Abstract
Cancer classification and feature (gene) selection plays an important role in knowledge discovery in genomic data. Although logistic regression is one of the most popular classification methods, it does not induce feature selection. In this paper, we presented a new hybrid L1/2 +2 regularization (HLR) function, a linear combination of L1/2 and L2 penalties, to select the relevant gene in the logistic regression. The HLR approach inherits some fascinating characteristics from L1/2 (sparsity) and L2 (grouping effect where highly correlated variables are in or out a model together) penalties. We also proposed a novel univariate HLR thresholding approach to update the estimated coefficients and developed the coordinate descent algorithm for the HLR penalized logistic regression model. The empirical results and simulations indicate that the proposed method is highly competitive amongst several state-of-the-art methods.
1. Introduction
With advances in high-throughput molecular techniques, the researchers can study the expression of tens of thousands of genes simultaneously. Cancer classification based on gene expression levels is one of the central problems in genome research. Logistic regression is a popular classification method and has an explicit statistical interpretation which can obtain probabilities of classification regarding the cancer phenotype. However, in most gene expression studies, the number of genes typically far exceeds the number of the sample size. This situation is called high-dimensional and low sample size problem, and the normal logistic regression method cannot be directly used to estimate the regression parameters.
To deal with the problem of high dimensionality, one of the popular techniques is the regularization method. A well-known regularization method is the L1 penalty [1], which is the least absolute shrinkage and selection operator (Lasso). It is performing continuous shrinkage and gene selection at the same time. Other L1 norm type regularization methods typically include the smoothly-clipped-absolute-deviation (SCAD) penalty [2], which is symmetric, nonconcave, and has singularities at the origin to produce sparse solutions. The adaptive Lasso [3] penalizes the different coefficients with the dynamic weights in the L1 penalty. However, the L1 type regularization may yield inconsistent feature selections in some situations [3] and often introduces extra bias in the estimation of the parameters in the logistic regression [4]. Xu et al. [5] proposed the L1/2 penalty, a method that can be taken as a representative of Lq (0 <q < 1) penalties in both sparsity and computational efficiency, and has demonstrated many attractive properties, such as unbiasedness, and oracle properties [5–7]. However, similar to most of the regularization methods, the L1/2 penalty ignores the correlation between features, and consequently unable to analyze data with dependent structures. If there is a group of variables among which the pair-wise correlations are very high, then the L1/2 method tends to select only one variable to represents the corresponding group. In gene expression study, genes are often highly correlated if they share the same biological pathway [8]. Some efforts had been made to deal with the problem of highly correlated variables. Zhou and Hastie proposed Elastic net penalty [9] which is a linear combination of L1 and L2 (the ridge technique) penalties, and such method emphasizes a grouping effect, where strongly correlated genes tend to be in or out of the model together. Becker et al. [10] proposed the Elastic SCAD (SCAD − L2), a combination of SCAD and L2 penalties. By introducing the L2 penalty term, Elastic SCAD also works for the groups of predictors.
In this article, we proposed the HLR (Hybrid L1/2 + 2 Regularization) approach to fit the logistic regression models for gene selection, where the regularization is a linear combination of the L1/2 and L2 penalties. The L1/2 penalty achieves feature selection. In theory, a strictly convex penalty function provides a sufficient condition for the grouping effect of variables and the L2 penalty guarantees strict convexity [11]. Therefore, the L2 penalty induces the grouping effect simultaneously in the HLR approach. Experimental results on artificial and real gene expression data in this paper demonstrate that our proposed method is very promising.
The rest of the article is organized as follows. In Section 2, we first defined the HLR approach and presented an efficient algorithm for solving the logistic regression model with the HLR penalty. In Section 3, we evaluated the performance of our proposed approach on the simulated data and five public gene expression datasets. We presented a conclusion of the paper in Section 4.
2. Methods
2.1 Regularization
Suppose that dataset D has n samples D = {(X1, y1), (X2, y2),…,(Xn, yn)}, where Xi = (xi1, xi2, …, xip) is ith sample with p dimensional and yi is the corresponding dependent variable.
For any non-negative λ, the normal regularization form is:
| (1) |
where P(β) represents the regularization term. There are many regularization methods proposed in recent years. One of the popular methods is the L1 regularization (Lasso), where . The others L1 type regularizations include SCAD, the adaptive Lasso, Elastic net, Stage wise Lasso [12], Dantzig selector [13] and Elastic SCAD. However, in genomic research, the result of the L1 type regularization may not sparse enough for interpretation. Actually, a typical microarray or RNA-seq data set has many thousands of predictors (genes), and researchers often desire to select fewer but informative genes. Beside this, the L1 regularization is asymptotically biased [14,15]. Although the L0 regularization, where , yields the sparsest solutions, it has to deal with NP-hard combinatory optimization problem. To gain a more concise solution and improve the predictive accuracy of the classification model, we need to think beyond the L1 and L0 regularizations to the Lq (0<q<1) regularization. The L1/2 regularization can be taken as a representative of the Lq (0<q<1) penalties and has permitted an analytically expressive thresholding representation [5]. With the thresholding representation, solving the L1/2 regularization is much easier than solving the L0 regularization. Moreover, the L1/2 penalty is unbiasedness and has oracle properties [5–7]. These characteristics are making the L1/2 penalty became an efficient tool for high dimensional problems [16,17]. However, due to the insensitivity of the highly correlated data, the L1/2 penalty tends to select only one variable to represent the correlated group. This drawback may deteriorate the performance of the L1/2 method.
2.2 Hybrid L1/2 +2 Regularization (HLR)
For any fixed non-negative λ1 and λ2, we define the hybrid L1/2 +2 regularization (HLR) criterion:
| (2) |
where β = (β1, …, βp) are the coefficients to be estimated and
The HLR estimator is the minimizer of Eq (2):
| (3) |
Let α = λ1/(1 + λ2), then solving in Eq (3) is equivalent to the optimization problem:
| (4) |
We call the function α|β|1/2 + (1 − α)|β|2 as the HLR, which is a combination of the L1/2 and L2 penalties. When α = 0, the HLR penalty becomes ridge regularization. When α = 1, the HLR becomes L1/2 regularization. The L2 penalty is enjoying the grouping effect and the L1/2 penalty induces sparse solutions. This combination of the both penalties makes the HLR approach not only capable of dealing with the correlation data, but also able to generate a succinct result.
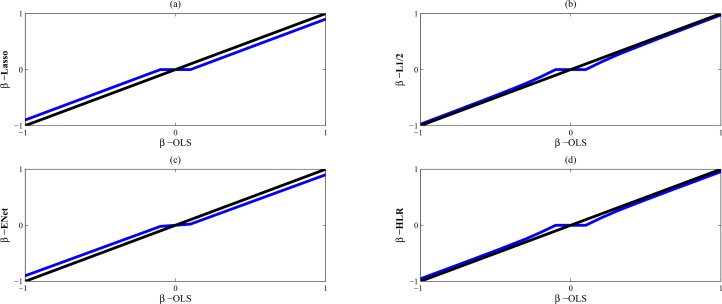
Fig 1 shows four regularization methods: Lasso, L1/2, Elastic net and HLR penalties with an orthogonal design matrix in the regression model. The estimators of Lasso and Elastic net are biased, whereas the L1/2 penalty is asymptotically unbiased. Similar to the L1/2 method, the HLR approach also performs better than Lasso and Elastic net in the property of unbiasedness.
Fig 1.
Exact solutions of (a) Lasso, (b) L1/2, (c) Elastic net, and (d) HLR in an orthogonal design. The regularization parameters are λ = 0.1 and α = 0.8 for Elastic net and HLR. (β-OLS is the ordinary least-squares (OLS) estimator).
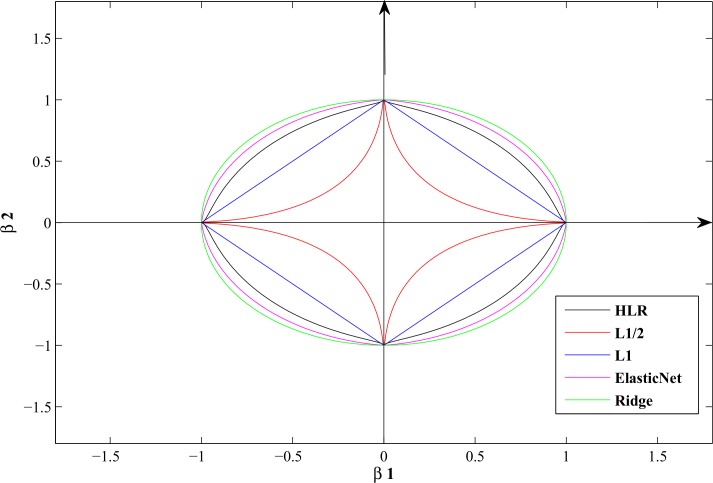
Fig 2 describes the contour plots on two-dimensional for the penalty functions of Lasso, Elastic net, L1/2 and HLR approaches. It is suggest that the L1/2 penalty is non-convex, whereas the HLR is convex for the given α. The following theorem will show how the HLR strengthens the L1/2 regularization.
Fig 2. Contour plots (two-dimensional) for the regularization methods.
The regularization parameters are λ = 1 and α = 0.2 for the HLR method.
Theorem 1
Given dataset (y, X) and (λ1, λ2), then the HLR estimates are given by
| (5) |
The L1/2 regularization can be rewritten as
| (6) |
The proof of Theorem 1 can be found in S1 File. Therorem1 shows the HLR approach is a stabilized version of the L1/2 regularization. Note that is a sample version of the correlation matrix Σ and
where δ = λ2/(1 + λ2) shrinks that towards the identity matrix. The classification accuracy can often be enhanced by replacing by a more shrunken estimate in linear discriminate analysis [18,19]. In other word, the HLR improves the L1/2 technique by regularizing in Eq (6).
2.3 The sparse logistic regression with the HLR method
Suppose that dataset D has n samples D = {(X1, y1), (X2, y2), …, (Xn, yn)}, where Xi = (xi1, xi2, …, xip) is ith sample with p genes and yi is the corresponding dependent variable that consist of a binary value with 0 or 1. Define a classifier f(x) = ex / (1 + ex) and the logistic regression is defined as:
| (7) |
Where β = (β1, …, βp) are the coefficients to be estimated. With a simple algebra, the regression model can be presented as:
| (8) |
In this paper, we apply the HLR approach to the logistic regression model. For any fixed non-negative λ and α, the sparse logistic regression model based on the HLR approach is defined as:
| (9) |
2.4 Solving algorithm for the sparse logistic regression with the HLR approach
The coordinate descent algorithm [20] is an efficient method for solving regularization models because its computational time increases linearly with the dimension of the problems. Its standard procedure can be showed as follows: for every βj (j = 1,2,…,p), to partially optimize the target function with respect to coefficient with the remaining elements of β fixed at their most recently updated values, iteratively cycling through all coefficients until meet converged. The specific form of renewing coefficients is associated with the thresholding operator of the penalty.
Suppose that dataset D has n samples D = {(X1, y1), (X2, y2), …, (Xn, yn)}, where Xi = (xi1, xi2, …, xip) is ith sample with p dimensional and yi is the corresponding dependent variable. The variables are standardized: .
Following Friedman et al. [20] and Liang et al. [16], in this paper, we present the original coordinate-wise update form for the HLR approach:
| (10) |
where , and as the partial residual for fitting βj. is the L1/2 thresholding operator
| (11) |
where , π = 3.14.
The Eq (9) can be linearized by one-term Taylor series expansion:
| (12) |
where is the estimated response, is the weight for the estimated response. is the evaluated value under the current parameters. Thus, we can redefine the partial residual for fitting current as and . The procedure of the coordinate descent algorithm for the HLR penalized logistic model is described as follows.
Algorithm: The coordinate descent approach for the HLR penalized logistic model
Step 1: Initialize all βj(m) ← 0 (j = 1, 2,…,p) and X, y,
set m ← 0, λ and α are chosen by cross-validation;
Step 2: Calculate Z(m) and W(m) and approximate the loss function (12) based on the current β(m);
Step 3: Update each βj(m), and cycle over j = 1,…, p;
Step 3.1: Compute and ;
Step 3.2: Update
Step 4: Let m ← m + 1, β(m + 1) ← β(m);
If β(m) dose not convergence, then repeat Steps 2, 3;
3. Results and Discussion
3.1 Analyzes of simulated data
The goal of this section is to evaluate the performance of the logistic regression with the HLR approach in the simulation study. Four approaches are compared with our proposed method: logistic regression with the Lasso regularization, L1/2 regularization, SCAD − L2 and Elastic net regularization respectively. We simulate data from the true model
where X ∼ N(0, 1), ϵ is the independent random error and σ is the parameter that controls the signal to noise. Four scenarios are presented here. In every example, the dimension of predictors is 1000. The notation. /. was represented the number of observations in the training and test sets respectively, e.g. 100/100. Here are the details of the four scenarios.
- In scenario 1, the dataset consists of 100/100 observations, we set σ = 0.3 and
, we simulated a grouped variable situation
where ρ is the correlation coefficient of the grouped variables. - The scenario 2 was defined similarly to the scenario 1, except that we considered the case when there are other independent factors also contributes to the corresponding classification variable y,
- In scenario 3, we set σ = 0.4 and the dataset consist of 200/200 observations, and
, we defined two grouped variables - In scenario 4, the true features were added up to 20% of the total features, σ = 0.4 and the dataset consist of 400/400 observations, and
, we defined three grouped variables
In this example, there were three groups of the correlated features and some single independent features. An ideal sparse regression method would select only the 200 true features and set the coefficients of the 800 noise features to zero.
In our experiment, we set the correlation coefficient ρ of features are 0.3, 0.6, 0.9 respectively. The Lasso and Elastic net were conducted by Glmnet (a Matlab package, version 2014-04-28, download at http://web.stanford.edu/~hastie/glmnet_matlab/). The optimal regularization parameters or tuning parameters (balance the tradeoff between data fit and model complexity) of the Lasso, L1/2, SCAD − L2, Elastic net and the HLR approaches were tuned by the 10-fold cross-validation (CV) approach in the training set. Note that, the Elastic net and HLR methods were tuned by the 10-CV approach on the two-dimensional parameter surfaces. The SCAD − L2 were tuned by the 10-CV approach on the three-dimensional parameter surfaces. Then, the different classifiers were built by these sparse logistic regressions with the estimated tuning parameters. Finally, the obtained classifiers were applied to the test set for classification and prediction.
We repeated the simulations 500 times for each penalty method and computed the mean classification accuracy on the test sets. To evaluate the quality of the selected features for the regularization approaches, the sensitivity and specificity of the feature selection performance [21] were defined as the follows:
where the .* is the element-wise product, and |.|0 calculates the number of non-zero elements in a vector, and are the logical “not” operators on the vectors β and .
As showed in Table 1, for all scenarios, our proposed HLR procedure generally gave higher or comparable classification accuracy than the Lasso, SCAD − L2, Elastic net and L1/2 methods. Also, the HLR approach results in much higher sensitivity for identifying true features compared to the other four algorithms. For example, in the scenario 1 with ρ = 0.9, our proposed method gained the impressive performance (accuracy 99.87% with perfect sensitivity and specificity). The specificity of the HLR approach is somewhat decreased, but not greatly as compared to the achieved in sensitivity.
Table 1. Mean results of the simulation.
In bold–the best performance amongst all the methods.
| Scenario | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ρ | Method | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 |
| Sensitivity of feature selection | Specificity of feature selection | Accuracy of classification (test set) | |||||||||||
| Lasso | 0.966 | 0.798 | 0.344 | 0.361 | 0.996 | 0.968 | 0.967 | 0.966 | 89.26% | 81.47% | 84.76% | 80.26% | |
| L1/2 | 0.971 | 0.888 | 0.411 | 0.355 | 0.998 | 0.974 | 0.975 | 0.970 | 92.05% | 82.22% | 85.11% | 81.45% | |
| 0.3 | SCAD − L2 | 1.000 | 0.913 | 0.722 | 0.674 | 0.995 | 0.928 | 0.890 | 0.723 | 93.21% | 82.90% | 84.51% | 82.51% |
| EN | 0.997 | 0.916 | 0.737 | 0.662 | 0.994 | 0.926 | 0.886 | 0.735 | 91.03% | 81.34% | 84.47% | 80.27% | |
| HLR | 1.000 | 0.924 | 0.791 | 0.708 | 0.999 | 0.931 | 0.892 | 0.769 | 95.27% | 82.66% | 84.99% | 85.05% | |
| Lasso | 0.887 | 0.723 | 0.351 | 0.270 | 0.995 | 0.975 | 0.981 | 0.923 | 94.24% | 84.10% | 91.88% | 85.88% | |
| L1/2 | 0.755 | 0.630 | 0.275 | 0.220 | 1.000 | 0.974 | 0.988 | 0.928 | 95.90% | 86.50% | 90.20% | 84.20% | |
| 0.6 | SCAD − L2 | 1.000 | 0.866 | 0.800 | 0.629 | 1.000 | 0.949 | 0.929 | 0.849 | 96.33% | 86.43% | 89.20% | 93.03% |
| EN | 1.000 | 0.854 | 0.795 | 0.621 | 1.000 | 0.953 | 0.939 | 0.837 | 96.22% | 86.41% | 92.12% | 91.01% | |
| HLR | 1.000 | 0.875 | 0.816 | 0.636 | 1.000 | 0.968 | 0.942 | 0.841 | 99.53% | 87.16% | 92.71% | 92.82% | |
| Lasso | 0.548 | 0.548 | 0.174 | 0.145 | 0.938 | 0.972 | 0.987 | 0.934 | 96.05% | 86.79% | 93.22% | 91.15% | |
| L1/2 | 0.337 | 0.495 | 0.159 | 0.139 | 0.999 | 0.977 | 0.991 | 0.944 | 97.89% | 87.90% | 93.70% | 92.70% | |
| 0.9 | SCAD − L2 | 1.000 | 0.872 | 0.809 | 0.636 | 1.000 | 0.954 | 0.952 | 0.861 | 97.28% | 88.60% | 93.70% | 93.19% |
| EN | 1.000 | 0.856 | 0.818 | 0.622 | 0.995 | 0.951 | 0.949 | 0.875 | 98.22% | 88.14% | 93.52% | 93.82% | |
| HLR | 1.000 | 0.897 | 0.824 | 0.645 | 1.000 | 0.966 | 0.956 | 0.880 | 99.87% | 89.40% | 94.76% | 94.40% | |
Mean results are based on 500 repeats. The sensitivity and specificity are both dedicated to measures the quality of the selected features, the accuracy evaluates the classification performance of the different regularization approaches on the test sets.
3.2 Analyzes of real data
To further evaluate the effectiveness of our proposed method, in this section, we used several publicly available datasets: Prostate, DLBCL and Lung cancer. The prostate and DLBCL datasets were both downloaded from http://ico2s.org/datasets/microarray.html, and the lung cancer dataset can be downloaded at http://www.ncbi.nlm.nih.gov/geo with access number [GSE40419].
More information on these datasets is given in Table 2.
Table 2. Real datasets used in this paper.
| Dataset | No. of Samples (Total) | No. of Genes | Classes |
|---|---|---|---|
| Prostate | 102 | 12600 | Normal/Tumor |
| Lymphoma | 77 | 7129 | DLBCL/FL |
| Lung cancer | 164 | 22401 | Normal/Tumor |
Prostate
This dataset was originally proposed by Singh et al. [22]; it is contains the expression profiles of 12,600 genes for 50 normal tissues and 52 prostate tumor tissues.
Lymphoma
This dataset (Shipp et al. [23]) contains 77 microarray gene expression profiles of the two most prevalent adult lymphoid malignancies: 58 samples of diffuse large B-cell lymphomas (DLBCL) and 19 follicular lymphomas (FL). The original data contains 7,129 gene expression values.
Lung cancer
As RNA- sequencing (RNA-seq) technique widely used, therefore, it is important to test the proposed method whether it has the ability to handle the RNA-seq data. To verify it, one dataset that used the next-generation sequencing was involved in our analysis. This dataset [24] contains 164 samples with 87 lung adenocarcinomas and 77 adjacent normal tissues.
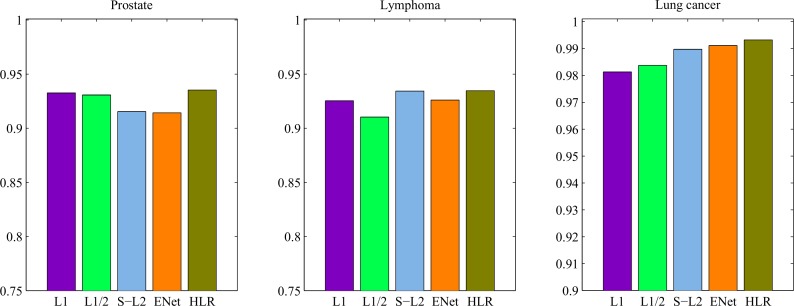
We evaluate the performance of the HLR penalized logistic regression models using the random partition. This means that we divide the datasets at random such that approximate 75% of the datasets becomes the training samples and the other 25% as the test samples. The optimal tuning parameters were found by using the 10-fold cross-validation in the training set. Then, the classification model was built by the sparse logistic regression with the estimated tuning parameters. Finally, application of the classifier to the test set provides the prediction characteristics such as classification accuracy, AUC under the receiver operating characteristic (ROC) analysis. The above procedures were repeated 500 times with different random dataset partitions. The mean number of the selected genes, the training and the testing classification accuracies, were summarized in Table 3 and the averaged AUC performances were showed in Fig 3.
Table 3. Mean results of empirical datasets.
In bold–the best performance.
| Dataset | Method | Training accuracy (10-CV) | Accuracy (testing) | No. of selected genes | ||||
|---|---|---|---|---|---|---|---|---|
| Lasso | 96.22% | 92.40% | 13.7 | |||||
| L1/2 | 96.13% | 92.18% | 8.2 | |||||
| Prostate | SCAD − L2 | 95.99% | 91.33% | 22 | ||||
| ElasticNet | 96.28% | 91.35% | 15.2 | |||||
| HLR | 97.61% | 93.68% | 12.6 | |||||
| Lymphom | Lasso | 96.03% | 91.11% | 13.2 | ||||
| L1/2 | 95.15% | 91.20% | 10.7 | |||||
| SCAD − L2 | 95.78% | 92.99% | 20.9 | |||||
| ElasticNet | 96.01% | 92.17% | 21.2 | |||||
| HLR | 96.55% | 94.23% | 15.1 | |||||
| Lung cancer | Lasso | 96.32% | 96.99% | 13.8 | ||||
| L1/2 | 97.17% | 97.20% | 11.5 | |||||
| SCAD − L2 | 97.95% | 98.17% | 25.1 | |||||
| ElasticNet | 97.21% | 98.38% | 28.9 | |||||
| HLR | 98.59% | 98.35% | 15.6 | |||||
Mean results are based on 500 repeats.
Fig 3. The performance of the AUC from ROC analyzes of each method on prostate, lymphoma and lung cancer datasets.
As showed in Table 3, for prostate dataset, the classifier with the HLR approach gives the average 10-fold CV accuracy of 97.61% and the average test accuracy of 93.68% with about 12.6 genes selected. The classifiers with Lasso, L1/2, SCAD − L2 and Elastic net methods give the average 10-fold CV accuracy of 96.22%, 96.13%, 95.99%, 96.28% and the average test accuracy of 92.4%, 92.18%, 91.33%, 91.35% with 13.7, 8.2, 22 and 15.2 genes selected respectively. For lymphoma datasets, it can be seen that the HLR method also achieves the best classification performances with the highest accuracy rates in the training and test sets. For lung cancer, our method gained the best training accuracy. The testing performance of Elastic net was slightly better than our method. However, the HLR method achieved its success using only about 15.6 predictors (genes), compared to 28.9 genes for the Elastic net method. Although the Lasso or L1/2 methods gained the sparsest solutions, the classification performance of these two approaches were worse than the HLR method. This is an important consideration for screening and diagnostic applications, where the goal is often to develop an accurate test using as few features as possible in order to control cost.
As showed in Fig 3, our proposed method achieved the best classification performances in these three real datasets amongst all the competitors. For example, the AUC from ROC analysis of the HLR method for datasets prostate, lymphoma and lung cancer datasets were estimated to be 0.9353, 0.9347 and 0.9932 respectively. AUC results of the Lasso method for the three datasets were calculated to be 0.9327, 0.9253 and 0.9813 respectively, which were worse than the proposed HLR method.
We summarized the top 10 ranked (most frequently) genes selected by the five regularization methods for the lung cancer gene expression dataset in Table 4, the information of top 10 ranked genes for the other datasets could be found in S2 File. Note that in Table 1, the proposed HLR method has the impressive performances to select the true features in the simulation data. It is implied that the genes selected by the HLR method in these three cancer datasets are valuable to the researchers who want to find out the key factors that associated with the cancer development. For example, in Table 4, the biomarkers selected by our HLR method include advanced glycosylation end product receptor (AGER), which is a member of the immunoglobulin superfamily predominantly expressed in the lung. AGER plays a role in epithelial organization, and decreased express of AGER in lung tumors may conduce to loss of epithelial tissue structure, potentially leading to malignant transformation [25]. The unique function of AGER in lung, making it could be used as an additional diagnostic tool for lung cancer [26], and even a target [27]. GATA2 (GATA binding protein 2) are expressed principally in hematopoietic lineages, and have essential roles in the development of multiple hematopoietic cells, including erythrocytes and megakaryocytes. It is crucial for the proliferation and maintenance of hematopoietic stem cells and multi-potential progenitors [28]. Kumar et al. [29] showed a strong relationship between GATA2 and RAS-pathway mutant lung tumor cells.
Table 4. The most frequently selected 10 genes found by the five sparse logistic regression methods from the lung cancer dataset.
| Rank | Lasso | L1/2 | SCAD − L2 | ElasticNet | HLR |
|---|---|---|---|---|---|
| 1 | STX11 | A2M | ABCA8 | CCDC69 | ACADL |
| 2 | GABARAPL1 | ACADL | ADH1B | STX11 | CCDC69 |
| 3 | PDLIM2 | PNLIP | CAT | GABARAPL1 | STX11 |
| 4 | CAV1 | AAAS | CAV1 | TNXB | ABCA8 |
| 5 | ABCA8 | A4GALT | CCDC69 | PDLIM2 | PAEP |
| 6 | GPM6A | ABHD8 | GABARAPL1 | FAM13C | AGER |
| 7 | GRK5 | ADD2 | GPM6A | GPM6A | GATA2 |
| 8 | TNXB | SLN | GRK5 | SFTPC | PNLIP |
| 9 | ADH1B | ACTL7B | PDLIM2 | ARHGAP44 | A2M |
| 10 | PTRF | ADAR | PTRF | CAT | ACAN |
To further verify the biomarkers selected by our method, we had collected two independent lung cancer datasets for validation. The GSE19804 [30] contains 120 samples with 60 lung adenocarcinomas and 60 adjacent normal tissues. The GSE32863 [31] contains 116 samples include 58 lung adenocarcinomas and 58 healthy controls. These two datasets are available from the GEO series accession number [GSE19804] and [GSE32863].
We used the support vector machine (SVM) approach to build the classifiers based on the first two, first five and first ten genes selected by the different regularization approaches from the lung cancer dataset (Table 4), and were trained on the lung cancer dataset (Table 2) respectively. These classifiers then were applied to the two independent lung cancer datasets, GSE19804 and GSE32863, respectively.
It is known that the obtained prediction models may be only applicable to samples from the same platform, cell type, environmental conditions and experimental procedure. However, interestingly, as demonstrated in Table 5, we can see that all the classification accuracies predicted by the classifiers with the selected genes by the HLR approach, are higher than 90%. Especially the classification accuracy on the GSE32863 dataset is 97.41% with the classifier based on the first ten genes. Such performances are better than the genes selected by other methods. For example, the accuracy of the classifier with the first two genes selected by Elastic net, for GSE19804, was estimated to be 86.67% that was worse than the classifier with the genes selected by our method, 90.83%. The performance of the classifier with the first five genes selected by SCAD − L2, for GSE32863, was calculated to be 92.24% that was worse than the classifier with the genes selected by our HLR method, 96.55%. The results indicate that the sparse logistic regression with the HLR approach can select powerful discriminatory genes.
Table 5. The validation results of the classifiers based on the top rank selected genes from lung cancer dataset.
In bold–the best performance.
| Dataset | Method | SVM with the top genes | ||||||
|---|---|---|---|---|---|---|---|---|
| 2 | 5 | 10 | ||||||
| GSE19804 | Lasso | 89.17% | 93.33% | 92.50% | ||||
| L1/2 | 85.83% | 90.83% | 91.67% | |||||
| SCAD − L2 | 89.17% | 89.17% | 93.33% | |||||
| ElasticNet | 86.67% | 87.50% | 89.17% | |||||
| HLR | 90.83% | 92.50% | 94.17% | |||||
| GSE32863 | Lasso | 93.10% | 95.69% | 93.97% | ||||
| L1/2 | 93.97% | 94.83% | 95.69% | |||||
| SCAD − L2 | 90.28% | 92.24% | 94.83% | |||||
| ElasticNet | 89.66% | 91.38% | 93.97% | |||||
| HLR | 94.83% | 96.55% | 97.41% | |||||
We used the SVM approach to build the classifiers based on the first two, first five and first ten genes selected by the different regularization approaches from the lung cancer dataset (Table 4), and were trained on the lung cancer dataset (Table 2) respectively. These classifiers then were applied to the two independent lung cancer datasets, GSE19804 and GSE32863, respectively.
In addition to comparing with the Lasso, L1/2, SCAD − L2 and Elastic net techniques, we also make a comparison with the results of other methods for datasets prostate and lymphoma published in the literature. Note that we only considered methods using the CV approach for evaluation, since approaches based on a mere training/test set partition are now widely known as unreliable [32]. Table 6 displays the best classification accuracy of other methods. In Table 6, classification accuracy achieved by the HLR approach is greater than other methods. Meanwhile, the number of selected genes is smaller than other methods except on the Lymphoma dataset.
Table 6. The result of the literature.
In bold–the best performance.
| Dataset | Author | Accuracy (CV) | No. of selected features |
|---|---|---|---|
| T.K. Paul et al. [33] | 96.60% | 48.5 | |
| Wessels et al. [34] | 93.40% | 14 | |
| Shen et al. [35] | 94.60% | unknown | |
| prostate | Lecocke et al. [36] | 90.10% | unknown |
| Dagliyan et al. [37] | 94.80% | unknown | |
| Glaab et al. [38] | 94.00% | 30 | |
| HLR | 97.61% | 12.6 | |
| Lymphoma | Wessels et al. [34] | 95.70% | 80 |
| Liu et al. [39] | 93.50% | 6 | |
| Shipp et al. [23] | 92.20% | 30 | |
| Goh et al. [40] | 91.00% | 10 | |
| Lecocke et al. [36] | 90.20% | unknown | |
| Hu et al. [41] | 87.01% | unknown | |
| Dagliyan et al. [37] | 92.25% | unknown | |
| Glaab et al. [38] | 95.00% | 30 | |
| HLR | 96.55% | 15.1 |
4. Conclusion
In this paper, we have proposed the HLR function, a new shrinkage and selection method. The HLR approach is inherited some valuable characteristics from the L1/2 (sparsity) and L2 (grouping effect where highly correlated variables are in or out a model together) penalties. We also proposed a novel univariate HLR thresholding function to update the estimated coefficients and developed the coordinate descent algorithm for the HLR penalized logistic regression model.
The empirical results and simulations show the HLR method was highly competitive amongst Lasso, L1/2, SCAD − L2 and Elastic net in analyzing high dimensional and low sample sizes data (microarray and RNA-seq data). Thus, logistic regression with the HLR approach is the promising tool for feature selection in the classification problem. Source code of sparse logistic regression with the HLR approach was provided in S3 File.
Supporting Information
(PDF)
Top-10 ranked genes selected by all the methods for prostate and lymphoma datasets.
(PDF)
MATLAB code of sparse logistic regression with the HLR approach.
(RAR)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the Macau Science and Technology Develop Funds (Grant No. 099/2013/A3) of Macau SAR of China.
References
- 1.Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc B. JSTOR; 1996; 267–288. [Google Scholar]
- 2.Fan J, Li R. Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Stat Assoc. Taylor & Francis; 2001;96: 1348–1360. [Google Scholar]
- 3.Zou H. The adaptive lasso and its oracle properties. J Am Stat Assoc. Taylor & Francis; 2006;101: 1418–1429. [Google Scholar]
- 4.Meinshausen N, Yu B. Lasso-type recovery of sparse representations for high-dimensional data. Ann Stat. JSTOR; 2009; 246–270. [Google Scholar]
- 5.Xu Z, Zhang H, Wang Y, Chang X, Liang Y. L1/2 regularization. Sci China Inf Sci. Springer; 2010;53: 1159–1169. [Google Scholar]
- 6.Zeng J, Lin S, Wang Y, Xu Z. Regularization: Convergence of Iterative Half Thresholding Algorithm. Signal Process IEEE Trans. IEEE; 2014;62: 2317–2329. [Google Scholar]
- 7.Xu Z, Chang X, Xu F, Zhang H. L1/2 regularization: a thresholding representation theory and a fast solver. IEEE Trans neural networks Learn Syst. United States; 2012;23: 1013–1027. 10.1109/TNNLS.2012.2197412 [DOI] [PubMed] [Google Scholar]
- 8.Segal MR, Dahlquist KD, Conklin BR. Regression approaches for microarray data analysis. J Comput Biol. Mary Ann Liebert, Inc.; 2003;10: 961–980. [DOI] [PubMed] [Google Scholar]
- 9.Zou H, Hastie T. Regularization and variable selection via the elastic net. J R Stat Soc Ser B (Statistical Methodol. Wiley Online Library; 2005;67: 301–320. [Google Scholar]
- 10.Becker N, Toedt G, Lichter P, Benner A. Elastic SCAD as a novel penalization method for SVM classification tasks in high-dimensional data. BMC Bioinformatics. German Cancer Research Center, Division Molecular Genetics, INF 280, 69120 Heidelberg, Germany. natalia.becker@dkfz.de; 2011;12: 138 10.1186/1471-2105-12-138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeng L, Xie J. Group variable selection via SCAD-L 2. Statistics (Ber). Taylor & Francis; 2014;48: 49–66. [Google Scholar]
- 12.Zhao P, Yu B. Stagewise lasso. J Mach Learn Res. JMLR. org; 2007;8: 2701–2726. [Google Scholar]
- 13.Candes E, Tao T. The Dantzig selector: statistical estimation when p is much larger than n. Ann Stat. JSTOR; 2007; 2313–2351. [Google Scholar]
- 14.Knight K, Fu W. Asymptotics for lasso-type estimators. Ann Stat. JSTOR; 2000; 1356–1378. [Google Scholar]
- 15.Malioutov D, Çetin M, Willsky AS. A sparse signal reconstruction perspective for source localization with sensor arrays. Signal Process IEEE Trans. IEEE; 2005;53: 3010–3022. [Google Scholar]
- 16.Liang Y, Liu C, Luan XZ, Leung KS, Chan TM, Xu ZB, et al. Sparse logistic regression with a L1/2 penalty for gene selection in cancer classification. BMC Bioinformatics. Faculty of Information Technology & State Key Laboratory of Quality Research in Chinese Medicines, Macau University of Science and Technology, Macau, China. yliang@must.edu.mo; 2013;14: 198 10.1186/1471-2105-14-198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang H-H, Liang Y, Liu X-Y. Network-Based Logistic Classification with an Enhanced Solver Reveals Biomarker and Subnetwork Signatures for Diagnosing Lung Cancer. Biomed Res Int. Hindawi Publishing Corporation; 2015;2015. [DOI] [PMC free article] [PubMed]
- 18.Friedman JH. Regularized discriminant analysis. J Am Stat Assoc. Taylor & Francis; 1989;84: 165–175. [Google Scholar]
- 19.Hastie T, Tibshirani R, Friedman J, Franklin J. The elements of statistical learning: data mining, inference and prediction. Math Intell. Springer; 2005;27: 83–85. [Google Scholar]
- 20.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. NIH Public Access; 2010;33: 1 [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Wan YW, Allen GI, Pang K, Anderson ML, Liu Z. Molecular pathway identification using biological network-regularized logistic models. BMC Genomics. England; 2013;14 Suppl 8: S7–2164–14–S8–S7. Epub 2013 Dec 9. 10.1186/1471-2164-14-S8-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Singh D, Febbo PG, Ross K, Jackson DG, Manola J, Ladd C, et al. Gene expression correlates of clinical prostate cancer behavior. Cancer Cell. Elsevier; 2002;1: 203–209. [DOI] [PubMed] [Google Scholar]
- 23.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RCT, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. Nature Publishing Group; 2002;8: 68–74. [DOI] [PubMed] [Google Scholar]
- 24.Seo JS, Ju YS, Lee WC, Shin JY, Lee JK, Bleazard T, et al. The transcriptional landscape and mutational profile of lung adenocarcinoma. Genome Res. Genomic Medicine Institute (GMI), Medical Research Center, Seoul National University, Seoul 110–799, Korea. jeongsun@snu.ac.kr; 2012;22: 2109–2119. 10.1101/gr.145144.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartling B, Hofmann HS, Weigle B, Silber RE, Simm A. Down-regulation of the receptor for advanced glycation end-products (RAGE) supports non-small cell lung carcinoma. Carcinogenesis. Clinic of Cardiothoracic Surgery, Martin Luther University Halle-Wittenberg, Ernst-Grube-Strasse 40, D-06120 Halle/Saale, Germany. babett.barling@medizin.uni-halle.de; 2005;26: 293–301. doi:bgh333 [pii]. [DOI] [PubMed] [Google Scholar]
- 26.Buckley ST, Ehrhardt C. The receptor for advanced glycation end products (RAGE) and the lung. J Biomed Biotechnol. School of Pharmacy and Pharmaceutical Sciences, Trinity College Dublin, Dublin 2, Ireland.; 2010;2010: 917108 10.1155/2010/917108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jing R, Cui M, Wang J, Wang H. Receptor for advanced glycation end products (RAGE) soluble form (sRAGE): a new biomarker for lung cancer. Neoplasma. Center of Laboratory Medicine, Affiliated Hospital of Nantong University, Affiliated Hospital of Nantong University, 20 Xi Si Road, Nantong 226001, PR China. jrjr2020@163.com; 2010;57: 55–61. [DOI] [PubMed] [Google Scholar]
- 28.Vicente C, Conchillo A, García-Sánchez MA, Odero MD. The role of the GATA2 transcription factor in normal and malignant hematopoiesis. Crit Rev Oncol Hematol. Elsevier; 2012;82: 1–17. 10.1016/j.critrevonc.2011.04.007 [DOI] [PubMed] [Google Scholar]
- 29.Kumar MS, Hancock DC, Molina-Arcas M, Steckel M, East P, Diefenbacher M, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. Elsevier; 2012;149: 642–655. [DOI] [PubMed] [Google Scholar]
- 30.Lu TP, Tsai MH, Lee JM, Hsu CP, Chen PC, Lin CW, et al. Identification of a novel biomarker, SEMA5A, for non-small cell lung carcinoma in nonsmoking women. Cancer Epidemiol Biomarkers Prev. Graduate Institute of Biomedical Electronics and Bioinformatics, National Taiwan University, Taipei, Taiwan.: AACR; 2010;19: 2590–2597. 10.1158/1055-9965.EPI-10-0332 [DOI] [PubMed] [Google Scholar]
- 31.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, et al. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. Department of Surgery, Department of Biochemistry and Molecular Biology, Norris Comprehensive Cancer Center, Keck School of Medicine, University of Southern California, Los Angeles, CA 90089–9176, USA.; 2012;22: 1197–1211. 10.1101/gr.132662.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambroise C, McLachlan GJ. Selection bias in gene extraction on the basis of microarray gene-expression data. Proc Natl Acad Sci U S A. Laboratoire Heudiasyc, Unite Mixte de Recherche/Centre National de la Recherche Scientifique 6599, 60200 Compiegne, France.; 2002;99: 6562–6566. 10.1073/pnas.102102699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paul TK, Iba H. Extraction of informative genes from microarray data. Proceedings of the 7th annual conference on Genetic and evolutionary computation. ACM; 2005. pp. 453–460.
- 34.Wessels LF, Reinders MJ, Hart AA, Veenman CJ, Dai H, He YD, et al. A protocol for building and evaluating predictors of disease state based on microarray data. Bioinformatics. Department of Mediamatics, Faculty of Electrical Engineering, Mathematics and Computer Science, Delft University of Technology Mekelweg 4, 2628 CD Delft, The Netherlands. l.f.a.wessels@ewi.tudelft.nl; 2005;21: 3755–3762. doi:bti429 [pii]. [DOI] [PubMed] [Google Scholar]
- 35.Shen L, Tan EC. Dimension reduction-based penalized logistic regression for cancer classification using microarray data. IEEE/ACM Trans Comput Biol Bioinforma. IEEE Computer Society Press; 2005;2: 166–175. [DOI] [PubMed] [Google Scholar]
- 36.Lecocke M, Hess K. An empirical study of univariate and genetic algorithm-based feature selection in binary classification with microarray data. Cancer Inform. Department of Mathematics, St. Mary’s University, San Antonio, Texas 78228, USA.; 2007;2: 313–327. [PMC free article] [PubMed] [Google Scholar]
- 37.Dagliyan O, Uney-Yuksektepe F, Kavakli IH, Turkay M. Optimization based tumor classification from microarray gene expression data. PLoS One. Department of Chemical and Biological Engineering, Koc University, Istanbul, Turkey.; 2011;6: e14579 10.1371/journal.pone.0014579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Glaab E, Bacardit J, Garibaldi JM, Krasnogor N. Using rule-based machine learning for candidate disease gene prioritization and sample classification of cancer gene expression data. PLoS One. Public Library of Science; 2012;7: e39932 10.1371/journal.pone.0039932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu J, Zhou H. Tumor classification based on gene microarray data and hybrid learning method. Machine Learning and Cybernetics, 2003 International Conference on. IEEE; 2003. pp. 2275–2280.
- 40.Goh L, Song Q, Kasabov N. A novel feature selection method to improve classification of gene expression data. Proceedings of the second conference on Asia-Pacific bioinformatics-Volume 29. Australian Computer Society, Inc.; 2004. pp. 161–166. [Google Scholar]
- 41.Hu Y, Kasabov N. Ontology-based framework for personalized diagnosis and prognosis of cancer based on gene expression data. Neural Information Processing. Springer; 2008. pp. 846–855. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
Top-10 ranked genes selected by all the methods for prostate and lymphoma datasets.
(PDF)
MATLAB code of sparse logistic regression with the HLR approach.
(RAR)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.