immediately followed by a cleavage site for a furin protein convertase that precedes the carboxy-terminal mature TGF-β family polypeptide. In the case of the three TGF-β isoforms, the prosegments are also known as latency-associated peptides (LAPs). Following secretion, the LAPs remain noncovalently associated with the mature TGF-β dimer and keep the TGF-βs latent, that is, unable to bind and activate the receptors, in complexes that are called “small latent complexes” (SLCs). Whereas these prosegments are likely to play important roles in folding and transport of the TGF-β family proteins, they were shown to remain associated following secretion with a number of mature TGF-β family proteins, such as with BMP-4, BMP-7, BMP-9 (also known as GDF-2), BMP-10, GDF-5, GDF-11, myostatin (also known as GDF-8), and Müllerian-inhibiting substance (MIS, also known as anti-Müllerian hormone, AMH) (Wilson et al. 1993; Wolfman et al. 2003; Brown et al. 2005; Ge et al. 2005; Sengle et al. 2008). However, only some of the TGF-β family members are produced as latent forms that are unable to bind the receptors. With the exception of the TGF-β1 complex, the roles of the prosegments for other TGF-β family proteins have been poorly characterized, even for the closely related TGF-β2 and -β3.
The latency and activation mechanisms that result in the release of active TGF-β protein have been primarily studied in the case of TGF-β1, with additional knowledge gained on the activation of myostatin (Wolfman et al. 2003). The prosegment of TGF-β1 contains a proline-rich loop, or latency lasso, which surrounds a TGF-β1 monomer like a straitjacket (Shi et al. 2011). Comparison between TGF-β1 LAP and the BMP-9 prosegment, which does not confer latency, suggests conformational differences (Mi et al. 2015). Further analysis of the prosegments of other TGF-β family members, especially nonlatent ones, will reveal functional conservation among them.
For TGF-β to bind to its cell-surface receptors, the LAP needs to be released from the mature peptide. Both the TGF-β1 LAP and -β3 LAP contain an integrin recognition motif Arg-Gly-Asp, or RGD, and bind to several different integrins, for example, αvβ6 and αvβ8. Biochemical and structural studies revealed that contractile force is necessary for the release of mature TGF-β1 after complex formation between integrin αvβ6 and TGF-β1 LAP (Annes et al. 2004; Shi et al. 2011). In agreement with these findings, mice with RGD-to-RGE mutation in TGF-β1 LAP (Yang et al. 2007) and integrin β6-deficient mice (Itgb6−/−) (Munger et al. 1999) recapitulate aspects of the phenotype of Tgfb1−/− mice, indicating the importance of the interaction between LAP and integrins in latent TGF-β1 activation. The SLC covalently attaches the large latent TGF-β-binding protein (LTBP) through LAP, thus, forming the large latent complex (LLC). The association of LTBP with the SLC is also important for proper TGF-β1 function. Mice expressing a mutant TGF-β1 precursor (C33S) with a prosegment that is unable to bind LTBP, instead of the wild-type TGF-β1 precursor, are hypomorphic for TGF-β1; Tgfb1C33S/C33S mice show decreased levels of active TGF-β1, decreased TGF-β signaling, and formation of gastrointestinal tumors (Yoshinaga et al. 2008). LTBPs are efficiently incorporated in the extracellular matrix (ECM) (Taipale et al. 1994; Dallas et al. 1995), where latent TGF-β, as a part of LLC, is stored for future mobilization and activation. Thus, the regulation of latency of TGF-β has substantial biological significance, although it has received little attention. The control of TGF-β latency has been reviewed elsewhere (Horiguchi et al. 2012).
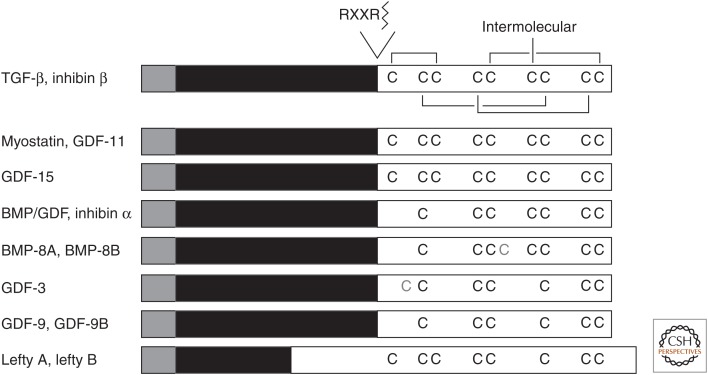
The mammalian genome encodes three different TGF-β isoforms, TGF-β1, -β2, and -β3. The mature proteins are highly conserved in sequence and are marked by the presence of nine cysteine residues. The three-dimensional structure of TGF-β2 reveals that four cysteine pairs are formed intramolecularly, but the sixth cysteine at position 77 forms the single intermolecular disulfide bridge that results in dimer formation (Fig. 1) (Daopin et al. 1992; Schlunegger and Grütter 1992).
Figure 1.
Comparison of the polypeptide organization of TGF-β family proteins. The precursor protein is cleaved by a furin protein convertase, which mainly recognizes the Arg-X-X-Arg (RXXR) motif, but also the Arg-X-Lys/Arg-Arg (RXK/RR) motif. The mature polypeptides (white) contain seven or nine cysteines conserved in most TGF-β family members. These cysteines engage in inter- and intramolecular disulfide bonds. Human BMP-8A, BMP-8B, and GDF-3 have an additional cysteine residue, which is shown in gray, whereas five members lack the cysteine that mediates the disulfide bond in ligand dimerization. Signal peptide, gray; prosegment, black; mature polypeptide, white; TGF-β, transforming growth factor β; BMP, bone morphogenetic protein; GDF, growth and differentiation factor.
Another distinct subfamily is the inhibin β (or activin β) family. Inhibins are heterodimeric proteins consisting of an inhibin α chain and an inhibin β chain, whereas activins are homodimers of inhibin/activin β chains. The four known β chains are closely related to each other and have the corresponding set of nine cysteines, which is apparent in the TGF-βs. Inhibin α, in contrast, is more divergent and has seven cysteines, corresponding to the seven carboxy-terminal cysteines seen in all TGF-β proteins.
All other ligands in the TGF-β family, with five exceptions discussed further below, have a characteristic seven-cysteine pattern that corresponds to the cysteine pattern observed in TGF-β and inhibin β, but without its amino-terminal two cysteines (Fig. 1). In these cases, the disulfide-linked dimerization is mediated by the fourth cysteine in each monomer. Many of these proteins have been named BMP or GDF followed by a number, based on their sequence relationship to other BMPs or GDFs, which were often identified in the same laboratories.
Within this large group of ligands, there is a “core” BMP/GDF subfamily of proteins with similar or related activities. These are structurally closely related to each other and are shown as the top 12 ligands in Figure 2. These BMPs and GDFs signal through heteromeric complexes that comprise the BMP type I receptors, including ALK-1 (activin receptor-like kinase 1), ALK-2, ALK-3/BMPRIA, or ALK-6/BMPRIB. In contrast, some ligands that have the seven-cysteine pattern, like the BMPs and GDFs, do not signal through the same BMP/GDF receptor complexes and intracellular mediators as BMPs and GDFs. Most notably, nodal signals through ALK-4, the major type I receptor for activins, which is also known as ActRIB, whereas myostatin signals through ALK-4/ActRIB or ALK-5, the TGF-β type I receptor also known as TβRI.
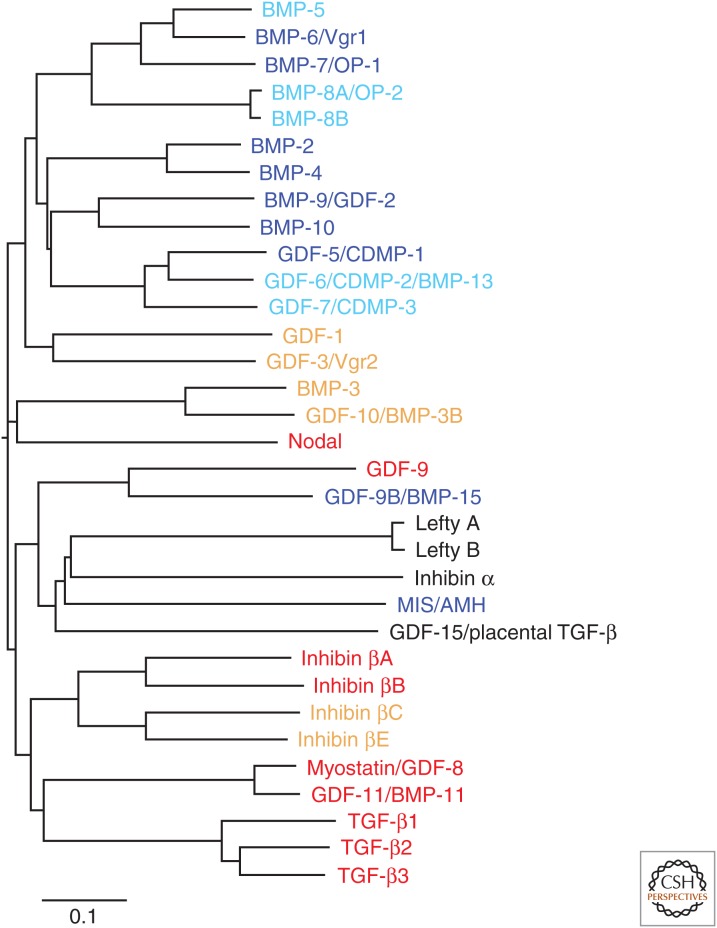
Figure 2.
Phylogenetic tree of the 33 TGF-β family polypeptides in human. The amino acid sequences of 33 TGF-β family polypeptides, encoded by their corresponding human genes, were obtained from NCBI’s protein database (www.ncbi.nlm.nih.gov), and the carboxy-terminal mature polypeptides were assigned based on validated or predicted furin cleavage sites. The sequences were aligned by Clustal Omega (www.clustal.org/omega) (Sievers et al. 2011) and the phylogenetic tree was illustrated using FigTree v1.4.2 (tree.bio.ed.ac.uk/software/figtree). Ligands that signal through activin- or TGF-β-activated R-Smads or BMP-activated R-Smads are shown in red or blue, respectively. Ligands that may activate these two types of R-Smads, but whose receptors and Smad-signaling pathways have not been fully determined, are shown in orange or light blue, respectively. BMP, Bone morphogenetic protein; OP, osteogenic protein; GDF, growth and differentiation factor; CDMP, cartilage-derived morphogenetic protein; MIS/AMH, Müllerian-inhibiting substance/anti-Müllerian hormone; TGF-β, transforming growth factor β.
As with the heterodimeric inhibins, BMP and/or GDF heterodimers have been identified and shown to be fully active. In mesodermal differentiation, BMP-4/-7 and -2/-7 heterodimers potently induce mesoderm in frog (Suzuki et al. 1997; Eimon and Harland 1999) and zebrafish (Little and Mullins 2009). In cultured cells, BMP-2/-7, -4/-7, and -2/-6 heterodimers are more active than the corresponding homodimers in ectopic bone-formation assays (Aono et al. 1995; Israel et al. 1996). BMP-2/GDF-6 heterodimer is also able to regulate ectodermal cell fate determination (Chang and Hemmati-Brivanlou 1999), and BMP-7/GDF-7 heterodimer is more potent than the corresponding homodimers in neuronal differentiation (Butler and Dodd 2003).
Finally, some TGF-β family proteins, lefty A and lefty B, GDF-3, and GDF-9 and GDF-9B (also known as BMP-15), lack the fourth cysteine in the seven-cysteine pattern and thus have eight or six cysteines (Fig. 1). As this missing cysteine mediates in other ligands the disulfide bond dimerization, one should assume that these TGF-β family proteins do not form disulfide-bonded dimers and have alternative means of interacting with other TGF-β family proteins. Lefty is found to associate with nodal and to inhibit nodal signaling through a dual mechanism involving its interaction with nodal and the EGF-CFC coreceptors, Cripto-1, FRL-1, or Cryptic, which are required for nodal signaling (Chen and Shen 2004; Tabibzadeh and Hemmati-Brivanlou 2006). GDF-3, which also lacks the cysteine required for intermolecular disulfide formation, may also function as an inhibitor of TGF-β/BMP signaling, similarly to lefty. Indeed, GDF-3 was reported to function as a BMP antagonist (Levine and Brivanlou 2006), whereas it was also reported to act as a nodal-like ligand (Chen et al. 2006; Levine et al. 2009) and to signal possibly through ALK-7, a type I receptor for activins (Andersson et al. 2008). GDF-9B has been reported to bind to BMP type I receptor ALK-6/BMPRIB and phosphorylate Smad1, Smad5, and Smad8, whereas GDF-9 activates Smad2 through ALK-5/TβRI (Moore et al. 2003; Mazerbourg et al. 2004; Peng et al. 2013). In addition, a GDF-9/-9B heterodimer is more bioactive for ovarian functions (Peng et al. 2013). Further characterization of the functions of this subfamily of TGF-β family proteins is expected.
COMPLEXITY OF NOMENCLATURE
The name “TGF-β family” derives from the identification of TGF-β1 as a founder member and its activity in the initial phenotypic transformation assay. However, this name may be somewhat misleading. The three TGF-β isoforms regulate a wide variety of cellular processes, both in development and in the adult organisms, and their effects are context-dependent. The three TGF-β isoforms have been established as some of the most potent growth inhibitors, and inhibit proliferation of most types of cells.
The identification of the BMP/GDF subfamily was based on the isolation and characterization of BMP-2 and BMP-4 with their ability to elicit ectopic bone formation and affect skeletal morphogenesis. However, most BMP/GDF proteins were assigned their names because of their sequence similarities to the other BMPs/GDFs, whereas their activities remain largely uncharacterized. It requires substantial caution to assume their biological functions merely because they share a name, especially for BMPs or GDFs. Recently, several BMP/GDF ligands were rediscovered as critical regulators of distinct physiological and pathological processes. For example, BMP-9 and BMP-10, which were identified as ligands of the ALK-1 receptor (David et al. 2007), play essential roles in the maintenance of homeostasis of vascular system and heart development (David et al. 2009; Pardali et al. 2010). Additionally, BMP-6 was identified as an endogenous ligand that regulates hepcidin expression and iron metabolism in vivo (Andriopoulos et al. 2009; Meynard et al. 2009). GDF-9 and the closely related GDF-9B are secreted by oocytes and regulate female fertility in several mammals.
Myostatin/GDF-8 (encoded by the Mstn gene) is now established as a key regulator of skeletal muscle mass; it inhibits muscle differentiation and regeneration, and induces muscle atrophy through receptor complexes of the ActRII or ActRIIB type II receptors with ALK-4 or -5 type I receptors that activate the Smad2/3 pathway (McPherron et al. 1997; Lee and McPherron 2001). As shown in Mstn-null mice and in loss-of-function mutations in cattle with double-muscling phenotype, mutations in the Mstn gene result in increased muscle mass with a combination of muscle cell hyperplasia and hypertrophy (Grobet et al. 1997; Kambadur et al. 1997; McPherron and Lee 1997; McPherron et al. 1997). GDF-11, which is highly related to myostatin, also signals through ActRII and/or ActRIIB (Oh et al. 2002). Similar to myostatin, GDF-11 has been reported to inhibit myoblast differentiation and regulate the same target genes (Egerman et al. 2015). It is worth noting that soluble ActRIIB-Fc, a ligand trap for myostatin, activin, and GDF-11, prevents muscle wasting and reverses loss of skeletal muscle and cardiac atrophy in tumor-bearing mice with cachexia (Zhou et al. 2010). Recently, GDF-11 has been identified as a circulating factor in young mice, which reverses age-related dysfunction in mouse cardiac and skeletal muscle (Loffredo et al. 2013; Sinha et al. 2014). Whereas these reports claimed that serum levels of GDF-11 decrease with age and that treating older mice with GDF-11 improved muscle regeneration, a contradictory report claims that serum levels of GDF-11 increase with age (Egerman et al. 2015). Further work on the biological activity of GDF-11 is required to confirm the significance of GDF-11 as a potential youth-promoting factor. In contrast, GDF-5 expression was shown to be associated with muscle hypertrophy through activation of BMP signaling (Sartori et al. 2013). Another TGF-β family protein, MIS/AMH, is secreted by Sertoli cells in the testis, and plays important roles in early gonadal development and sex differentiation through inhibition of the Müllerian duct development. MIS signals through its type II receptor, MISRII, and BMPRIA/ALK-3 in vivo, which subsequently activates the Smad1 pathway (Jamin et al. 2002). Mutations in the gene encoding MIS (Behringer et al. 1994) or MISRII (Imbeaud et al. 1995) result in persistent Müllerian duct syndrome. Intriguingly, the sex-determining roles of MIS/MISRII signaling are also conserved in fish, non-mammalian vertebrates that lack a Müllerian duct (Morinaga et al. 2007).
Figure 2 and Table 1 show the 33 known human TGF-β family polypeptides with their evolutionary relationship calculated based on sequence similarities. Because multiple and often parallel approaches have led to the identification of TGF-β family proteins, it is not surprising that several ligands became known with multiple names (Table 1). Now, most investigators have settled on a common name for each individual TGF-β family protein; this nomenclature is used in Figure 2 and Table 1.
With genome-wide and transcriptome analyses gaining in popularity, much research has involved the control of TGF-β family members. However, some discrepancies between the “official gene symbols” and the commonly used names for TGF-β family proteins have caused confusion for researchers and students who enter the field. For example, BMP-9 is the accepted and commonly used name for the protein, but its official gene symbol is GDF2. Lefty A and lefty B are widely used names for the proteins, but the corresponding official gene symbols are Lefty2 and Lefty1, respectively.
TGF-β SIGNALING IN THE CONTROL OF GENE EXPRESSION
More than 30 years of research on these multifunctional ligands has revealed different and sometimes opposite responses to TGF-β, which strongly depend on the cellular context. Most information on the activities of TGF-β ligands derives from studies using cells cultured in media that are supplemented with cytokine-containing serum, thus allowing for signaling cross talk. Although Smads directly target and bind regulatory gene sequences, they either activate or repress gene expression in cooperation with high affinity DNA-binding transcription factors and transcription coregulators that are controlled by and/or dependent on other signaling pathways, therefore providing scenarios for highly context-dependent transcription responses controlled by signaling cross talk. Furthermore, Smad activation and Smad activities are regulated by other kinase and ubiquitylation pathways. Additionally, Smad activation in response to TGF-β ligand-induced receptor activation is complemented by TGF-β/BMP-induced activation of non-Smad-signaling pathways that may be separately controlled. The combination of this signaling cross talk at multiple levels, together with differences in the physiology of different cell types, provides the basis for the high context dependence of the responses to TGF-β.
Several genes, such as the SMAD7 gene encoding the inhibitory Smad7, SERPINE1 encoding plasminogen activator inhibitor 1, or CDKN1A encoding the cyclin-dependent kinase inhibitor p21Cip1/Waf1, have been established as direct Smad target genes that are generally and rapidly induced after TGF-β treatment in many cell types (Lund et al. 1987; Datto et al. 1995b; Nakao et al. 1997; Koinuma et al. 2009b). The development of DNA microarrays, which allows simultaneous monitoring of thousands of transcripts, has revealed “synexpression groups” of genes that are coregulated (Niehrs and Pollet 1999). FoxO and C/EBPβ proteins were identified as some of the first examples of transcription factors that control the expression of TGF-β/Smad-regulated synexpression groups (Gomis et al. 2006a,b). Furthermore, chromatin immunoprecipitation (ChIP) with promoter array analysis (ChIP-chip) and ChIP followed by sequencing (ChIP-seq) revealed chromatin-binding landscapes of Smad proteins (Chen et al. 2008; Koinuma et al. 2009a,b; Qin et al. 2009). Some of these are defined through Smad binding with lineage-specific transcription factors at corresponding enhancer regions (Chen et al. 2008; Lee et al. 2011; Morikawa et al. 2011; Mullen et al. 2011; Trompouki et al. 2011). Forced expression of different lineage-specific transcription factors then redirects Smads to novel binding regions of different lineages (Mullen et al. 2011; Trompouki et al. 2011). These findings suggest that the binding of Smad complexes to target DNA sequences is defined through cooperation with other DNA-binding transcription cofactors in two different ways: (1) cell-type- or lineage-specific transcription factors, or pioneer factors, open up local chromatin structure to make it accessible for Smads, and (2) DNA-binding cofactors that are induced and activated in a context-dependent manner strengthen the interaction between Smads and DNA. The control of gene expression by Smads has been reviewed elsewhere (Morikawa et al. 2013; Gaarenstroom and Hill 2014).
In addition to genes that encode proteins, TGF-β also regulates expression of noncoding RNAs, such as microRNAs (miRNAs) and long noncoding RNAs (lncRNAs). miRNAs are ∼19–25 nucleotides long (with an average of 22 nucleotides), and are transcribed as long primary transcripts with hairpin structures (pri-miRNAs). Most pri-miRNAs derive from introns of coding genes or are independently transcribed as noncoding transcripts. In the nucleus, a microprocessor complex containing the RNase III enzyme Drosha and the double-stranded RNA (dsRNA)-binding protein DGCR8 (DiGeorge syndrome critical region 8) specifically recognizes and cleaves pri-miRNAs to generate precursor miRNAs (pre-miRNAs), which are exported to the cytoplasm, where another RNase III Dicer generates a duplex of 19–22 nucleotides long. One strand of the duplex is incorporated into the RNA-induced silencing complex, which posttranscriptionally regulates hundreds of target mRNAs (Blahna and Hata 2013).
TGF-β regulates directly or indirectly the expression of pri-miRNA transcript and the generation of mature miRNAs. Among them, miRNA-200 family (miR-200a, -200b, -200c, -141, and -429) and miR-205 are markedly down-regulated in cells that have undergone epithelial-mesenchymal transition (EMT) in response to TGF-β. TGF-β indirectly represses miR-200 family members through induction of ZEB1 (also known as δEF1) and ZEB2 (also known as Smad-interacting protein 1, or SIP1) transcription factors, and miR-200 family members target and inhibit the expression of ZEB proteins, which stabilize regulatory networks for TGF-β-induced EMT (Gregory et al. 2008; Park et al. 2008). In addition, TGF-β activates processing of pri-miRNAs. R-Smads interact with RNA helicase p68 (also known as DDX5), a component of the Drosha microprocessor complex, to promote pri-miRNA processing by Drosha (Davis et al. 2008). The role of TGF-β signaling in the control of miRNA generation is more extensively discussed elsewhere (Butz et al. 2012; Hata and Lieberman 2015).
Similarly to miRNAs, the emerging physiological and pathological roles of lncRNAs have started to attract the interest of investigators. LncRNAs are noncoding RNAs usually longer than 200 nucleotides. Several lncRNAs have been reported to play essential roles in TGF-β-regulated biological processes. Future work will confirm their biological significance.
CONTROL OF CELL PROLIFERATION BY TGF-β
TGF-β ligands convey strong growth inhibitory activity in most of the cell types, and this inhibition is reversible after ligand removal (Tucker et al. 1984; Roberts et al. 1985; Ohta et al. 1987; Kimchi et al. 1988). Although the antiproliferative response is observed in many cell types, including epithelial, endothelial, hematopoietic, and glial cells, most knowledge about the molecular mechanisms comes from studies in epithelial cells and keratinocytes (Siegel and Massagué 2003, 2008). The extent of the growth inhibitory response to TGF-β varies depending on the cell type and the particular cell system studied, and reaches a growth arrest in some cell types (Tucker et al. 1984). Confirming observations in cell culture, evidence for an antiproliferative effect of TGF-β1 in vivo was first obtained with polymer beads impregnated with TGF-β1 and implanted near the epithelial end buds of immature mammary glands (Silberstein and Daniel 1987). Also, intravenous injection of TGF-β1 or -β2 inhibits proliferation of the regenerating rat liver (Russell et al. 1988). The cytostatic effects by TGF-β are mediated mainly through two sets of mechanisms: induction of cyclin-dependent kinase inhibitors and elimination of proliferative drivers. TGF-β induces expression of p15Ink4b, p21Cip1/Waf1, and p27Kip1 (Hannon and Beach 1994; Polyak et al. 1994; Datto et al. 1995a), and inhibits the expression of mediators that contribute to cell proliferation, such as c-Myc (Pietenpol et al. 1990), Cdc25A (Iavarone and Massagué 1997), and Id proteins (Kang et al. 2003; Anido et al. 2010). In addition to direct effects of TGF-β signaling on cell-cycle control, TGF-β opposes the action of specific mitogens, such as EGF in keratinocytes (Coffey et al. 1988), or platelet-derived growth factor (PDGF) in rat fibroblast and embryonic cells (Roberts et al. 1985; Anzano et al. 1986). How TGF-β signaling interferes with the growth stimulation resulting from growth factor-induced receptor tyrosine kinase signaling has remained largely unexplored.
In contrast to the growth inhibition in many cell types, TGF-β can also promote cell proliferation in other cell types under certain conditions, as initially reported (Roberts et al. 1981). TGF-β has been shown to stimulate proliferation of several cell types, including chondrocytes, osteoblasts under some conditions, mesenchymal stem cells (MSCs), some fibroblasts, and endothelial cells under some conditions (Roberts et al. 1981; Goumans et al. 2002; Jian et al. 2006). As mentioned, the effects are often context-dependent because TGF-β can induce both growth promotion and growth inhibition in the same cells depending on the cell culture conditions (Roberts et al. 1985). Thus, not only may different cell types respond differently to TGF-β, but also the same cell type may show opposite responses under different experimental conditions. The molecular mechanisms responsible for inducing proliferation are less defined than those that lead to inhibition of proliferation. In some contexts, the growth-promoting effect seems to be secondary to induction of other cytokines such as PDGF (Leof et al. 1986; Matsuyama et al. 2003; Bruna et al. 2007). The control of cell proliferation has been reviewed elsewhere (Siegel and Massagué 2003, 2008).
CONTROL OF CELL DIFFERENTIATION AND STEMNESS BY TGF-β PROTEINS
TGF-β family members control the development, differentiation, and function of diverse cell types. For example, activin A, which uses similar repertoires of receptors and intracellular signaling pathways as TGF-β, was identified as a critical mesoderm-inducing factor in vivo (Asashima et al. 1990) as well as an erythroid differentiation factor (EDF) (Eto et al. 1987; Yu et al. 1987) and a nerve cell survival factor (Schubert et al. 1990). Additionally, the Spemann’s organizer in early embryonic development secretes extracellular antagonists, such as follistatin, which prevents activin signaling, and the BMP inhibitor noggin (Smith and Harland 1992; Hemmati-Brivanlou et al. 1994), which are critical for the establishment of morphogen gradients of activins and BMPs.
Similar to the developmental roles of activin, TGF-β ligands are implicated in differentiation toward a wide variety of lineages, including immune cells (Li and Flavell 2008), blood cells (Blank and Karlsson 2015), and neural/neuronal cells (Krieglstein et al. 2011). Consistently, TGF-β2-deficient mice show defects in multiple organs, including heart, lung, craniofacial, limb, spinal cord, eye, inner ear, and urogenital tracts (Sanford et al. 1997), and TGF-β3-deficient mice display defects during pulmonary and palate development, suggesting its critical roles in EMT (Kaartinen et al. 1995), whereas TGF-β1-deficient mice show a phenotype in the immune system (Shull et al. 1992). In addition, mice deficient in TGF-β2 and -β3 expression reveal defects in the central nervous system (Vogel et al. 2010).
Among a wide variety of effects on many lineages, TGF-β ligands play essential roles in determining the direction and extent of mesenchymal differentiation, as first shown in ectoderm explants from frog embryos (Kimelman and Kirschner 1987; Rosa et al. 1988). In cultured cells, TGF-β1 inhibits differentiation toward adipocytes (Ignotz and Massagué 1985) and skeletal myocytes (Florini et al. 1986; Massagué et al. 1986). In contrast, TGF-β1 and -β2 enhance differentiation toward chondrocytes, consistent with their independent identification and purification as cartilage-inducing factors A and B (CIF-A and CIF-B), respectively (Seyedin et al. 1985). Additionally, TGF-β promotes or represses the expression of differentiation markers by bone matrix-depositing osteoblasts, depending on their extent of differentiation. Inhibition of endogenous TGF-β and activin signaling in human MSCs by pharmacological inhibition of the ALK-4, -5, and -7 type I receptor kinases, induces osteoblast maturation (Maeda et al. 2004), and transcriptome analysis further supports the notion that inhibition of ALK-5/TβRI-mediated TGF-β signaling enhances adipogenic and osteogenic differentiation, and prevents chondrogenic differentiation (Ng et al. 2008).
Complementing the control of MSC differentiation toward specific lineages, TGF-β helps maintain MSCs in an undifferentiated state (Jian et al. 2006). In serum-free media with basic fibroblast growth factor (FGF-2), PDGF-BB, and insulin, TGF-β1 enables multiple passages of MSCs, and inhibition of any of these signaling pathways decreases proliferation of MSCs, suggesting a requirement of these tyrosine kinase-activated pathways for MSC proliferation (Ng et al. 2008). Thus, the cooperation of TGF-β with other cytokines plays important roles in the maintenance and expansion of MSC populations before differentiation commitment, while also restricting their differentiation potential, inhibiting differentiation along the osteoblast, myoblast, and adipocyte lineages.
TGF-β signaling also controls the maintenance of the undifferentiated state and pluripotency of embryonic stem cells (ESCs). In mouse ESCs, leukemia inhibitory factor (LIF), an interleukin 6 (IL-6) class cytokine, and BMP-4 play essential roles in the maintenance of pluripotency. However, in this model of “naïve pluripotency,” the role of TGF-β and activin signaling remains to be better defined. Whereas kinase inhibitors for TGF-β/activin type I receptors were reported to help maintain the naïve pluripotent state (Hassani et al. 2012), nodal/activin signaling was also shown to enhance self-renewal of mouse ESCs (Ogawa et al. 2007).
Human ESCs, which share characteristics with mouse epiblast stem cells derived from postimplantation embryos, represent a more mature stage in pluripotency, and are “primed” for differentiation. These cells are usually maintained with FGF-2 and activin A (Beattie et al. 2005; James et al. 2005; Vallier et al. 2005), which stabilize the primed pluripotent state. They no longer require LIF, which has essential roles in the naïve state of mouse ESCs. Defined culture conditions were reported to achieve a naïve pluripotent state for human ESCs and, in this context, two groups used TGF-β or activin ligands (Gafni et al. 2013; Theunissen et al. 2014) rather than their inhibitor(s). The roles of TGF-β family proteins in stem cells are reviewed elsewhere (Oshimori and Fuchs 2012; Sakaki-Yumoto et al. 2013).
Accumulating evidence also indicates essential roles of TGF-β/activin signaling in the maintenance of stem-cell-like properties of some cancer-initiating cell populations, such as glioma-initiating cells (Ikushima et al. 2009; Penuelas et al. 2009), mammary cancer stem cells (Mani et al. 2008), pancreatic cancer–initiating cells (Lonardo et al. 2011), and leukemia-initiating cells in chronic myeloid leukemia (Naka et al. 2010). Consistent with these observations, pharmacological inhibition of TGF-β/activin type I receptor kinases reduces cancer progression in some animal models (Ikushima et al. 2009; Penuelas et al. 2009; Naka et al. 2010; Lonardo et al. 2011). On the other hand, TGF-β was also shown to reduce the cancer-initiating cell populations in certain cancers, including breast cancer, pancreatic cancer, and diffuse-type gastric cancer (Tang et al. 2007; Ehata et al. 2011; Hoshino et al. 2015). These observations suggest that targeting the TGF-β/activin-signaling pathways could be an attractive therapeutic basis for some advanced cancers, although inhibition of these pathways in normal tissues may increase the risk for development of other tumors. The roles of TGF-β in cancer stem cells and cancer progression are reviewed elsewhere (Caja et al. 2012; Katsuno et al. 2013).
TGF-β CONTROLS WOUND HEALING
Normal wound healing is a complex process that involves (1) cell migration and inflammation, (2) proliferation of fibroblasts with formation of granulation tissue and ECM deposition, and (3) remodeling of scar tissue for an extended time period. TGF-βs have been shown to regulate these different steps through effects on multiple cell types, and to promote the wound healing process in vivo. Indeed, early studies, initiated shortly after the isolation and purification of TGF-β1, revealed that TGF-β1 strongly accelerates wound healing in vivo (Sporn et al. 1983).
TGF-β1 expression and activation are rapidly induced in response to injury, progressing outward from the site of injury (Kane et al. 1991). Platelets, which immediately after wounding facilitate the formation of the hemostatic plug, store large amounts of TGF-β1 (Assoian et al. 1983) that is then released at the site of injury. Because TGF-β1 acts as a potent chemoattractant for monocytes (Wahl et al. 1987) and fibroblasts (Postlethwaite et al. 1987), the platelet-derived TGF-β1 is able to attract both cell populations to sites of inflammation and repair. Accordingly, administration of TGF-β1 into wound chambers, or to incisional wounds, stimulates the accumulation of granulation tissue and the cellularization of the wound bed, and accelerates the wound healing response in experimental models (Sporn et al. 1983; Roberts et al. 1986; Mustoe et al. 1987). TGF-β1 is also a potent inducer of the expression of major ECM proteins, such as fibronectin and collagens (Ignotz and Massagué 1986; Roberts et al. 1986), thus promoting ECM deposition at the site of wound healing. Furthermore, TGF-β also induces inhibition of ECM degradation as a result of repressed expression of metalloproteinases and induction of tissue inhibitor of metalloproteinase synthesis (Edwards et al. 1987).
The repression of the epithelial phenotype with its apical-basal polarity and epithelial junction architecture in response to TGF-β, and concomitant induction of mesenchymal traits with increased cell motility also contribute to epithelial wound healing. At the site of injury, epithelial cells undergo partial or complete EMT as a result of increased TGF-β signaling, therefore promoting epithelial resurfacing, and then reacquire their epithelial phenotype (Kalluri and Weinberg 2009). TGF-β has also been shown to induce the expression of α-smooth muscle actin (Desmouliere et al. 1993), a marker of myofibroblasts. In addition, TGF-β treatment enhances acquisition of the myofibroblastic phenotype, and costimulation with FGF-2 is shown to inhibit the process (Shirakihara et al. 2011). This and other observations raise the possibility that increased TGF-β stimulation during wound healing may result in or contribute to the generation of myofibroblasts.
Although TGF-β1, -β2, and -β3 act through the same receptors and Smad2/3 pathway, they show differences in the control of wound healing that remain to be fully explained. Administration of TGF-β1 or -β2 promotes ECM deposition in the early stages of wound healing, but wounds treated with TGF-β1 or -β2 do not differ from control wounds in the final quality of scarring (Shah et al. 1995). However, inhibition of TGF-β/Smad3 signaling, for example, in the absence of Smad3 expression or in the presence of an antibody to TGF-β1 and -β2, enhances the quality of wound healing and reduces scarring (Ashcroft et al. 1999; Amendt et al. 2002). Additionally, administration of TGF-β3, but not TGF-β1 nor -β2, also reduces cutaneous scarring (Shah et al. 1995). Thus, recombinant TGF-β3 could be considered to promote scarless wound healing, which is supported by results from prophylactic use of recombinant TGF-β3 ligand in phase I/II trial (Ferguson et al. 2009).
TGF-β CONTROLS THE IMMUNE SYSTEM
TGF-β acts as a potent immunosuppressive cytokine through effects on both cell differentiation and cell proliferation. For example, TGF-β inhibits proliferation of T-lymphocytes (Kehrl et al. 1986) and thymocytes (Ristow 1986), and TGF-β2 was isolated as glioblastoma-derived T-cell suppressor factor, based on the observation that glioblastoma is frequently accompanied by immunosuppression (de Martin et al. 1987). The key roles of TGF-β1 in suppressing immune responses also explain the rapid neonatal development of overt inflammation in TGF-β1-deficient mice (Shull et al. 1992). In contrast, TGF-β2- and -β3-deficient mice display developmental defects (Kaartinen et al. 1995; Sanford et al. 1997), emphasizing their roles in development rather than in immune regulation. Consistent with the phenotype of Tgfb1−/− mice, T-cell-specific expression of a dominant-negative TGF-β type II receptor (TβRII) mutant, and T-cell-specific ablation of TβRII or TβRI/ALK-5 show that inhibition of TGF-β signaling in T cells causes neonatal lethal inflammatory disease.
Conversely, TGF-β1 promotes T-cell differentiation. Similarities in the autoimmune phenotype of mice lacking expression of TGF-β1 (Tgfb1−/−) or Foxp3, the transcription factor required for CD4+CD25+ regulatory T cells (Treg) (Brunkow et al. 2001), led to the discovery that TGF-β, in combination with IL-2, induces Treg cell differentiation through induction of Foxp3 expression in CD4+ T cells (Chen et al. 2003; Liu et al. 2008). TGF-β is also essential for the generation of IL-17-expressing proinflammatory T helper cells (TH17), in combination with IL-6 or -21 (Veldhoen et al. 2006; Korn et al. 2007). Thus, TGF-β can induce both regulatory/inhibitory and proinflammatory T cells, depending on whether proinflammatory cytokines such as IL-6 are present. The roles of TGF-β in the immune system are reviewed elsewhere (Li and Flavell 2008; Flavell et al. 2010).
ANOMALOUS TGF-β ACTIVATION IN CONNECTIVE TISSUE AND SKELETAL DISEASES
Gene mutations that result in dysregulation of TGF-β latency and confer increased release of activated TGF-β have been implicated in the pathogenesis of connective tissue diseases and skeletal diseases. Camurati–Engelmann disease (CED) is characterized by a progressive diaphyseal dysplasia with hyperostosis and sclerosis of the diaphyses of long bones. Most individuals in CED families have a mutation in the region of the TGFB1 gene locus that encodes LAP (Janssens et al. 2000; Kinoshita et al. 2000), which affects LAP dimerization and results in hyperactivation of TGF-β signaling (Saito et al. 2001; Tang et al. 2009), whereas no mutations are found in the domain encoding the mature TGF-β1 peptide. As an underlying mechanism, increased TGF-β may promote migration of bone marrow stromal cells to sites of bone resorption (Tang et al. 2009). Accordingly, a type I receptor kinase inhibitor partially rescues the uncoupled bone remodeling in a mouse model for CED with the mutant TGF-β1 precursor gene (Tang et al. 2009).
Mutations in the gene encoding fibrillin-1 have been causally linked to Marfan syndrome (Neptune et al. 2003), and their etiology highlights important roles of TGF-β in Marfan syndrome or Marfan-like connective tissue disorders. The fibrillins, which form extracellular microfibrils, associate with LTBP-1, which interacts with the LAP in the LLC, consequently linking this complex to elastic microfibrils (Isogai et al. 2003). The absence of fibrillin-1 in Fbn1-deficient mice results in excessive TGF-β activity in vivo, which could be at the basis of the pathogenesis of emphysema and other manifestations of Marfan syndrome (Neptune et al. 2003). Additionally, mutant fibrillin-1 disturbs the microfibril structure (Judge et al. 2004; Charbonneau et al. 2010) and activates TGF-β signaling, which may then over time generate the pathological features of Marfan syndrome.
Clinically overlapping conditions, which result in aortic aneurysms, for example, in Loeys–Dietz syndrome, familial thoracic aortic aneurysms and dissections, and aneurysms-osteoarthritis syndrome, are also caused by enhanced TGF-β signaling. Mutations that affect TGF-β-signaling pathway components have been reported; these include mutations in loci encoding TGFB2 (Boileau et al. 2012; Lindsay et al. 2012) or TGFB3 (Bertoli-Avella et al. 2015), as well as other signaling components or regulators, that is, TGFBR1 (Loeys et al. 2005, 2006), TGFBR2 (Mizuguchi et al. 2004; Loeys et al. 2005, 2006), SMAD3 (van de Laar et al. 2011), and SKI (Doyle et al. 2012). These syndromes share some clinical features in the aorta, including dilation of the aortic root or ascending aorta and degeneration of the medial layer of the aorta. Another common feature is paradoxical activation of TGF-β signaling in aortic lesions, although they harbor loss-of-function mutations in TGF-β-signaling components. The molecular mechanisms that explain the paradoxical activation have not been elucidated. The dysregulation of TGF-β signaling in connective tissue and skeletal diseases has been discussed elswhere by Gallo (Lindsay and Dietz 2011; Akhurst 2012).
INCREASED TGF-β SIGNALING DRIVES FIBROSIS
A pivotal role of TGF-β in fibrogenesis was first revealed when subcutaneous injection of purified TGF-β1 was seen to induce fibrotic lesions at the injection site (Roberts et al. 1986). This potent effect was further confirmed by the fibrotic response in lungs of rodents following intratracheal administration of adenovirus that expresses active TGF-β1 (Sime et al. 1997), whereas neutralization of TGF-β with antiserum ameliorated experimental fibrosis in the kidney, heart, and liver (Border et al. 1990; Denis 1994; Kuwahara et al. 2002). In addition, inhibitors or antagonists of the TGF-β/Smad pathway, such as soluble TβRII (George et al. 1999) or adenovirally expressed Smad7 (Nakao et al. 1999), were shown to prevent liver and lung fibrosis in mice, respectively. Most fibrogenic effects resulting from increased TGF-β signaling are thought to be mediated by the Smad pathway because Smad3-null mice are resistant to bleomycin-induced lung fibrosis (Zhao et al. 2002), radiation-induced skin fibrosis (Flanders et al. 2002), and tubulointerstitial fibrosis following unilateral ureteral obstruction (Sato et al. 2003). In addition, phenotypes of integrin αvβ6-deficient mice support a role of integrin-mediated TGF-β activation in progressive fibrosis because bleomycin-induced inflammation can occur without progression to fibrosis in these mice (Munger et al. 1999). These and other observations suggest that dysregulation and sustained activation of TGF-β/Smad3 signaling play essential roles in the initiation and maintenance of the fibrotic tissue phenotype.
The profibrotic effects of TGF-β involve a combination of mechanisms and cell types, including enhanced infiltration and/or proliferation of preexisting fibroblasts, generation of myofibroblasts, increased ECM synthesis, and inhibition of collagenolysis, all of which are also apparent in wound healing. In addition, lineage tracing strongly suggests important contributions of mesenchymal cells arising through TGF-β-induced EMT. For example, in a model of tubulointerstitial renal fibrosis, the use of the promoter region of the gene encoding γ-glutamyltranspeptidase, a proximal tubule marker, for cell lineage tracing suggests that 36% of renal fibroblasts derive through EMT from renal tubular epithelium (Iwano et al. 2002). Additionally, using the promoter of surfactant protein C, a type II alveolar epithelial cell marker, for lineage tracing highlights a substantial contribution of EMT of alveolar epithelial cells in TGF-β1-induced pulmonary fibrosis (Kim et al. 2006). Endothelial cells have also been identified as a source of fibroblasts in fibrosis. In heart fibrosis, for example, 27%–35% of all fibroblasts are estimated to originate from endothelial cells through endothelial-mesenchymal transition (Zeisberg et al. 2007).
Although TGF-β has been well established as a key profibrotic cytokine, its activity is attenuated or aggravated by several other cytokines. For example, IL-1 and tumor necrosis factor-α, which cross talk with TGF-β signaling at multiple levels, are well-characterized profibrotic cytokines. Conversely, some members of the nuclear receptor superfamily, such as the vitamin D receptor, PPAR-γ (peroxisome proliferator-activated receptor-γ), and NR4A1, directly repress fibroblast activation induced by TGF-β (Wu et al. 2009; Ding et al. 2013; Palumbo-Zerr et al. 2015).
TGF-β SIGNALING IN CANCER PROGRESSION
TGF-β induces an antiproliferative response in many cell types, including both normal epithelial cells and transformed cells. Initial reports showed that antisense inhibition of TGF-β enhances tumorigenicity in vivo (Wu et al. 1992), and that certain tumor cells can become unresponsive to TGF-β, which was primarily assessed by its effects on proliferation (Shipley et al. 1986; Kimchi et al. 1988). It was, therefore, assumed that disruption of TGF-β signaling is implicated in the pathogenesis of tumor development. In line with this hypothesis, inactivating mutations and deletions in the TGFBR2 and SMAD4 gene loci were found in cancers (Markowitz et al. 1995; Hahn et al. 1996) and thought to drive tumorigenesis (reviewed in Garraway and Lander 2013; Vogelstein et al. 2013).
As advocated by Vogelstein and colleagues, the multistep tumorigenesis model (Fearon and Vogelstein 1990; Vogelstein et al. 2013) involves the requirement of sequential genetic alterations over time, which enables the development of tumors and promotes cancer progression. In colon cancers, for example, gatekeeping mutations most often occur in the adenomatous polyposis coli (APC) gene, and second mutations in another gene, such as KRAS, then stimulate clonal growth and expansion. Mutations in genes such as PIK3CA, SMAD4, and TP53 follow this mutation process, resulting in more invasive and metastatic tumors. Conceptually similarly, TGF-β’s function as a suppressor of epithelial cell tumorigenesis may need to be alleviated at an early stage in tumor development, which does not preclude roles of TGF-β signaling at later stages.
On the other hand, tumor cells show increased expression of TGF-β, most commonly TGF-β1, when compared with normal surrounding tissue, and secrete TGF-β ligands. Additionally, elevated TGF-β expression correlates with tumor progression and poorer prognosis, indicating pro-oncogenic roles for TGF-β at late stages. Substantial evidence now supports the notion that the increased TGF-β expression by tumor cells promotes tumor progression by enhancing migration, invasion, and survival of tumor cells during later stages of tumorigenesis, through stimulating ECM deposition and tissue fibrosis, perturbing immune and inflammatory function, stimulating angiogenesis, promoting EMT that enables increased migration and invasion, and maintaining cancer stem cells. Platelets attached to tumor cells in the circulation provide an additional source of TGF-β that promotes an invasive mesenchymal-like phenotype and enhances metastasis in vivo through stimulating the TGF-β/Smad and NF-κB pathways in the tumor cells (Labelle et al. 2011).
With the sequential acquisition of genomic mutations, tumor cells were shown to display a range of mutations in different signaling pathways, which lead to changes in TGF-β responsiveness. This is illustrated by a model system consisting of a set of human breast epithelial MCF10A–derived cell lines, in which TGF-β functions as a tumor suppressor early in the carcinogenic process, but switches to a pro-oncogenic agent in the later stage. Intriguingly, in the most aggressive cell line, M-IV, which resembles a high-grade breast carcinoma, loss of TGF-β response does not affect primary tumorigenesis, but suppresses metastasis (Tang et al. 2003).
As illustrated with these observations, TGF-β signaling controls both the initial development of tumors as well as the cancer progression of a large variety of tumor types. Its roles in tumor suppression and its extensive roles in cancer progression through direct effects on the cancer cells themselves, as well as the microenvironment, are the subject of many reviews (Bierie and Moses 2006; Ikushima and Miyazono 2010; Pickup et al. 2013).
CONCLUSIONS
The TGF-β family comprises a large number of secreted and structurally related proteins with multiple roles in developmental patterning, tissue differentiation, and maintenance of homeostasis. As discussed, the biological effects of TGF-β family proteins are contextual, and even the same cell type may show different or opposite responses to the ligand under different biological contexts. Some TGF-β family members are made available to cells with concentration gradients in vivo, resulting in graded effects of cell differentiation and function by the responsive cells. The activation of distinctive sets of genes by the ligand in a dosage-dependent manner plays essential roles in the specification of cell fates during normal development. Cells also respond to TGF-β family proteins in combination with other external stimuli. Costimulation by other cytokines modifies the responsiveness to the ligand, as observed in the immune system. In addition, the complexity of the TGF-β family may reflect some functional redundancy. It is, therefore, possible that some family members could play redundant roles in the regulation of some biological processes. Therefore, detailed profiling of expression and activation of all TGF-β family ligands in each context could lead to a better understanding of the complex nature of TGF-β family members.
Recent advances in single-cell assays have revealed intertumor heterogeneity between tumors of the same histopathological subtype, and intratumor genetic and epigenetic heterogeneity within individual cancer masses. In addition, characterization of the transcriptome and histone modification markers in normal cells, such as mouse ESCs, similarly suggests substantial cell-to-cell variability even in cells with identical genetic backgrounds. It is, therefore, possible that the heterogeneity could contribute to different responsiveness to TGF-β family members. Thus, spatial and temporal profiling of signaling at the single-cell level will help us to understand the complex mechanisms of TGF-β family members.
Footnotes
Editors: Rik Derynck and Kohei Miyazono
Additional Perspectives on The Biology of the TGF-β Family available at www.cshperspectives.org
REFERENCES
- Akhurst RJ. 2012. The paradoxical TGF-β vasculopathies. Nat Genet 44: 838–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendt C, Mann A, Schirmacher P, Blessing M. 2002. Resistance of keratinocytes to TGFβ-mediated growth restriction and apoptosis induction accelerates re-epithelialization in skin wounds. J Cell Sci 115: 2189–2198. [DOI] [PubMed] [Google Scholar]
- Andersson O, Korach-Andre M, Reissmann E, Ibanez CF, Bertolino P. 2008. Growth/differentiation factor 3 signals through ALK7 and regulates accumulation of adipose tissue and diet-induced obesity. Proc Natl Acad Sci 105: 7252–7256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriopoulos B Jr, Corradini E, Xia Y, Faasse SA, Chen S, Grgurevic L, Knutson MD, Pietrangelo A, Vukicevic S, Lin HY, et al. 2009. BMP6 is a key endogenous regulator of hepcidin expression and iron metabolism. Nat Genet 41: 482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anido J, Saez-Borderias A, Gonzalez-Junca A, Rodon L, Folch G, Carmona MA, Prieto-Sanchez RM, Barba I, Martinez-Saez E, Prudkin L, et al. 2010. TGF-β receptor inhibitors target the CD44high/Id1high glioma-initiating cell population in human glioblastoma. Cancer Cell 18: 655–668. [DOI] [PubMed] [Google Scholar]
- Annes JP, Chen Y, Munger JS, Rifkin DB. 2004. Integrin αVβ6-mediated activation of latent TGF-β requires the latent TGF-β binding protein-1. J Cell Biol 165: 723–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzano MA, Roberts AB, Sporn MB. 1986. Anchorage-independent growth of primary rat embryo cells is induced by platelet-derived growth factor and inhibited by type-β transforming growth factor. J Cell Physiol 126: 312–318. [DOI] [PubMed] [Google Scholar]
- Aono A, Hazama M, Notoya K, Taketomi S, Yamasaki H, Tsukuda R, Sasaki S, Fujisawa Y. 1995. Potent ectopic bone-inducing activity of bone morphogenetic protein-4/7 heterodimer. Biochem Biophys Res Commun 210: 670–677. [DOI] [PubMed] [Google Scholar]
- Asashima M, Nakano H, Shimada K, Kinoshita K, Ishii K, Shibai H, Ueno N. 1990. Mesodermal induction in early amphibian embryos by activin A (erythroid differentiation factor). Rouxs Arch Dev Biol 198: 330–335. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Yang X, Glick AB, Weinstein M, Letterio JL, Mizel DE, Anzano M, Greenwell-Wild T, Wahl SM, Deng C, et al. 1999. Mice lacking Smad3 show accelerated wound healing and an impaired local inflammatory response. Nat Cell Biol 1: 260–266. [DOI] [PubMed] [Google Scholar]
- Assoian RK, Komoriya A, Meyers CA, Miller DM, Sporn MB. 1983. Transforming growth factor-β in human platelets. Identification of a major storage site, purification, and characterization. J Biol Chem 258: 7155–7160. [PubMed] [Google Scholar]
- Beattie GM, Lopez AD, Bucay N, Hinton A, Firpo MT, King CC, Hayek A. 2005. Activin A maintains pluripotency of human embryonic stem cells in the absence of feeder layers. Stem Cells 23: 489–495. [DOI] [PubMed] [Google Scholar]
- Behringer RR, Finegold MJ, Cate RL. 1994. Müllerian-inhibiting substance function during mammalian sexual development. Cell 79: 415–425. [DOI] [PubMed] [Google Scholar]
- Bertoli-Avella AM, Gillis E, Morisaki H, Verhagen JM, de Graaf BM, van de Beek G, Gallo E, Kruithof BP, Venselaar H, Myers LA, et al. 2015. Mutations in a TGF-β ligand, TGFB3, cause syndromic aortic aneurysms and dissections. J Am Coll Cardiol 65: 1324–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bierie B, Moses HL. 2006. TGFβ: The molecular Jekyll and Hyde of cancer. Nat Rev Cancer 6: 506–520. [DOI] [PubMed] [Google Scholar]
- Blahna MT, Hata A. 2013. Regulation of miRNA biogenesis as an integrated component of growth factor signaling. Curr Opin Cell Biol 25: 233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank U, Karlsson S. 2015. TGF-β signaling in the control of hematopoietic stem cells. Blood 125: 3542–3550. [DOI] [PubMed] [Google Scholar]
- Boileau C, Guo DC, Hanna N, Regalado ES, Detaint D, Gong L, Varret M, Prakash SK, Li AH, d’Indy H, et al. 2012. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of Marfan syndrome. Nat Genet 44: 916–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Border WA, Okuda S, Languino LR, Sporn MB, Ruoslahti E. 1990. Suppression of experimental glomerulonephritis by antiserum against transforming growth factor β1. Nature 346: 371–374. [DOI] [PubMed] [Google Scholar]
- Brown MA, Zhao Q, Baker KA, Naik C, Chen C, Pukac L, Singh M, Tsareva T, Parice Y, Mahoney A, et al. 2005. Crystal structure of BMP-9 and functional interactions with pro-region and receptors. J Biol Chem 280: 25111–25118. [DOI] [PubMed] [Google Scholar]
- Bruna A, Darken RS, Rojo F, Ocana A, Penuelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, et al. 2007. High TGFβ-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell 11: 147–160. [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. 2001. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet 27: 68–73. [DOI] [PubMed] [Google Scholar]
- Butler SJ, Dodd J. 2003. A role for BMP heterodimers in roof plate-mediated repulsion of commissural axons. Neuron 38: 389–401. [DOI] [PubMed] [Google Scholar]
- Butz H, Racz K, Hunyady L, Patocs A. 2012. Crosstalk between TGF-β signaling and the microRNA machinery. Trends Pharmacol Sci 33: 382–393. [DOI] [PubMed] [Google Scholar]
- Caja L, Kahata K, Moustakas A. 2012. Context-dependent action of transforming growth factor β family members on normal and cancer stem cells. Curr Pharm Des 18: 4072–4086. [DOI] [PubMed] [Google Scholar]
- Chang C, Hemmati-Brivanlou A. 1999. Xenopus GDF6, a new antagonist of noggin and a partner of BMPs. Development 126: 3347–3357. [DOI] [PubMed] [Google Scholar]
- Charbonneau NL, Carlson EJ, Tufa S, Sengle G, Manalo EC, Carlberg VM, Ramirez F, Keene DR, Sakai LY. 2010. In vivo studies of mutant fibrillin-1 microfibrils. J Biol Chem 285: 24943–24955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Shen MM. 2004. Two modes by which Lefty proteins inhibit nodal signaling. Curr Biol 14: 618–624. [DOI] [PubMed] [Google Scholar]
- Chen W, Jin W, Hardegen N, Lei KJ, Li L, Marinos N, McGrady G, Wahl SM. 2003. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-β induction of transcription factor Foxp3. J Exp Med 198: 1875–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Ware SM, Sato A, Houston-Hawkins DE, Habas R, Matzuk MM, Shen MM, Brown CW. 2006. The Vg1-related protein Gdf3 acts in a Nodal signaling pathway in the pre-gastrulation mouse embryo. Development 133: 319–329. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. 2008. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133: 1106–1117. [DOI] [PubMed] [Google Scholar]
- Coffey RJ Jr, Bascom CC, Sipes NJ, Graves-Deal R, Weissman BE, Moses HL. 1988. Selective inhibition of growth-related gene expression in murine keratinocytes by transforming growth factor β. Mol Cell Biol 8: 3088–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallas SL, Miyazono K, Skerry TM, Mundy GR, Bonewald LF. 1995. Dual role for the latent transforming growth factor-β binding protein in storage of latent TGF-β in the extracellular matrix and as a structural matrix protein. J Cell Biol 131: 539–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daopin S, Piez KA, Ogawa Y, Davies DR. 1992. Crystal structure of transforming growth factor-β2: An unusual fold for the superfamily. Science 257: 369–373. [DOI] [PubMed] [Google Scholar]
- Datto MB, Li Y, Panus JF, Howe DJ, Xiong Y, Wang XF. 1995a. Transforming growth factor β induces the cyclin-dependent kinase inhibitor p21 through a p53-independent mechanism. Proc Natl Acad Sci 92: 5545–5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datto MB, Yu Y, Wang XF. 1995b. Functional analysis of the transforming growth factor β responsive elements in the WAF1/Cip1/p21 promoter. J Biol Chem 270: 28623–28628. [DOI] [PubMed] [Google Scholar]
- David L, Mallet C, Mazerbourg S, Feige JJ, Bailly S. 2007. Identification of BMP9 and BMP10 as functional activators of the orphan activin receptor-like kinase 1 (ALK1) in endothelial cells. Blood 109: 1953–1961. [DOI] [PubMed] [Google Scholar]
- David L, Feige JJ, Bailly S. 2009. Emerging role of bone morphogenetic proteins in angiogenesis. Cytokine Growth Factor Rev 20: 203–212. [DOI] [PubMed] [Google Scholar]
- Davis BN, Hilyard AC, Lagna G, Hata A. 2008. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 454: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martin R, Haendler B, Hofer-Warbinek R, Gaugitsch H, Wrann M, Schlusener H, Seifert JM, Bodmer S, Fontana A, Hofer E. 1987. Complementary DNA for human glioblastoma-derived T cell suppressor factor, a novel member of the transforming growth factor-β gene family. EMBO J 6: 3673–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denis M. 1994. Neutralization of transforming growth factor-β1 in a mouse model of immune-induced lung fibrosis. Immunology 82: 584–590. [PMC free article] [PubMed] [Google Scholar]
- Derynck R, Jarrett JA, Chen EY, Eaton DH, Bell JR, Assoian RK, Roberts AB, Sporn MB, Goeddel DV. 1985. Human transforming growth factor-β complementary DNA sequence and expression in normal and transformed cells. Nature 316: 701–705. [DOI] [PubMed] [Google Scholar]
- Derynck R, Lindquist PB, Lee A, Wen D, Tamm J, Graycar JL, Rhee L, Mason AJ, Miller DA, Coffey RJ, et al. 1988. A new type of transforming growth factor-β, TGF-β3. EMBO J 7: 3737–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmouliere A, Geinoz A, Gabbiani F, Gabbiani G. 1993. Transforming growth factor-β1 induces α-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J Cell Biol 122: 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Yu RT, Subramaniam N, Sherman MH, Wilson C, Rao R, Leblanc M, Coulter S, He M, Scott C, et al. 2013. A vitamin D receptor/SMAD genomic circuit gates hepatic fibrotic response. Cell 153: 601–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle AJ, Doyle JJ, Bessling SL, Maragh S, Lindsay ME, Schepers D, Gillis E, Mortier G, Homfray T, Sauls K, et al. 2012. Mutations in the TGF-β repressor SKI cause Shprintzen–Goldberg syndrome with aortic aneurysm. Nat Genet 44: 1249–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DR, Murphy G, Reynolds JJ, Whitham SE, Docherty AJ, Angel P, Heath JK. 1987. Transforming growth factor β modulates the expression of collagenase and metalloproteinase inhibitor. EMBO J 6: 1899–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerman MA, Cadena SM, Gilbert JA, Meyer A, Nelson HN, Swalley SE, Mallozzi C, Jacobi C, Jennings LL, Clay I, et al. 2015. GDF11 increases with age and inhibits skeletal muscle regeneration. Cell Metab 22: 164–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehata S, Johansson E, Katayama R, Koike S, Watanabe A, Hoshino Y, Katsuno Y, Komuro A, Koinuma D, Kano MR, et al. 2011. Transforming growth factor-β decreases the cancer-initiating cell population within diffuse-type gastric carcinoma cells. Oncogene 30: 1693–1705. [DOI] [PubMed] [Google Scholar]
- Eimon PM, Harland RM. 1999. In Xenopus embryos, BMP heterodimers are not required for mesoderm induction, but BMP activity is necessary for dorsal/ventral patterning. Dev Biol 216: 29–40. [DOI] [PubMed] [Google Scholar]
- Eto Y, Tsuji T, Takezawa M, Takano S, Yokogawa Y, Shibai H. 1987. Purification and characterization of erythroid differentiation factor (EDF) isolated from human leukemia cell line THP-1. Biochem Biophys Res Commun 142: 1095–1103. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. 1990. A genetic model for colorectal tumorigenesis. Cell 61: 759–767. [DOI] [PubMed] [Google Scholar]
- Ferguson MW, Duncan J, Bond J, Bush J, Durani P, So K, Taylor L, Chantrey J, Mason T, James G, et al. 2009. Prophylactic administration of avotermin for improvement of skin scarring: Three double-blind, placebo-controlled, phase I/II studies. Lancet 373: 1264–1274. [DOI] [PubMed] [Google Scholar]
- Flanders KC, Sullivan CD, Fujii M, Sowers A, Anzano MA, Arabshahi A, Major C, Deng C, Russo A, Mitchell JB, et al. 2002. Mice lacking Smad3 are protected against cutaneous injury induced by ionizing radiation. Am J Pathol 160: 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell RA, Sanjabi S, Wrzesinski SH, Licona-Limon P. 2010. The polarization of immune cells in the tumour environment by TGFβ. Nat Rev Immunol 10: 554–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florini JR, Roberts AB, Ewton DZ, Falen SL, Flanders KC, Sporn MB. 1986. Transforming growth factor-β. A very potent inhibitor of myoblast differentiation, identical to the differentiation inhibitor secreted by Buffalo rat liver cells. J Biol Chem 261: 16509–16513. [PubMed] [Google Scholar]
- Gaarenstroom T, Hill CS. 2014. TGF-β signaling to chromatin: How Smads regulate transcription during self-renewal and differentiation. Semin Cell Dev Biol 32: 107–118. [DOI] [PubMed] [Google Scholar]
- Gafni O, Weinberger L, Mansour AA, Manor YS, Chomsky E, Ben-Yosef D, Kalma Y, Viukov S, Maza I, Zviran A, et al. 2013. Derivation of novel human ground state naive pluripotent stem cells. Nature 504: 282–286. [DOI] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. 2013. Lessons from the cancer genome. Cell 153: 17–37. [DOI] [PubMed] [Google Scholar]
- Ge G, Hopkins DR, Ho WB, Greenspan DS. 2005. GDF11 forms a bone morphogenetic protein 1–activated latent complex that can modulate nerve growth factor-induced differentiation of PC12 cells. Mol Cell Biol 25: 5846–5858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George J, Roulot D, Koteliansky VE, Bissell DM. 1999. In vivo inhibition of rat stellate cell activation by soluble transforming growth factor β type II receptor: A potential new therapy for hepatic fibrosis. Proc Natl Acad Sci 96: 12719–12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, He W, Wang Q, Seoane J, Lash A, Massagué J. 2006a. A FoxO-Smad synexpression group in human keratinocytes. Proc Natl Acad Sci 103: 12747–12752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomis RR, Alarcon C, Nadal C, Van Poznak C, Massagué J. 2006b. C/EBPβ at the core of the TGFβ cytostatic response and its evasion in metastatic breast cancer cells. Cancer Cell 10: 203–214. [DOI] [PubMed] [Google Scholar]
- Goumans MJ, Valdimarsdottir G, Itoh S, Rosendahl A, Sideras P, ten Dijke P. 2002. Balancing the activation state of the endothelium via two distinct TGF-β type I receptors. EMBO J 21: 1743–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. 2008. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 10: 593–601. [DOI] [PubMed] [Google Scholar]
- Grobet L, Martin LJ, Poncelet D, Pirottin D, Brouwers B, Riquet J, Schoeberlein A, Dunner S, Menissier F, Massabanda J, et al. 1997. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat Genet 17: 71–74. [DOI] [PubMed] [Google Scholar]
- Hahn SA, Schutte M, Hoque AT, Moskaluk CA, da Costa LT, Rozenblum E, Weinstein CL, Fischer A, Yeo CJ, Hruban RH, et al. 1996. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 271: 350–353. [DOI] [PubMed] [Google Scholar]
- Hannon GJ, Beach D. 1994. p15INK4B is a potential effector of TGF-β-induced cell cycle arrest. Nature 371: 257–261. [DOI] [PubMed] [Google Scholar]
- Hassani SN, Totonchi M, Farrokhi A, Taei A, Larijani MR, Gourabi H, Baharvand H. 2012. Simultaneous suppression of TGF-β and ERK signaling contributes to the highly efficient and reproducible generation of mouse embryonic stem cells from previously considered refractory and non-permissive strains. Stem Cell Rev 8: 472–481. [DOI] [PubMed] [Google Scholar]
- Hata A, Lieberman J. 2015. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci Signal 8: re3. [DOI] [PubMed] [Google Scholar]
- Hemmati-Brivanlou A, Kelly OG, Melton DA. 1994. Follistatin, an antagonist of activin, is expressed in the Spemann organizer and displays direct neuralizing activity. Cell 77: 283–295. [DOI] [PubMed] [Google Scholar]
- Horiguchi M, Ota M, Rifkin DB. 2012. Matrix control of transforming growth factor-β function. J Biochem 152: 321–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Y, Nishida J, Katsuno Y, Koinuma D, Aoki T, Kokudo N, Miyazono K, Ehata S. 2015. Smad4 decreases the population of pancreatic cancer-initiating cells through transcriptional repression of ALDH1A1. Am J Pathol 185: 1457–1470. [DOI] [PubMed] [Google Scholar]
- Iavarone A, Massagué J. 1997. Repression of the CDK activator Cdc25A and cell-cycle arrest by cytokine TGF-β in cells lacking the CDK inhibitor p15. Nature 387: 417–422. [DOI] [PubMed] [Google Scholar]
- Ignotz RA, Massagué J. 1985. Type β transforming growth factor controls the adipogenic differentiation of 3T3 fibroblasts. Proc Natl Acad Sci 82: 8530–8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignotz RA, Massagué J. 1986. Transforming growth factor β stimulates the expression of fibronectin and collagen and their incorporation into the extracellular matrix. J Biol Chem 261: 4337–4345. [PubMed] [Google Scholar]
- Ikushima H, Miyazono K. 2010. TGFβ signalling: A complex web in cancer progression. Nat Rev Cancer 10: 415–424. [DOI] [PubMed] [Google Scholar]
- Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. 2009. Autocrine TGF-β signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell 5: 504–514. [DOI] [PubMed] [Google Scholar]
- Imbeaud S, Faure E, Lamarre I, Mattei MG, di Clemente N, Tizard R, Carre-Eusebe D, Belville C, Tragethon L, Tonkin C, et al. 1995. Insensitivity to anti-Müllerian hormone due to a mutation in the human anti-Müllerian hormone receptor. Nat Genet 11: 382–388. [DOI] [PubMed] [Google Scholar]
- Isogai Z, Ono RN, Ushiro S, Keene DR, Chen Y, Mazzieri R, Charbonneau NL, Reinhardt DP, Rifkin DB, Sakai LY. 2003. Latent transforming growth factor β-binding protein 1 interacts with fibrillin and is a microfibril-associated protein. J Biol Chem 278: 2750–2757. [DOI] [PubMed] [Google Scholar]
- Israel DI, Nove J, Kerns KM, Kaufman RJ, Rosen V, Cox KA, Wozney JM. 1996. Heterodimeric bone morphogenetic proteins show enhanced activity in vitro and in vivo. Growth Factors 13: 291–300. [DOI] [PubMed] [Google Scholar]
- Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. 2002. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest 110: 341–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- James D, Levine AJ, Besser D, Hemmati-Brivanlou A. 2005. TGFβ/activin/nodal signaling is necessary for the maintenance of pluripotency in human embryonic stem cells. Development 132: 1273–1282. [DOI] [PubMed] [Google Scholar]
- Jamin SP, Arango NA, Mishina Y, Hanks MC, Behringer RR. 2002. Requirement of Bmpr1a for Müllerian duct regression during male sexual development. Nat Genet 32: 408–410. [DOI] [PubMed] [Google Scholar]
- Janssens K, Gershoni-Baruch R, Guanabens N, Migone N, Ralston S, Bonduelle M, Lissens W, Van Maldergem L, Vanhoenacker F, Verbruggen L, et al. 2000. Mutations in the gene encoding the latency-associated peptide of TGF-β1 cause Camurati–Engelmann disease. Nat Genet 26: 273–275. [DOI] [PubMed] [Google Scholar]
- Jian H, Shen X, Liu I, Semenov M, He X, Wang XF. 2006. Smad3-dependent nuclear translocation of β-catenin is required for TGF-β1-induced proliferation of bone marrow-derived adult human mesenchymal stem cells. Genes Dev 20: 666–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judge DP, Biery NJ, Keene DR, Geubtner J, Myers L, Huso DL, Sakai LY, Dietz HC. 2004. Evidence for a critical contribution of haploinsufficiency in the complex pathogenesis of Marfan syndrome. J Clin Invest 114: 172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaartinen V, Voncken JW, Shuler C, Warburton D, Bu D, Heisterkamp N, Groffen J. 1995. Abnormal lung development and cleft palate in mice lacking TGF-β3 indicates defects of epithelial-mesenchymal interaction. Nat Genet 11: 415–421. [DOI] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. 2009. The basics of epithelial-mesenchymal transition. J Clin Invest 119: 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambadur R, Sharma M, Smith TP, Bass JJ. 1997. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res 7: 910–916. [DOI] [PubMed] [Google Scholar]
- Kane CJ, Hebda PA, Mansbridge JN, Hanawalt PC. 1991. Direct evidence for spatial and temporal regulation of transforming growth factor β1 expression during cutaneous wound healing. J Cell Physiol 148: 157–173. [DOI] [PubMed] [Google Scholar]
- Kang Y, Chen CR, Massagué J. 2003. A self-enabling TGFβ response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell 11: 915–926. [DOI] [PubMed] [Google Scholar]
- Katsuno Y, Lamouille S, Derynck R. 2013. TGF-β signaling and epithelial-mesenchymal transition in cancer progression. Curr Opin Oncol 25: 76–84. [DOI] [PubMed] [Google Scholar]
- Kawabata M, Inoue H, Hanyu A, Imamura T, Miyazono K. 1998. Smad proteins exist as monomers in vivo and undergo homo- and hetero-oligomerization upon activation by serine/threonine kinase receptors. EMBO J 17: 4056–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrl JH, Wakefield LM, Roberts AB, Jakowlew S, Alvarez-Mon M, Derynck R, Sporn MB, Fauci AS. 1986. Production of transforming growth factor β by human T lymphocytes and its potential role in the regulation of T cell growth. J Exp Med 163: 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Kugler MC, Wolters PJ, Robillard L, Galvez MG, Brumwell AN, Sheppard D, Chapman HA. 2006. Alveolar epithelial cell mesenchymal transition develops in vivo during pulmonary fibrosis and is regulated by the extracellular matrix. Proc Natl Acad Sci 103: 13180–13185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi A, Wang XF, Weinberg RA, Cheifetz S, Massagué J. 1988. Absence of TGF-β receptors and growth inhibitory responses in retinoblastoma cells. Science 240: 196–199. [DOI] [PubMed] [Google Scholar]
- Kimelman D, Kirschner M. 1987. Synergistic induction of mesoderm by FGF and TGF-β and the identification of an mRNA coding for FGF in the early Xenopus embryo. Cell 51: 869–877. [DOI] [PubMed] [Google Scholar]
- Kinoshita A, Saito T, Tomita H, Makita Y, Yoshida K, Ghadami M, Yamada K, Kondo S, Ikegawa S, Nishimura G, et al. 2000. Domain-specific mutations in TGFB1 result in Camurati-Engelmann disease. Nat Genet 26: 19–20. [DOI] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Imamura T, Aburatani H, Miyazono K. 2009a. Promoter-wide analysis of Smad4 binding sites in human epithelial cells. Cancer Sci 100: 2133–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma D, Tsutsumi S, Kamimura N, Taniguchi H, Miyazawa K, Sunamura M, Imamura T, Miyazono K, Aburatani H. 2009b. Chromatin immunoprecipitation on microarray analysis of Smad2/3 binding sites reveals roles of ETS1 and TFAP2A in transforming growth factor β signaling. Mol Cell Biol 29: 172–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T, Bettelli E, Gao W, Awasthi A, Jager A, Strom TB, Oukka M, Kuchroo VK. 2007. IL-21 initiates an alternative pathway to induce proinflammatory TH17 cells. Nature 448: 484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieglstein K, Zheng F, Unsicker K, Alzheimer C. 2011. More than being protective: Functional roles for TGF-β/activin signaling pathways at central synapses. Trends Neurosci 34: 421–429. [DOI] [PubMed] [Google Scholar]
- Kuwahara F, Kai H, Tokuda K, Kai M, Takeshita A, Egashira K, Imaizumi T. 2002. Transforming growth factor-β function blocking prevents myocardial fibrosis and diastolic dysfunction in pressure-overloaded rats. Circulation 106: 130–135. [DOI] [PubMed] [Google Scholar]
- Labelle M, Begum S, Hynes RO. 2011. Direct signaling between platelets and cancer cells induces an epithelial-mesenchymal-like transition and promotes metastasis. Cancer Cell 20: 576–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagna G, Hata A, Hemmati-Brivanlou A, Massagué J. 1996. Partnership between DPC4 and SMAD proteins in TGF-β signalling pathways. Nature 383: 832–836. [DOI] [PubMed] [Google Scholar]
- Lee SJ, McPherron AC. 2001. Regulation of myostatin activity and muscle growth. Proc Natl Acad Sci 98: 9306–9311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KL, Lim SK, Orlov YL, Yit le Y, Yang H, Ang LT, Poellinger L, Lim B. 2011. Graded Nodal/Activin signaling titrates conversion of quantitative phospho-Smad2 levels into qualitative embryonic stem cell fate decisions. PLoS Genet 7: e1002130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leof EB, Proper JA, Goustin AS, Shipley GD, DiCorleto PE, Moses HL. 1986. Induction of c-sis mRNA and activity similar to platelet-derived growth factor by transforming growth factor β: A proposed model for indirect mitogenesis involving autocrine activity. Proc Natl Acad Sci 83: 2453–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine AJ, Brivanlou AH. 2006. GDF3, a BMP inhibitor, regulates cell fate in stem cells and early embryos. Development 133: 209–216. [DOI] [PubMed] [Google Scholar]
- Levine AJ, Levine ZJ, Brivanlou AH. 2009. GDF3 is a BMP inhibitor that can activate Nodal signaling only at very high doses. Dev Biol 325: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MO, Flavell RA. 2008. TGF-β: A master of all T cell trades. Cell 134: 392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Dietz HC. 2011. Lessons on the pathogenesis of aneurysm from heritable conditions. Nature 473: 308–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay ME, Schepers D, Bolar NA, Doyle JJ, Gallo E, Fert-Bober J, Kempers MJ, Fishman EK, Chen Y, Myers L, et al. 2012. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nat Genet 44: 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little SC, Mullins MC. 2009. Bone morphogenetic protein heterodimers assemble heteromeric type I receptor complexes to pattern the dorsoventral axis. Nat Cell Biol 11: 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Zhang P, Li J, Kulkarni AB, Perruche S, Chen W. 2008. A critical function for TGF-β signaling in the development of natural CD4+CD25+Foxp3+ regulatory T cells. Nat Immunol 9: 632–640. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Chen J, Neptune ER, Judge DP, Podowski M, Holm T, Meyers J, Leitch CC, Katsanis N, Sharifi N, et al. 2005. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet 37: 275–281. [DOI] [PubMed] [Google Scholar]
- Loeys BL, Schwarze U, Holm T, Callewaert BL, Thomas GH, Pannu H, De Backer JF, Oswald GL, Symoens S, Manouvrier S, et al. 2006. Aneurysm syndromes caused by mutations in the TGF-β receptor. N Engl J Med 355: 788–798. [DOI] [PubMed] [Google Scholar]
- Loffredo FS, Steinhauser ML, Jay SM, Gannon J, Pancoast JR, Yalamanchi P, Sinha M, Dall’Osso C, Khong D, Shadrach JL, et al. 2013. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153: 828–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonardo E, Hermann PC, Mueller MT, Huber S, Balic A, Miranda-Lorenzo I, Zagorac S, Alcala S, Rodriguez-Arabaolaza I, Ramirez JC, et al. 2011. Nodal/Activin signaling drives self-renewal and tumorigenicity of pancreatic cancer stem cells and provides a target for combined drug therapy. Cell Stem Cell 9: 433–446. [DOI] [PubMed] [Google Scholar]
- Lund LR, Riccio A, Andreasen PA, Nielsen LS, Kristensen P, Laiho M, Saksela O, Blasi F, Dano K. 1987. Transforming growth factor-β is a strong and fast acting positive regulator of the level of type-1 plasminogen activator inhibitor mRNA in WI-38 human lung fibroblasts. EMBO J 6: 1281–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda S, Hayashi M, Komiya S, Imamura T, Miyazono K. 2004. Endogenous TGF-β signaling suppresses maturation of osteoblastic mesenchymal cells. EMBO J 23: 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. 1995. Inactivation of the type II TGF-β receptor in colon cancer cells with microsatellite instability. Science 268: 1336–1338. [DOI] [PubMed] [Google Scholar]
- Massagué J, Cheifetz S, Endo T, Nadal-Ginard B. 1986. Type β transforming growth factor is an inhibitor of myogenic differentiation. Proc Natl Acad Sci 83: 8206–8210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews LS, Vale WW. 1991. Expression cloning of an activin receptor, a predicted transmembrane serine kinase. Cell 65: 973–982. [DOI] [PubMed] [Google Scholar]
- Matsuyama S, Iwadate M, Kondo M, Saitoh M, Hanyu A, Shimizu K, Aburatani H, Mishima HK, Imamura T, Miyazono K, et al. 2003. SB-431542 and Gleevec inhibit transforming growth factor-β-induced proliferation of human osteosarcoma cells. Cancer Res 63: 7791–7798. [PubMed] [Google Scholar]
- Mazerbourg S, Klein C, Roh J, Kaivo-Oja N, Mottershead DG, Korchynskyi O, Ritvos O, Hsueh AJ. 2004. Growth differentiation factor-9 signaling is mediated by the type I receptor, activin receptor-like kinase 5. Mol Endocrinol 18: 653–665. [DOI] [PubMed] [Google Scholar]
- McPherron AC, Lee SJ. 1997. Double muscling in cattle due to mutations in the myostatin gene. Proc Natl Acad Sci 94: 12457–12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron AC, Lawler AM, Lee SJ. 1997. Regulation of skeletal muscle mass in mice by a new TGF-β superfamily member. Nature 387: 83–90. [DOI] [PubMed] [Google Scholar]
- Meynard D, Kautz L, Darnaud V, Canonne-Hergaux F, Coppin H, Roth MP. 2009. Lack of the bone morphogenetic protein BMP6 induces massive iron overload. Nat Genet 41: 478–481. [DOI] [PubMed] [Google Scholar]
- Mi LZ, Brown CT, Gao Y, Tian Y, Le VQ, Walz T, Springer TA. 2015. Structure of bone morphogenetic protein 9 procomplex. Proc Natl Acad Sci 112: 3710–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi T, Collod-Beroud G, Akiyama T, Abifadel M, Harada N, Morisaki T, Allard D, Varret M, Claustres M, Morisaki H, et al. 2004. Heterozygous TGFBR2 mutations in Marfan syndrome. Nat Genet 36: 855–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RK, Otsuka F, Shimasaki S. 2003. Molecular basis of bone morphogenetic protein-15 signaling in granulosa cells. J Biol Chem 278: 304–310. [DOI] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Tsutsumi S, Vasilaki E, Kanki Y, Heldin CH, Aburatani H, Miyazono K. 2011. ChIP-seq reveals cell type-specific binding patterns of BMP-specific Smads and a novel binding motif. Nucleic Acids Res 39: 8712–8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikawa M, Koinuma D, Miyazono K, Heldin CH. 2013. Genome-wide mechanisms of Smad binding. Oncogene 32: 1609–1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga C, Saito D, Nakamura S, Sasaki T, Asakawa S, Shimizu N, Mitani H, Furutani-Seiki M, Tanaka M, Kondoh H. 2007. The hotei mutation of medaka in the anti-Müllerian hormone receptor causes the dysregulation of germ cell and sexual development. Proc Natl Acad Sci 104: 9691–9696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. 2011. Master transcription factors determine cell-type-specific responses to TGF-β signaling. Cell 147: 565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger JS, Huang X, Kawakatsu H, Griffiths MJ, Dalton SL, Wu J, Pittet JF, Kaminski N, Garat C, Matthay MA, et al. 1999. The integrin αvβ6 binds and activates latent TGFβ1: A mechanism for regulating pulmonary inflammation and fibrosis. Cell 96: 319–328. [DOI] [PubMed] [Google Scholar]
- Mustoe TA, Pierce GF, Thomason A, Gramates P, Sporn MB, Deuel TF. 1987. Accelerated healing of incisional wounds in rats induced by transforming growth factor-β. Science 237: 1333–1336. [DOI] [PubMed] [Google Scholar]
- Naka K, Hoshii T, Muraguchi T, Tadokoro Y, Ooshio T, Kondo Y, Nakao S, Motoyama N, Hirao A. 2010. TGF-β-FOXO signalling maintains leukaemia-initiating cells in chronic myeloid leukaemia. Nature 463: 676–680. [DOI] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin CH, et al. 1997. Identification of Smad7, a TGFβ-inducible antagonist of TGF-β signalling. Nature 389: 631–635. [DOI] [PubMed] [Google Scholar]
- Nakao A, Fujii M, Matsumura R, Kumano K, Saito Y, Miyazono K, Iwamoto I. 1999. Transient gene transfer and expression of Smad7 prevents bleomycin-induced lung fibrosis in mice. J Clin Invest 104: 5–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neptune ER, Frischmeyer PA, Arking DE, Myers L, Bunton TE, Gayraud B, Ramirez F, Sakai LY, Dietz HC. 2003. Dysregulation of TGF-β activation contributes to pathogenesis in Marfan syndrome. Nat Genet 33: 407–411. [DOI] [PubMed] [Google Scholar]
- Ng F, Boucher S, Koh S, Sastry KS, Chase L, Lakshmipathy U, Choong C, Yang Z, Vemuri MC, Rao MS, et al. 2008. PDGF, TGF-β, and FGF signaling is important for differentiation and growth of mesenchymal stem cells (MSCs): Transcriptional profiling can identify markers and signaling pathways important in differentiation of MSCs into adipogenic, chondrogenic, and osteogenic lineages. Blood 112: 295–307. [DOI] [PubMed] [Google Scholar]
- Niehrs C, Pollet N. 1999. Synexpression groups in eukaryotes. Nature 402: 483–487. [DOI] [PubMed] [Google Scholar]
- Ogawa K, Saito A, Matsui H, Suzuki H, Ohtsuka S, Shimosato D, Morishita Y, Watabe T, Niwa H, Miyazono K. 2007. Activin-nodal signaling is involved in propagation of mouse embryonic stem cells. J Cell Sci 120: 55–65. [DOI] [PubMed] [Google Scholar]
- Oh SP, Yeo CY, Lee Y, Schrewe H, Whitman M, Li E. 2002. Activin type IIA and IIB receptors mediate Gdf11 signaling in axial vertebral patterning. Genes Dev 16: 2749–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Greenberger JS, Anklesaria P, Bassols A, Massagué J. 1987. Two forms of transforming growth factor-β distinguished by multipotential haematopoietic progenitor cells. Nature 329: 539–541. [DOI] [PubMed] [Google Scholar]
- Oshimori N, Fuchs E. 2012. The harmonies played by TGF-β in stem cell biology. Cell Stem Cell 11: 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palumbo-Zerr K, Zerr P, Distler A, Fliehr J, Mancuso R, Huang J, Mielenz D, Tomcik M, Furnrohr BG, Scholtysek C, et al. 2015. Orphan nuclear receptor NR4A1 regulates transforming growth factor-β signaling and fibrosis. Nat Med 21: 150–158. [DOI] [PubMed] [Google Scholar]
- Pardali E, Goumans MJ, ten Dijke P. 2010. Signaling by members of the TGF-β family in vascular morphogenesis and disease. Trends Cell Biol 20: 556–567. [DOI] [PubMed] [Google Scholar]
- Park SM, Gaur AB, Lengyel E, Peter ME. 2008. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev 22: 894–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Li Q, Wigglesworth K, Rangarajan A, Kattamuri C, Peterson RT, Eppig JJ, Thompson TB, Matzuk MM. 2013. Growth differentiation factor 9:bone morphogenetic protein 15 heterodimers are potent regulators of ovarian functions. Proc Natl Acad Sci 10: E776–E785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penuelas S, Anido J, Prieto-Sanchez RM, Folch G, Barba I, Cuartas I, Garcia-Dorado D, Poca MA, Sahuquillo J, Baselga J, et al. 2009. TGF-β increases glioma-initiating cell self-renewal through the induction of LIF in human glioblastoma. Cancer Cell 15: 315–327. [DOI] [PubMed] [Google Scholar]
- Pickup M, Novitskiy S, Moses HL. 2013. The roles of TGFβ in the tumour microenvironment. Nat Rev Cancer 13: 788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietenpol JA, Stein RW, Moran E, Yaciuk P, Schlegel R, Lyons RM, Pittelkow MR, Munger K, Howley PM, Moses HL. 1990. TGF-β1 inhibition of c-myc transcription and growth in keratinocytes is abrogated by viral transforming proteins with pRB binding domains. Cell 61: 777–785. [DOI] [PubMed] [Google Scholar]
- Polyak K, Kato JY, Solomon MJ, Sherr CJ, Massagué J, Roberts JM, Koff A. 1994. p27Kip1, a cyclin-Cdk inhibitor, links transforming growth factor-β and contact inhibition to cell cycle arrest. Genes Dev 8: 9–22. [DOI] [PubMed] [Google Scholar]
- Postlethwaite AE, Keski-Oja J, Moses HL, Kang AH. 1987. Stimulation of the chemotactic migration of human fibroblasts by transforming growth factor β. J Exp Med 165: 251–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H, Chan MW, Liyanarachchi S, Balch C, Potter D, Souriraj IJ, Cheng AS, Agosto-Perez FJ, Nikonova EV, Yan PS, et al. 2009. An integrative ChIP-chip and gene expression profiling to model SMAD regulatory modules. BMC Syst Biol 3: 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery LA, Twombly V, Wharton K, Gelbart WM. 1995. Genetic screens to identify elements of the decapentaplegic signaling pathway in Drosophila. Genetics 139: 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow HJ. 1986. BSC-1 growth inhibitor/type β transforming growth factor is a strong inhibitor of thymocyte proliferation. Proc Natl Acad Sci 83: 5531–5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Lamb LC, Newton DL, Sporn MB, De Larco JE, Todaro GJ. 1980. Transforming growth factors: Isolation of polypeptides from virally and chemically transformed cells by acid/ethanol extraction. Proc Natl Acad Sci 77: 3494–3498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Anzano MA, Lamb LC, Smith JM, Sporn MB. 1981. New class of transforming growth factors potentiated by epidermal growth factor: Isolation from non-neoplastic tissues. Proc Natl Acad Sci 78: 5339–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Anzano MA, Meyers CA, Wideman J, Blacher R, Pan YC, Stein S, Lehrman SR, Smith JM, Lamb LC, et al. 1983. Purification and properties of a type β transforming growth factor from bovine kidney. Biochemistry 22: 5692–5698. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Anzano MA, Wakefield LM, Roche NS, Stern DF, Sporn MB. 1985. Type β transforming growth factor: A bifunctional regulator of cellular growth. Proc Natl Acad Sci 82: 119–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AB, Sporn MB, Assoian RK, Smith JM, Roche NS, Wakefield LM, Heine UI, Liotta LA, Falanga V, Kehrl JH, et al. 1986. Transforming growth factor type β: Rapid induction of fibrosis and angiogenesis in vivo and stimulation of collagen formation in vitro. Proc Natl Acad Sci 83: 4167–4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa F, Roberts AB, Danielpour D, Dart LL, Sporn MB, Dawid IB. 1988. Mesoderm induction in amphibians: The role of TGF-β2-like factors. Science 239: 783–785. [DOI] [PubMed] [Google Scholar]
- Russell WE, Coffey RJ Jr, Ouellette AJ, Moses HL. 1988. Type β transforming growth factor reversibly inhibits the early proliferative response to partial hepatectomy in the rat. Proc Natl Acad Sci 85: 5126–5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito T, Kinoshita A, Yoshiura K, Makita Y, Wakui K, Honke K, Niikawa N, Taniguchi N. 2001. Domain-specific mutations of a transforming growth factor (TGF)-β1 latency-associated peptide cause Camurati–Engelmann disease because of the formation of a constitutively active form of TGF-β1. J Biol Chem 276: 11469–11472. [DOI] [PubMed] [Google Scholar]
- Sakaki-Yumoto M, Katsuno Y, Derynck R. 2013. TGF-β family signaling in stem cells. Biochim Biophys Acta 1830: 2280–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanford LP, Ormsby I, Gittenberger-de Groot AC, Sariola H, Friedman R, Boivin GP, Cardell EL, Doetschman T. 1997. TGFβ2 knockout mice have multiple developmental defects that are non-overlapping with other TGFβ knockout phenotypes. Development 124: 2659–2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori R, Schirwis E, Blaauw B, Bortolanza S, Zhao J, Enzo E, Stantzou A, Mouisel E, Toniolo L, Ferry A, et al. 2013. BMP signaling controls muscle mass. Nat Genet 45: 1309–1318. [DOI] [PubMed] [Google Scholar]
- Sato M, Muragaki Y, Saika S, Roberts AB, Ooshima A. 2003. Targeted disruption of TGF-β1/Smad3 signaling protects against renal tubulointerstitial fibrosis induced by unilateral ureteral obstruction. J Clin Invest 112: 1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage C, Das P, Finelli AL, Townsend SR, Sun CY, Baird SE, Padgett RW. 1996. Caenorhabditis elegans genes sma-2, sma-3, and sma-4 define a conserved family of transforming growth factor β pathway components. Proc Natl Acad Sci 93: 790–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlunegger MP, Grütter MG. 1992. An unusual feature revealed by the crystal structure at 2.2 Å resolution of human transforming growth factor-β2. Nature 358: 430–434. [DOI] [PubMed] [Google Scholar]
- Schubert D, Kimura H, LaCorbiere M, Vaughan J, Karr D, Fischer WH. 1990. Activin is a nerve cell survival molecule. Nature 344: 868–870. [DOI] [PubMed] [Google Scholar]
- Sekelsky JJ, Newfeld SJ, Raftery LA, Chartoff EH, Gelbart WM. 1995. Genetic characterization and cloning of mothers against dpp, a gene required for decapentaplegic function in Drosophila melanogaster. Genetics 139: 1347–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengle G, Charbonneau NL, Ono RN, Sasaki T, Alvarez J, Keene DR, Bachinger HP, Sakai LY. 2008. Targeting of bone morphogenetic protein growth factor complexes to fibrillin. J Biol Chem 283: 13874–13888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedin SM, Thomas TC, Thompson AY, Rosen DM, Piez KA. 1985. Purification and characterization of two cartilage-inducing factors from bovine demineralized bone. Proc Natl Acad Sci 82: 2267–2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M, Foreman DM, Ferguson MW. 1995. Neutralisation of TGF-β1 and TGF-β2 or exogenous addition of TGF-β3 to cutaneous rat wounds reduces scarring. J Cell Sci 108: 985–1002. [DOI] [PubMed] [Google Scholar]
- Shi M, Zhu J, Wang R, Chen X, Mi L, Walz T, Springer TA. 2011. Latent TGF-β structure and activation. Nature 474: 343–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipley GD, Childs CB, Volkenant ME, Moses HL. 1984. Differential effects of epidermal growth factor, transforming growth factor, and insulin on DNA and protein synthesis and morphology in serum-free cultures of AKR-2B cells. Cancer Res 44: 710–716. [PubMed] [Google Scholar]
- Shipley GD, Pittelkow MR, Wille JJ Jr, Scott RE, Moses HL. 1986. Reversible inhibition of normal human prokeratinocyte proliferation by type β transforming growth factor-growth inhibitor in serum-free medium. Cancer Res 46: 2068–2071. [PubMed] [Google Scholar]
- Shirakihara T, Horiguchi K, Miyazawa K, Ehata S, Shibata T, Morita I, Miyazono K, Saitoh M. 2011. TGF-β regulates isoform switching of FGF receptors and epithelial-mesenchymal transition. EMBO J 30: 783–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shull MM, Ormsby I, Kier AB, Pawlowski S, Diebold RJ, Yin M, Allen R, Sidman C, Proetzel G, Calvin D, et al. 1992. Targeted disruption of the mouse transforming growth factor-β1 gene results in multifocal inflammatory disease. Nature 359: 693–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Massagué J. 2003. Cytostatic and apoptotic actions of TGF-β in homeostasis and cancer. Nat Rev Cancer 3: 807–821. [DOI] [PubMed] [Google Scholar]
- Siegel PM, Massagué J. 2008. Growth control by TGF-β: Mechanisms controlling cell cycle progression and apoptosis. The TGF-β family (eds. Derynck R, Miyazono K), pp. 333–362. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberstein GB, Daniel CW. 1987. Reversible inhibition of mammary gland growth by transforming growth factor-β. Science 237: 291–293. [DOI] [PubMed] [Google Scholar]
- Sime PJ, Xing Z, Graham FL, Csaky KG, Gauldie J. 1997. Adenovector-mediated gene transfer of active transforming growth factor-β1 induces prolonged severe fibrosis in rat lung. J Clin Invest 100: 768–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha M, Jang YC, Oh J, Khong D, Wu EY, Manohar R, Miller C, Regalado SG, Loffredo FS, Pancoast JR, et al. 2014. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science 344: 649–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WC, Harland RM. 1992. Expression cloning of noggin, a new dorsalizing factor localized to the Spemann organizer in Xenopus embryos. Cell 70: 829–840. [DOI] [PubMed] [Google Scholar]
- Sporn MB, Roberts AB, Shull JH, Smith JM, Ward JM, Sodek J. 1983. Polypeptide transforming growth factors isolated from bovine sources and used for wound healing in vivo. Science 219: 1329–1331. [DOI] [PubMed] [Google Scholar]
- Suzuki A, Kaneko E, Maeda J, Ueno N. 1997. Mesoderm induction by BMP-4 and -7 heterodimers. Biochem Biophys Res Commun 232: 153–156. [DOI] [PubMed] [Google Scholar]
- Tabibzadeh S, Hemmati-Brivanlou A. 2006. Lefty at the crossroads of “stemness” and differentiative events. Stem Cells 24: 1998–2006. [DOI] [PubMed] [Google Scholar]
- Taipale J, Miyazono K, Heldin CH, Keski-Oja J. 1994. Latent transforming growth factor-β1 associates to fibroblast extracellular matrix via latent TGF-β binding protein. J Cell Biol 124: 171–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Vu M, Booker T, Santner SJ, Miller FR, Anver MR, Wakefield LM. 2003. TGF-β switches from tumor suppressor to prometastatic factor in a model of breast cancer progression. J Clin Invest 112: 1116–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, Yoo N, Vu M, Mamura M, Nam JS, Ooshima A, Du Z, Desprez PY, Anver MR, Michalowska AM, et al. 2007. Transforming growth factor-β can suppress tumorigenesis through effects on the putative cancer stem or early progenitor cell and committed progeny in a breast cancer xenograft model. Cancer Res 67: 8643–8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Wu X, Lei W, Pang L, Wan C, Shi Z, Zhao L, Nagy TR, Peng X, Hu J, et al. 2009. TGF-β1-induced migration of bone mesenchymal stem cells couples bone resorption with formation. Nat Med 15: 757–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theunissen TW, Powell BE, Wang H, Mitalipova M, Faddah DA, Reddy J, Fan ZP, Maetzel D, Ganz K, Shi L, et al. 2014. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 15: 471–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trompouki E, Bowman TV, Lawton LN, Fan ZP, Wu DC, DiBiase A, Martin CS, Cech JN, Sessa AK, Leblanc JL, et al. 2011. Lineage regulators direct BMP and Wnt pathways to cell-specific programs during differentiation and regeneration. Cell 147: 577–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker RF, Shipley GD, Moses HL, Holley RW. 1984. Growth inhibitor from BSC-1 cells closely related to platelet type β transforming growth factor. Science 226: 705–707. [DOI] [PubMed] [Google Scholar]
- Vallier L, Alexander M, Pedersen RA. 2005. Activin/nodal and FGF pathways cooperate to maintain pluripotency of human embryonic stem cells. J Cell Sci 118: 4495–4509. [DOI] [PubMed] [Google Scholar]
- van de Laar IM, Oldenburg RA, Pals G, Roos-Hesselink JW, de Graaf BM, Verhagen JM, Hoedemaekers YM, Willemsen R, Severijnen LA, Venselaar H, et al. 2011. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet 43: 121–126. [DOI] [PubMed] [Google Scholar]
- Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. 2006. TGFβ in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 24: 179–189. [DOI] [PubMed] [Google Scholar]
- Vogel T, Ahrens S, Buttner N, Krieglstein K. 2010. Transforming growth factor β promotes neuronal cell fate of mouse cortical and hippocampal progenitors in vitro and in vivo: Identification of Nedd9 as an essential signaling component. Cereb Cortex 20: 661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA Jr, Kinzler KW. 2013. Cancer genome landscapes. Science 339: 1546–1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl SM, Hunt DA, Wakefield LM, McCartney-Francis N, Wahl LM, Roberts AB, Sporn MB. 1987. Transforming growth factor type β induces monocyte chemotaxis and growth factor production. Proc Natl Acad Sci 84: 5788–5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CA, di Clemente N, Ehrenfels C, Pepinsky RB, Josso N, Vigier B, Cate RL. 1993. Müllerian inhibiting substance requires its N-terminal domain for maintenance of biological activity, a novel finding within the transforming growth factor-β superfamily. Mol Endocrinol 7: 247–257. [DOI] [PubMed] [Google Scholar]
- Wolfman NM, McPherron AC, Pappano WN, Davies MV, Song K, Tomkinson KN, Wright JF, Zhao L, Sebald SM, Greenspan DS, et al. 2003. Activation of latent myostatin by the BMP-1/tolloid family of metalloproteinases. Proc Natl Acad Sci 100: 15842–15846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SP, Theodorescu D, Kerbel RS, Willson JK, Mulder KM, Humphrey LE, Brattain MG. 1992. TGF-β1 is an autocrine-negative growth regulator of human colon carcinoma FET cells in vivo as revealed by transfection of an antisense expression vector. J Cell Biol 116: 187–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Melichian DS, Chang E, Warner-Blankenship M, Ghosh AK, Varga J. 2009. Rosiglitazone abrogates bleomycin-induced scleroderma and blocks profibrotic responses through peroxisome proliferator-activated receptor-γ. Am J Pathol 174: 519–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Mu Z, Dabovic B, Jurukovski V, Yu D, Sung J, Xiong X, Munger JS. 2007. Absence of integrin-mediated TGFβ1 activation in vivo recapitulates the phenotype of TGFβ1-null mice. J Cell Biol 176: 787–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinaga K, Obata H, Jurukovski V, Mazzieri R, Chen Y, Zilberberg L, Huso D, Melamed J, Prijatelj P, Todorovic V, et al. 2008. Perturbation of transforming growth factor (TGF)-β1 association with latent TGF-β binding protein yields inflammation and tumors. Proc Natl Acad Sci 105: 18758–18763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Shao LE, Lemas V, Yu AL, Vaughan J, Rivier J, Vale W. 1987. Importance of FSH-releasing protein and inhibin in erythrodifferentiation. Nature 330: 765–767. [DOI] [PubMed] [Google Scholar]
- Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, et al. 2007. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med 13: 952–961. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Feng X, We R, Derynck R. 1996. Receptor-associated Mad homologues synergize as effectors of the TGF-β response. Nature 383: 168–172. [DOI] [PubMed] [Google Scholar]
- Zhao J, Shi W, Wang YL, Chen H, Bringas P Jr, Datto MB, Frederick JP, Wang XF, Warburton D. 2002. Smad3 deficiency attenuates bleomycin-induced pulmonary fibrosis in mice. Am J Physiol Lung Cell Mol Physiol 282: L585–L593. [DOI] [PubMed] [Google Scholar]
- Zhou X, Wang JL, Lu J, Song Y, Kwak KS, Jiao Q, Rosenfeld R, Chen Q, Boone T, Simonet WS, et al. 2010. Reversal of cancer cachexia and muscle wasting by ActRIIB antagonism leads to prolonged survival. Cell 142: 531–543. [DOI] [PubMed] [Google Scholar]