Abstract
Study objective
To determine the onset of Gametocyte Specific Factor (Gtsf1) expression in embryogenesis and its relation to Nobox, and determine its localization during gonadal development and gametocyte maturation
Design
Developmental Animal Study
Setting
University Reproductive Biology Laboratory
Animals
Mice ranging in age from embryonic day 12.5 to 8 weeks old
Interventions
Polymerase Chain Reaction and Quantitative Polymerase Chain Reaction were performed to determine the onset of and relative mRNA expression. Western Blot was done to confirm protein expression and antibody specificity. In-situ hybridization & Immunohistochemistry were used determine localization of expression.
Main outcome measure(s)
Gtsf1 mRNA expression levels during embryogenesis through adulthood in wild-type mice and in newborn Nobox knockout mice, GTSF1 expression and localization in postnatal mice
Results
Gtsf1 functions downstream of Nobox and is highly expressed in embryonic male and female gonads, localizing to germ cells throughout development. GTSF1 expression is confined to the cytoplasm in all stages of postnatal oocyte maturation, and to pre-spermatogonia during early postnatal testicular development.
Conclusions
The expression pattern of Gtsf1 and its high conservation suggests that it may play important role in germ cell development. Further characterization of Gtsf1 may elucidate mechanisms involved in Premature Ovarian Failure.
Keywords: Nobox, Premature Ovarian Failure, Folliculogenesis, Germ-cell, Gametes
Introduction
In mammals, oocyte maturation is a complex process involving the interaction of many genes during embryogenesis (1). In mice, 45 primordial germ cells (PGCs) appear at embryonic day 7 after conception (E7.5) and give rise to the germ cell lineage (2,3). From E9.5 to E11.5, PGCs migrate from the hindgut epithelium to the urogenital ridge to form germ cell clusters called cysts (4–8). Primordial germ cells within the cysts then undergo mitosis with cytokinesis to form oocyte clusters (1). At E13.5, the oocytes initiate meiosis and become arrested in the diplotene phase of meiosis I until ovulation (9). Shortly after birth these oocyte clusters separate and become associated with pre-granulosa cells to form primordial follicles (1) with most germ cell cysts dissipating by postnatal day 7 (5). At postnatal day 3 some primordial follicles begin maturing into primary follicles (10). During this transition, numerous oocyte-specific genes critical to folliculogenesis such as Nobox, Figla, Gdf-9 and BMP-15 (10) are expressed.
Nobox, first isolated using in silico screening techniques to identify expressed sequence tags (EST) specific to the gonads (11), encodes a homeobox transcription factor critical for the transition of primordial to primary follicles (12,13). Since Nobox deficiency in mice leads to a rapid postnatal loss of oocytes, it serves as a model for Premature Ovarian Failure (POF). Persistent expression of Nobox in the adult ovary throughout folliculogenesis suggests that it may also be responsible for maintaining the expression of genes in growing oocytes (10). Gtsf1 (MGI:1921424) was identified on an Affymetrix 430 2.0 microarray chip as having a 13-fold reduction in mRNA expression in Nobox deficient as compared to wild-type mice (14). It is highly conserved among 27 species, including humans, with evolutionary conservation extending to Euteleostomi (bony vertebrates). We aim to further characterize Gtsf1 to determine its involvement in folliculogenesis and spermatogenesis, and therefore describe its expression during embryogenesis and early postnatal development in the ovary and testis.
Materials and Methods
Experimental Animals
Animals used in this research were maintained in accordance with the Institutional Animal Care and Use Committee (IACUC) at Baylor College of Medicine. Postnatal days were determined by counting the number of days after birth (PD 0.5). All tissues used in RT-PCR or immunochemistry were derived from C57BL/6/129S5 SvEvBrd hybrid background.
Determination of Relative mRNA expression levels of Gtsf1 using real time quantitative polymerase chain reaction
Complimentary DNA was derived from ovarian messenger RNA isolated from newborn ovaries using RNeasy Mini Kit (Qiagen, MD). RT-QPCR was performed using Sybr Green with oligonucleotide primers corresponding to Gtsf1 (forward primer 5′ tgc cct cct tgt gat gaa gac 3′, reverse primer 5′ gaa ctc gca tgc ctg aag c 3′), and Gapdh (forward primer 5′ caa tgt gtc cgt cgt gga tct 3′, reverse primer 5′ gcc tgc ttc acc acc ttc tt 3′) as a control. RT-QPCR was performed three times, as previously described (10), to verify results. Student’s t-test was used to calculate level of significance of the change in mRNA levels.
Determination of Tissue Specificity and Onset of Gtsf1 mRNA Expression with reverse transcriptase polymerase chain reaction
Complimentary DNA was derived from messenger RNA isolated from multiple adult tissues as well as the ovaries and testis of both embryonic and adult mice using RNeasy Mini Kit (Qiagen, Maryland). Equal amounts of messenger RNA were reverse transcribed using Jumpstart Taq polymerase (Sigma, St. Louis, Missouri). Reverse transcriptase PCR (RT-PCR) was performed using Gtsf1 primers (5′-GCC AAG CTT CTG TGC ATT TCC ATT GTT TTT CCA T-3′ and 5′-CCG GAT CCG ATG GAA GAC ACT TAC ATC GAC TCC CT-3′) to amplify a 501 base pair product. RT-PCR was performed for 29 cycles at 94 degrees Celsius for 30 seconds (denaturation), 55 degrees Celsius for 30 seconds (annealing) and 72 degrees for 30 seconds (extension). Amplification was performed using actin-specific primers as a positive control for equivalent mRNA levels.
Confirmation of GTSF1 Expression in Gonadal Tissue
Tissue from the ovaries, testis and liver of postnatal day 1 to 3 mice were homogenized in Buffer C [20mM Hepes, 25% Glycerol, 0.42 NaCl, 1.5mM MgCl2, 0.2 mM EDTA]. One hundred micrograms of protein extract from each specific tissue was loaded onto a 4–12% Bis-Tris Gel (Invitrogen; Carlsbad, CA), subjected to electrophoresis and then the protein was transferred onto a nitrocellulose membrane [Whatman GmbH; Dassel, Germany]. Nitrocellulose membranes were blocked in a 5% dry non-fat milk, 0.1% Tween in TBS solution [50 mM Tris base, pH 7.5, 150 mM NaCl, and 0.1% (w/v) Tween 20] for one hour at room temperature. The membranes were incubated at 4 degrees overnight with affinity-purified anti-GTSF1 antibodies at 1:100 dilution. The membranes were then incubated with a rabbit anti-guinea pig peroxidase-conjugated secondary antibody at 1:10,000 for one hour at room temperature. The enzyme products were visualized using enhanced chemiluminescence (ECL Western Blotting Analysis System, GE Healthcare, Buckinghamshire, UK).
In situ hybridization of Gtsf1
In situ hybridization was performed as previously described (15). In summary ovaries were initially fixed in 4% paraformaldehyde and then embedded in parafin. Five micrometer sections were hybridized to 35Slabeled sense and antisense riboprobes as previously described (11). The signal was detected by using NTB-2 emulsion autoradiography (Eastman Kodak, Rochester, NY) and tissue was counterstained with hematoxylin.
Localization of GTSF1 Expression with Immunohistochemistry
Antibodies against GTSF1 were created by expressing the full length protein in the pET-23b vector and immunizing guinea pigs at Cocalico Biologicals (Lancaster, PA). The anti-GTSF1 antibodies were immunoaffinity purified over Affi-Gel 10 (Bio-Rad, CA) and used in immunohistochemistry as previously described (16).
Results
Gtsf1 localizes to mouse chromosome 15 and its human orthologue localizes to chromosome 12q13.2. Murine GTSF1 (XP_900203) protein shares 100% and 92% homology with rat and human orthologues, respectively. According to publicly available databases (www.ensembl.org), Gtsf1 is a highly conserved gene, showing 1 to 1 conservation with twenty seven species including zebrafish and frogs (www.ensembl.org, http://www.ncbi.nlm.nih.gov/sites/entrez). It belongs to a functionally uncharacterized protein family (UPF0224) with an unknown functional domain (http://www.bmm.icnet.uk/servers/3djigsaw/). Use of publicly available prediction models for subcellular localization demonstrated different locations including nuclear (http://psort.nibb.ac.jp/form2.html), cytoplasmic (http://www.cs.ualberta.ca/~bioinfo/PA/Sub/) and extracellular (http://www.bioinfo.tsinghua.edu.cn/SubLoc/cgi-bin/eu_subloc. cgi) regions.
To further evaluate earlier Afymetrix findings, real time quantitative polymerase chain reaction (RT-QPCR) was performed on three independent samples to determine relative expression levels of Gtsf1 mRNA in wild-type and Nobox deficient newborn ovaries (Figure 1). Relative expression levels of Gtsf1 in wild-type ovaries and Nobox deficient ovaries were 0.38 and 0.017, 0.71 and 0.055, 0.84 and 0.022 for the first, second and third time that QPCR was performed. The average relative expression of Gtsf1 for all three experiments was 0.64 ± 0.24 in wild-type newborn ovaries and 0.031 ± 0.02 in Nobox deficient ovaries (Figure 1). Gtsf1 expression in Nobox deficient ovaries is 4.8% of the level of Gtsf1 expression in wild-type mice, representing a 20-fold reduction in expression, a significant difference (p ≤ 0.011).
Figure 1.
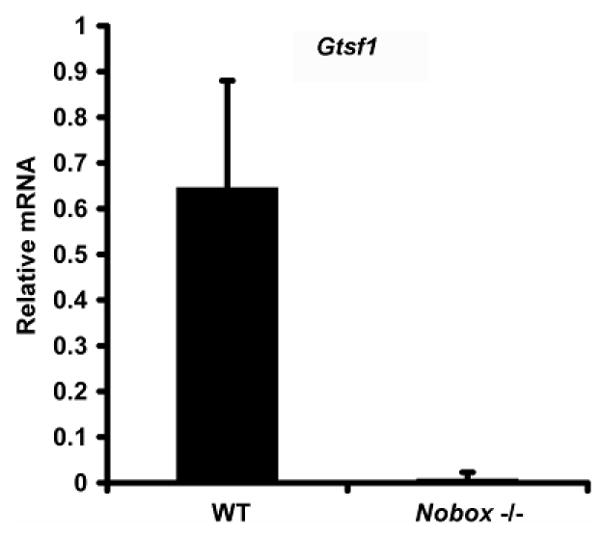
Gtsf1 expression in Wild-type and Nobox deficient female mice. Gtsf1 expression is downregulated 20-fold in Nobox deficient female mice compared to Wild-type female mice (p ≤ 0.011).
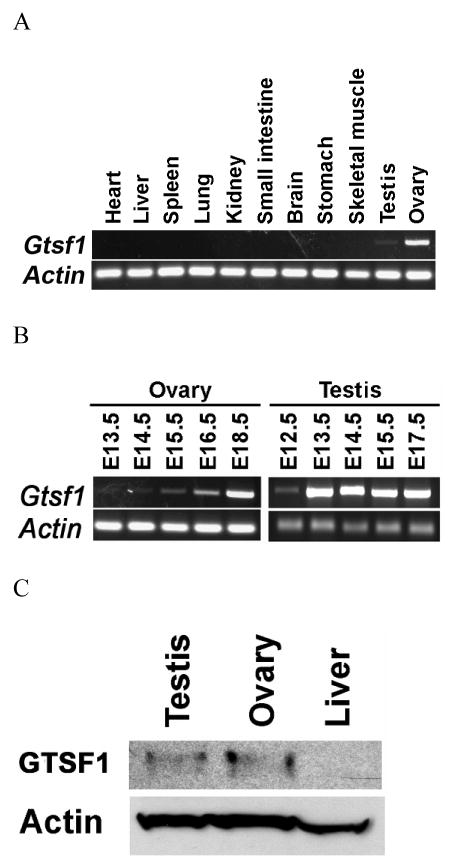
Multitissue reverse-transcriptase polymerase chain reaction (RT-PCR) was performed on adult murine tissue using primers to Gtsf1 and actin. Actin, serving as a positive control, was detected in all tissues. Gtsf1 expression was detected primarily in in adult ovaries and testis (Figure 2A). RT-PCR was performed on embryonic female gonadal tissue from embryonic day 12.5 (E12.5) to E18.5 with actin as a positive control. Gtsf1 mRNA expression was detected as early as E13.5 (data from E12.5 not shown) and demonstrated a relative increase throughout embryonic development (Figure 2B). RT-PCR was performed on embryonic male gonadal tissue from embryonic day 12.5(E12.5) to E18.5 with actin as a positive control as well. Expression of Gtsf1 was weaker at E12.5 but remained relatively constant from E13.5 to E18.5.
Figure 2.
(A–C). Gtsf1 RNA and protein expression. Equal amounts of total RNA from 11 different adult tissues as indicated above (A) and from various embryonic time points (B) were reverse transcribed. Oligonucleotides complementary to Gtsf1 were used to amplify the corresponding RNA sequences. (C) Western blot with affinity-purified anti-GTSF1 antibodies were used to detect a 29 kDa band in Postnatal Day 3 ovaries and testis, but not liver.
Western blotting was performed to confirm specificity of binding of polyclonal antibodies derived against GTSF1 protein in gonadal tissue. Antibodies to actin bound to testis, ovary and liver extracts which served as a positive control. Anti-GTSF1 antibodies bound to testis and ovary extracts, but not liver extracts, at the expected band size of 29 kD (Figure 2C). The 29kD band was determined to be GTSF1 from prior coomasie gels used to verify GTSF1 expression from the pET-23b expression system.
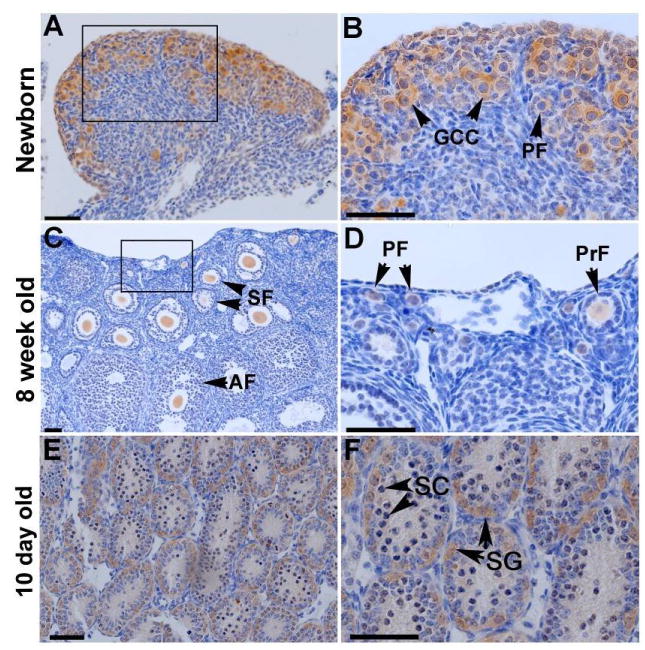
In situ hybridization was performed on adult murine ovarian tissue and demonstrated localization the highest degree of localization to the cytoplasm of primary follicles and primordial follicles to a lesser degree (not shown). To further characterize localization in the ovaries, immunohistochemistry was performed on ovarian tissue from newborn and eight-week old female mice using the same polyclonal antibodies as in western blotting (Figure 3). In the newborn ovary, anti-GTSF1 staining occurred predominantly in the cytoplasm of oocytes within germ cell cysts and in primordial follicles (panel A and B). In the eight week adult ovary, anti-GTSF1 staining was specific to the cytoplasm of oocytes of secondary and antral follicles (panel C). Addtionally, anti-GTSF1 cytoplasmic staining was noted in the oocytes of primordial and primary follicles (see panel D) in the eight week old ovary. This demonstrates that GTSF1 expression occurs throughout all stages of folliculogenesis. Immunohistochemistry performed on postnatal day 10 testis shows anti-GTSF1 staining of spermatogonia but not spermatocytes (Figure 3, panel E & F).
Figure 3.
GTSF1 expression in the ovaries (Panels A-D). Immunohistochemistry was performed on Newborn (Panel A & B) and 8 week old ovaries (Panel C & D). GTSF1 can be clearly detected (Panel B) in the cytoplasm of germ cell clusters (GCC) and primordial follicles (PF) in Newborn ovaries. At 8 weeks, GTSF1 can be clearly seen in all stages of developing oocytes, from primordial (PF) and primary follicles (PrF) (Panel D) to secondary (SF) and antral follicles (AF) (Panel C). GTSF1 expression in the testis (Panels E & F). Immunohistochemistry is performed on Postnatal day 10 (P10) testis. At Postnatal day 10, GTSF1 expression can be seen in primary spermatogonia (SG) but not in primary spermatocytes (SC). Scale bars, 50 μm
Discussion
In this study, our aim was to further characterize the expression and localization of Gtsf1. Gtsf1 was originally identified on an Affymetrix 430 2.0 microarray chip as one of 35 genes downregulated at least 5-fold in Nobox deficient murine ovaries (14). Further investigation into its protein sequence suggested a high level of conservation with 1 to 1 alignment with 27 other species. Since GTSF1 belongs to a novel family (UPF0224) with an uncharacterized functional domain, it is an ideal candidate for further investigation.
RT-QPCR was initially performed to confirm microarray chip findings and demonstrated a 20-fold decrease in Gtsf1 expression, suggesting that Gtsf1 is downstream of Nobox. Multitissue RT-PCR demonstrates that Gtsf1 expression first occurs at E13.5 in the ovaries when meiosis is initiated and continues to increase throughout embryonic development into adulthood. The onset of Gtsf1 expression in the testis occurs at E12.5, when testis cords patterns develop and become the foundation of adult seminiferous tubules. Expression becomes more pronounced at E13.5 when the testis are enlarging secondary to the proliferation of germ cells and pre-Sertoli cells, and remains at a high level until birth.
Western blotting of postnatal day 1–3 ovaries, testis and liver, performed when the highest percentage of gonadal tissue is made up of gametes, confirmed that GTSF1 expression was specific to ovaries and testis. Since in situ hybridization yielded only preliminary information on localization, immunohistochemistry was performed at different stages of folliculogenesis. Immunohistochemistry in the newborn and adult ovary demonstrated that GTSF1 was expressed at all stages of follicle maturation, from germ cell cysts to antral follicles. Since Gtsf1 and Nobox are expressed throughout folliculogenesis and Gtsf1 is downstream of Nobox, it may in part be responsible for the phenotype demonstrated in Nobox deficient mice. Antibody staining performed on postnatal day 10 mice showing localization to the spermatogonia suggests that GTSF1 may play a role in spermatogenesis, although any potential role at this time is unclear. The failure of online models to predict protein localization consistently is likely secondary to the lack of characterization of members of the UPF0224 family (FamB112).
Conclusion
This study has shown that Gtsf1 is a highly conserved and novel gene specific to the ovaries and testis. It functions downstream of Nobox with expression first occuring with the formation of gonadal tissue during embryogenesis. Its presence throughout folliculogenesis and into adulthood, and localization to the ooplasm, suggests it may have a critical role throughout this process. Detection of its presence in the pre-spermatogonia of the testis suggests that further evaluation of its role in male reproduction is warranted. Knockout models of Gtsf1 in the future will allow further elucidation of its role in folliculogenesis and spermatogenesis, and may provide further insight into the process of Premature Ovarian Failure.
Acknowledgments
Research supported by a National Institutes of Health grant (HD44858; Bethesda, Maryland) and a March of Dimes (White Plains, New York) Basil O’Connor Award (5-FY02-266; both grants, Aleksandar Rajkovic)
We would like to thank Dr. Stephanie Pangas for her assistance in the analysis of the RT-QPCR results. Financial support for this research has been provided to Dr. Aleksandar Rajkovic by the National Institutes of Health (Bethesda, Maryland) and the March of Dimes.
Footnotes
None of the above authors has any potential conflict of interest, financial or otherwise.
Oral Presentation, 63rd Annual Meeting of the American Society for Reproductive Medicine, Washington, D.C. October 2007. Expression and Localization of the Novel and Highly-Conserved Germ-Cell Specific Factor 1 During Early Oogenesis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Choi Y, Rajkovic A. Genetics of early mammalian folliculogenesis. Cell Mol Life Sci. 2006;63:579–90. doi: 10.1007/s00018-005-5394-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lawson KA, Hage WJ. Clonal analysis of the origin of primordial germ cells in the mouse. Ciba Found Symp. 1994;182:68–84. doi: 10.1002/9780470514573.ch5. discussion 84–91. [DOI] [PubMed] [Google Scholar]
- 3.McClellan KA, Gosden R, Taketo T. Continuous loss of oocytes throughout meiotic prophase in the normal mouse ovary. Dev Biol. 2003;258:334–348. doi: 10.1016/s0012-1606(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 4.Elvin JA, Yan C, Matzuk MM. Oocyte-expressed TGF-beta superfamily members in female fertility. Mol Cell Endocrinol. 2000;159:1–5. doi: 10.1016/s0303-7207(99)00185-9. [DOI] [PubMed] [Google Scholar]
- 5.Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol. 2001;234:339–51. doi: 10.1006/dbio.2001.0269. [DOI] [PubMed] [Google Scholar]
- 6.Clark JM, Eddy EM. Fine structural observations on the origin and associations of primordial germ cells of the mouse. Dev Biol. 1975;47:136–55. doi: 10.1016/0012-1606(75)90269-9. [DOI] [PubMed] [Google Scholar]
- 7.Donovan PJ, Stott D, Cairns LA, Heasman J, Wylie CC. Migratory and postmigratory mouse primordial germ cells behave differently in culture. Cell. 1986;44:831–8. doi: 10.1016/0092-8674(86)90005-x. [DOI] [PubMed] [Google Scholar]
- 8.Anderson R, Copeland TK, Scholer H, Heasman J, Wylie C. The onset of germ cell migration in the mouse embryo. Mech Dev. 2000;91:61–8. doi: 10.1016/s0925-4773(99)00271-3. [DOI] [PubMed] [Google Scholar]
- 9.Pedersen T, Peters H. Proposal for a classification of oocytes and follicles in the mouse ovary. J Reprod Fertil. 1968;17:555–7. doi: 10.1530/jrf.0.0170555. [DOI] [PubMed] [Google Scholar]
- 10.Pangas SA, Choi Y, Ballow DJ, Zhao Y, Westphal H, Matzuk MM, et al. Oogenesis requires germ cell-specific transcriptional Sohlh1 and Lhx8. Proc Natl Acad Sci U S A. 2006;103:8090–5. doi: 10.1073/pnas.0601083103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajkovic A, Yan MSC, Klysik M, Matzuk M. Discovery of germ cell-specific transcripts by expressed sequence tag database analysis. Fertil Steril. 2001;76:550–4. doi: 10.1016/s0015-0282(01)01966-5. [DOI] [PubMed] [Google Scholar]
- 12.Suzumori N, Yan C, Matzuk MM, Rajkovic A. Nobox is a homeobox-encoding gene preferentially expressed in primordial and growing oocytes. Mech Dev. 2002;111:137–41. doi: 10.1016/s0925-4773(01)00620-7. [DOI] [PubMed] [Google Scholar]
- 13.Rajkovic A, Pangas SA, Ballow D, Suzumori N, Matzuk MM. Nobox deficiency disrupts early folliculogenesis and oocyte-specific gene expression. Science. 2004;305:1157–9. doi: 10.1126/science.1099755. [DOI] [PubMed] [Google Scholar]
- 14.Choi Y, Qin Y, Berger MF, Ballow DJ, Bulyk ML, Rajkovic A. Microarray analyses of newborn mouse ovaries lacking Nobox. Biol Reprod. 2007;77:312–9. doi: 10.1095/biolreprod.107.060459. [DOI] [PubMed] [Google Scholar]
- 15.Elvin JA, Yan C, Wang P, Nishimori K, Matzuk MM. Molecular characterization of the follicle defects in the growth differentiation factor-9-deficient ovary. Mol Endocrinol. 1999;13:1018–1034. doi: 10.1210/mend.13.6.0309. [DOI] [PubMed] [Google Scholar]
- 16.Pangas SA, Rademaker AW, Fishman DA, Woodruff TK. Localization of the activin signal transduction components in normal human ovarian follicles: implications for autocrine and paracrine signaling in the ovary. J Clin Endocrinol Metab. 2002;87:2644–57. doi: 10.1210/jcem.87.6.8519. [DOI] [PubMed] [Google Scholar]