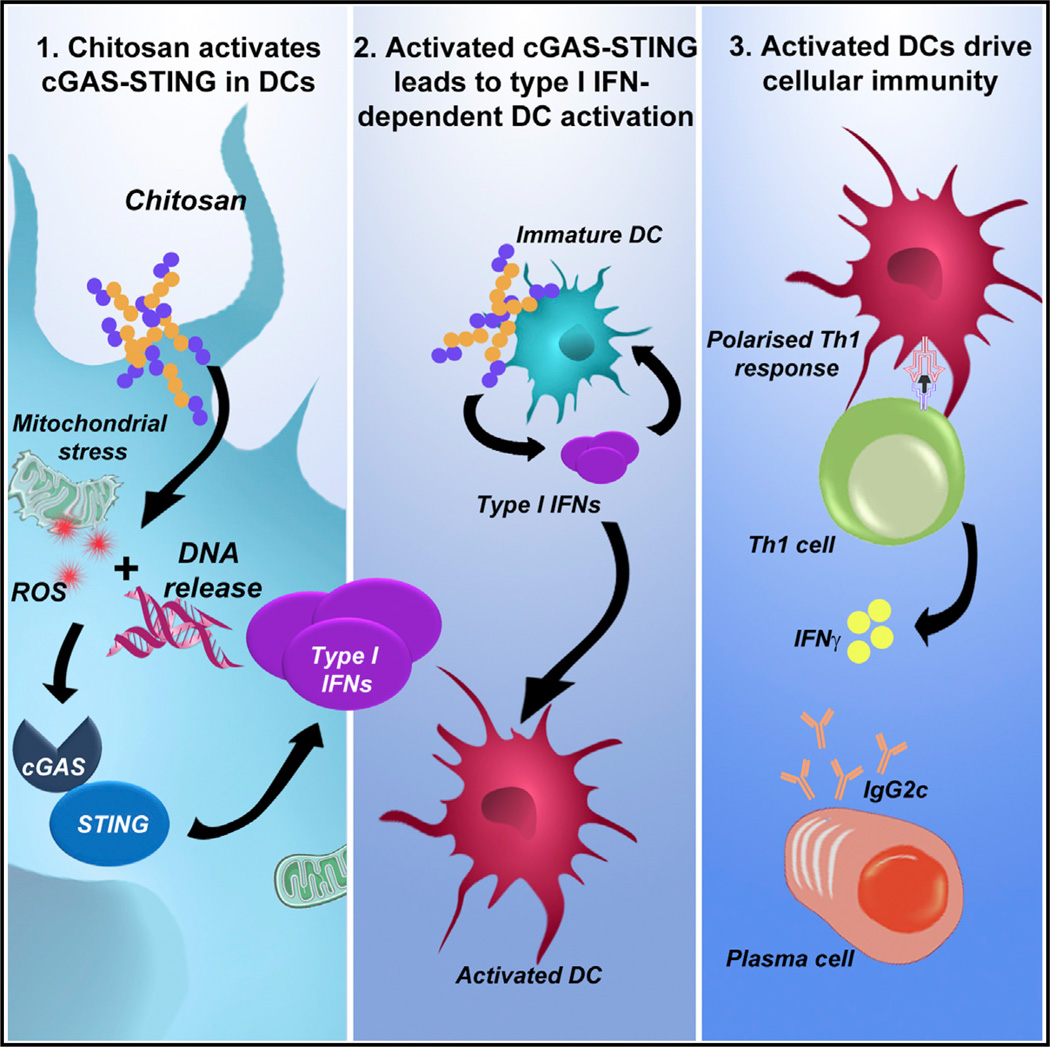
SUMMARY
The cationic polysaccharide chitosan is an attractive candidate adjuvant capable of driving potent cell-mediated immunity, but the mechanism by which it acts is not clear. We show that chitosan promotes dendritic cell maturation by inducing type I interferons (IFNs) and enhances antigen-specific T helper 1 (Th1) responses in a type I IFN receptor-dependent manner. The induction of type I IFNs, IFN-stimulated genes and dendritic cell maturation by chitosan required the cytoplasmic DNA sensor cGAS and STING, implicating this pathway in dendritic cell activation. Additionally, this process was dependent on mitochondrial reactive oxygen species and the presence of cytoplasmic DNA. Chitosan-mediated enhancement of antigen specific Th1 and immunoglobulin G2c responses following vaccination was dependent on both cGAS and STING. These findings demonstrate that a cationic polymer can engage the STING-cGAS pathway to trigger innate and adaptive immune responses.
Graphical Abstract
INTRODUCTION
There is a significant impetus to develop new and improved vaccine strategies for challenging diseases including HIV, tuberculosis, and malaria (Rappuoli and Aderem, 2011). A major obstacle is the lack of adjuvants that can safely drive potent cellular immunity against intracellular pathogens. Aluminum salts (alum) find wide clinical application and promote humoral immunity and T helper type 2 (Th2) cell responses (Brewer et al., 1999). However, a major disadvantage of alum is its limited ability to efficiently drive T helper 1 (Th1) responses. The chitin derivative chitosan is an attractive alternative to alum, being biocompatible, flexible in terms of formulation and degree of deacetylation, and efficacious when administered mucosally (Vasiliev, 2015). We found that chitosan is superior to alum in promoting Th1 responses (Mori et al., 2012). However, like alum, the mechanism underlying the adjuvanticity of chitosan is not fully understood.
A deeper understanding of adjuvanticity is critical for the rational design of new and improved vaccines. Activation of innate immunity is essential for effective induction of protective, antigen-specific responses (Pulendran and Ahmed, 2011). Modulation of dendritic cells (DCs) by adjuvants is a major focus due to their superior ability to present antigen to naive T cells. DC activation is characterized by enhanced surface expression of costimulatory molecules, including CD40 and CD86, in a process referred to as maturation (Reis e Sousa, 2006). Additionally, in the case of pattern-recognition receptors (PRRs) such as Toll-like receptors (TLRs), engagement of pathogen associated molecular patterns (PAMPs) by DCs also results in increased secretion of inflammatory and immunomodulatory cytokines. Given the pivotal role of these sentinels as a bridge between innate and adaptive immunity, understanding how adjuvants regulate DC-mediated adaptive immunity is crucial (Palucka et al., 2010).
The innate sensing of nucleic acids as either PAMPs or danger associated molecular patterns (DAMPs) has been postulated to underlie the efficacy of live attenuated vaccines, alum-adjuvanted vaccines, and DNA vaccines (Desmet and Ishii, 2012). A range of PRRs involved in nucleic-acid sensing have been identified in recent years. Cytosolic DNA triggers robust immune responses including the activation of the absent in melanoma 2 (AIM2) inflammasome and the induction of type I interferons (IFNs). Stimulator of IFN genes (STING) has been identified as a central adaptor protein mediating intracellular signaling events in response to cytosolic DNA, by directing the activation of the transcription factors, nuclear factor kappa B (NF-κB) and interferon regulatory factor 3 (IRF3) through the kinases IκB kinase (IKK) and Tank-binding kinase-1 (TBK1), respectively. Recently, the enzyme cyclic-di-GMP-AMP synthase (cGAS) has been identified as the main DNA sensor upstream of STING, essential for DNA-mediated immune responses irrespective of DNA sequence. cGAS binds to DNA to catalyze the synthesis of cyclic-di-GMP-AMP (cGAMP) from ATP and GTP, which subsequently binds to and activates STING (Cai et al., 2014). STING is also capable of binding cyclic dinucleotides (CDNs) and bacterial second messenger molecules such as cyclic di-GMP (c-di-GMP) and cyclic di-AMP (c-di-AMP; Burdette et al., 2011) to induce a similar gene induction profile to cytosolic DNA (McWhirter et al., 2009). Interestingly, these STING-activating molecules hold significant potential for use as vaccine adjuvants in a mucosal setting (Dubensky et al., 2013).
Type I IFNs belong to a family of cytokines that are widely recognized for their roles in antiviral immunity. They consist of 16 members, 12 IFN-α subtypes, IFN-β, IFN-ε, IFN-κ, and IFN-ω. These cytokines signal through a common heterodimeric receptor, the IFN-α:IFN-β receptor (IFNAR), which consists of two subunits: IFNAR1 and IFNAR2 (González-Navajas et al., 2012). Type I IFNs have important roles in the induction of adaptive immunity, they promote the generation of cytotoxic T cell responses as well as a Th1 biased CD4+ T cell phenotype (Tough, 2012). Furthermore, type I IFNs promote the activation and functional maturation of DCs, increasing their migration and facilitating antigen presentation to CD4+ T cells as well as cross priming of CD8+ T cells (Longhi et al., 2009).
We have demonstrated the potential of chitosan to serve as an adjuvant capable of driving potent cell-mediated immunity (Mori et al., 2012). More recently, the mechanisms involved in chitosan-mediated inflammasome activation have been addressed (Bueter et al., 2014). Despite the widespread use of chitosan in vaccine research and for other biomedical applications, its immunomodulatory effects are not well understood. Herein, we describe the mechanism by which chitosan promoted DC activation and induction of cellular immunity. Chitosan stimulated the intracellular release of DNA, which engaged the cGAS-STING pathway to mediate the selective production of type I IFN and interferon-stimulated genes (ISGs). These cytokines were then responsible for mediating the activation of DCs and induction of cellular immunity.
RESULTS
Chitosan-Induced Cellular Immunity and IgG2 Production Is Dependent on Signaling through the Type I Interferon Receptor, IFNAR
We previously highlighted the potential of chitosan as an adjuvant to promote cellular immunity. Unlike alum, chitosan does not inhibit production of the Th1 driving cytokine interleukin-12 (IL-12). Furthermore, when combined with the TLR9 agonist CpG, chitosan promotes antigen-specific Th1 and Th17 responses (Mori et al., 2012). This study focused on the potential of chitosan to drive cellular immunity and elucidate the mechanism underlying its adjuvanticity. The antigen used, Hybrid-1 (H1), is a fusion protein based on two immunodominant antigens: antigen 85B (Ag85B) and early secreted antigenic target (ESAT-6) from Mycobacterium tuberculosis, and has shown promise in clinical trials (van Dissel et al., 2010, 2014). Given the requirement for cellular responses in conferring protection against M. tuberculosis (Agger et al., 2008), the identification of effective safe adjuvants that drive cellular immunity is a priority.
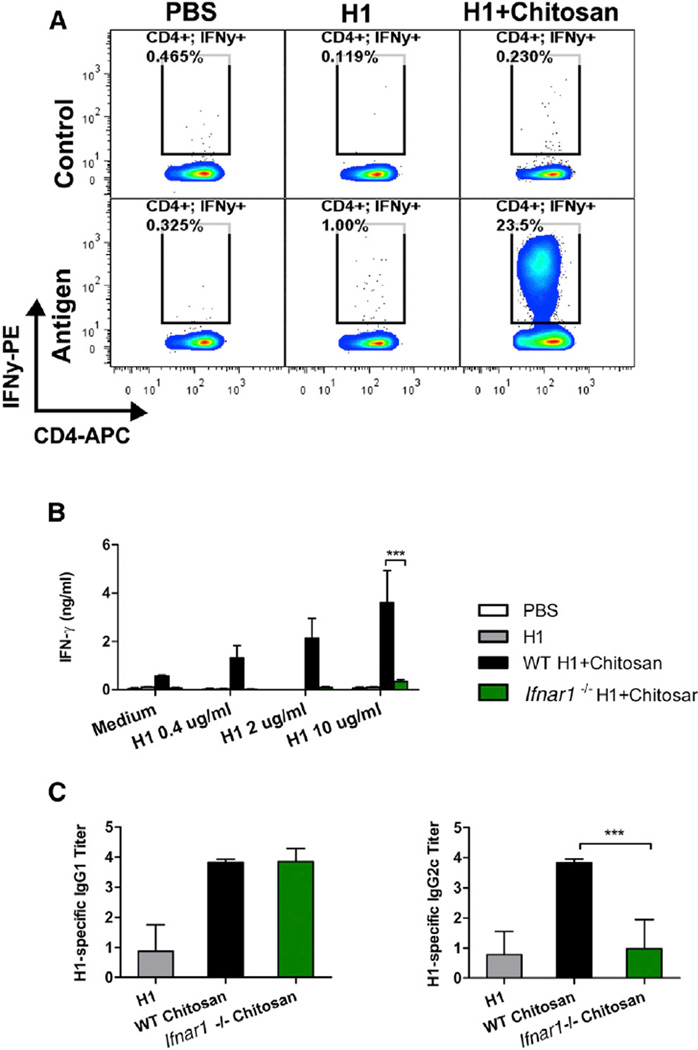
Immunization of mice with H1 and chitosan promoted antigen-specific production of the Th1-associated cytokine IFN-γ from CD3+CD4+ T cells (Figure 1A, Figure S1A). IFN-γ secretion after antigen stimulation remained unchanged in CD8+ T cells and γδ-T cells (Figures S1B and S1C). Limited production of Th2-associated cytokines (data not shown) and IL-17 (Figure S1D) was detected, indicating a Th1 polarized response. As type I IFNs can promote Th1 polarized responses in other contexts (Tough, 2012), a role for this family of cytokines in the adjuvanticity of chitosan was investigated. Wild-type (WT) C57BL/6 mice and mice deficient in the IFNAR1 chain of IFNAR (Ifnar1−/−) were immunized with chitosan and H1. The ability of chitosan to drive antigen-specific IFN-γ secretion by draining mediastinal lymph node (mLNs) cells was significantly decreased in Ifnar1−/− mice (Figure 1B). Although a limited response, chitosan-mediated enhancement of H1-specific IL-17 was also impaired in the mLNs of Ifnar1−/− mice compared to WT mice (Figure S1D). Furthermore, injection of mice with chitosan and H1 increased IFN-γ and IL-17 secretion by mLNs in response to anti-CD3 stimulation, and this effect was abrogated in Ifnar1−/− mice (Figures S1E and S1F). Co-injection of chitosan enhanced H1-specific immunoglobulin G1 (IgG1) and IgG2c antibody responses when compared to immunization with antigen alone. Chitosan-mediated enhancement of IgG2c but not IgG1 was compromised in the absence of IFNAR signaling (Figure 1C). In summary, chitosan, when combined with the clinically relevant antigen H1, promoted robust Th1 and to a lesser extent Th17 responses which were dependent on signaling through IF-NAR. In line with these observations, antigen-specific induction of the Th1-associated antibody isotype IgG2c was compromised in Ifnar1−/− mice. These data highlight a crucial role for type I IFNs in the adjuvanticity of chitosan, in particular for its ability to promote cell-mediated immune responses.
Figure 1. Chitosan Promotes Robust IFNAR-Dependent Th1 Responses and IgG2c Production.
(A) H1-specific IFN-γ production by CD4+ T cells in PECs of WT mice after vaccination with H1 alone or in combination with chitosan. Results are shown as representative dot plots or bar graphs.
(B) Antigen-specific IFN-γ production in mLNs isolated from WT and Ifnar1−/− mice immunized with H1 alone or with chitosan.
(C) H1-specific IgG1 and IgG2c in serum of WT and Ifnar1−/− mice immunized with H1 alone or with chitosan. Data shown as mean ± SEM of three independent experiments, n = 5 per group. WT versus Ifnar1−/−, ***p < 0.001. See also Figure S1.
Chitosan Promoted the Upregulation of Costimulatory Molecules on the Surface of DCs in the Absence of Proinflammatory Cytokine Production
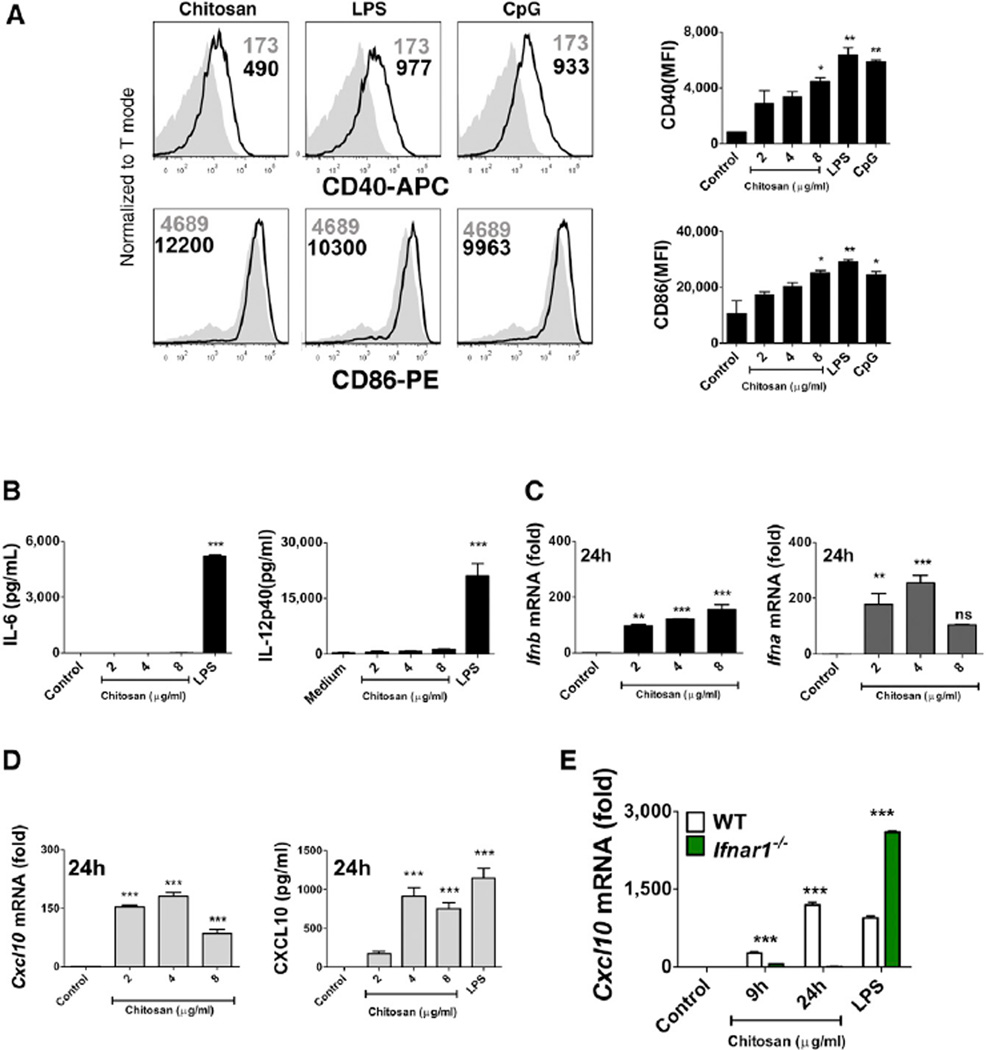
The activation of DCs is an essential requirement for effective adjuvanticity (Palucka et al., 2010). To determine whether chitosan promoted DC maturation, we studied its ability to modulate surface expression of the costimulatory molecules CD40 and CD86 by bone-marrow-derived DCs (BMDCs). The TLR agonists LPS and CpG served as positive controls to promote DC maturation and as expected, both strongly enhanced CD40 and CD86 surface expression. Chitosan also enhanced surface expression of CD40 and CD86 by DCs. This upregulation reached significance at the highest concentration of chitosan tested when compared to the control (Figure 2A). However, chitosan-induced DC maturation did not coincide with the secretion of pro-inflammatory cytokines such as IL-6 and IL-12p40 by DCs. This was in sharp contrast to stimulation with LPS, which induced robust secretion of both (Figure 2B). The involvement of TLR4 in chitosan-induced DC maturation was ruled out, as demonstrated by intact upregulation of CD40 in DCs generated from C3H/HeJ mice, which are defective in TLR4 signaling (Figure S2). Thus, chitosan promoted DC maturation by a mechanism other than that involving TLR4.
Figure 2. Chitosan Induces DC Maturation and Type I IFN Signaling in the Absence of Proinflammatory Cytokine Production.
(A) CD40 and CD86 expression in WT DCs incubated for 24 hr with medium (shaded histogram), chitosan (8 µg/ml), LPS, or CpG (black line). Data are represented as mean ± SEM of MFI values representative of three independent experiments.
(B) IL-6 and IL-12p40 in supernatants of DCs incubated with indicated concentrations of chitosan or LPS for 24 hr.
(C) qPCR analysis of Ifnb and Ifna. mRNA expression by DCs stimulated for the indicated times with chitosan.
(D) Cxcl10 mRNA expression and CXCL10 secreted into the supernatant of DCs incubated with chitosan or LPS for 24 hr.
(E) qPCR analysis of Cxcl10 mRNA expression in WT (black bars) and Ifnar1−/− DCs (green bars) incubated with chitosan for the indicated time points and LPS for 4 hr. mRNA levels calculated with respect to β-actin and fold increase calculated relative to untreated cells. Results are expressed as the mean ± SEM. Asterisks represent significant differences for *p < 0.05, **p < 0.01, ***p < 0.001 for medium versus treated in (A), and, WT versus Ifnar1−/− in (B)–(D).
Chitosan Promoted the Transcription and Secretion of Type I Interferons and ISGs by DCs
Signaling through IFNAR was shown to be crucial for the ability of chitosan to promote antigen-specific Th1 responses. As a result, we addressed the ability of chitosan to promote type I IFN production by DCs. The capacity of DCs to express Ifnb and Ifna messenger RNA (mRNA) in response to increasing concentrations of chitosan was studied. Primers were designed to amplify the single Ifnb gene and all of the murine Ifna subtypes described. Additionally, DCs were either treated with LPS or transfected with the double-stranded (ds) DNA analog poly(dA:dT) as positive controls. LPS induced robust expression of Ifnb but not Ifna, while poly(dA:dT) stimulated robust induction of Ifna mRNA compared to Ifnb mRNA (Figures S3A and S3B). We found that chitosan was capable of inducing the expression of both Ifnb and Ifna mRNA; this response was detectable after 9 hr and peaked after 24 hr (Figure 2C, Figures S3C and S3D).
In response to viral infection, type I IFN production in DCs occurs in two phases; an initial induction phase, followed by an IFN feedback loop whereby type I IFN signaling, in an autocrine or paracrine manner, amplifies the response (Tailor et al., 2007). To determine whether this was the case with chitosan-mediated type I IFN production, we tested IFN transcript induction in DCs from Ifnar1−/− mice that are incapable of generating this feedback loop. The initial induction of Ifnb mRNA, 9 hr following chitosan treatment, was independent of signaling through IFNAR. However, after 24 hr, Ifnb mRNA transcripts were no longer detected in Ifnar1−/− DCs (Figure S3C). We concluded that IFNAR signaling contributed to a feed-forward loop of IFN-β production in DCs in response to chitosan. While the initial induction of Ifnb mRNA in response to chitosan was independent of signaling through IFNAR, Ifna mRNA transcription was not (Figure S3D). This suggested that chitosan-induced Ifna transcription first required priming by IFN-β.
Type I IFNs are known to induce the transcription of several ISGs, including the chemokine CXCL10 (IP-10) (Holm et al., 2012). Indeed, we observed induction of Cxcl10 transcripts in response to chitosan treatment (Figure 2D). To further confirm the ability of chitosan to promote CXCL10 production, we detected CXCL10 protein in supernatants of DCs incubated with chitosan (Figure 2D). The transcription of Cxcl10 in response to chitosan, but not LPS, was dependent on autocrine IFN-β signaling through IFNAR and was absent in Ifnar1−/− mice, thus confirming that in our experimental setting, CXCL10 is an ISG induced in response to chitosan-mediated IFN-β production (Figure 2E).
Previously, the immunostimulatory properties of malarial hemozoin were shown to be the consequence of contaminating malarial DNA (Parroche et al., 2007). To eliminate this possibility, we subjected chitosan to nuclease digestion. DNase I digestion of chitosan did not impair its ability to induce Cxcl10 transcription or secretion, but did abolish poly(dA:dT)-induced secretion of CXCL10 (Figures S4A and S4B). Together, these data indicate that chitosan directly promoted IFN-β responses in DCs resulting in the transcription of ISGs. Furthermore, the induction of type I IFN and associated genes in response to chitosan was not the consequence of contaminating DNA.
Chitosan-Induced DC Maturation Is Dependent on Signaling through IFNAR
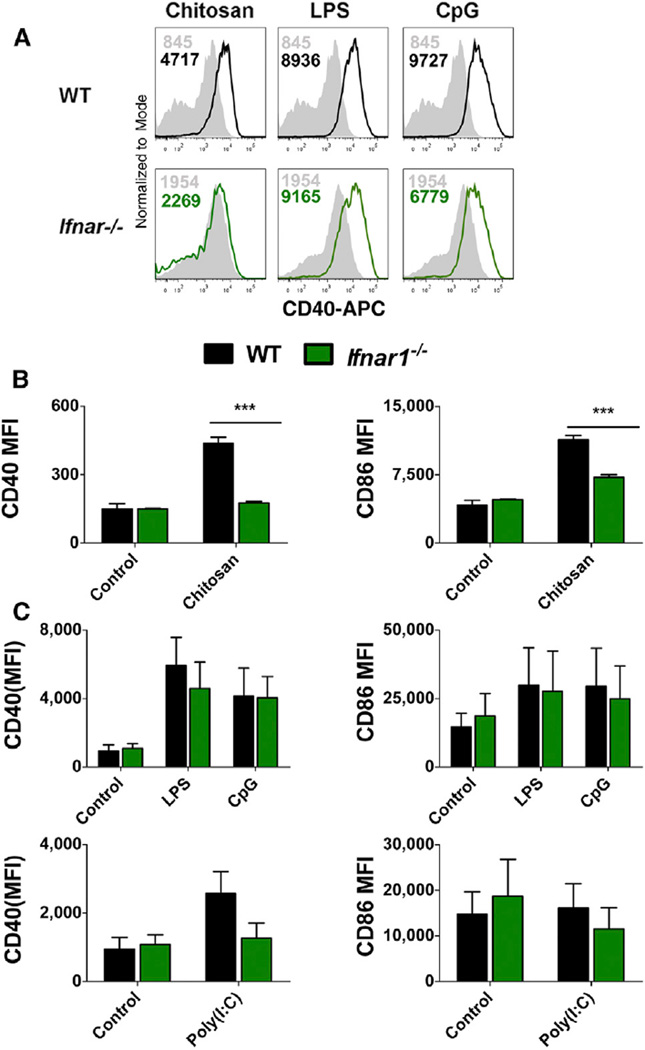
Type I IFNs exert a multitude of effects on both the innate and adaptive immune systems. In particular, these cytokines are implicated in the phenotypic and functional maturation of DCs in response to dsRNA and viral infection (Honda et al., 2003). With this in mind, the role of signaling through IFNAR in chitosan-induced DC maturation was investigated by measuring chitosan-induced upregulation of CD40 and CD86 in WT and Ifnar1−/− DCs. The ability of chitosan to enhance CD40 expression was abrogated in the absence of signaling through IFNAR (Figure 3A). Median fluorescence intensity (MFI) values for both CD40 and CD86 were markedly lower in Ifnar1−/− DCs treated with the highest concentration of chitosan when compared to WT DCs (Figure 3B). In contrast, MFI values for both CD40 and CD86 were comparable in WT and Ifnar1−/− DCs treated with the positive control CpG. Upregulation of costimulatory molecules by dsRNA and LPS is reported to be entirely dependent on type I IFN receptor signaling (Hoebe et al., 2003). While we did not observe a substantial increase in CD86 expression in response to the TLR3 ligand poly(I:C) in WT DCs, the upregulation of CD40 in response to poly(I:C) was compromised in the absence of IFNAR. In contrast, we noted that LPS-induced DC maturation was IFNAR-independent (Figure 3C). Thus chitosan but not LPS or CpG requires IFNAR to promote DC maturation.
Figure 3. Chitosan Induced Upregulation of CD40 and CD86 on DCs Is Compromised in the Absence of Type I IFN Receptor Signaling.
(A) CD40 expression in WT (black) or Ifnar1−/− (green) DCs unstimulated (shaded histogram) or stimulated with chitosan (8 µg/ml), LPS, or CpG for 24 hr.
(B) CD40 and CD86 expression in WT or Ifnar1−/− DCs stimulated with chitosan and represented as MFI values.
(C) CD40 and CD86 expression in WT or Ifnar1−/− DCs stimulated with medium, LPS, or CpG and represented as MFI values. Results expressed as mean ± SEM of three independent experiments. WT versus Ifnar1−/−, ***p < 0.001.
The cGAS-STING Pathway Is Required for Chitosan-Induced Type I IFN Production
Chitosan is phagocytosed by macrophages and this action is important for its ability to activate the NLRP3 inflammasome (Bueter et al., 2011). We hypothesized that uptake of chitosan by DCs and subsequent engagement with an intracellular sensing pathway induced the production of type I IFNs. Indeed, we confirmed the uptake of fluorescein-5-isothiocyanate (FITC)-conjugated chitosan by DCs by flow cytometry and confocal microscopy (Figures S5A–S5C).
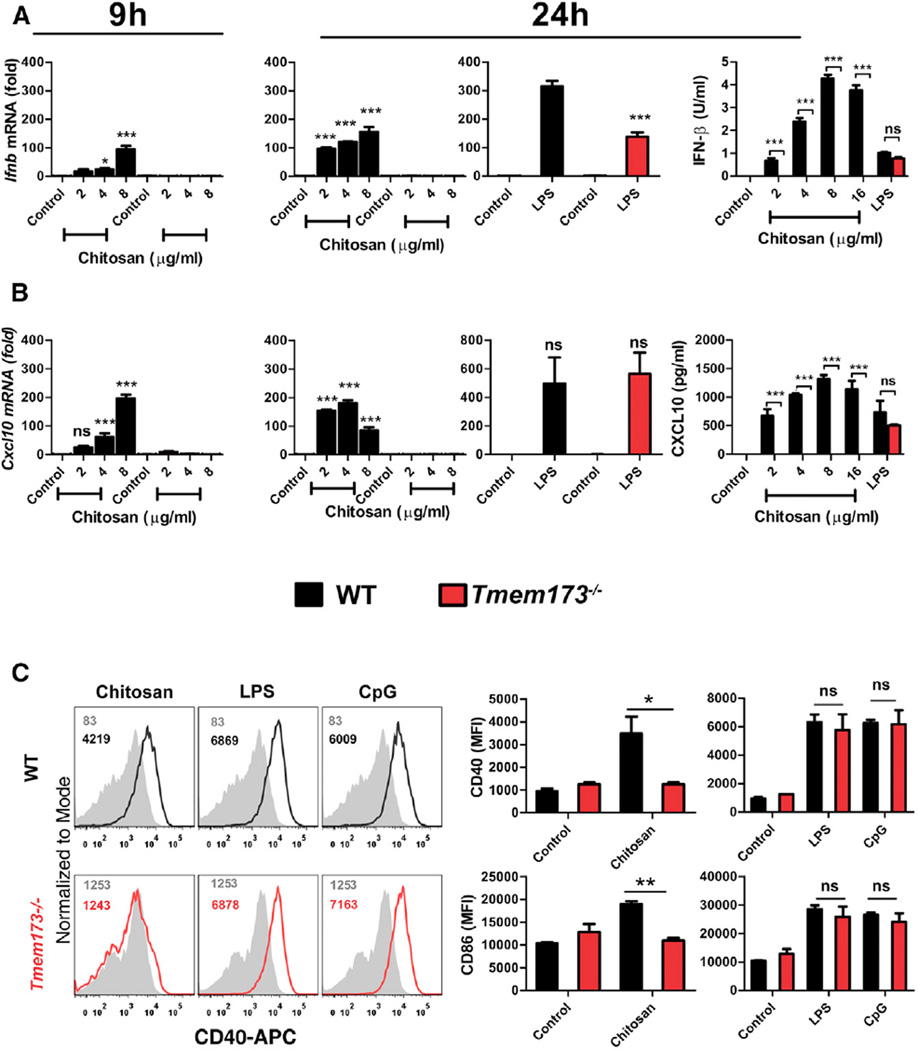
The induction of type I IFN can be mediated by the engagement of a range of PRRs, in particular by intracellular nucleic acid sensors. STING has been identified as a central player in mediating type I IFN induction in response to cytosolic DNA (Cai et al., 2014), with a number of intracellular type I IFN activating pathways converging at the level of STING. It was hypothesized that chitosan might be engaging with this adaptor to mediate type I IFN production and DC maturation. The expression of Ifnb, Ifna, and Cxcl10 mRNA in response to chitosan was studied in WT and STING-deficient DCs (STING is encoded by Tmem173). Induction of Ifnb in response to chitosan was completely abolished in Tmem173−/− DCs, and this impairment was observed 9 hr and 24 hr following treatment, suggesting that STING was crucial for the initial induction of Ifnb (Figure 4A). Moreover, chitosan-induced Ifna (Figure S3E) and Cxcl10 expression, in addition to the secretion of IFN-β and CXCL10, was also absent in Tmem173−/− DCs (Figures 4A and 4B). We noted that LPS-induced Ifnb mRNA transcription was significantly decreased in Tmem173−/− DCs compared to WT cells (Figure 4A). However, LPS-induced Cxcl10 mRNA expression was comparable between WT and Tmem173−/− cells (Figure 4B). Additionally, there were no significant differences in LPS-induced secretion of IFNβ protein and CXCL10 proteins between WT and Tmem173−/− DCs (Figures 4A and 4B), which demonstrated an intact capacity to respond to STING-independent ligands.
Figure 4. STING Is Required for Chitosan-Induced Type I IFN Responses and Maturation of DCs.
mRNA expression (9 and 24 hr) and protein levels (24 hr) of (A) IFN-β or (B) CXCL10 produced by WT (black) or Tmem173−/− (red) DCs stimulated with chitosan or LPS. RNA levels were calculated with respect to Actb and results are presented as fold increase compared to untreated cells.
(C) CD40 and CD86 expression on WT and Tmem173−/− DCs which were either untreated (shaded histogram) or stimulated with chitosan (8 µg/ml), LPS or CpG. Numbers indicate MFI values. Results are expressed as the mean ± SEM of two independent experiments. Asterisks indicate *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S3.
The ability of chitosan to promote DC maturation was also abolished in the absence of STING. Specifically, treatment of DCs from STING-deficient mice with chitosan failed to upregulate CD40 or CD86 surface expression, whereas LPS and CpG-induced upregulation of CD40 and CD86 was intact in Tmem173−/− DCs (Figure 4C). These data are in accordance with our earlier observations that chitosan-induced DC maturation was dependent on signaling through IFNAR.
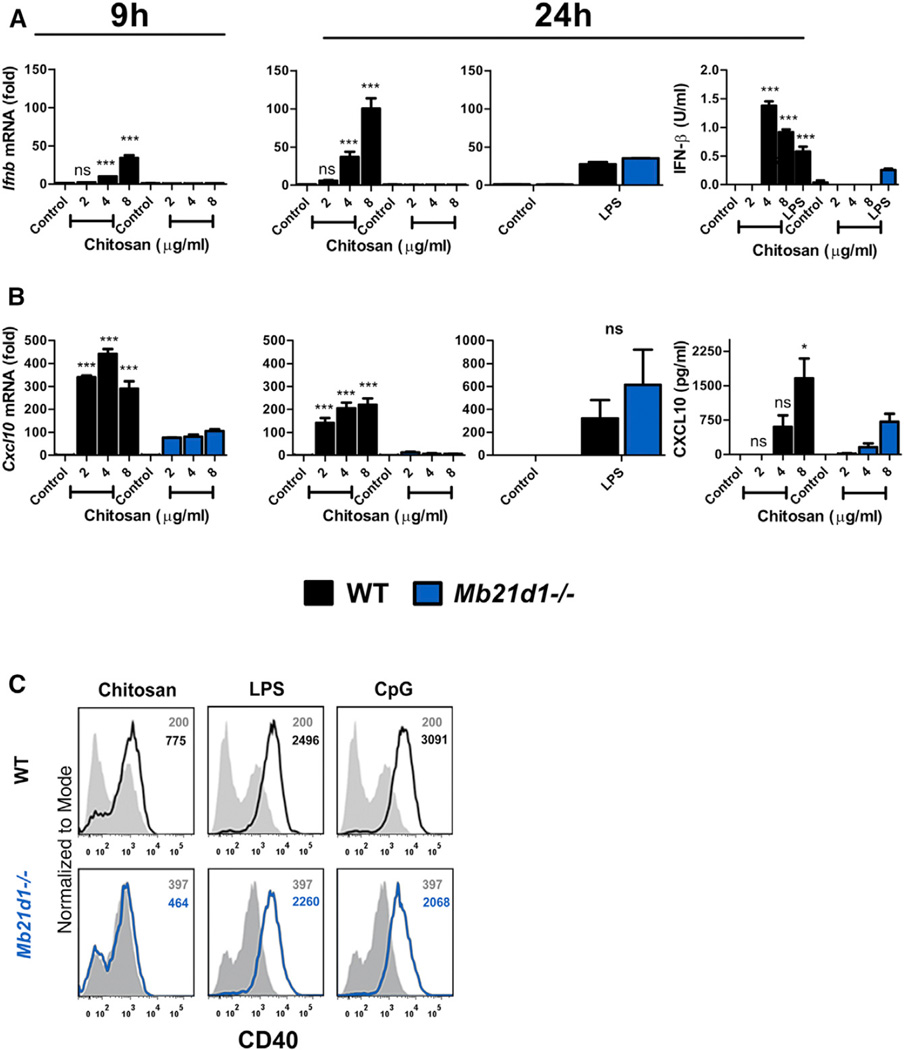
We next sought to identify what components upstream of STING were involved in mediating the activation of the chitosan-STING-type I IFN pathway. The enzyme cGAS is identified as the main DNA sensor upstream of STING (Cai et al., 2014). The possibility that chitosan engaged the DNA-sensing cGAS-STING pathway was therefore explored in DCs from cGAS-deficient mice (cGAS is encoded by Mb21d1).We found that induction of Ifnb mRNA and Cxcl10 expression in response to chitosan was abrogated in Mb21d1−/− DCs (Figures 5A and 5B) whereas LPS-induced expression of both Ifnb and Cxcl10 mRNA was completely intact. Furthermore, the secretion of IFN-β and CXCL10 in response to chitosan was compromised in the absence of cGAS (Figures 5A and 5B). Importantly, Mb21d1−/− DCs were still capable of producing IFN-β in response to LPS, although it was noted to be significantly lower in Mb21d1−/− DCs compared to WT cells. Finally, chitosan-mediated enhancement of CD40 expression on DCs was abolished in the absence of cGAS while LPS- and CpG-induced CD40 expression was intact (Figure 5C). In summary, the ability of chitosan to promote type I interferons and DC maturation was dependent on both cGAS and STING.
Figure 5. Chitosan-Induced DC Maturation and Type I IFN Production Requires cGAS.
(A) qPCR analysis of Ifnb mRNA expression by WT (black) or Mb21d1−/− (blue) DCs which were either untreated or stimulated with chitosan or LPS for 9 or 24 hr.
(B) Transcript (9 and 24 hr) and protein (24 hr) levels for CXCL10 in WT and Mb21d1−/− DCs after treatment with chitosan or LPS. mRNA levels were normalized with respect to Actb and results are presented as fold increase compared to untreated cells. Data are shown as mean ± SEM for triplicate samples. Results are representative of two independent experiments. WT versus Mb21d1−/−, *p < 0.05, ***p < 0.001.
(C) Representative histograms for CD40 expression on WT and Mb21d1−/− DCs which were either untreated (shaded) or stimulated (solid lines) with chitosan (8 µg/ml), LPS, or CpG.
Chitosan-Induced Cellular Immunity and IgG2c Production Required Both cGAS and STING
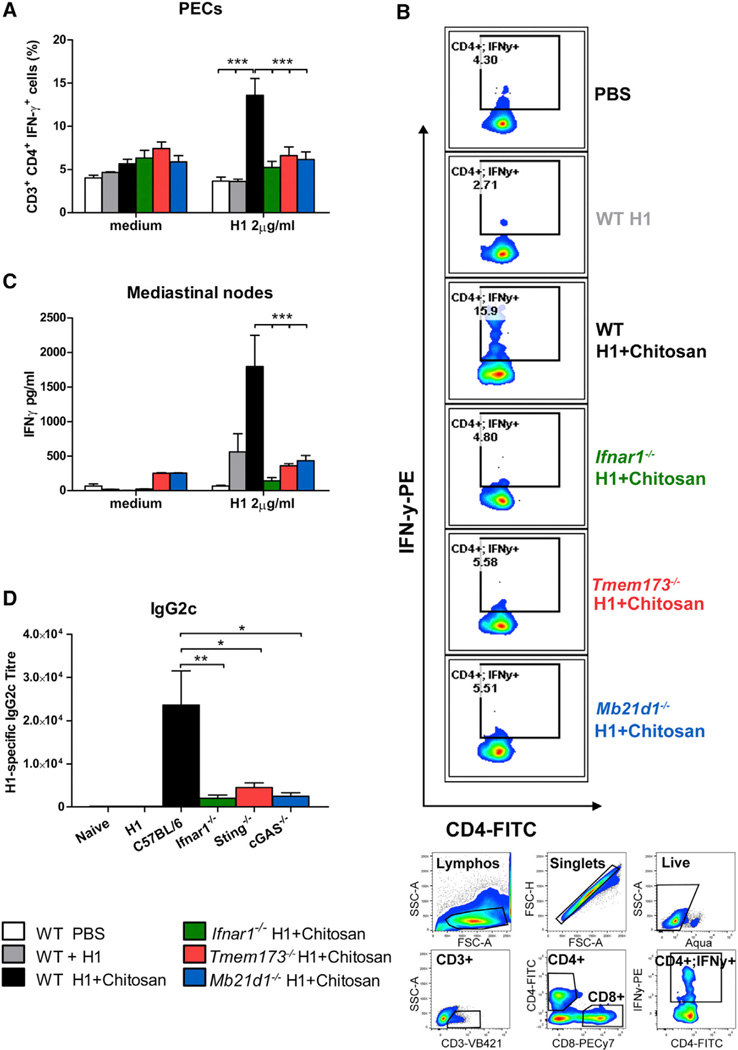
The previous section highlighted the critical role of both cGAS and STING in chitosan-induced type I IFN responses in DCs in vitro. Because we had established that type I IFNs were essential for the ability of chitosan to drive cellular immunity (Figure 1), we set out to address whether cGAS and STING were required for chitosan-induced cellular immune responses in vivo. The enhanced H1-specific IFN-γ response in CD3+CD4+ cells from peritoneal exudates (PECs) taken from WT mice immunized with chitosan and H1 was significantly compromised in Tmem173−/− mice (Figures 6A and 6B). Similarly, the chitosan-induced enhancement of antigen-specific IFN-γ secretion by mLNs was markedly decreased in Tmem173−/− mice compared to WT mice (Figure 6C). Further supporting a role for STING in the adjuvanticity of chitosan, H1-specific IgG2c (Figure 6D), but not IgG1 (data not shown), titers induced by chitosan were compromised in the absence of STING.
Figure 6. Chitosan Promotes cGAS-STING-IFNAR1-Dependent Th1 Responses and IgG2c Production.
(A) H1-specific intracellular IFN-γ production in CD3+CD4+ isolated from PECs of WT (white, PBS; gray, antigen alone; black, antigen+adjuvant), Ifnar1−/− (green), Tmem173−/− (red), and Mb21d1−/− (blue) mice immunized with H1 alone or in combination with chitosan.
(B) Representative dot plots showing levels of H1 specific intracellular IFN-γ in CD3+CD4+ (upper panel). Gating strategy used for analysis of intracellular IFN-γ production (lower panel).
(C) Antigen-specific IFN-γ production in mLNs isolated from WT, Ifnar1−/−, Tmem173−/−, and Mb21d1−/− mice immunized with H1 alone or with chitosan.
(D) H1-specific IgG2c in serum of WT, Ifnar1−/−, Tmem173−/−, and Mb21d1−/− mice immunized with H1 alone or with chitosan. Representative of two independent experiments for WT, Ifnar1−/−, Tmem173−/−, and one experiment for WT, Ifnar1−/−, Tmem173−/−, and Mb21d1−/−, n = 5 per group. Data shown as mean ± SEM. WT versus Ifnar1−/−, WT versus Tmem173−/−, WT versus Mb21d1−/−,*p < 0.05, ***p < 0.001. See also Figure S7.
An almost identical requirement for cGAS in chitosan-induced cellular immunity was demonstrated. The chitosan-induced enhancement of H1-specific IFN-γ by CD3+CD4+ PECs was significantly decreased in Mb21d1−/− mice in comparison to WT mice (Figures 6A and 6B). In line with this observation, antigen-specific IFN-γ secretion in mLNs was markedly lower in Mb21d1−/− than WT mice (Figure 6C). Finally, as in the case of Ifnar1−/− and Tmem173−/− mice, chitosan-mediated induction of H1-specific IgG2c was compromised in the absence of cGAS (Figure 6D).
Because chitosan has been shown to activate the NLRP3 inflammasome, the ability of chitosan to induce cellular immunity in Nlrp3−/− mice was analyzed. The capacity of chitosan to promote antigen-specific Th1 responses was markedly reduced in mLNs from Nlrp3−/− mice compared to WT mice (Figure S6A). Additionally, the enhanced production of IFN-γ by mLNs of mice immunized with H1 and chitosan in response to anti-CD3 stimulation was also reduced in Nlrp3−/− mice (Figure S6B).However, in contrast to the findings in Ifnar1−/− mice, antigen-specific IgG2c responses were not significantly reduced in Nlrp3−/− mice compared to WT mice (Figure S6C) as was the case for IgG1 (Figure S6D). These data indicated complementary roles for the cGAS-STING pathway and the NLRP3 inflammasome in chitosan-mediated Th1 responses and IgG2c antibody induction.
Mucosal vaccination is an attractive alternative to delivery by injection. To address whether the role of STING in the induction of cellular immunity by chitosan was specific to the intraperitoneal (i.p.) route or more broadly applicable,WTandTmem173−/− mice were immunized intranasally with pneumococcal surface protein A (PspA) and chitosan. Vaccination with antigen and chitosan enhanced PspA specific IFN-γ production by restimulated lung cells in WT mice, and this was significantly reduced in Tmem173−/− mice (Figure S7D). Furthermore, while chitosan enhanced PspA-specific serum IgG1 and IgG2c responses in WT mice, the IgG2c response was reduced in Tmem173−/− mice while the IgG1 response remained intact (Figures S7A and S7B). These findings demonstrated that the requirement for STING in the adjuvanticity of chitosan was applicable to both injected and mucosally delivered vaccines and applied to two distinct vaccine antigens.
Chitosan Enhanced Mitochondrial Reactive Oxygen Species Production and Endogenous DNA Release, Which Are Required for Subsequent Secretion of Type I IFNs
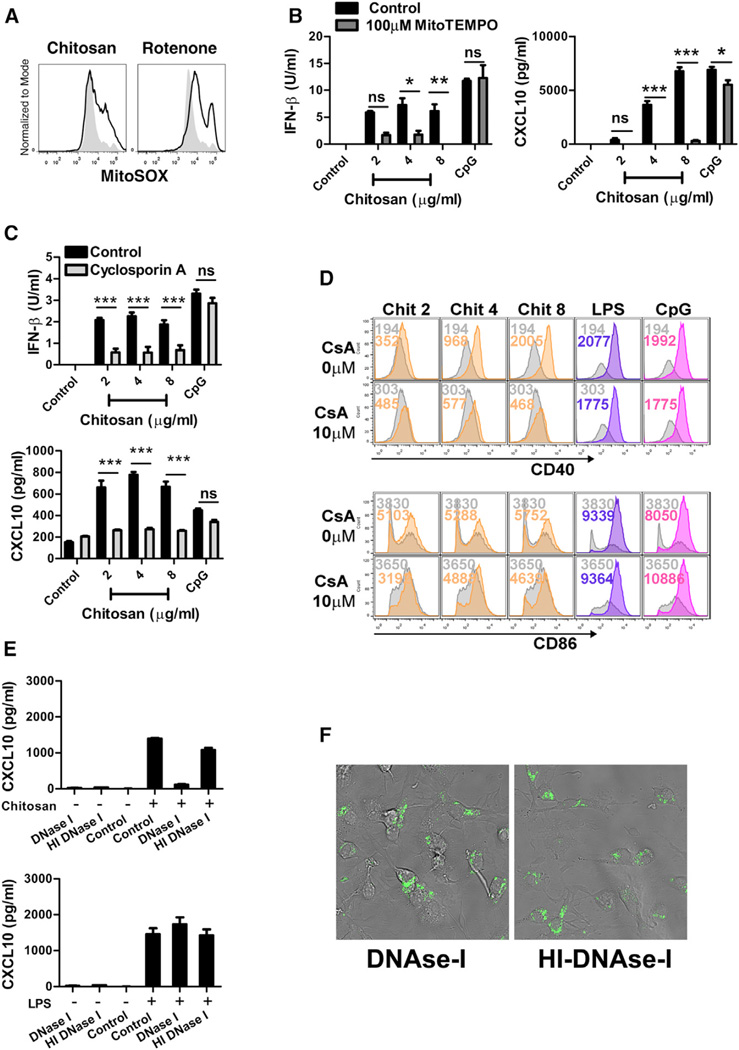
Mitochondrial stress can trigger the release of mitochondrial DNA (mtDNA) into the cytosol, culminating in the induction of innate inflammatory responses (Shimada et al., 2012; West et al., 2015; Zhou et al., 2011). The generation of mitochondrial ROS is indicative of mitochondrial stress. We speculated that chitosan might be inducing some degree of mitochondrial stress, sufficient to induce the production of ROS. The mitochondrial-specific ROS indicator MitoSOX was used to selectively detect superoxide in the mitochondria of live cells after stimulation with chitosan. Rotenone, the mitochondrial complex I inhibitor known to induce the accumulation of ROS (Li et al., 2003), was used as a positive control and, as expected, promoted an increase in MitoSOX fluorescence. A similar enhancement in MitoSOX fluorescence was seen in WT DCs incubated with chitosan. The production of mitochondrial-specific ROS was observed as early as 3 hr following treatment and sustained for up to 24 hr (Figure 7A).
Figure 7. Chitosan Induces Mitochondrial Damage, Characterized by the Generation of Mitochondrial-Derived ROS and Release of DNA into the Cytosol.
(A) DCs left untreated (control, shaded) or treated with chitosan (4 µg/ml, black line) for 3 hr stained with MitoSOX. As a positive control cells treated with rotenone (5 µM) for 6 hr were used.
(B) IFN-β and CXCL10 in supernatants of WT DCs treated for 24 hr with chitosan or CpG in the presence (dark gray) or absence (black) of Mito-TEMPO (500 µM).
(C) IFN-β and CXCL10 in supernatants of WT DCs treated for 24 hr or 16 hr with chitosan or CpG in the presence (light gray) or absence (black) of CsA (20 µM).
(D) CD40 and CD86 expression in WT DCs stimulated with chitosan (orange), LPS (violet) or CpG (pink) or left unstimulated (gray) in the presence or absence of CsA (10 µM); colored numbers indicate MFI values for the corresponding treatment.
(E) CXCL10 in supernatants of DCs incubated for 24 hr after delivery of 1.5 µg DNase I or HI-DNase I into the cytosol, followed by stimulation with chitosan or LPS.
(F) Representative confocal micrograph showing DCs transfected with fluorescent DNase I (left panel) or heat inactivated DNase I (right panel). Data are shown as mean ± SEM for triplicate samples. Results are representative of two independent experiments. Adjuvant (control) versus adjuvant (MitoTEMPO/CsA/DNase I), *p < 0.05, **p < 0.01, ***p < 0.001.
Mitochondrial ROS can positively regulate RLR-mediated type I IFN production (Tal et al., 2009). We postulated that chitosan-induced mitochondrial ROS was important for the subsequent induction of type I IFN. Indeed, pre-treatment of DCs with the mitochondria-targeted antioxidant MitoTEMPO, suppressed chitosan-induced secretion of IFN-β and CXCL10 (Figure 7B). MitoTEMPO-induced suppression of IFN-β and CXCL10 was overcome at the highest concentration of chitosan; it is possible that the magnitude of superoxide induced by high concentrations of chitosan is too great for the concentration of MitoTEMPO used in these experiments to scavenge. The secretion of IFN-β and CXCL10 in response to LPS or CpG was not affected by pre-treatment with Mito-TEMPO, indicating that this pathway was specific for chitosan activation.
Cellular stress events, including oxidative stress can result in the opening of the mitochondrial permeability transition (MPT) pore, making the inner mitochondrial membrane permeable to a range of solutes. This event leads to the release of mitochondrial factors into the cytosol, including mtDNA (García et al., 2005). Cyclosporin A (CsA) is identified as a potent inhibitor of MPT, capable of blocking the release of mtDNA fragments into the cytosol (García et al., 2005; Nakahira et al., 2011; Patrushev et al., 2004; Shimada et al., 2012). Prior incubation of WT DCs with CsA abrogated the ability of chitosan to induce the secretion of IFN-β and CXCL10 (Figure 7C), suggesting that CsA was blocking the release of a mitochondrial factor into the cytosol which was essential for chitosan-induced type I IFN responses. Furthermore, chitosan induced upregulation of CD40 on DCs was suppressed in cells pre-treated with CsA; CD86 expression was also affected although to a lesser extent (Figure 7D). In contrast, CpG-induced upregulation of CD40 remained intact. Based on these observations, we hypothesized that treatment of DCs with chitosan mediated the release of mtDNA into the cytosol, which activated the cGAS-STING pathway to result in type I IFN production. However, we cannot discount other sources of DNA contributing to the response.
To determine whether the release of endogenous cytosolic DNA was involved in chitosan–induced type I IFN production, we delivered DNase I, or heat-inactivated (HI)-DNase I as a control into DCs using the transfection agent DOTAP. Transfection was confirmed using fluorescently labeled active or inactivated DNase I by confocal microscopy (Figure 7F) and cells were stimulated with medium, chitosan, or LPS. Transfection of DNase I or HI-DNase I did not elicit CXCL10 production, and costimulatory molecules were not upregulated in response to DNase I or HI-DNase I treatment alone (data not shown). Secretion of CXCL10 was significantly compromised in cells transfected with DNase I when compared to cells exposed to HI-DNase. In contrast, LPS-induced CXCL10 secretion was not impaired in DCs transfected with either DNase I or HI-DNase I (Figure 7E). Together, these data suggested that chitosan induced mitochondrial damage, characterized by the generation of mitochondrial ROS and release of endogenous DNA into the cytosol. These events culminated in the activation of the cGAS-STING pathway and the induction of type I IFN-dependent DC maturation. Thus, chitosan induced cellular immunity can be explained by its activation of cGAS-STING signaling.
DISCUSSION
The rational development of new and improved vaccines requires a comprehensive understanding of innate immune activation and how this translates into the induction of specific adaptive immune responses. The inclusion of adjuvants as immune potentiating components in vaccine formulations enhances the response to co-administered antigen and an understanding of their mode of action will facilitate the development of formulations capable of eliciting an adaptive immune response of the desired phenotype for the pathogen in question. Chitosan is an attractive adjuvant for injectable and mucosal vaccines and this work set out to elucidate its mechanism of action. We identified type I IFNs as critical mediators of chitosan’s adjuvanticity. Type I IFN production is known to strongly influence the development of Th1 type immunity (Coffman et al., 2010). Our work established a critical role for IFNAR-mediated signaling in chitosan-induced Th1 responses. The ability of chitosan to drive strong antigen-specific IFN-γ was likely to be the factor responsible for promoting IgG2c production in this system, given that IFN-γ is important for IgG2c class switching (Bossie and Vitetta, 1991; Snapper et al., 1988).
DC activation is pivotal in the capacity of vaccines to induce protective adaptive immunity. This activation is characterized by enhanced surface expression of costimulatory molecules, including CD40 and CD86, in a process referred to as maturation (Reis e Sousa, 2006), in addition to an enhanced capacity to secrete cytokines. We demonstrated that chitosan promotes the expression of costimulatory molecules and the selective production of type I IFNs and ISGs, but not proinflammatory cytokines. This observation was in agreement with a recent publication reporting chitosan to function as an “adjuvant-like substrate” to enhance DC activation and anti-tumor immunity (Lin et al., 2014). We demonstrated that type I IFNs are crucial for the adjuvanticity of chitosan, both in its ability to promote DC activation in vitro and its ability to enhance cell-mediated immunity in vivo. This observation contrasts with a previous report which proposes a role for TLR4 in chitosan-mediated DC activation (Villiers et al., 2009). We and others have demonstrated the inflammasome-activating potential of chitosan (Bueter et al., 2011, 2014; Mori et al., 2012). Furthermore, the inability of chitosan to promote proinflammatory cytokine secretion in unprimed innate immune cells is documented (Bueter et al., 2014). However, we have now shown that chitosan can directly promote the induction of type I IFNs in DCs.
In contrast to an earlier report (Hoebe et al., 2003), we found that LPS-induced DC maturation was IFNAR-independent. This might be due to the difference in mice strains used in our study and that published by Hoebe et al. The Ifnar1−/− mice used in the latter study were generated on a 129 background, whereas in this work the mice are on a C57BL/6 background.
We demonstrated that chitosan engaged the cGAS-STING pathway, inducing type I IFN production and robust Th1 cellular immune responses. We provided evidence that chitosan exposure led to mitochondrial stress, evidenced by the generation of mitochondrial-specific ROS, necessary for subsequent secretion of IFN-β and the ISG, CXCL10. Mitochondrial stress can trigger MPT pore formation and the release of mitochondrial DNA into the cytosol, which can subsequently engage the NLRP3 inflammasome. In this setting, inhibition of mitochondrial DNA release through the MPT pore compromises inflammasome activation (Shimada et al., 2012). In the current study, inhibition of MPT formation abrogated the capacity of chitosan to promote type I IFN production and upregulate expression of costimulatory molecules on DCs. The physiological relevance of the cGAS-STING pathway in a number of different contexts has been highlighted recently. Apoptotic caspases are shown to prevent cGAS-STING signaling in response to mitochondrial DNA, suppressing the inflammation induced by dying cells (Rongvaux et al., 2014; White et al., 2014). More recently, herpes virus-induced mitochondrial stress is shown to promote the release of mitochondrial DNA into the cytosol where it engages cGAS-STING-IRF3-dependent signaling to enhance type I IFN responses (West et al., 2015). We observed that delivery of DNase I into the cytosol of DCs compromised their ability to produce CXCL10 in response to chitosan, indicating that release of DNA into the cytosol was an important prerequisite for chitosan-induced type I IFN responses. Given the capacity of cGAS to be activated in response to DNA of host origin, we suggest that chitosan promoted the release of mitochondrial DNA, which is then responsible for triggering the cGAS-STING pathway, mediating type I IFN production. Mitochondrial ROS generation can result in the formation of oxidized mitochondrial DNA. Oxidized cytosolic DNA can enhance STING-dependent type I IFN induction due to its increased resistance to TREX1-mediated degradation but not to DNase I or DNase II-mediated digestion (Gehrke et al., 2013). It is possible that chitosan might promote the release of oxidized mitochondrial DNA into the cytosol of DCs where it engages the cGAS-STING pathway, owing to its enhanced resistance to TREX1-mediated degradation. While our data point to a mitochondrial origin for the chitosan-induced cytoplasmic DNA, we cannot exclude a contribution from other sources, including the nucleus or endosomes.
Chitosan-induced Th1 responses and IgG2c induction, following both parenteral and mucosal vaccination, were highly dependent on cGAS and STING. Therefore, the cGAS-STING pathway was a critical mediator of the adjuvanticity of chitosan both in vitro and in vivo. However, there is likely more than one mechanism contributing to chitosan’s adjuvant activity and our data show that in addition to the central role for cGAS-STING, the NLRP3 inflammasome also contributes to chitosan induced Th1 responses. The ability of chitosan to promote IgG2c secreting B cells was dependent on type I IFNs but largely independent of the NLRP3 inflammasome. This might suggest that while IFN-γ is essential, type I IFN might enhance class switching to IgG2c both by promoting Th1 responses and potentially by enhancing IFN-γ production by other cell types or via direct effects on B cells (Swanson et al., 2010). It will be important to establish whether chitosan-induced type I IFN similarly contributes to IgG2c+ memory B cell induction and maintenance (Wang et al., 2012).
Proposed mechanisms underlying the adjuvanticity of chitosan include antigen protection, depot formation, enhanced antigen uptake and presentation and direct modulation of immune responses (Vasiliev, 2015). Another plausible suggestion is that chitosan-induced cell death might promote the release of DAMPs. The observations presented here provide a novel mechanism by which chitosan promotes innate and adaptive cell-mediated immunity. We conclude that chitosan engages the cGAS-STING pathway to induce type I IFNs, which in turn mediates DC activation and induction of cellular immunity with a distinct Th1-associated profile. This indicates that a cationic polymer can engage the cGAS-STING DNA sensing pathway. This work emphasizes the unique potential of chitosan as a tool in rational vaccine design given its innate immune modulating properties via the cGAS-STING pathway to promote cellular immunity.
EXPERIMENTAL PROCEDURES
Mice
Female C57BL/6 mice were obtained from Harlan Olac and used at 8–16 weeks of age. Animals were maintained according to the regulations of the European Union and the Irish Department of Health or Albany Medical College. All animal studies were approved by the Trinity College Dublin Animal Research Ethics Committee (Reference Number 091210) or carried out in accordance with the regulations and approval of Albany Medical College and the Institutional Animal Care and Use Committee. Ifnar1−/− mice were generously provided by Professor Paul Hertzog (Centre for Innate Immunity and Infectious Diseases MIMR-PHI and Monash University). C3H/HeN and C3H/HeJ mice were from Harlan Olac. C3H/HeN, C3H/HeJ, Ifnar1−/−, and Nlrp3−/− mice were bred in TBSI Comparative Medicines Unit. Tmem173−/− mice were a gift from Dr. Lei Jin (Centre for Immunology and Microbial Disease, Albany Medical Centre). Professor Kate Fitzgerald (Program in Innate Immunity, Division of Infectious Diseases and Immunology, Department of Medicine, University of Massachusetts Medical School) kindly provided Mb21d1−/− mice.
Reagents
Protasan ultrapure (endotoxins ≤ 100EU/g) chitosan salt (CL213) was obtained from Novamatrix. LPS from E. coli, Serotype R515, TLR grade was purchased from Enzo Life Sciences. CpG oligonucleotide 1826 was supplied by Oligos etc. Mito-TEMPO, rotenone, cyclosporine A, and poly(dA:dT) were purchased from Sigma. Poly(I:C) was purchased from Invitrogen. Recombinant DNase I and DOTAP liposomal transfection reagent were obtained from Roche. Alexa-488 DNase I was obtained from Fisher. Flow cytometry antibodies and ELISA kits were obtained from BD, eBiosciences, or BioLegend as indicated in the corresponding sections. Reagents for confocal microscopy were obtained from DAKO or Life Technologies.
Cell Culture
BMDCs were generated as described previously (Lutz et al., 1999). Cells were plated on day 10 at concentrations of either 1 × 106 cells/ml or 0.625 × 106 cells/ml and stimulated 2 hr later (unless stated otherwise).
DNase I Transfection
DCs were transfected with 1.5 µg of DNase I (Roche) or HI-DNase I (75°C for 10 min/U) using DOTAP (Roche) transfection reagent according to manufacturer’s instructions. After incubating the cells with DOTAP-DNase I or DOTAP-HI-DNase I for 40 min, cells were washed to remove excess of transfection reagent and enzyme and were then stimulated with chitosan (4 µg/ml) or LPS (10 ng/ml) and incubated for 24 hr. IFN-β and CXCL10 were measured by ELISA. Transfection of DNase I or HI-DNase I was confirmed by confocal microscopy in cells transfected with active or HI Alexa Fluor 488-DNase I (Fisher) as described below.
Cyclosporine A Treatment
DCs were treated with CsA (20 µM) 40 min prior to treatment with chitosan (2 µg/ml, 4 µg/ml, and 8 µg/ml), and CpG (4 µg/ml) for 16 hr and 24 hr. Concentrations of CXCL10 and IFN-β were subsequently measured by ELISA.
Flow Cytometry
For the analysis of costimulatory molecule expression, cells were incubated with AQUA LIVE/DEAD fluorescent dye (0.5 µl) (Invitrogen) for 30 min. Cells were subsequently stained with 0.3 µg/ml of anti-CD11c (HL3), 1.25 µg/ml of anti-CD86 (24F) both obtained from BD Biosciences and 2.5 µg/ml of anti-CD40 (3.23) from eBiosciences. Mitochondrial ROS were measured in cells by MitoSOX (Invitrogen) staining (1 µM for 15 min at 37°C) (Nakahira et al., 2011; Tal et al., 2009). Samples were acquired on BD FACSCanto II or Cyan using FACSDiva (BD) or Summit (Dako) software and the data analyzed using FlowJo software (Treestar).
RNA Analysis
RNA was collected with a High Pure RNA Isolation Kit (Roche) and reverse transcribed into complementary DNA (cDNA) with an M-MLV reverse transcriptase, RNase H minus, point mutant (Promega). Quantitative PCR was performed using KAPA SYBR® FAST qPCR Kit Master Mix (2X) (KAPA Biosystems) in accordance with the instructions provided by the manufacturer using Aligent Technologies Stratagene Mx3005P technology. The following primers were used (5′ → 3′). Actb forward, TCCAGCCTTTCTTGGT, Actb reverse, GCACTGTGTTGGCATAGAGGTC, Ifnb forward, ATGGTGGTCCGA GCAGAGAT, Ifnb reverse, CCACCACTCATTCTGAGGCA, Ifna forward, ATGGCTAGGCTCTGTGCTTTCCT, Ifna reverse, AGGGCTCTCCAGAGTTCT GCTCTG, Cxcl10 forward, TCTGAGTGGGACTCAAGGGAT, Cxcl10 reverse, TCGTGGCAATGATCTCAACACG. RNA expression was normalized to β-actin expression and to expression in the relevant untreated control sample.
Measurement of Cytokine Levels by ELISA
Concentrations of IL-6 and IL-12p40 were measured using antibodies obtained from BD Biosciences. IFN-β was detected using antibodies from Santa Cruz and Interferome. CXCL10 ELISA kits were purchased from R&D systems.
Adaptive Immunity to Antigen Injected with Chitosan
Female C57BL/6, Ifnar1−/−, Tmem173−/−, or Mb21d1−/− mice were immunized i.p. on days 0 and 14 with H1 (2 µg/mouse) alone or in combination with chitosan (1 mg/mouse). Mice were sacrificed 7 days after the last immunization. Mediastinal lymph nodes were cultured for 3 days with medium, H1 or anti-CD3 (0.1 µg/ml; BD Biosciences). IFN-γ and IL-17 concentrations were determined by ELISA (R&D systems and BioLegend, respectively). Antigen-specific IgG1 and IgG2c in serum were quantified using isotype-specific antibodies from BD Biosciences and AbD Serotec, respectively. PECs were restimulated for 6 hr in cRPMI with or without antigen. Brefeldin A (10 µg/ml, Sigma) was added to the culture after 1 hr. 6 hr later cells were labeled with anti-CD3 (17A2) and anti-CD4 (H29.19) fluorophore-conjugated antibodies purchased from eBiosciences and BD Biosciences respectively. Cells were fixed and permeabilized (FIX&PERM; ADg Bioresearch GMBH) and intracellular IFN-γ labeled using anti-IFN-γ (XMG1.2) from BD Biosciences. Data were acquired using summit software (Dako) and analyzed using FlowJo software (Treestar).
For intranasal (i.n.) vaccination, mice were immunized on days 0 and 14 with PspA (2 µg, BEI Resources) alone, or in combination with chitosan (50 µg), as previously described (Blaauboer et al., 2015). Briefly, animals were anaesthetized using isoflurane in an E-Z Anesthesia system (Euthanex Corp). PspA, with or without chitosan, was administrated i.n. in 25 µl saline (InvivoGen). Ag-specific Ab production was determined 14 days after the last immunization. Briefly, mice were sacrificed by CO2 asphyxiation and lungs were lavaged with 0.8 ml ice-cold PBS. The lavage fluid was centrifuged at 2,000 g for 1 min. Collected BALF was analyzed by ELISA for antibody production, using anti-mouse anti-IgG1-, IgG2C-, and IgA-HRP antibodies (Southern Biotech).
To determine Ag-specific Th1 response, we excised and digested lungs from immunized mice in DMEM contain 200 µg/ml DNase I (Roche), 25 µg/ml Liberase (Roche) at 37°C for 3 hr. Single lung cell suspension were seeded in 96-well U-bottom plate at 5×107 cells/ml and stimulated with 5 µg/ml PspA for 4 days. IFN-γ production in the culture supernatant was determined by ELISA (eBioscience,) according to the manufacturer’s instructions.
Statistical Analysis
Statistical analysis was performed using Graphpad Prism 5 software. The means for three or more groups were compared by one-way ANOVA. Where significant differences were found, the Tukey-Kramer multiple comparisons test was used to identify differences between individual groups.
Supplementary Material
In Brief.
Adjuvants are essential in many vaccines to promote innate and adaptive immunity. However, our limited understanding of their mode(s) of action is an obstacle to progress. Lavelle and colleagues demonstrate that the cationic polysaccharide chitosan activates dendritic cells and promotes Th1 responses by engaging the DNA sensor cGAS-STING pathway.
Highlights.
The adjuvant chitosan promotes DC maturation by inducing type I interferons
Chitosan-triggered type I interferons and DC maturation requires cGAS and STING
ROS and cytoplasmic DNA are necessary for chitosan to drive type I interferons
Promotion of Th1 responses by chitosan requires IFNAR, cGAS, and STING
Acknowledgments
We thank Dr. Gavin McManus for experimental advice with the confocal microscopy and Barry Moran M.Sc. for technical assistance with flow cytometry experiments. This work was funded by a Science Foundation Ireland Investigator Award 12/1A/1421 (E.C.L.) and an EMBARK postgraduate scholarship awarded by the Irish Research Council (E.C.C.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes seven figures and Supplemental Experimental Procedures and can be found with this article online at http://dx.doi.org/10.1016/j.immuni.2016.02.004.
AUTHOR CONTRIBUTION
Conceptualization: E.C.L., E.O., A.M., A.G.B.; Methodology: E.C.C., L.J., N.M.-W., A.G.B., E.C.L.; Investigation: E.C.C., A.M., N.M.-W., E.O., H.B.T.M., S.M., C.P.M.; Formal Analysis: E.C.C., A.M., N.M.-W., S.M., C.P.M., E.C.L.; Resources: L.J., E.M.A., P.A., C.C., P.H., K.A.F.; Writing – Original Draft: E.C.C., E.L.; Writing – Review & Editing: E.C.C., N.M.-W., C.P.M., A.B., J.L., E.C.L.; Funding Acquisition: E.C.L.; Supervision: E.C.L.
REFERENCES
- Agger EM, Cassidy JP, Brady J, Korsholm KS, Vingsbo-Lundberg C, Andersen P. Adjuvant modulation of the cytokine balance in Mycobacterium tuberculosis subunit vaccines; immunity, pathology and protection. Immunology. 2008;124:175–185. doi: 10.1111/j.1365-2567.2007.02751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaauboer SM, Mansouri S, Tucker HR, Wang HL, Gabrielle VD, Jin L. The mucosal adjuvant cyclic di-GMP enhances antigen uptake and selectively activates pinocytosis-efficient cells in vivo. eLife. 2015;4:1–25. doi: 10.7554/eLife.06670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossie A, Vitetta ES. IFN-γ enhances secretion of IgG2a from IgG2a-committed LPS-stimulated murine B cells: implications for the role of IFN-γ in class switching. Cell. Immunol. 1991;135:95–104. doi: 10.1016/0008-8749(91)90257-c. [DOI] [PubMed] [Google Scholar]
- Brewer JM, Conacher M, Hunter CA, Mohrs M, Brombacher F, Alexander J. Aluminium hydroxide adjuvant initiates strong antigen-specific Th2 responses in the absence of IL-4- or IL-13-mediated signaling. J. Immunol. 1999;163:6448–6454. [PubMed] [Google Scholar]
- Bueter CL, Lee CK, Rathinam VA, Healy GJ, Taron CH, Specht CA, Levitz SM. Chitosan but not chitin activates the inflammasome by a mechanism dependent upon phagocytosis. J. Biol. Chem. 2011;286:35447–35455. doi: 10.1074/jbc.M111.274936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueter CL, Lee CK, Wang JP, Ostroff GR, Specht CA, Levitz SM. Spectrum and mechanisms of inflammasome activation by chitosan. J. Immunol. 2014;192:5943–5951. doi: 10.4049/jimmunol.1301695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Monroe KM, Sotelo-Troha K, Iwig JS, Eckert B, Hyodo M, Hayakawa Y, Vance RE. STING is a direct innate immune sensor of cyclic di-GMP. Nature. 2011;478:515–518. doi: 10.1038/nature10429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Chiu Y-H, Chen ZJ. The cGAS-cGAMP-STING pathway of cytosolic DNA sensing and signaling. Mol. Cell. 2014;54:289–296. doi: 10.1016/j.molcel.2014.03.040. [DOI] [PubMed] [Google Scholar]
- Coffman RL, Sher A, Seder RA. Vaccine adjuvants: putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmet CJ, Ishii KJ. Nucleic acid sensing at the interface between innate and adaptive immunity in vaccination. Nat. Rev. Immunol. 2012;12:479–491. doi: 10.1038/nri3247. [DOI] [PubMed] [Google Scholar]
- Dubensky TW, Jr, Kanne DB, Leong ML. Rationale, progress and development of vaccines utilizing STING-activating cyclic dinucleotide adjuvants. Ther. Adv. Vaccines. 2013;1:131–143. doi: 10.1177/2051013613501988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García N, García JJ, Correa F, Chávez E. The permeability transition pore as a pathway for the release of mitochondrial DNA. Life Sci. 2005;76:2873–2880. doi: 10.1016/j.lfs.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Gehrke N, Mertens C, Zillinger T, Wenzel J, Bald T, Zahn S, Tüting T, Hartmann G, Barchet W. Oxidative damage of DNA confers resistance to cytosolic nuclease TREX1 degradation and potentiates STING-dependent immune sensing. Immunity. 2013;39:482–495. doi: 10.1016/j.immuni.2013.08.004. [DOI] [PubMed] [Google Scholar]
- González-Navajas JM, Lee J, David M, Raz E. Immunomodulatory functions of type I interferons. Nat. Rev. Immunol. 2012;12:125–135. doi: 10.1038/nri3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoebe K, Janssen EM, Kim SO, Alexopoulou L, Flavell RA, Han J, Beutler B. Upregulation of costimulatory molecules induced by lipopolysaccharide and double-stranded RNA occurs by Trif-dependent and Trif-independent pathways. Nat. Immunol. 2003;4:1223–1229. doi: 10.1038/ni1010. [DOI] [PubMed] [Google Scholar]
- Holm CK, Jensen SB, Jakobsen MR, Cheshenko N, Horan KA, Moeller HB, Gonzalez-Dosal R, Rasmussen SB, Christensen MH, Yarovinsky TO, et al. Virus-cell fusion as a trigger of innate immunity dependent on the adaptor STING. Nat. Immunol. 2012;13:737–743. doi: 10.1038/ni.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda K, Sakaguchi S, Nakajima C, Watanabe A, Yanai H, Matsumoto M, Ohteki T, Kaisho T, Takaoka A, Akira S, et al. Selective contribution of IFN-alpha/beta signaling to the maturation of dendritic cells induced by double-stranded RNA or viral infection. Proc. Natl. Acad. Sci. USA. 2003;100:10872–10877. doi: 10.1073/pnas.1934678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Ragheb K, Lawler G, Sturgis J, Rajwa B, Melendez JA, Robinson JP. Mitochondrial complex I inhibitor rotenone induces apoptosis through enhancing mitochondrial reactive oxygen species production. J. Biol. Chem. 2003;278:8516–8525. doi: 10.1074/jbc.M210432200. [DOI] [PubMed] [Google Scholar]
- Lin Y-C, Lou P-J, Young T-H. Chitosan as an adjuvant-like substrate for dendritic cell culture to enhance antitumor effects. Biomaterials. 2014;35:8867–8875. doi: 10.1016/j.biomaterials.2014.07.014. [DOI] [PubMed] [Google Scholar]
- Longhi MP, Trumpfheller C, Idoyaga J, Caskey M, Matos I, Kluger C, Salazar AM, Colonna M, Steinman RM. Dendritic cells require a systemic type I interferon response to mature and induce CD4+ Th1 immunity with poly IC as adjuvant. J. Exp. Med. 2009;206:1589–1602. doi: 10.1084/jem.20090247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz MB, Kukutsch N, Ogilvie AL, Rössner S, Koch F, Romani N, Schuler G. An advanced culture method for generating large quantities of highly pure dendritic cells from mouse bone marrow. J. Immunol. Methods. 1999;223:77–92. doi: 10.1016/s0022-1759(98)00204-x. [DOI] [PubMed] [Google Scholar]
- McWhirter SM, Barbalat R, Monroe KM, Fontana MF, Hyodo M, Joncker NT, Ishii KJ, Akira S, Colonna M, Chen ZJ, et al. A host type I interferon response is induced by cytosolic sensing of the bacterial second messenger cyclic-di-GMP. J. Exp. Med. 2009;206:1899–1911. doi: 10.1084/jem.20082874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori A, Oleszycka E, Sharp FA, Coleman M, Ozasa Y, Singh M, O’Hagan DT, Tajber L, Corrigan OI, McNeela EA, Lavelle EC. The vaccine adjuvant alum inhibits IL-12 by promoting PI3 kinase signaling while chitosan does not inhibit IL-12 and enhances Th1 and Th17 responses. Eur. J. Immunol. 2012;42:2709–2719. doi: 10.1002/eji.201242372. [DOI] [PubMed] [Google Scholar]
- Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palucka K, Banchereau J, Mellman I. Designing vaccines based on biology of human dendritic cell subsets. Immunity. 2010;33:464–478. doi: 10.1016/j.immuni.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parroche P, Lauw FN, Goutagny N, Latz E, Monks BG, Visintin A, Halmen KA, Lamphier M, Olivier M, Bartholomeu DC, et al. Malaria hemozoin is immunologically inert but radically enhances innate responses by presenting malaria DNA to Toll-like receptor 9. Proc. Natl. Acad. Sci. USA. 2007;104:1919–1924. doi: 10.1073/pnas.0608745104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrushev M, Kasymov V, Patrusheva V, Ushakova T, Gogvadze V, Gaziev A. Mitochondrial permeability transition triggers the release of mtDNA fragments. Cell. Mol. Life Sci. 2004;61:3100–3103. doi: 10.1007/s00018-004-4424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Ahmed R. Immunological mechanisms of vaccination. Nat. Immunol. 2011;12:509–517. doi: 10.1038/ni.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappuoli R, Aderem A. A 2020 vision for vaccines against HIV, tuberculosis and malaria. Nature. 2011;473:463–469. doi: 10.1038/nature10124. [DOI] [PubMed] [Google Scholar]
- Reis e Sousa C. Dendritic cells in a mature age. Nat. Rev. Immunol. 2006;6:476–483. doi: 10.1038/nri1845. [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Jackson R, Harman CC, Li T, West AP, de Zoete MR, Wu Y, Yordy B, Lakhani SA, Kuan CY, et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell. 2014;159:1563–1577. doi: 10.1016/j.cell.2014.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K, Crother TR, Karlin J, Dagvadorj J, Chiba N, Chen S, Ramanujan VK, Wolf AJ, Vergnes L, Ojcius DM, et al. Oxidized mitochondrial DNA activates the NLRP3 inflammasome during apoptosis. Immunity. 2012;36:401–414. doi: 10.1016/j.immuni.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper CM, Peschel C, Paul WE. IFN-gamma stimulates IgG2a secretion by murine B cells stimulated with bacterial lipopolysaccharide. J. Immunol. 1988;140:2121–2127. [PubMed] [Google Scholar]
- Swanson CL, Wilson TJ, Strauch P, Colonna M, Pelanda R, Torres RM. Type I IFN enhances follicular B cell contribution to the T cell-independent antibody response. J. Exp. Med. 2010;207:1485–1500. doi: 10.1084/jem.20092695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailor P, Tamura T, Kong HJ, Kubota T, Kubota M, Borghi P, Gabriele L, Ozato K. The feedback phase of type I interferon induction in dendritic cells requires interferon regulatory factor 8. Immunity. 2007;27:228–239. doi: 10.1016/j.immuni.2007.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tal MC, Sasai M, Lee HK, Yordy B, Shadel GS, Iwasaki A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA. 2009;106:2770–2775. doi: 10.1073/pnas.0807694106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tough DF. Modulation of T-cell function by type I interferon. Immunol. Cell Biol. 2012;90:492–497. doi: 10.1038/icb.2012.7. [DOI] [PubMed] [Google Scholar]
- van Dissel JT, Arend SM, Prins C, Bang P, Tingskov PN, Lingnau K, Nouta J, Klein MR, Rosenkrands I, Ottenhoff THM. Ag85B-ESAT-6 adjuvanted with IC31 promotes strong and long-lived Mycobacterium tuberculosis specific T cell responses in naïve human volunteers. Vaccine. 2010;28:3571–3581. doi: 10.1016/j.vaccine.2010.02.094. [DOI] [PubMed] [Google Scholar]
- van Dissel JT, Joosten SA, Hoff ST, Soonawala D, Prins C, Hokey DA, O’Dee DM, Graves A, Thierry-Carstensen B, Andreasen LV, et al. A novel liposomal adjuvant system, CAF01, promotes long-lived Mycobacterium tuberculosis-specific T-cell responses in human. Vaccine. 2014;32:7098–7107. doi: 10.1016/j.vaccine.2014.10.036. [DOI] [PubMed] [Google Scholar]
- Vasiliev YM. Chitosan-based vaccine adjuvants: incomplete characterization complicates preclinical and clinical evaluation. Expert Rev. Vaccines. 2015;14:37–53. doi: 10.1586/14760584.2015.956729. [DOI] [PubMed] [Google Scholar]
- Villiers C, Chevallet M, Diemer H, Couderc R, Freitas H, Van Dorsselaer A, Marche PN, Rabilloud T. From secretome analysis to immunology: chitosan induces major alterations in the activation of dendritic cells via a TLR4-dependent mechanism. Mol. Cell. Proteomics. 2009;8:1252–1264. doi: 10.1074/mcp.M800589-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang NS, McHeyzer-Williams LJ, Okitsu SL, Burris TP, Reiner SL, McHeyzer-Williams MG. Divergent transcriptional programming of class-specific B cell memory by T-bet and RORα. Nat. Immunol. 2012;13:604–611. doi: 10.1038/ni.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AP, Khoury-Hanold W, Staron M, Tal MC, Pineda CM, Lang SM, Bestwick M, Duguay BA, Raimundo N, MacDuff DA, et al. Mitochondrial DNA stress primes the antiviral innate immune response. Nature. 2015;520:553–557. doi: 10.1038/nature14156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ, Mcarthur K, Metcalf D, Lane RM, Cambier JC, Herold MJ, Van Delft MF, Bedoui S, Lessene G, Ritchie ME, et al. Apoptotic Caspases Suppress Type I IFN Production. Cell. 2014;159:1549–1562. doi: 10.1016/j.cell.2014.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Yazdi AS, Menu P, Tschopp J. A role for mitochondria in NLRP3 inflammasome activation. Nature. 2011;469:221–225. doi: 10.1038/nature09663. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.