Abstract
The clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated (Cas) system has been broadly adopted for highly efficient genome editing in a variety of model organisms and human cell types. Unlike previous genome editing technologies such as Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs), the CRISPR/Cas technology does not require complex protein engineering and can be utilized by any researcher proficient in basic molecular biology and cell culture techniques. Here we describe protocols for design and cloning of vectors expressing single or multiplex gRNAs, for transient transfection of human cell lines, and for quantitation of mutation frequencies by T7 Endonuclease I assay. These protocols also include guidance for using two improvements that increase the specificity of CRISPR/Cas nucleases: truncated gRNAs and dimeric RNA-guided FokI nucleases.
Keywords: CRISPR, Cas9, genome editing, human cells, tru-gRNA, RNA-guided FokI nucleases, FokI-dCas9
INTRODUCTION
The advant of engineered Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)/CRISPR- associated (Cas) nucleases has transformed our ability to introduce targeted modifications into the genome of human cells and a wide variety of organisms (Sander and Joung, 2014). Previous genome editing technologies like Zinc Finger Nucleases (ZFNs) or Transcription Activator-Like Effector Nucleases (TALENs) (Reyon et al., 2012) required laborious and expensive protein engineering strategies. By contrast, engineered CRISPR/Cas nucleases can be easily practiced in any laboratory and thereby make genome editing accessible to any scientist. Many of the plasmids for CRISPR-based genome editing have been deposited with the non-profit plasmid distribution service Addgene (http://www.addgene.org/crispr-cas), which makes the technology easily available to the academic research community.
Engineered CRISPR/Cas technology comprises two components, a short guide RNA (gRNA) and the Cas9 nuclease, that work together to recognize and cleave a specific target DNA site (Jinek et al., 2012) (see Figure 1a). The target site consists of a 20 bp protospacer sequence, which is complementary to the first 20 nucleotides of the gRNA (counted from the 5’ end), and the protospacer-adjacent motif (PAM). The PAM sequence of the most frequently used Cas9 from Streptococcus pyogenes matches the form 5’-NGG-3’. Hence, Cas9 nuclease can be used to efficiently induce double stranded breaks into any genomic DNA locus bearing a 5’-N20NGG-3’ sequence by co-expression of an appropriately designed gRNA. Cas9-induced double-stranded breaks (DSBs) are generally repaired by one of two major pathways: non-homologous end joining (NHEJ) and homology-directed repair (HDR). The NHEJ pathway is characterized by imprecise re-joining of genomic DNA ends, resulting in the creation of variable-length insertions and deletions (indels) at the DSB site. Indel mutations can disrupt the translational reading frame and therefore, if introduced into coding sequence, may result in knockout of the target gene. The HDR pathway can be used to precisely repair a DSB in the presence of an exogenously added DNA donor template that bears homology to the DNA sequences upstream and downstream of the target site.
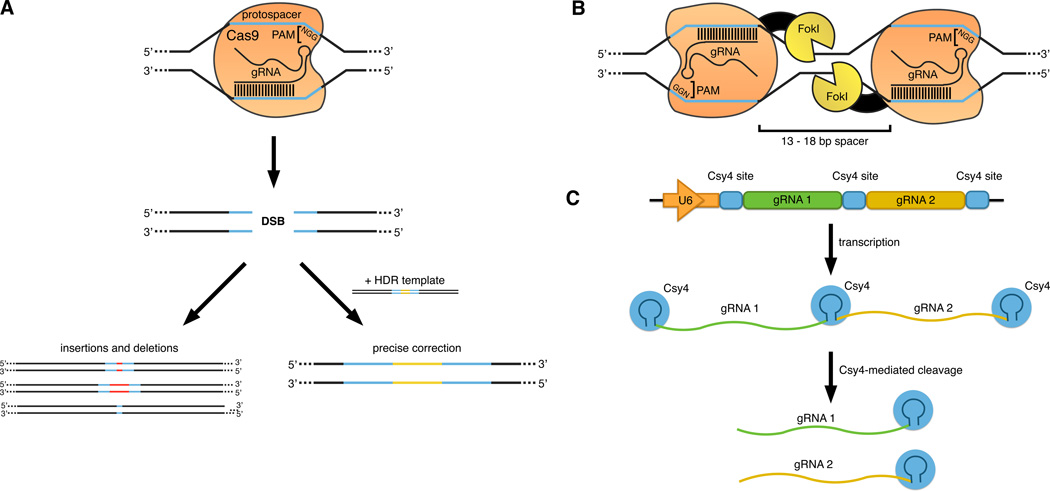
Figure 1. Cas9, RFNs and Csy4-based gRNA processing.
(a) Cas9-mediated DSB formation and repair by NHEJ and HDR pathways. The Cas9 target site is specified by the protospacer sequence, which is recognized by the most 5’ nucleotides of the gRNA, and the adjacent 5’-NGG-3’ PAM sequence. Upon target site recognition, the Cas9 nuclease induces a double-stranded break (DSB). It can be repaired by either imprecise re-joining of genomic DNA ends (NHEJ) or by precise repair according to an exogenously added DNA template (HDR).
(b) Dimeric CRISPR RNA-guided FokI nucleases. Dimeric RFNs consist of two gRNA/FokI-dCas9 complexes recognizing opposite DNA strands of target sites that are separated by a 13 – 18 bp spacer. Dimerization of FokI nuclease domains is required for efficient DNA cleavage.
(c) Csy4-mediated gRNA processing. Both Csy4 site-flanked gRNA sequences are transcribed as part of a single polymerase II transcript. The ribonuclease Csy4 specifically recognizes Csy4 cleavage sites and releases individual gRNAs.
PAM, Protospacer-adjacent motif; NHEJ, non-homologous end joining; HDR, homology-directed repair.
Soon after the development of the CRISPR/Cas system as a highly efficient genome editing technology, the realization that Cas9 can induce high-frequency off-target mutagenesis has suggested potential limitations for use for high-fidelity research and therapeutic applications (Fu et al., 2013; Hsu et al., 2013; Mali et al., 2013; Pattanayak et al., 2013). These studies showed that Cas9 can cleave off-target sites bearing mismatches at as many as five nucleotides and that off-target NHEJ-mediated mutagenesis rates in some cases even exceeded those at the on-target site. In order to overcome these limitations, various improvements to the CRISPR/Cas nuclease platform designed to increase its specificity have been described. Two of the improvements described by our group are the truncated gRNAs (tru-gRNAs) (Fu et al., 2014) and the dimeric CRISPR RNA-guided FokI nucleases (RFNs) (Guilinger et al., 2014; Tsai et al., 2014).
Truncating gRNAs by two or three nucleotides can reduce off-target mutagenesis by 5,000 fold and more while generally maintaining full on-target activity (Fu et al., 2014). One potential explanation for this somewhat counterintuitive observation is that the full-length gRNA/Cas9 complex may have excess DNA binding affinity and might therefore tolerate mismatches in the target sequence. Truncation of gRNAs may reduce binding affinity and therefore might increase sensitivity for mismatches within the target site. Besides the significant specificity improvement, one major advantage of the tru-gRNA technology is that it can easily be implemented with any gRNA expression vector.
Dimeric RFNs are another recent approach to re-engineer the Cas9 platform for improved specificity. These fusion proteins combine the ease of CRISPR-based targeting with the high precision of dimerization-dependent genome editing tools like TALENs and ZFNs. RFNs are chimeric proteins consisting of the dimerization-dependent FokI nuclease domain fused to the amino-terminal end of a catalytically “dead” Cas9 protein (FokI-dCas9, Figure 1b). Two FokI-dCas9 fusion proteins can be recruited to adjacent target sites by two different gRNAs to enable FokI dimerization and efficient DNA cleavage of a “spacer” sequence in between. This approach essentially doubles the length of the target site, potentially making dimeric RFNs one of the most specific CRISPR/Cas-based genome editing platforms to date. Because of the need to express two gRNAs in each cell, Tsai et al. developed a novel strategy for multiplex gRNA expression in which both gRNAs are transcribed as part of a single transcript and subsequently processed and cleaved out of that RNA by the ribonuclease Csy4 (Figure 1c). The Csy4 ribonuclease is expressed on the same plasmid as the FokI-dCas9 protein and both gRNAs are transcribed from a single gRNA expression vector. Hence, the RFN technology, like the conventional CRISPR/Cas, is based on a two-plasmid expression system.
The decision of whether to use the conventional CRISPR/Cas system, tru-gRNAs or RFNs largely depends on the level of specificity required for a particular experiment. If the main objective is to induce high mutation rates, while off-target mutagenesis might be tolerated, we recommend the conventional CRISPR/Cas system. If, however, a higher degree of on-target specificity is desirable, the tru-gRNA technology or RFNs will likely have more favorable specificity profiles.
We have developed an easy-to-use, streamlined and scalable protocol for efficient genome editing in human cell lines that is applicable to the conventional CRISPR/Cas9 system as well as to the tru-gRNA and RFN platforms. The protocol can be conceptually divided into three steps: (a) target site identification and gRNA vector cloning, (b) transfection of a human cell line and (c) quantification of genomic DNA mutation frequencies by T7 Endonuclease I assay. The entire procedure requires only basic skills in molecular biology techniques and human cell line handling and can be performed in about two weeks.
BASIC PROTOCOL 1
Target site identification and single (tru-) gRNA expression vector cloning for the conventional CRISPR/Cas9 and tru-gRNA technologies
Unlike previous genome editing technologies, such as ZFNs and TALENs, no laborious protein engineering is required for targeting the CRISPR/Cas9 complex to novel genomic sites. While the Cas9 plasmid pSQT817 remains constant, a single DNA oligo cloning step is required to modify the single gRNA cloning vector pMLM3636 for a particular target site (Figure 2a). We have developed the online platform ZiFiT (zifit.partners.org) for designing DNA oligos needed to construct an expression vector encoding a gRNA to a given target DNA sequence. To build this vector, a pair of DNA oligos is annealed and directly ligated into the purified single gRNA vector backbone (Figure 2 b).
Figure 2. Plasmids used in this protocol and DNA oligo cloning strategy for single and multiplex gRNA expression vectors.
(a) Vectors used in this protocol. For the conventional CRISPR/Cas and the tru-gRNA technologies, the single gRNA cloning vector pMLM3636 and the Cas9 vector pSQT817 are used. RFNs require the multiplex gRNA cloning vector pSQT1313 and the Csy4-T2A-FokI-dCas9 expression vector SQT1601.
(b) Cloning strategy for the single (tru-) gRNA expression vector. Oligos identified by ZiFiT are annealed and subsequently ligated into the BsmBI-cut single gRNA cloning vector pMLM3636.
(c) Cloning strategy for the multiplex gRNA expression vector. Two pairs of oligos recommended by ZiFiT and the “middle RFN oligos” are annealed and directly ligated into the BsmBI-cut multiplex gRNA cloning vector pSQT1313.
T2A, self-cleaving T2A peptide; NLS, nuclear localisation signal.
Materials
For target site identification:
Genomic DNA sequence of genomic target region
Computer with internet access
For single gRNA vector cloning:
Plasmid pMLM3636 (Addgene plasmid 43860)
Plasmid pSQT817 (Addgene plasmid 53373)
Agar plates with 50 µg/ml carbenicillin
LB media with 50 µg/ml carbenicillin
QIAprep Spin Miniprep Kit (Qiagen)
BsmBI restriction enzyme (NEB)
10 × NEBuffer 3.1 (NEB)
Nuclease-free H2O
1% agarose gel
QIAquick Gel Extraction Kit (Qiagen)
10 × STE buffer (see recipe)
T4 Ligase (NEB)
T4 Polynucleotide Kinase (NEB)
10 × T4 Ligase Buffer (NEB)
Chemically competent XL1-Blue cells (Stratagene)
SOC media
Sequencing primer OS280: 5’-CAGGGTTATTGTCTCATGAGCGG-3’
Identifying Cas9 target sites
-
1.Visit zifit.partners.org, click on “ZiFiT” on the website menu and proceed to ZiFiT. Choose “CRISPR/Cas Nucleases” in the “Design Genome Editing Nucleases/Nickases“ category.
- For conventional gRNAs, choose a target site length of 20 bp and the U6 promoter.
- For tru-gRNAs, choose a target site length of 17 or 18 bp and the U6 promoter.
Paste the sequence of your target region into the text box and hit “Identify target site”. ZiFiT ignores all numbers and characters in the sequence that are not G, A, T, or C. Choose a preferred DSB site by framing it with brackets (e.g., [A] or [G]).Step annotation: It is highly recommended to only use human genomic sequences for target site identification by ZiFiT. Target sites in cDNA sequences may not be present in the genome if they contain junction sequences that are formed after splicing. -
2.
Using the ‘Identify potential off-target sites’ feature in ZiFit, choose target sites with minimal numbers of potential off-target sites with 1–3 bp mismatches. Ideally, targets would have no potential off-target sites in the genome with 1–2 mismatches.
-
1.
Verify that the chosen target site does not contain any polymorphisms with respect to the human genome reference in your cell line of interest. If it does, adjust target sequences to match. Order the corresponding pair of DNA oligos designed by ZiFiT.
Single gRNA expression vector cloning
Streak bacteria from pMLM3636 and pSQT817 Addgene stocks onto LB agar plates containing 50 µg/ml carbenicillin with sterile pipet tips to obtain single bacterial colonies. Incubate at 37°C for 16 hours, then store plate pSQT817 at 4°C and proceed with plate pMLM3636. Pick a single colony of pMLM3636-transformed bacteria with a sterile pipet tip and inoculate 4 ml of LB media supplemented with 50 µg/ml carbenicillin. Shake and incubate at 37°C for 16 hours.
Use the QIAprep Spin Miniprep kit according to the manufacturer’s instructions to purify plasmid pMLM3636 DNA. Elute DNA in 50 µl 0.1 × Elution Buffer.
- Digest plasmid pMLM3636 using the conditions listed below. Incubate the reaction overnight at 55°C, heat inactivate for 20 minutes at 80°C.
- 3 µg plasmid pMLM3636
- 2 µl 10 × NEBuffer 3.1
- 2 µl 10 U/µl BsmBI
- to 20 µl dH2O
Step annotation: It is not recommended to significantly shorten digest time, since an incomplete digest of the pMLM3636 backbone will result in a large proportion of clones not bearing the desired gRNA sequence after gRNA expression vector cloning and transformation. Transfer the restriction digest product of step 3 to a 1% agarose gel, perform gel electrophoresis and excise the ~ 2.25 kb band. Purify DNA from excised agarose gel band according to the manual provided with the QIAquick Gel Extraction kit. Elute DNA in 50 µl 0.1 × Elution Buffer. Adjust DNA concentration to 10 ng/µl.
Resuspend synthesized DNA oligos for gRNA expression vector cloning in nuclease-free water for a concentration of 100 µM.
- Anneal resuspended DNA oligos using the conditions listed below. Incubate the DNA oligo annealing mixture at 95°C for 5 minutes in a thermocycler, then ramp down temperature to 25°C at 1°C per 30 seconds.
- 10 µl 100 µM DNA oligo 1
- 10 µl 100 µM DNA oligo 2
- 10 µl 10 × STE buffer
- 70 µl nuclease-free water
Dilute DNA oligoduplexes 1:1,000 in nuclease-free water for a final concentration of 0.01 µM.
- Prepare a ligation mixture of BsmBI-cut plasmid pMLM3636 from step 4 and the diluted oligoduplexes using the conditions listed below. Incubate the ligation mixture at a temperature of 16°C for 1 hour.
- 2 µl 0.01 µM oligoduplex
- 2 µl 10 ng/µl BsmBI-cut plasmid pMLM3636
- 1 µl 10 × T4 Ligase Buffer
- 0.5 µl T4 Ligase
- 0.5 µl Polynucleotide Kinase
- 4 µl nuclease-free water
- Transform ligation product into chemically competent XL1-Blue bacteria:
- Thaw a 50 µl aliquot of XL1-Blue bacteria on ice.
- Carefully add 5 µl of ligation product.
- Incubate for 20 minutes on ice.
- Heat shock at 42°C for 45 seconds.
- Incubate on ice for 5 minutes.
- Add 250 µl of SOC media, then incubate in a thermoshaker at 37°C for one hour.
- Plate on LB agar plate containing 100 µg/ml carbenicillin.
Incubate agar plate for 16 hours at 37°C, then pick 1 – 2 colonies with sterile pipet tips and inoculate 4 ml LB media supplemented with 50 µg/ml carbenicillin. Shake and incubate at 37°C for 16 hours.
Isolate plasmid DNA of bacterial cultures using QIAprep Spin Miniprep kit. Save an aliquot of each bacterial culture for step 13 and store at 4°C.
Sequence plasmids using the gRNA sequencing primer OS280 and align sequencing results with reference sequence 1 (When cloning tru-gRNAs remove two or three nucleotides from the variable segment of the reference sequence 1 for proper alignment. The number of Ns (17 or 18) should correspond to the length of the tru-gRNA target sequence.)
Inoculate 50 ml of LB media supplemented with 50 µg/ml carbenicillin with a bacterial culture bearing a sequence-verified single gRNA expression vector (form step 11). Also, pick a single bacterial colony from plate pSQT817 (from step 1) with a sterile pipet tip and inoculate 50 ml of LB media with 50 µg/ml carbenicillin. Shake and incubate at 37°C for 16 hours and purify plasmid DNA using the HiSpeed Plasmid Midi kit.
ALTERNATE PROTOCOL 1
Target site identification and multiplex gRNA vector cloning for RFNs
Similar to the conventional CRISPR/Cas system, dimeric CRISPR RNA-guided FokI nucleases can be expressed from two plasmids. While the Csy4-T2A-FokI-dCas9 expression plasmid pSQT1601 remains constant, a single cloning step is required to modify the multiplex gRNA cloning vector pSQT1313 for a given dimeric target site. After annealing of two pairs of DNA oligos recommended by ZiFiT and of a pair of constant “middle DNA oligos”, these oligoduplexes are directly ligated into BsmBI-cut pSQT1313 backbone to build a multiplex gRNA expression plasmid.
Additional Materials
Plasmid pSQT1313 (Addgene plasmid 53370) instead of plasmid pMLM3636
Plasmid pSQT1601 (Addgene plasmid 53369) instead of plasmid pSQT817
oSQT875 middle RFN oligo 1: /5Phos/AGCTAGAAATAGCAAGTTAAAATAAGGC
TAGTCCGTTATCAACTTGAAAAAGTGGCACCGAGTCGGTGCGTTCACTGCCGTATA
(20 nmole Ultramer DNA, IDT)
oSQT876 middle RFN oligo 2: /5Phos/TGCCTATACGGCAGTGAACGCACCGACT
CGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCT
(20 nmole Ultramer DNA, IDT)
Identification of RFN target sites
-
2.Visit zifit.partners.org, click on “ZiFiT” on the website menu and proceed to ZiFiT. Choose “CRISPR RFNs (RNA-guided FokI Nucleases)” in the “Design Genome Editing Nucleases/Nickases“ category. Paste the sequence of your target region into the text box and hit “Identify target site”. ZiFiT ignores all numbers and characters in the sequence that are not G, A, T, or C. Choose a preferred DSB site by framing it with brackets (e.g., [A] or [G]). Alternately, target sites can also be picked manually and should be of the form CCN(N20)(N13–18)(N20)NGG.Step annotation: It is highly recommended to only use human genomic sequences for target site identification by ZiFiT. Target sites in cDNA sequences may not be present in the genome if they contain junction sequences that are formed after splicing.
-
3.
Choose one or more target sites, preferably with a spacer length of 16 bp, and perform a UCSC Genome Blat Search. Verify that the chosen target site does not contain any polymorphisms with respect to the human genome reference in your cell line of interest. If it does, adjust target sequences to match.
-
4.
If a target site can only be found once in the human genome, order the corresponding two pairs (left/right) of DNA oligos designed by ZiFiT.
Multiplex gRNA vector cloning
Streak bacteria from pSQT1313 and pSQT1601 Addgene stocks onto LB agar plates containing 50 µg/ml carbenicillin with sterile pipet tips to obtain single bacterial colonies. Incubate at 37°C for 16 hours, then store plate pSQT1601 at 4°C and proceed with plate pSQT1313. Pick a single colony of pSQT1313-transformed bacteria with a sterile pipet tip and inoculate 4 ml of LB media supplemented with 50 µg/ml carbenicillin. Shake and incubate at 37°C for 16 hours.
Use the QIAprep Spin Miniprep kit according to the manufacturer’s instructions to purify plasmid pSQT1313 DNA. Elute DNA in 50 µl 0.1 × Elution Buffer.
- Digest plasmid pSQT1313 using the conditions listed below. Incubate the reaction overnight at 55°C, heat inactivate for 20 minutes at 80°C.
- 3 µg plasmid pSQT1313
- 2 µl 10 × NEBuffer 3.1
- 2 µl 10 U/µl BsmBI
- to 20 µl nuclease-free water
Step annotation: It is not recommended to significantly shorten digest time, since an incomplete digest of the pSQT1313 backbone will result in a large proportion of clones not bearing the desired gRNA sequence after gRNA expression vector cloning and transformation. Transfer the restriction digest product of step 3 to a 1% agarose gel, perform gel electrophoresis and excise the ~ 2.3 kb band. Purify DNA from excised agarose gel band according to the manual provided with the QIAquick Gel Extraction kit. Elute DNA in 50 µl 0.1 × Elution Buffer. Adjust DNA concentration to 10 ng/µl.
- Prepare 100 µM DNA oligo solutions in nuclease-free water for the following DNA oligos:
- left RFN oligo 1
- left RFN oligo 2
- oSQT875 middle RFN oligo 1
- oSQT876 middle RFN oligo 2
- right RFN oligo 1
- right RFN oligo 2
-
Anneal the following pairs of resuspended DNA oligos separately:
- Left oligoduplex: left RFN oligo 1 and left RFN oligo 2
- Middle oligoduplex: oSQT875 middle RFN oligo 1 and oSQT876 middle RFN oligo 2
- Right oligoduplex: right RFN oligo 1 and right RFN oligo 2
For each pair, incubate the DNA oligo annealing mixture at 95°C for 5 minutes in a thermocycler, then ramp down temperature to 25°C at 1°C per 30 seconds.- 10 µl 100 M DNA oligo 1
- 10 µl 100 M DNA oligo 2
- 10 µl 10 × STE buffer
- 70 µl nuclease-free water
Dilute oligoduplexes 1:1,000 in nuclease-free water for a final concentration of 0.01 µM.
- Prepare a ligation mixture of BsmBI-cut plasmid pSQT1313 from step 4 and diluted oligoduplexes on ice. Incubate at 16°C for 30 minutes, then keep at 4°C over night.
- 2 µl 0.01 µM left oligoduplex
- 2 µl 0.01 µM middle oligoduplex
- 2 µl 0.01 µM right oligoduplex
- 2 µl 10 ng/µl BsmBI-cut plasmid pSQT1313
- 1 µl 10 × T4 Ligase Buffer
- 0.5 µl 10 U/µl T4 Polynucleotide Kinase
- 0.5 µl 400 U/µl T4 Ligase
Step annotation: It is critical that the ligation mixture is cooled throughout the procedure to reduce the likelihood of false ligation products. It is recommended to prepare the ligation mixture in a PCR tube and to perform overnight ligation in a thermocycler. - Transform 5 µl ligation product into chemically competent XL1-Blue bacteria:
- Thaw a 50 µl aliquot of XL1-Blue bacteria on ice.
- Carefully add 5 µl of ligation product.
- Incubate for 20 minutes on ice.
- Heat shock at 42°C for 45 seconds.
- Incubate on ice for 5 minutes.
- Add 250 ul of SOC media, then incubate in a thermoshaker at 37°C for one hour.
- Plate on an LB agar plate containing 100 µg/ml carbenicillin.
Incubate agar plate for 16 hours at 37°C, then pick 3 – 6 colonies with sterile pipet tips and inoculate 4 ml of LB media with 50 µg/ml carbenicillin. Shake and incubate at 37°C for 16 hours.
Isolate plasmid DNA of bacterial cultures using QIAprep Spin Miniprep kit. Save an aliquot of each bacterial culture for step 13 and store at 4°C.
Sequence plasmids using the gRNA sequencing primer OS280 and align sequencing results with reference sequence 2.
Inoculate 50 ml of LB media supplemented with 50 µg/ml carbenicillin with a bacterial culture bearing a sequence-verified single gRNA expression vector (form step 11). Also, pick a single bacterial colony from plate pSQT1601 (from step 1) with a sterile pipet tip and inoculate 50 ml of LB media with 50 µg/ml carbenicillin. Shake and incubate at 37°C for 16 hours and purify plasmid DNA using the HiSpeed Plasmid Midi kit.
BASIC PROTOCOL 2
Transient transfection of human HEK-293 and U2OS cell lines
In this protocol, we describe transient transfection of Cas9 or Csy4-T2A-FokI-dCas9 plasmids along with single or multiplex gRNA vectors into Human Embryonic Kidney (HEK-293) cells or human osteosarcoma (U2OS) cells. This protocol may be used to study genomic mutations in these two cell types, but it might also be valuable for screening multiple gRNAs targeting the same genomic locus for maximal activity before proceeding to more demanding cell types.
Materials
Media for both HEK-293 and U2OS cell lines:
Advanced DMEM (Life Technologies)
10% Fetal Calf Serum (FCS, Life Technologies)
2 mM GlutaMax (Life Technologies)
Penicillin/Streptomycin (Life Technologies)
For transfection of HEK-293 cells:
HEK-293 cells
Cell culture media
Trypsin (Life Technologies)
Phosphate-Buffered Saline (PBS, Life Technologies)
Midiprep of nuclease expression vector (pSQT817 or pSQT1601)
Midiprep of single or multiplex gRNA expression vector
ptdTomato-N1 plasmid (Clontech)
Opti-MEM reduced serum medium (Life Technologies)
Lipofectamine LTX reagent (Life Technologies)
For transfection of U2OS cells:
U2OS cells
Cell culture media
Trypsin (Life Technologies)
Phosphate-Buffered Saline (PBS, Life Technologies)
Falcon Cell Strainers (nylon, 100 µm mesh, Fisher Scientific)
Midiprep of nuclease expression vector (pSQT817 or pSQT1601)
Midiprep of single or multiplex gRNA expression vector
ptdTomato-N1 plasmid (Clontech)
SE Cell Line 4D-Nucleofector X Kit S (Lonza)
4D-Nucleofector System (Lonza)
Transfection of HEK-293 cells
24 hours prior to transfection, remove cell culture media from cells and wash with PBS. Trypsinize HEK-293 cells for 5 – 10 minutes until they are completely detached from the plate. Stop trypsin treatment and separate cells by adding cell culture media and pipetting up and down 5 – 10 times. Count cells and plate 0.5 × 106 cells per well in 24-well plate(s).
-
For each sample, dilute the following DNA amounts in 50 µl of OptiMEM:
For conventional CRISPR/Cas9 or tru-gRNAs:- 375 ng pSQT817
- 125 ng single (tru-) gRNA vector
- 10 ng ptdTomato-N1 plasmid
For RFNs:- 750 ng pSQT1601
- 250 ng multiplex gRNA vector
- 10 ng ptdTomato-N1 plasmid
For each sample, dilute 2.5 µl of Lipofectamine LTX reagent in 50 µl OptiMEM. Add diluted Lipofectamine LTX to diluted DNA, mix and incubate at room temperature for 30 minutes to allow for the formation of DNA:Lipofectamine LTX complexes.
Add 100 µl of DNA:Lipofectamine LTX mixtures directly to the 24-well plate(s) containing HEK-293 cells. Gently shake plate horizontally to ensure even distribution of DNA:Lipofectamine complexes. Put 24-well plate(s) back into the incubator for 72 hours. Evaluate transfection efficiency after 24 hours by estimating the percentage of TD tomato-positive cells under the fluorescence microscope.
Transfection of U2OS cells
24 hours prior to transfection, split U2OS cells and plate at 40% confluency.
Prepare 24-well-plate(s) with 500 µl aliquots of cell culture media and pre-equilibrate at 37°C under 5% CO2 in incubator for at least 30 minutes before transfection. Make up complete nucleofection solution SE (150 ul Supplement, 675 ul solution SE) and keep at room temperature.
- Mix plasmids for each transfection in individual PCR tubes or a 96-well V-bottom tissue culture plate, in a total volume of 2.3 µl per sample:
- For conventional CRISPR/Cas9 or tru-gRNAs:
- 750 ng pSQT817
- 250 ng single (tru-) gRNA vector
- 10 ng ptdTomato-N1 plasmid
- For RFNs:
- 975 ng pSQT1601
- 325 ng multiplex gRNA vector
- 10 ng ptdTomato-N1 plasmid
- Also include negative control samples transfected with ptdTomato-N1 plasmid only.
Remove cell culture media from cells and wash with PBS. Trypsinize U2OS cells for ~ 10 minutes until they are completely detached from the plate. Stop trypsin activity and separate cells by adding cell culture media and pipetting up and down 5 – 10 times. Filter cells through cell strainer to remove cell clumps.
Count cells and transfer cell suspension containing n × 2.5 × 105 cells (n = number of transfections) into a clean falcon tube. Centrifuge at 3,000 rpm for 5 minutes at room temperature and remove media. Gently resuspend cell pellet in n × 25 µl complete nucleofection solution SE. Add 23 µl aliquots of cell suspension to DNA aliquots and mix by pipetting up and down 5 times.
Transfer 20 µl of DNA/cell mixtures into 16-well Nucleocuvette strip and gently tap to remove bubbles. Insert Nucleocuvette strip into the 4D-Nucleofector System and transfect using the DN-100 program.
After transfection, wait for 10 minutes until adding 80 µl of cell culture media. Resuspend cells by gently pipetting up and down 5 times, and transfer 80 µl of cell suspensions to pre-equilibrated media in 24-well plate(s). Gently shake plate(s) with short horizontal side-to-side and front-to-back motion to ensure even distribution of transfected cells. Put 24-well plate(s) back into the incubator for 72 hours. Evaluate transfection efficiency after 24 hours by estimating the percentage of TD tomato-positive cells under the fluorescence microscope.
BASIC PROTOCOL 3
Quantification of genomic DNA mutation frequencies by T7 Endonuclease I assay
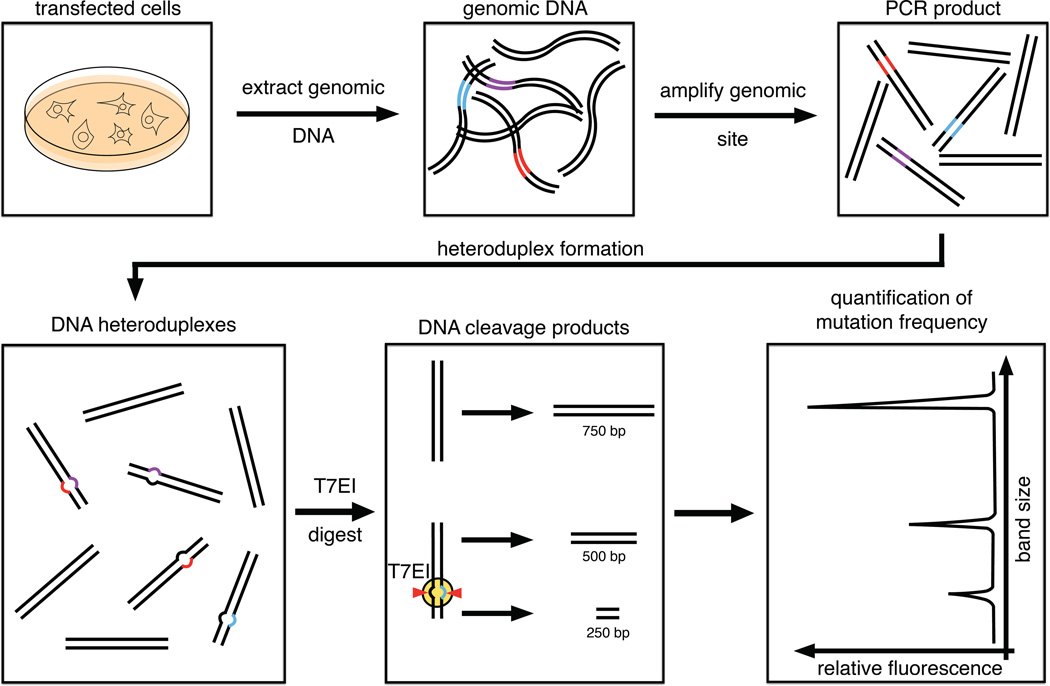
The T7EI assay is a simple and cost-effective assay for quantification of genomic mutation frequencies (Figure 3). The protocol consists of five main steps: (1) Isolation of genomic DNA from transfected cells. This genomic DNA is generally a mixture of modified and wild type genomes. (2) In a second step, the locus comprising the Cas9/RFN target site is amplified by PCR. (3) Next, PCR amplicons are melted and re-annealed, resulting in the formation of DNA heteroduplexes. (4) Subsequently, these DNA heteroduplexes are recognized and digested by the T7EI enzyme. The digest product is comprised of DNA fragments of three different lengths, representing the full-length amplicon and both cleavage products. (5) Lastly, restriction fragments are quantified and NHEJ rates are calculated. The complete procedure can be performed in one or two days. For high-throughput applications, the protocol can be scaled up to a 96-well format.
Figure 3.
T7 Endonuclease I assay for quantification of NHEJ mutation frequencies. The protocol consists of five main steps: (1) Genomic DNA isolation from human HEK-293 or U2OS cells, (2) PCR amplification of the genomic target site, (3) formation of DNA heteroduplexes via melting and annealing of PCR amplicons, (4) T7EI digest of DNA heteroduplexes and (5) quantification of mutation frequencies by restriction fragment analysis.
Materials
Agencourt DNAdvance gDNA isolation kit (Beckmann Coulter)
70% Ethanol
Nuclease-free water
Phusion Hot-start FLEX DNA polymerase (NEB)
5 × Phusion Polymerase HF buffer (NEB)
10 mM dNTPs
Agencourt AMPure XP - PCR Purification kit (Beckmann Coulter)
1% and 2% agarose gels
T7 Endonuclease I (NEB)
10 × NEBuffer 2 (NEB)
0.25 M EDTA
QIAxcel Advanced Instrument (Qiagen)
QIAxcel DNA High Resolution Kit (Qiagen)
T7EI assay, protocol steps
Purify genomic DNA from transfected cells using the Agencourt DNAdvance kit or any other genomic DNA purification kit. Elute DNA from magnetic beads in 50 µl 0.1 × Elution Buffer per sample. Measure DNA concentrations with a spectrophotometer and adjust to 10 ng/µl.
Design and order a pair of DNA primers for each target locus. Optimally, the amplicon should be about 750 bp in length and the Cas9 or RFN target site should be around the 250th or 500th base pair.
-
For each target site and each gDNA sample, perform three PCR reactions:
- Nuclease-treated sample: Use gDNA of cells transfected with Cas9 or RFNs as PCR template
- Negative transfection control: Use gDNA of cells transfected with plasmid ptdTomato-N1 only as PCR template
- Negative PCR control: Use 0.1 × Elution Buffer as PCR template. (Only one set of negative controls is required per target site.)
Mix the following PCR reagents in PCR tubes or a 96-well PCR plate on ice. Touchdown PCR thermocycling conditions can be used for a large variety of primer melting temperatures and generally produce a clean PCR product.- 10 µl 5 × Phusion HF buffer
- 1 µl 10 mM dNTPs
- 2.5 µl 10 mM forward primer
- 2.5 µl 10 mM reverse primer
- 10 µl PCR template (e.g. 10 ng/µl gDNA of nuclease-treated cells)
- 24 µl nuclease-free water
Touchdown PCR thermocycling conditions:Number of cycles Denature Anneal Extend 1 98°C, 3 min - - 10 98°C, 10 s 72°C, ramp down to
62°C at 1°C/cycle, 15 s72°C, 30 s 25 98°C, 10 s 62°C, 15 s 72°C, 30 s 1 - - 72°C, 3 min - Test whether PCR was successful by running 5 µl of each PCR product on a 1% agarose gel. If “negative PCR control” samples show no bands, proceed to step 5 with the “nuclease-treated sample” and “negative transfection control” samples only.Step annotation: It is critical that “nuclease-treated samples” and “negative transfection control” samples only show a single, sharp band. If there are more than one or no bands consider alternate thermocycling conditions for standard PCR.Step annotation: Negative PCR control samples should not show any bands on agarose gel. Possible explanations for a PCR product in these samples include DNA contamination of PCR reagents or self-complementary primers.
Clean up PCR reactions by using the Agencourt AMPure XP kit or any other PCR purification kit. Elute DNA in 30 µl 0.1 × Elution Buffer. Measure DNA concentrations using a spectrophotometer.
- Use the following conditions for the formation of DNA heteroduplexes:
- 200 ng purified PCR product
- 2 µl 10 × NEBuffer 2
- to 19 µl nuclease-free water
Step Denature Hybridize 1 95°C for 5 min - 2 - 95°C, ramp down at 2°C/s, 5 s 3 - 85°C ramp down at 0.1°C/s, 10 min Step annotation: When analyzing a large number of samples, it is recommended to pipet volumes containing 200 ng into a 96-well PCR plate, followed by complete removal of water by using a vacuum concentrator. Subsequently, dried DNA can be resuspended in 19 µl of 1 × NEBuffer 2. - Put samples back on ice and add 1 µl of T7 Endonuclease I to each sample. Incubate at 37°C for 15 minutes in a thermocycler and immediately put samples back on ice. Stop T7EI digest by adding 2 µl of 0.25 M EDTA.Step annotation: It is not recommended to significantly extend T7EI digestion times beyond 15 minutes. Longer incubation at 37°C might result in T7EI star activity.
Purify products of T7EI digest using the Agencourt AMPure XP kit, analyze samples with the Qiaxcel capillary electrophoresis instrument using program OM500 and an injection time of 20 s. Alternatively, samples can be analyzed on a 2% agarose gel.
- Quantify T7EI restriction fragment bands, then calculate T7EI cleavage rate and NHEJ mutation rate using the following formulas:
f
REAGENTS AND SOLUTIONS
10 × STE buffer (pH 8, store at 4°C)
| Component | Concentration | Amount | Final concentration |
|---|---|---|---|
| NaCl | 5 M | 10 ml | 1 M |
| Tris HCL | 1 M | 5 ml | 100 mM |
| EDTA | 0.5 M | 1 ml | 10 mM |
| dH2O | 34 ml |
COMMENTARY
Background Information
All plasmids used in this protocol have been designed and codon-optimized for expression in human cells and high genome editing efficiencies have been observed in both HEK-293 and U2OS cell lines (Fu et al., 2014; Tsai et al., 2014). Fu et al. were able to show that tru-gRNAs dramatically reduce off-target cleavage while maintaining high NHEJ or HDR rates at most on-target sites. However, at a minority of sites, some tru-gRNAs exhibited reduced on-target mutation frequencies compared to full-length gRNAs. Therefore, it might be beneficial to screen multiple tru-gRNAs targeting the same locus for highest activity. Tsai et al. have found that RFNs can induce NHEJ mutations at lower frequencies than conventional CRISPR/Cas but at similar rates as the recently described Cas9 nickase technology (Mali et al., 2013). In the same study, Tsai et al. used deep sequencing to look for mutations at the 8 most closely matched genomic off-target sites of each of the three tested genes, but found no evidence of RFN-mediated off-target activity. However, a more sensitive, unbiased genome-wide approach is needed to fully evaluate the specificity profiles of conventional CRISPR/Cas, tru-gRNAs and RFNs.
Critical Parameters and troubleshooting
It is important to exclusively submit human genomic DNA sequences to ZiFiT for target site identification. Target sites found in cDNA may not be present in the human genome if they contain sequences that are separated by introns. In case a gRNA does not show any activity, it may be beneficial to sequence the target site in the cell line and to compare it with the reference target site used for gRNA design. Repetitive genomic sequences should also be excluded using Repeat Masker software.
It is critical that the target site is only present in the human genome once. Multiple target sites in the genome might result in more than one high efficiency DSB in a cell, potentially leading to chromosomal translocations, deletions, or other undesirable large-scale genomic rearrangements (Maddalo et al., 2014).
If a large number of bacterial clones also appear on the backbone-only control agar plate of the oligo cloning procedure, the BsmBI digestion of the single or multiple gRNA cloning vector (pMLM3636 or pSQT1313) has likely not been complete. It is critical to dilute oligoduplexes before ligation into the BsmBI-cleaved backbone to reduce the likelihood of ligation of multiple oligoduplexes into a single backbone. The multi-piece ligation step for multiplex gRNA expression vector cloning is relatively less efficient compared to single-target ligations and will result in a high number of false ligation products unless performed at 4°C overnight.
Transfection efficiency can be impaired by impurities in vector preparations. It is therefore recommended to only transfect plasmid DNA that has been prepared by midi- or maxi-prep. Ethanol precipitation after midiprep may be used to ensure high purity of plasmid preparation.
It is critical to obtain a clean PCR product when amplifying the target locus from genomic DNA for the T7EI assay. Additional bands on the agarose gel often are a consequence of mis-annealing of primers at off-target sites. Increasing annealing temperature or using alternative primer pairs often resolves this issue.
Anticipated Results
Single oligo cloning is generally very efficient and accurate. Hence, the frequency of false single gRNA expression vector sequences comes close to the oligo synthesis error rate. Under standard ligation conditions, multiplex gRNA expression vector cloning is much less accurate than single oligo cloning. However, using the optimized cloning procedure, we generally obtain the correct desired sequences in > 50% of tested clones. The genome editing efficiency strongly depends on the transfection efficiency. Under optimal conditions (i.e. in cell lines like HEK-293 or U2OS that can be transfected with up to ~ 90% efficiency) NHEJ-mediated mutagenesis frequencies can be as high as 30% to 50% or more with the conventional CRISPR/Cas system. Tsai et al. have tested RFNs at twelve different genomic loci with average NHEJ rates of 11% and 19% in HEK-293 and U2OS cells, respectively. Mutation rates measured by the T7EI assay correlate well with deep sequencing analyses. Note that T7E1 is a relatively insensitive assay for detecting mutation rates of less than 2% to 5%.
Time Considerations
The complete procedure can be performed in approximately two weeks: gRNA expression vector design, cloning and sequence validation takes ~ 1 week, genome editing of human cells requires ~ 4 days and the T7EI assay can be performed in ~ 1 or 2 days. Moreover, gRNA expression vector cloning and the T7EI assay can easily be scaled up to a 96-well format.
Acknowledgments
This work was funded by a National Institutes of Health (NIH) Director’s Pioneer Award (DP1 GM105378), NIH R01 GM088040, NIH P50 HG005550, and the Jim and Ann Orr Massachusetts General Hospital (MGH) Research Scholar Award. S.Q.T. was supported by NIH F32 GM105189. This material is based upon work supported by, or in part by, the US Army Research Laboratory and the US Army Research Office under grant number W911NF-11-2-0056.
Footnotes
Competing financial interests
J.K.J. has financial interests in Editas Medicine and Transposagen Biopharmaceuticals. J.K.J. is a consultant for Horizon Discovery. J.K.J.'s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies. J.K.J. has filed a patent application on the tru-gRNA/tru-RGN technology. J.K.J. and S.Q.T. are inventors on patent applications describing the FokI-dCas9 technology and the multiplex gRNA expression method.
INTERNET RESOURCES
The online resource ZiFiT (http://zifit.partners.org/ZiFiT/) provides assistance in finding target sites for conventional CRISPR/Cas nucleases as well as for the tru-gRNA and RFN technologies. It also identifies DNA oligo sequences compatible with our single and gRNA expression vector cloning protocols.
Reference sequences:
Reference sequence 1 (first 20 nucleotides of gRNA sequence colored in  , for tru-gRNAs remove two or three nucleotides):
, for tru-gRNAs remove two or three nucleotides):
LITERATURE CITED
- Fu Y, Foden JA, Khayter C, Maeder ML, Reyon D, Joung JK, Sander JD. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013;31:822–826. doi: 10.1038/nbt.2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat. Biotechnol. 2014;32:279–284. doi: 10.1038/nbt.2808. Compares genome editing efficiency and specificity of the tru-gRNA technology with the conventional CRISPR/Cas system in human cells.
- Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat. Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han Y-C, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014 doi: 10.1038/nature13902. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat. Biotechnol. 2013;31:839–843. doi: 10.1038/nbt.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nat. Biotechnol. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. Comprehensive review of the CRISPR/Cas9 technology.
- Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat. Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. Describes the dimeric RFN platform in human cells and compares its genome editing efficiency and specificity with the conventional CRISPR/Cas system.