Abstract
Background
An attractive approach in the study of human cancers is the use of transparent zebrafish (Danio rerio) embryos, which enable the visualization of cancer progression in a living animal.
Materials and methods
We implanted mixtures of fluorescently labeled glioblastoma (GBM) cells and bonemarrow-derived mesenchymal stem cells (MSCs) into zebrafish embryos to study the cellular pathways of their invasion and the interactions between these cells in vivo.
Results
By developing and applying a carbocyanine-dye-compatible clearing protocol for observation of cells in deep tissues, we showed that U87 and U373 GBM cells rapidly aggregated into tumor masses in the ventricles and midbrain hemispheres of the zebrafish embryo brain, and invaded the central nervous system, often using the ventricular system and the central canal of the spinal cord. However, the GBM cells did not leave the central nervous system. With co-injection of differentially labeled cultured GBM cells and MSCs, the implanted cells formed mixed tumor masses in the brain. We observed tight associations between GBM cells and MSCs, and possible cell-fusion events. GBM cells and MSCs used similar invasion routes in the central nervous system.
Conclusions
This simple model can be used to study the molecular pathways of cellular processes in GBM cell invasion, and their interactions with various types of stromal cells in double or triple cell co-cultures, to design anti-GBM cell therapies that use MSCs as vectors.
Key words: brain tumors, tumor microenvironment, animal models, xenotransplantation
Introduction
Glioblastoma multiformae (GBM) is the most aggressive type of glioma and also the most frequent and fatal among brain tumors.1 An essential hallmark of GBM is its diffuse invasion into the brain parenchyma, which prevents successful surgical removal.2 Understanding the mechanisms and the pathways of GBM cell invasion is therefore of crucial importance for the treatment of the aggressive spread of GBM.1-3 The process of GBM cell infiltration into the brain parenchyma4 differs from carcinoma cells invading the more compact extracellular matrix and the basal membranes of blood vessels.5,6 The recently recognized important role of the tumor microenvironment in cancer cell invasion7 has become an important topic and the subject of intensive research.8-10 The effects of the microenvironment also include the impact of the different types of cells comprising the stroma within a tumor mass. Infiltrating and tumor-associated mesenchymal stem cells (MSCs) may significantly affect tumor progression and resistance to treatment, as reviewed in.8,11 MSCs are known to be recruited by tumor-secreted signaling molecules via the blood circulation, and to become part of the tumor-supporting stoma, where they have a role that remains poorly understood.10,12 Several studies have addressed this issue in vitro and in vivo, also by using genetically modified MSCs.13,14 Previously, we studied the molecular mechanisms that support the observed phenotype changes in GBM cells and MSCs upon co-culture in vitro, including decreased U87 GBM cell proliferation and invasion, and increased U373 GBM cell invasion in vitro.15,16 By investigating the GBM-MSC interactions in a mouse model, Behan et al.17 recently showed that cells with an MSC-like phenotype can infiltrate the stoma of the mouse GBM and have important roles in tumor cell growth. Moreover, their data demonstrated an alteration in GBM cell marker expression upon the encounter with MSCs in vivo. In the present study, we aimed to use an alternative and simpler in vivo GBM xenotransplantation model in zebrafish embryos18, to study human GBM cell invasion and their interactions with MSCs at the cellular level.
The zebrafish (Danio rerio) is the major vertebrate model in developmental biology and genetics.19 There are several technologies available in the zebrafish that have made it a unique model in cancer research.20 In particular, cancers can be studied throughout the life cycle of zebrafish, with each zebrafish developmental stage offering its own experimental advantages. This makes zebrafish a powerful complement to other more traditional model systems.21 As well as their high fecundity and ease of maintenance, the major advantage of zebrafish is the transparency of their embryos and larvae, which allows in vivo visualization of cellular processes related to cancer growth and progression at single-cell resolution.20,22,23 Xenotransplantation of either dyelabeled or fluorescent-protein-expresing human cells in zebrafish embryos is becoming an increasingly used tool to study cancers of the central nervous system (CNS).18,24-26
The aim of the present study was to investigate the interactions of GBM cells with the brain matrix components and MSCs in the zebrafish embryonic brain by coimplantation of fluorescently labeled GBM cells and MSCs. To deepen our understanding of GBM cell behavior within the brain of zebrafish embryos, we combined in vivo imaging of GBM progression with imaging of fluorescently counterstained whole-mount preparations that allowed the visualization of the anatomical context of the implanted cells. As chemical fixation leads to loss of transparency of the embryonic tissues, it necessitated the clearing of the embryos, which was achieved with the use of clearing agents.27-39 To this end, we optimized and applied protocols to clear fixed tissues while preserving the fluorescent protein signal over a period of several weeks.
Materials and methods
Ethical statement
The experimental procedures were approved by the Republic of Slovenia National Medical Ethics Committee, approval No. 92/06/12. All of the procedures were performed according to the relevant regulations.
Zebrafish husbandry
Wild-type AB zebrafish (Danio rerio) were maintained under conditions according to the Organisation for Economic Cooperation and Development guidelines.40 The zebrafish embryos were collected and incubated in dilution water (ISO 7346-3:1996) with 0.005% phenylthiourea, to inhibit pigment formation after 36 h of age.
Human cells
The U373 and U87-MG human GBM cell lines were from American Type Culture Collection (USA), and the BM-MSC2 human bone-marrow-derived MSC cell line was from Lonza Bioscience (USA). The cells were cultured in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich, USA) supplemented with 10% fetal bovine serum (Gibco, USA), 100 U/mL penicillin (Sigma-Aldrich), 100 mg/mL streptomycin (Sigma-Aldrich), 2 mM L-glutamine (Sigma-Aldrich), 1 mM Na-pyruvate (Gibco), and non-essential amino acids (Sigma-Aldrich).
Xenotransplantation procedures
The U373 GBM cells were transfected with the pEGFP-N1 plasmid to stably express enhanced green fluorescent protein (GFP). The U87 GBM cells were transfected for the expression of the red fluorescent protein DsRed, as described previously.16 Cells were transfected using the Superfect Transfection Reagent (Qiagen, Germany) by 3 h pre-incubation at 37°C, 5% CO2. The transfection mix was then removed and upon washing with phosphate-buffered saline (PBS), fresh culture medium was added to the cells. Transfected cells were selected for by adding 0.8 mg/ml Geneticin (G418, Gibco, USA) to the medium. The uniformity of emitted fluorescence was confirmed with flow cytometry. The stability of fluorescent protein expression was verified with repeated flow cytometry analyses after 10 and 20 passages and proved to be stable (>99% of fluorescent protein-expressing cells). Prior to implantation, MSCs were labeled with Vybrant DiI or DiO (Molecular Probes, USA) for co-implantation with the U373 and U87 cells, respectively, according to the manufacturer instructions. For injection into the embryos, suspensions of GBM cells prepared in PBS were mixed with labeled MSCs in a 1:1 ratio. Embryos at 52 h after fertilization were injected either with 50 to 100 GBM cells or 100 to 200 cells of the GBM/MSC mixture (i.e., maintaining 50–100 GBM cells), using a borosilicate glass capillary and a MICROINJECTOR system (Tritech Research, USA). After cell implantation, the embryos were incubated at 31°C in 48-well plates for 3 days.
Clearing agents
The clearing agents used were SeeDB, a near-saturated solution of fructose with α-thioglycerol34, sRIMS, a buffered solution of sorbitol39, and two aqueous solutions of urea and glycerol known as ScaleA2 and ScaleU2.33 An overview of the compositions of these clearing agents is shown in Table 1.
Table 1.
Compositions of the selected optical clearing agents
| Optical clearing agent | Composition | Reference |
|---|---|---|
| SeeDB | 80% (w/w) fructose, 0.5% (w/w) α-thioglycerol in water | Ke et al.34 |
| sRIMS | 70% (w/v) sorbitol, 0.01% (w/v) sodium azide, 0.1% (w/v) Tween 20 in 0.02 M phosphate buffer (pH 7.5) | Yang et al.39 |
| ScaleA2 | 4 M urea, 0.1% (w/v) Triton X-100, 10% (w/w) glycerol in water | Hama et al.33 |
| ScaleU2 | 4 M urea, 0.1% (w/v) Triton X-100, 30% (w/w) glycerol in water | Hama et al.33 |
Analysis of clearing efficiency and fluorescence preservation
At 3 days after fertilization (for measurements of transparency and size) and 5 days after fertilization (for fluorescence imaging of implanted cells), the zebrafish embryos were fixed in 4% paraformaldehyde at 4°C overnight, after which time the fixative was washed off with PBS. The fixed embryos were then embedded in 2% low-melting-point agarose in PBS in 50-mm Petri dishes. For the optical clearing agent SeeDB, the embryos were transferred to SeeDB through a graded series of sucrose, as described by Ke et al.34, and maintained at room temperature. For sRIMS and Scale, the embryos were immersed in the clearing agents and kept at 4°C in the dark. In all cases, the clearing agents were replaced every 3 days. The control embryos were incubated in parallel in PBS. For analysis of the whole-embryo clearing, transmitted light images were obtained at constant (maximum) illumination at 32× magnification. For imaging and quantification of GFP fluorescence, the images were captured at 80× magnification using the GFP filter set. Micrographs were obtained using a fluorescence stereomicroscope (Leica MZ FLIII). The imaging was carried out over a period of 21 days. The analysis of fluorescence preservation was performed on embryos with implanted U373 cells. Fluorescence intensity, embryo transparency, and embryo size were quantified with the image analysis software ImageJ.41 For determination of embryo transmittance, the parameter ‘Integrated density’ was measured. The relative integrated densities and areas were determined by dividing the values of these parameters at the given time points with their values at the beginning of the observation on day 0. As integrated density represents the product of pixel intensities and area, the changes in embryo size that were caused by some of the clearing agents were accounted for by dividing the integrated density of the structure analyzed with its area, thus obtaining the relative embryo transparency. Fluorescence intensity was analyzed by thresholding the fluorescent images with a constant threshold and measuring the integrated density of the selection. For the analysis of changes in embryo size and transparency, 8 embryos per treatment were measured. For the analysis of GFP fluorescence intensity changes, 24 embryos per treatment were measured. Measurements were compared with ANOVA in GraphPad Prism.
Confocal microscopy
The nuclei of the embryos that had been fixed, embedded in agarose, and cleared as described above were counterstained by addition of 0.004% methyl green to the individual clearing agent.42 Confocal zstacks of the embryos were obtained using a spectral confocal laser scanning microscope (Leica TCS SPE) at 10× magnification after 7 days of treatment.
Results
Clearing efficiencies of the different clearing agents
To evaluate the applicability of the different optical clearing agents (Table 1) to zebrafish embryo imaging, we treated embryos fixed at 3 days after fertilization with SeeDB, sRIMS, ScaleA2, and ScaleU2 for 3 weeks, with regular imaging. The tissues were cleared best by ScaleU2 and ScaleA2 with no statistically significant difference between them at any time point. These agents performed significantly better than SeeDB on all treatment days. The least effective agent was sRIMS (Figure 1A). After approximately 1 week of incubation, fructose began to crystallize from the SeeDB solution, preventing further analyses. With regard to the preservation of GFP fluorescence, ScaleU2 was the optimal clearing agent, resulting in a several-fold increase in fluorescence intensity during the first 3 days of incubation and no demonstrable reduction in its intensity during 3 weeks of observation (Figure 1B). These changes were statistically significant when compared with the control. Complete loss of fluorescence occurred in the case of See DB within a week and no statistically significant increases in intensity compared to the control were demonstrated in the cases of ScaleA2 and s RIMS. The loss of GFP fluorescence in SeeDB was accompanied by the appearance of strong auto fluorescence of the yolk and eye (Figure 1C).
Figure 1.
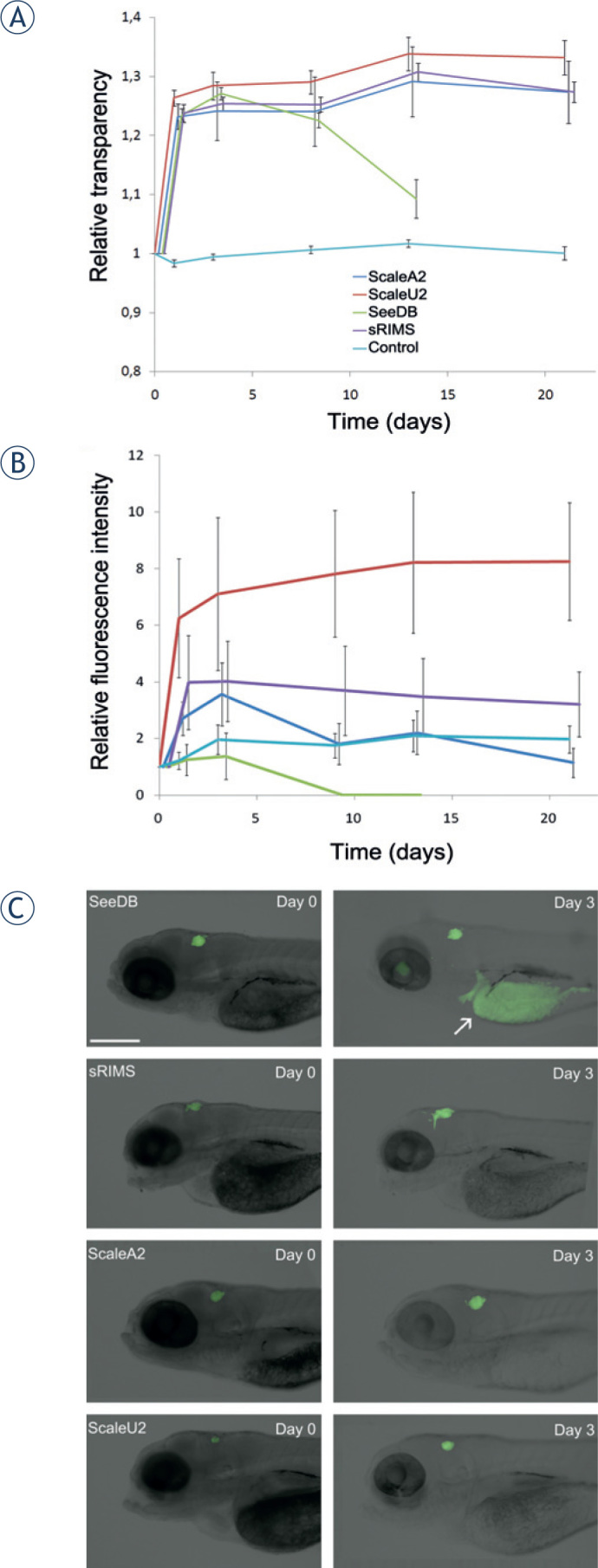
(A) Clearing of zebrafish embryos. Embryos were fixed 3 days after fertilization, exposed to the different clearing agents SeeDB, sRIMS, ScaleA2, and ScaleU2 for 21 days, and imaged regularly. Time courses of changes in relative transparency are shown, which represents the value of integrated density relative to day 0 divided by the embryo area relative to day 0. Differences between treatments are statistically significant in all cases except between ScaleA2 and Scale B2 (on all days) and between ScaleA2 and sRIMS on day 21. Green, SeeDB; purple, sRIMS; dark blue, ScaleA2; red, ScaleU2; light blue, PBS control. Data for the different clearing agents are displaced horizontally for improved clarity. Means ± SE of eight embryos per treatment are shown. (B) Preservation of GFP fluorescence during clearing. Changes in the detected GFP fluorescence intensity of glioblastoma cells implanted in the brain of zebrafish embryos during treatment with different clearing agents of SeeDB, sRIMS, ScaleA2, and ScaleU2, measured over 21 days of the treatment. The integrated density of GFP-expressing cells relative to day 0 was measured. Fluorescence intensity was significantly increased compared to control in the case of ScaleU2 on all days except on day 3, but not in the case of other clearing agents. Means ±SE of 24 embryos per treatment are shown. (C) Fluorescence of U373-GFP cells in the brain of zebrafish embryos. Representative images show embryos treated with the different clearing agents obtained at the beginning of observation (Day 0, left) and after 3 days of clearing (Day 3, right). The appearance of autofluorescence of the yolk (arrow) is evident in the case of SeeDB. Scale bar: 400 μm.
Localization of implanted glioblastoma cells and pathways of GBM cell invasion in the zebrafish embryonic brain
After selecting ScaleU2 as the optimal clearing agent for the visualization of the fluorescent-protein-labeled cells, it was applied to visualize tumor progression in the whole-mount preparations of the zebrafish embryos. We implanted U373-GFP cells into the brain of embryos 2 days after fertilization, and monitored these over the following 3 days.
By imaging the GBM cells in zebrafish embryos in vivo, we demonstrated that implanted cells aggregated and formed tumors in the zebrafish brain (Figure 2A). In some embryos, individual cells moved posteriorly in the embryo at great speed, as they progressed at several hundreds of micrometers per day (Figure 2B). This rapid invasion outside of the brain in the posterior direction was observed more frequently for the U87 cells (35 ± 5% of the embryos, as 3 experimental repeats, and 20 embryos per repeat) than for the U373 cells (20 ± 5% of the embryos, as 3 experimental repeats, and 20 embryos per repeat).
Figure 2.
In vivo imaging of glioblastoma cells in the brain of zebrafish embryos. (A) Embryo 3 days after the implantation of U87DsRed cells in the brain (visible as red fluorescence). Compact tumors have formed in the midbrain and for brain. (B) An embryo with implanted U87-DsRed cells 2 days after implantation, with a string of U87-DsRed cells rapidly invading from the tumor in the brain in the posterior direction (arrow). Scale bars: 300 μm (A); 50 μm (B).
Tumors were seen to form predominantly in the midbrain hemispheres and in the ventricles of the midbrain and hindbrain (Figure 3A-C). Individual cells, or small strands of cells, invaded the ventricular system and the brain tissue using pseudopodal movement (Figure 3A,B). In particular, cells present in the midbrain hemispheres formed elongated pseudopodia and invaded the neighboring brain areas dorsally, most likely along axonal tracts that connect the hemispheres (Figure 3A). Whole mount imaging of embryos with cells invading posteriorly in the body revealed that the rapidly invading cells invaded the spinal cord via the central canal (Figure 3D,E). We did not observe GBM cells outside of the brain and spinal cord, indicating that GBM cell invasion in zebrafish embryos was limited to the CNS, and that the cells did not spread via the circulatory system.
Figure 3.
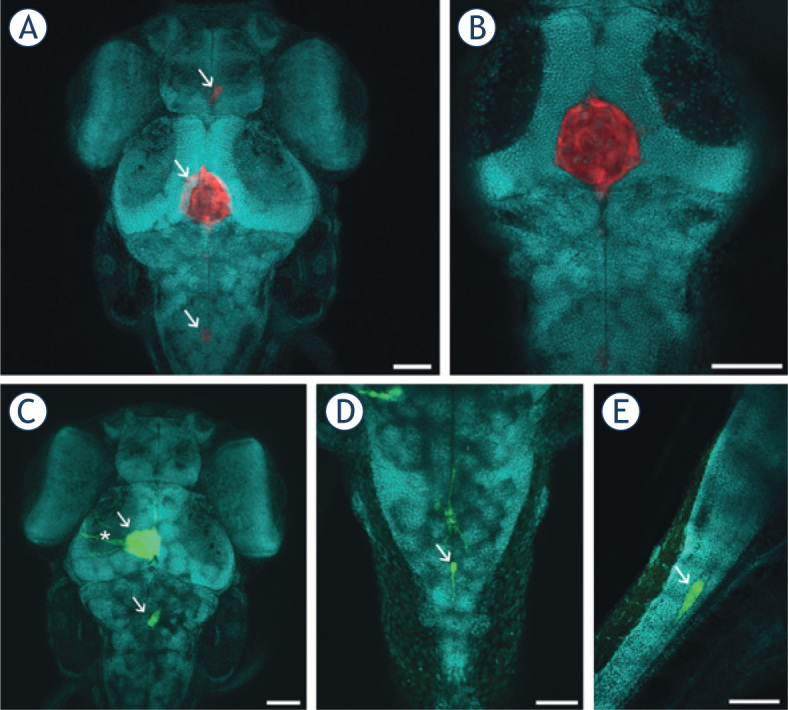
Visualization of GBM cells in cleared zebrafish embryos, counterstained with methyl green (presented in blue). Embryos with U373-GFP and U87-DsRed cells implanted in the brain were cleared with ScaleU2, counterstained with methyl green, and imaged with confocal microscopy. (A) U87-DsRed cells (arrows) in the brain of a zebrafish embryo 3 days after implantation. (B) An optical section through the tumor in (A), demonstrating that the tumor is a compact mass of U87 cells in the midbrain ventricle. (C) U373-GFP cells (arrows) in the brain of a zebrafish embryo 3 days after implantation. Elongated U373-GFP cells are visible invading from the tumor (asterisk). (D) A U373-GFP cell invading along the central canal of the spinal cord (arrow), in dorsal view. (E) A U373-GFP cell invading along the central canal of the spinal cord (arrow), in lateral view. Scale bars: 70 μm (A, B, C); 50 μm (D, E).
Imaging of xenotransplanted GBM and MSC co-cultures
To study the interactions between the GBM cells and MSCs in vivo, we implanted co-cultures of U373-GFP cells and carbocyanine-dye-labeled human MSCs into the brain of zebrafish embryos 2 days after fertilization. As labeling with carbocyanine dyes relies on the hydrophobic nature of these dyes, they can be washed out with organic sol vents and detergents. Thus, we prepared ScaleU2 without Triton X-100, with which we successfully visualized dye-labeled cells in whole-mount preparations. In implanted mixtures of GBM cells and MSCs, the MSCs preferentially associated with the implanted GBM cells instead of being interspersed individually or as separate clusters in the brain (Figure 4A–C). The MSCs often used similar invasion routes as the GBM cells, as they moved along the ventricles and the central canal of the spinal cord (Figure 4B,C).
Figure 4.
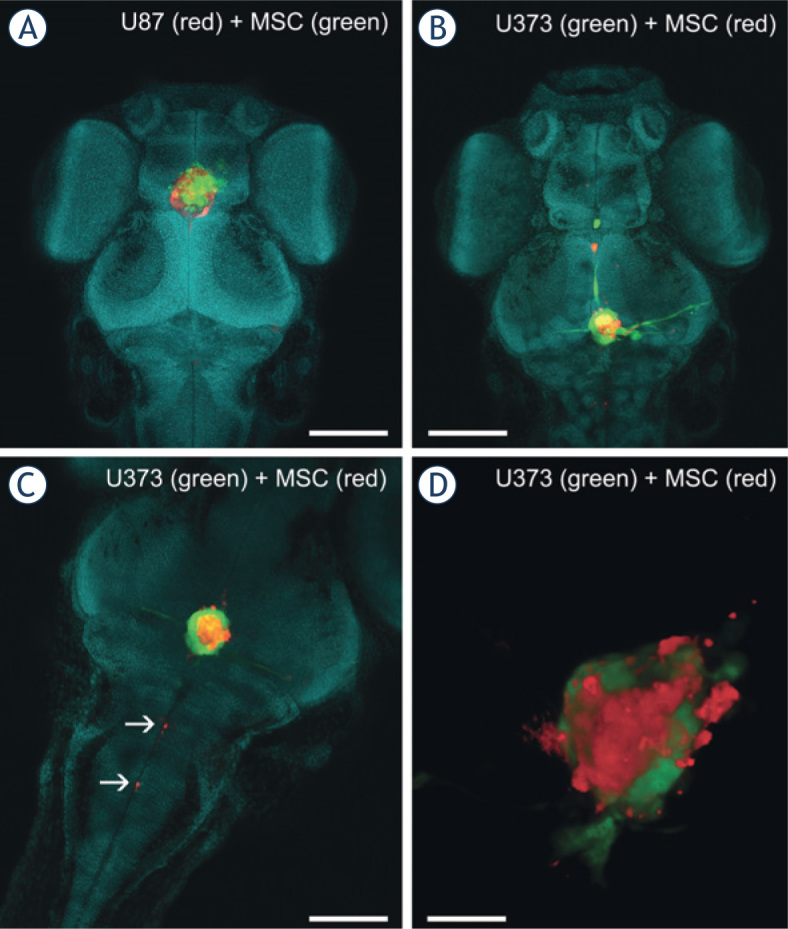
Imaging of co-cultures of GBM cells and MSCs in the brain of zebrafish embryos. A mixture of fluorescent-protein-expressing GBM cells and carbocyaninedye-labeled MSCs was implanted into the brain of the zebrafish embryos. Three days after implantation, the embryos were fixed, cleared in ScaleU2 without the addition of Triton X-100, and imaged with confocal microscopy. (A) The head of a zebrafish embryo with a co-culture of U87-DsRed cells (red) and DiO-labeled MSCs (green) implanted in the brain. (B) The head of a zebrafish embryo with a co-culture of U373-GFP cells (green) and DiI-labeled MSCs (red) implanted in the brain. (C) Invasion of DiI-labeled MSCs (red) along the central canal of the spinal cord. (D) Three-dimensional rendering of a mixed mass of U373 cells (green) and MSCs (red) in a brain obtained from a cleared embryo. Nuclei are stained with methyl green (presented in blue). Scale bars: 250 μm (A, B); 100 μm (C); 50 μm (D).
Confocal imaging of U373 and MSC co-cultures in situ revealed that the GBM cells and MSCs formed mixed tumor masses that consisted of both of these cell types in similar locations as for the GBM cells alone; i.e., in the ventricles and midbrain hemispheres (Figure 4A,B). These two cell types were interspersed in these tumors, and interacted closely with each other (Figure 4D). In some cases, cells simultaneously emitting the fluorescence of proteins and the carbocyanine dye were observed, which indicated possible cell fusion between the GBM cells and MSCs (Figure 5).
Figure 5.
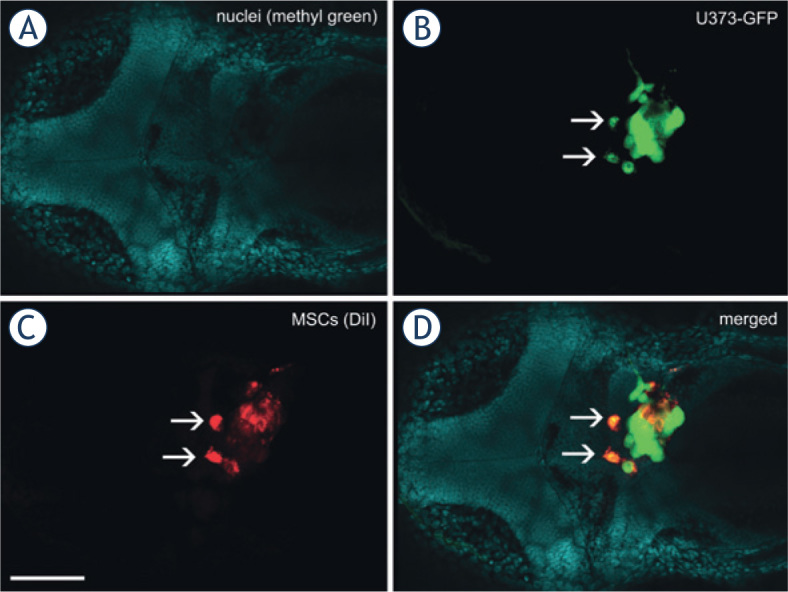
Fusion between GBM cells and MSCs in the zebrafish brain. A mixture of U373-GFP cells and DiI-labeled MSCs was implanted in the brain of the embryos, which were fixed, cleared and imaged 3 days after implantation of the cells. Two cells (arrows) emit green GFP fluorescence as well as red DiI fluorescence, which strongly indicates that the U373-GFP cells and MSCs have fused after implantation.Nuclei of embryonic tissues labeled with methyl green. (B) Green fluorescent protein fluorescence of U373 cells. (C) Red fluorescence of DiI, used to label the MSCs. (D) Merged image of all of the fluorescent channels. Scale bar: 200 μm.
Discussion
The major advantage of the zebrafish model is the transparency of their embryos and larvae, which allows in vivo visualization of cellular processes related to cancer progression at single-cell resolution. We identified ScaleU2 as the optimal clearing agent for zebrafish embryos among the agents tested. Furthermore, we were able to modify it to be compatible with the labeling of cells with carbocyanine dyes. The superiority of ScaleU2 in fluorescence preservation appears to result from its high glycerol concentration, and thus might be linked to the protein-stabilizing effects of glycerol in aqueous solution.43,44
The proposed protocols were used to study the invasion of the U87 and U373 human GBM cell lines alone and in co-cultures with bone-marrow derived MSCs in the zebrafish embryo. By combining the in vivo imaging with confocal microscopy of fluorescently counterstained whole-mount preparations, we demonstrated that GBM cells aggregate in the brain of zebrafish embryos and form tumors predominantly in the ventricles. This indicates that GBM cells have tropism towards each other upon implantation to form tumors, which are preferentially formed in the ventricles and dorsal areas of the midbrain. The tendency of the GBM cells to aggregate in these areas might be linked to the ease of dislodging the embryonic brain tissues in anatomical structures such as the ventricles, which are fluid-filled spaces where the brain cells are not in direct contact. The localization of GBM cells to the ventricular system has previously not been reported. Eden et al.45 recently reported that mouse xenografts in the brain of juvenile (30-day-old) zebrafish reproduced the histology and gene expression profiles of the murine tumors of their origin within the tissues of the brain. Thus, the observed localization of the GBM cells to the ventricular system might be limited to embryos and larval stages.
We were able to identify the central canal of the spinal cord as the major route of GBM invasion in zebrafish embryos that has not been reported before. Furthermore, GBM cells did not leave the central nervous system, which is similar to their behavior in humans.46 In a previous study on murine tumor xenografts in the zebrafish brain, tumor masses developed 1 day to 2 days after the implantation in the spinal cord. Histological analysis confirmed that these masses were independent, distant tumors, rather than direct extensions of the main tumor mass, thus demonstrating that the implanted GBM cells disseminated in the CNS of the zebrafish.45 This is in agreement with our observations of single-cell invasion along the central canal of the spinal cord, which may result in separate tumor formation within the spinal cord. The underlying molecular mechanisms of this rapid invasion remain to be established. The process might be facilitated by the low resistance of the central canal to cell invasion, as the GBM cells are believed to generally invade through structures that have low resistance to cell movement.5, 6,46
One of the preferential pathways of GBM invasion in the human brain is the white matter, where GBM cells invade along axons.46 It is likely that this preference is recapitulated in zebrafish embryos, as we observed pseudopodal invasion in the midbrain area that is rich in axonal connections between the two hemispheres (Figure 3C). This is reminiscent of tumor invasion via the corpus callosum in human patients.2
Pseudopodia and cell elongation characterize the mesenchymal type of cell invasion, which is typical for gliomas and depends on cell-matrix adhesion.5,6 The observed strand migration in the spinal cord as well as the midbrain (Figure 2B,4B) is linked to proteolytic matrix remodeling and is characteristic of cancer cells belonging to the mesenchymal type.5 A mesenchymal molecular fingerprint has recently been established for U373 GBM cells.47 This invasion pattern has also previously been observed for U87 cells in mouse models, together with elevated cathepsin B expression at the tumor periphery.48 In a study on zebrafish larvae, invasion of U87 GBM cells along the abluminal surface of blood vessels has also been demonstrated.25 As the basal lamina of blood vessels is a known invasion pathway in the human brain46, this further strengthens the view that the invasion of GBM cells in the zebrafish model resembles this process in mammals.
Confocal imaging of GBM and MSC co-cultures in situ revealed that both of these types of cells form mixed tumor masses at similar locations as for the GBM cells alone; i.e., in the ventricles and midbrain hemisphere. The GBM and MSC cells used similar invasion routes along the ventricles and the central canal of the spinal cord, but did not invade other tissues. The association of GBM cells and MSCs into such mixed tumors in the brain of these zebrafish embryos suggests a strong intercellular interaction between them, which would appear to also have a role in the human GBM microenvironment. Indeed, MSC tropism towards GBM cells has previously been described10, and a set of cytokines has been shown to mediate the interactions between these cells in vitro.49,50 Among these, the monocyte chemoattractant protein (MCP-1) has been suggested as the major trigger of various molecular pathways that can enhance MSC proliferation and invasion, whereas a different set of genes has been shown to impair U87 cell invasion and proliferation, and to even induce their senescence.49 When mixtures of U87 or U373 GBM cells and MSCs were implanted into the zebrafish embryos, these established very close contacts and apparently also formed a structural syncytium in vivo. This finding confirms our previous in-vitro studies of direct co-cultures using U373 GBM cells and MSCs. These studies showed the formation of gap junctions between these cell types (i.e., formation of a functional syncytium), as demonstrated by fluorescein transmission and connexin 43 expression. Furthermore, there was membrane fusion between these cells (i.e., formation of a structural syncytium), as demonstrated by the co-localization of different carbocyanine dyes.15 Direct membrane fusion between the GBM cells and MSCs affects gene expression and the cell phenotype, which might lead to enhanced invasion of hybrid cells, which is of relevance to tumor progression.15
Taken together, our enhanced clearing methodology has enabled us to study GBM cell localization in the brain of zebrafish embryos and to observe their interaction with MSCs at single-cell resolution. This allowed us to identify the invasion patterns of GBM cells in the zebrafish brain and identify the central canal of the spinal cord as a major invasion route. As the frequency of this single cell invasion of the spinal cord is also quantifiable, the observation of this process in high-throughput screening can now be developed as a fast and objective methodology. Thus it can not only to be used to study basic mechanisms in terms of the differences among heterogeneous GBM populations, but also for diagnostic purposes with patient biopsies. This study is also the first to address co-culture implantation in the zebrafish brain model in order to define the interactions between GBM cells and MSCs. We demonstrated that these two cell types invade the surrounding brain tissue along similar invasion roots as the GBM cells alone, whereby they often moved along the central canal of the spinal cord, but did not leave the CNS.
In perspective, the zebrafish xenotransplantation model of GBM has many benefits in terms of cost, simplicity, possibility for single-cell visualization in vivo, and high throughput.18 As demonstrated by the present study, this simple model can be used to study cell processes involved in GBM cell invasion and the interactions of GBM cells with many other cells of the stroma in double or triple cell co-cultures.
Acknowledgments
The authors wish to thank Prof. Dr. Cornelis J.F. van Noorden (Academic Medical Centre, Amsterdam) for his introduction to the CLARITY technique. We also thank Dr. David Dobnik (National Institute of Biology, Ljubljana) for his help with confocal microscopy. This work was funded by the INTERREG EC Project 2011, Ref. No. 42 (GLIOMA) and the Reserch Programme P1-0245, awarded by the Slovenian Research Agency.
Disclosure: No potential conflicts of interest were disclosed.
References
- 1.Ohgaki H, Kleihues P. The definition of primary and secondary glioblastoma. Clin Cancer Res 2013; 19: 764-72. [DOI] [PubMed]; Ohgaki H, Kleihues P.. The definition of primary and secondary glioblastoma. Clin Cancer Res. 2013;19:764–72. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 2.Claes A, Idema A, Wesseling P. Diffuse glioma growth: a guerilla war. Acta Neuropathol 2007; 114: 443-58. [DOI] [PMC free article] [PubMed]; Claes A, Idema A, Wesseling P.. Diffuse glioma growth: a guerilla war. Acta Neuropathol. 2007;114:443–58. doi: 10.1007/s00401-007-0293-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gupta MK, Jayaram S, Reddy DN, Polisetty RV, Sirdeshmukh R. Transcriptomic and proteomic data integration and two-dimensional molecular maps with regulatory and functional linkages: application to cell proliferation and invasion networks in glioblastoma. J Proteome Res 2015; 14: 5017-27 [DOI] [PubMed]; Gupta MK, Jayaram S, Reddy DN, Polisetty RV, Sirdeshmukh R.. Transcriptomic and proteomic data integration and two-dimensional molecular maps with regulatory and functional linkages: application to cell proliferation and invasion networks in glioblastoma. J Proteome Res. 2015;14:5017–27. doi: 10.1021/acs.jproteome.5b00765. [DOI] [PubMed] [Google Scholar]
- 4.Klekner Á, Hutóczki G, Virga J, Reményi-Puskár J, Tóth J, Scholtz B, et al. Expression pattern of invasion-related molecules in the peritumoral brain. Clin Neurol Neurosurg 2015; 139: 138-43. [DOI] [PubMed]; Klekner Á, Hutóczki G, Virga J, Reményi-Puskár J, Tóth J, Scholtz B. et al. Expression pattern of invasion-related molecules in the peritumoral brain. Clin Neurol Neurosurg. 2015;139:138–43. doi: 10.1016/j.clineuro.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Friedl P, Wolf K. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer 2003; 3: 362-74. [DOI] [PubMed]; Friedl P, Wolf K.. Tumour-cell invasion and migration: diversity and escape mechanisms. Nat Rev Cancer. 2003;3:362–74. doi: 10.1038/nrc1075. [DOI] [PubMed] [Google Scholar]
- 6.Friedl P, Wolf K. Plasticity of cell migration: a multiscale tuning model. J Cell Biol 2010; 188: 11-9. [DOI] [PMC free article] [PubMed]; Friedl P, Wolf K.. Plasticity of cell migration: a multiscale tuning model. J Cell Biol. 2010;188:11–9. doi: 10.1083/jcb.200909003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011; 144: 646-74. [DOI] [PubMed]; Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Grisendi G, Bussolari R, Veronesi E, Piccinno S, Burns JS, De Santis G, et al. Understanding tumor-stroma interplays for targeted therapies by armed mesenchymal stromal progenitors: the Mesenkillers. Am J Cancer Res 2011; 1: 787-805. [PMC free article] [PubMed]; Grisendi G, Bussolari R, Veronesi E, Piccinno S, Burns JS, De Santis G. et al. Understanding tumor stroma interplays for targeted therapies by armed mesenchymal stromal progenitors: the Mesenkillers. Am J Cancer Res. 2011;1:787–805. [PMC free article] [PubMed] [Google Scholar]
- 9.Kološa K, Motaln H, Herold-Mende C, Koršič M, Lah TT. Paracrine effects of mesenchymal stem cells induce senescence and differentiation of glioblastoma stem-like cells. Cell Transplant 2015; 24: 631-44. [DOI] [PubMed]; Kološa K, Motaln H, Herold Mende C, Koršič M, Lah TT.. Paracrine effects of mesenchymal stem cells induce senescence and differentiation of glioblastoma stem-like cells. Cell Transplant. 2015;24:631–44. doi: 10.3727/096368915X687787. [DOI] [PubMed] [Google Scholar]
- 10.Motaln H, Lah TT. Cytokines play a key role in communication between mesenchymal stem cells and brain cancer cells. Protein Pept Lett 2015; 22: 322-31. [DOI] [PubMed]; Motaln H, Lah TT.. Cytokines play a key role in communication between mesenchymal stem cells and brain cancer cells. Protein Pept Lett. 2015;22:322–31. doi: 10.2174/0929866522666150131123808. [DOI] [PubMed] [Google Scholar]
- 11.Barcellos-de-Souza P, Gori V, Bambi F, Chiarugi P. Tumor microenvironment: bone marrow-mesenchymal stem cells as key players. Biochim Biophys Acta 2013; 1836: 321-35. [DOI] [PubMed]; Barcellos-de-Souza P, Gori V, Bambi F, Chiarugi P.. Tumor microenvironment: bone marrow-mesenchymal stem cells as key players. Biochim Biophys Acta. 2013;1836:321–35. doi: 10.1016/j.bbcan.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 12.Arango-Rodriguez ML, Ezquer F, Ezquer M, Conget P. Could cancer and infection be adverse effects of mesenchymal stromal cell therapy? World J Stem Cells 2015; 7: 408-17. [DOI] [PMC free article] [PubMed]; Arango Rodriguez ML, Ezquer F, Ezquer M, Conget P.. Could cancer and infection be adverse effects of mesenchymal stromal cell therapy? World J Stem Cells. 2015;7:408–17. doi: 10.4252/wjsc.v7.i2.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fei S, Qi X, Kedong S, Guangchun J, Jian L, Wei Q. The antitumor effect of mesenchymal stem cells transduced with a lentiviral vector expressing cytosine deaminase in a rat glioma model. J Cancer Res Clin Oncol 2012; 138: 347-57. [DOI] [PubMed]; Fei S, Qi X, Kedong S, Guangchun J, Jian L, Wei Q.. The antitumor effect of mesenchymal stem cells transduced with a lentiviral vector expressing cytosine deaminase in a rat glioma model. J Cancer Res Clin Oncol. 2012;138:347–57. doi: 10.1007/s00432-011-1104-z. [DOI] [PubMed] [Google Scholar]
- 14.Kim SM, Woo JS, Jeong CH, Ryu CH, Lim JY, Jeun SS. Effective combination therapy for malignant glioma with TRAIL-secreting mesenchymal stem cells and lipoxygenase inhibitor MK886. Cancer Res 2012; 72: 4807-17. [DOI] [PubMed]; Kim SM, Woo JS, Jeong CH, Ryu CH, Lim JY, Jeun SS.. Effective combination therapy for malignant glioma with TRAIL-secreting mesenchymal stem cells and lipoxygenase inhibitor MK886. Cancer Res. 2012;72:4807–17. doi: 10.1158/0008-5472.CAN-12-0123. [DOI] [PubMed] [Google Scholar]
- 15.Schichor C, Albrecht V, Korte B, Buchner A, Riesenberg R, Mysliwietz J, et al. Mesenchymal stem cells and glioma cells form a structural as well as a functional syncytium in vitro. Exp Neurol 2012; 234: 208-19. [DOI] [PubMed]; Schichor C, Albrecht V, Korte B, Buchner A, Riesenberg R, Mysliwietz J. et al. Mesenchymal stem cells and glioma cells form a structural as well as a functional syncytium in vitro. Exp Neurol. 2012;234:208–19. doi: 10.1016/j.expneurol.2011.12.033. [DOI] [PubMed] [Google Scholar]
- 16.Pilat MM, Oliveira M, Motaln H, Breznik B, Glaser T, Lah TT, et al. Glioblastoma-mesenchymal stem cell communication modulates expression and activities of kinin receptors: possible involvement of bradykinin on information flow. Cytometry A 2016; DOI: 10.1002/cyto.a.22800 [DOI] [PubMed]; Pilat MM, Oliveira M, Motaln H, Breznik B, Glaser T, Lah TT, Cytometry A. 2016. Glioblastoma-mesenchymal stem cell communication modulates expression and activities of kinin receptors: possible involvement of bradykinin on information flow. DOI: [DOI] [PubMed] [Google Scholar]
- 17.Behnan J, Isakson P, Joel M, Cilio C, Langmoen I, Vik-Mo EO, et al. Recruited brain rumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells 2014; 32: 1110-23. [DOI] [PubMed]; Behnan J, Isakson P, Joel M, Cilio C, Langmoen I, Vik-Mo EO. et al. Recruited brain rumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells. 2014;32:1110–23. doi: 10.1002/stem.1614. [DOI] [PubMed] [Google Scholar]
- 18.Vittori M, Motaln H, Lah TT. The study of glioma by xenotransplantation in zebrafish early life stages. J Histochem Cytochem 2015; 63: 749-61. [DOI] [PMC free article] [PubMed]; Vittori M, Motaln H, Lah TT.. The study of glioma by xenotransplantation in zebrafish early life stages. J Histochem Cytochem. 2015;63:749–61. doi: 10.1369/0022155415595670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grunwald DJ, Eisen JS. Headwaters of the zebrafish – emergence of a new model vertebrate. Nat Rev Genet 2002; 3: 717-24. [DOI] [PubMed]; Grunwald DJ, Eisen JS.. Headwaters of the zebrafish – emergence of a new model vertebrate. Nat Rev Genet. 2002;3:717–24. doi: 10.1038/nrg892. [DOI] [PubMed] [Google Scholar]
- 20.Feng Y, Martin P. Imaging innate immune responses at tumour initiation: new insights from fish and flies. Nat Rev Cancer 2015; 15: 556-62. [DOI] [PubMed]; Feng Y, Martin P.. Imaging innate immune responses at tumour initiation: new insights from fish and flies. Nat Rev Cancer. 2015;15:556–62. doi: 10.1038/nrc3979. [DOI] [PubMed] [Google Scholar]
- 21.White R, Rose K, Zon L. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer 2013; 13: 624-36. [DOI] [PMC free article] [PubMed]; White R, Rose K, Zon L.. Zebrafish cancer: the state of the art and the path forward. Nat Rev Cancer. 2013;13:624–36. doi: 10.1038/nrc3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zon LI, Peterson RT. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov 2005; 4: 35-44. [DOI] [PubMed]; Zon LI, Peterson RT.. In vivo drug discovery in the zebrafish. Nat Rev Drug Discov. 2005;4:35–44. doi: 10.1038/nrd1606. [DOI] [PubMed] [Google Scholar]
- 23.Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 2008; 322: 1065-9. [DOI] [PubMed]; Keller PJ, Schmidt AD, Wittbrodt J, Stelzer EH.. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science. 2008;322:1065–9. doi: 10.1126/science.1162493. [DOI] [PubMed] [Google Scholar]
- 24.Geiger GA, Fu W, Kao GD. Temozolomide-mediated radiosensitization of human glioma cells in a zebrafish embryonic system. Cancer Res 2008; 68: 3396-404. [DOI] [PMC free article] [PubMed]; Geiger GA, Fu W, Kao GD.. Temozolomide-mediated radiosensitization of human glioma cells in a zebrafish embryonic system. Cancer Res. 2008;68:3396–404. doi: 10.1158/0008-5472.CAN-07-6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lal S, La Du J, Tanguay RL, Greenwood JA. Calpain 2 is required for the invasion of glioblastoma cells in the zebrafish brain microenvironment. J Neurosci Res 2012; 90: 769-81. [DOI] [PMC free article] [PubMed]; Lal S, La Du J, Tanguay RL, Greenwood JA.. Calpain 2 is required for the invasion of glioblastoma cells in the zebrafish brain microenvironment. J Neurosci Res. 2012;90:769–81. doi: 10.1002/jnr.22794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konantz M, Balci TB, Hartwig UF, Dellaire G, André MC, Berman JN, et al. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci 2012; 1266: 124-37. [DOI] [PubMed]; Konantz M, Balci TB, Hartwig UF, Dellaire G, André MC, Berman JN. et al. Zebrafish xenografts as a tool for in vivo studies on human cancer. Ann N Y Acad Sci. 2012;1266:124–37. doi: 10.1111/j.1749-6632.2012.06575.x. [DOI] [PubMed] [Google Scholar]
- 27.Sakhalkar HS, Dewhirst M, Oliver T, Cao Y, Oldham M. Functional imaging in bulk tissue specimens using optical emission tomography: fluorescence preservation during optical clearing. Phys Med Biol 2007; 52: 2035-54. [DOI] [PubMed]; Sakhalkar HS, Dewhirst M, Oliver T, Cao Y, Oldham M.. Functional imaging in bulk tissue specimens using optical emission tomography: fluorescence preservation during optical clearing. Phys Med Biol. 2007;52:2035–54. doi: 10.1088/0031-9155/52/8/001. [DOI] [PubMed] [Google Scholar]
- 28.Dodt HU, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods 2007; 4: 331-6. [DOI] [PubMed]; Dodt HU, Leischner U, Schierloh A, Jährling N, Mauch CP, Deininger K. et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4:331–6. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 29.Zukor KA, Kent DT, Odelberg SJ. Fluorescent whole-mount method for visualizing three-dimensional relationships in intact and regenerating adult newt spinal cords. Dev Dyn 2010; 239: 3048-57. [DOI] [PMC free article] [PubMed]; Zukor KA, Kent DT, Odelberg SJ.. Fluorescent whole-mount method for visualizing three-dimensional relationships in intact and regenerating adult newt spinal cords. Dev Dyn. 2010;239:3048–57. doi: 10.1002/dvdy.22441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oldham M, Sakhalkar H, Oliver T, Allan Johnson G, Dewhirst M. Optical clearing of unsectioned specimens for three-dimensional imaging via optical transmission and emission tomography. J Biomed Opt 2008; 13: 021113. [DOI] [PMC free article] [PubMed]; Oldham M, Sakhalkar H, Oliver T, Allan Johnson G, Dewhirst M.. Optical clearing of unsectioned specimens for three-dimensional imaging via optical transmission and emission tomography. J Biomed Opt. 2008;13:021113. doi: 10.1117/1.2907968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG, et al. Threedimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc 2012; 7: 1983-95. [DOI] [PubMed]; Ertürk A, Becker K, Jährling N, Mauch CP, Hojer CD, Egen JG. et al. Threedimensional imaging of solvent-cleared organs using 3DISCO. Nat Protoc. 2012;7:1983–95. doi: 10.1038/nprot.2012.119. [DOI] [PubMed] [Google Scholar]
- 32.Becker K, Jährling N, Saghafi S, Weiler R, Dodt H-U. Chemical clearing and dehydration of GFP expressing mouse brains. PLoS ONE 2012; 7: e33916 [DOI] [PMC free article] [PubMed]; Becker K, Jährling N, Saghafi S, Weiler R, Dodt H-U.. Chemical clearing and dehydration of GFP expressing mouse brains. PLoS ONE. 2012;7:e33916. doi: 10.1371/journal.pone.0033916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H, et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci 2011; 14: 1481-8. [DOI] [PubMed]; Hama H, Kurokawa H, Kawano H, Ando R, Shimogori T, Noda H. et al. Scale: a chemical approach for fluorescence imaging and reconstruction of transparent mouse brain. Nat Neurosci. 2011;14:1481–8. doi: 10.1038/nn.2928. [DOI] [PubMed] [Google Scholar]
- 34.Ke MT, Fujimoto S, Imai T. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci 2013; 16: 1154-61. [DOI] [PubMed]; Ke MT, Fujimoto S, Imai T.. SeeDB: a simple and morphology-preserving optical clearing agent for neuronal circuit reconstruction. Nat Neurosci. 2013;16:1154–61. doi: 10.1038/nn.3447. [DOI] [PubMed] [Google Scholar]
- 35.Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell 2014; 157: 726-39. [DOI] [PubMed]; Susaki EA, Tainaka K, Perrin D, Kishino F, Tawara T, Watanabe TM. et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–39. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 36.Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M, et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell 2014; 159: 911-24. [DOI] [PubMed]; Tainaka K, Kubota SI, Suyama TQ, Susaki EA, Perrin D, Ukai-Tadenuma M. et al. Whole-body imaging with single-cell resolution by tissue decolorization. Cell. 2014;159:911–24. doi: 10.1016/j.cell.2014.10.034. [DOI] [PubMed] [Google Scholar]
- 37.Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C. ClearT: a detergent- and solvent-free clearing method for neuronal and non-neuronal tissue. Development 2013; 140: 1364-8. [DOI] [PMC free article] [PubMed]; Kuwajima T, Sitko AA, Bhansali P, Jurgens C, Guido W, Mason C.. ClearT: a detergentand solvent-free clearing method for neuronal and non-neuronal tissue. Development. 2013;140:1364–8. doi: 10.1242/dev.091844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ, et al. Structural and molecular interrogation of intact biological systems. Nature 2013; 497: 332-7. [DOI] [PMC free article] [PubMed]; Chung K, Wallace J, Kim SY, Kalyanasundaram S, Andalman AS, Davidson TJ. et al. Structural and molecular interrogation of intact biological systems. Nature. 2013;497:332–7. doi: 10.1038/nature12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E, et al. Single-cell phenotyping within transparent intact tissue through wholebody clearing. Cell 2014; 158: 945-58. [DOI] [PMC free article] [PubMed]; Yang B, Treweek JB, Kulkarni RP, Deverman BE, Chen CK, Lubeck E. et al. Single cell phenotyping within transparent intact tissue through wholebody clearing. Cell. 2014;158:945–58. doi: 10.1016/j.cell.2014.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Organisation for Economic Cooperation and Development. OECD Guidelines For Testing of Chemicals. Test No. 236. Fish Embryo Acute Toxicity (FET) Test. Paris: OECD Publishing; 2013.; Organisation for Economic Cooperation and Development. OECD Guidelines For Testing of Chemicals Test No. 236. Fish Embryo Acute Toxicity (FET) Test. Paris: OECD Publishing; 2013. [Google Scholar]
- 41.Abramoff MD, Magalhães PJ, Ram SJ. Image processing with ImageJ. Biophotonics Intern 2004; 11: 36-42.; Abramoff MD, Magalhães PJ, Ram SJ.. Image processing with ImageJ. Biophotonics Intern. 2004;11:36–42. [Google Scholar]
- 42.Prieto D, Aparicio G, Morande PE, Zolessi FR. A fast, low-cost, and highly efficient fluorescent DNA labeling method using methyl green. Histochem Cell Biol 2014; 142: 335-45. [DOI] [PubMed]; Prieto D, Aparicio G, Morande PE, Zolessi FR.. A fast, low-cost, and highly efficient fluorescent DNA labeling method using methyl green. Histochem Cell Biol. 2014;142:335–45. doi: 10.1007/s00418-014-1215-0. [DOI] [PubMed] [Google Scholar]
- 43.Gekko, K, Timasheff SN. Mechanism of protein stabilization by glycerol: preferential hydration in glycerol-water mixtures. Biochemistry 1981; 20: 4667-76. [DOI] [PubMed]; Gekko K, Timasheff SN.. Mechanism of protein stabilization by glycerol: preferential hydration in glycerol-water mixtures. Biochemistry. 1981;20:4667–76. doi: 10.1021/bi00519a023. [DOI] [PubMed] [Google Scholar]
- 44.Street TO, Bolen DW, Rose GD. A molecular mechanism for osmolyteinduced protein stability. Proc Natl Acad Sci U S A 2006; 103: 13997-4002. [DOI] [PMC free article] [PubMed]; Street TO, Bolen DW, Rose GD.. A molecular mechanism for osmolyteinduced protein stability. Proc Natl Acad Sci U S A. 2006;103:13997–4002. doi: 10.1073/pnas.0606236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eden C, Ju B, Murugesan M, Phoenix T, Nimmervoll B, Tong Y, et al. Orthotopic models of pediatric brain tumors in zebrafish. Oncogene 2015; 34: 1736-42. [DOI] [PMC free article] [PubMed]; Eden C, Ju B, Murugesan M, Phoenix T, Nimmervoll B, Tong Y. et al. Orthotopic models of pediatric brain tumors in zebrafish. Oncogene. 2015;34:1736–42. doi: 10.1038/onc.2014.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gritsenko PG, Ilina O, Friedl P. Interstitial guidance of cancer invasion. J Pathol 2012; 226: 185-99. [DOI] [PubMed]; Gritsenko PG, Ilina O, Friedl P.. Interstitial guidance of cancer invasion. J Pathol. 2012;226:185–99. doi: 10.1002/path.3031. [DOI] [PubMed] [Google Scholar]
- 47.Motaln H, Koren A, Gruden K, Ramšak Ž, Schichor C, Lah TT. Heterogeneous glioblastoma cell cross-talk promotes phenotype alterations and enhanced drug resistance. Oncotarget 2015; 38: 40998-1017. [DOI] [PMC free article] [PubMed]; Motaln H, Koren A, Gruden K, Ramšak Ž, Schichor C, Lah TT.. Heterogeneous glioblastoma cell cross-talk promotes phenotype alterations and enhanced drug resistance. Oncotarget. 2015;38:40998–1017. doi: 10.18632/oncotarget.5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gole B, Huszthy PC, Popović M, Jeruc J, Ardebili YS, Bjerkvig R, Lah TT. The regulation of cysteine cathepsins and cystatins in human gliomas. Int J Cancer 2012; 131: 1779-89. [DOI] [PubMed]; Gole B, Huszthy PC, Popović M, Jeruc J, Ardebili YS, Bjerkvig R, Lah TT.. The regulation of cysteine cathepsins and cystatins in human gliomas. Int J Cancer. 2012;131:1779–89. doi: 10.1002/ijc.27453. [DOI] [PubMed] [Google Scholar]
- 49.Motaln H, Gruden K, Hren M, Schichor C, Primon M, Rotter A, et al. Human mesenchymal stem cells exploit the immune response mediating chemokines to impact the phenotype of glioblastoma. Cell Transplant 2012; 21: 1529-45. [DOI] [PubMed]; Motaln H, Gruden K, Hren M, Schichor C, Primon M, Rotter A. et al. Human mesenchymal stem cells exploit the immune response mediating chemokines to impact the phenotype of glioblastoma. Cell Transplant. 2012;21:1529–45. doi: 10.3727/096368912X640547. [DOI] [PubMed] [Google Scholar]
- 50.Tajnšek U, Motaln H, Levičar N, Rotter A, Lah TT. The duality of stem cell: double edged sword in tumour evolution and treatment. In: Resende RR, Ulrich H, editor. Trends in Stem Cell Proliferation and Cancer Research. Dordrecht: Springer Netherlands; 2013. p. 391-434.; Tajnšek U, Motaln H, Levičar N, Rotter A, Lah TT. Resende RR, Ulrich H. Trends in Stem Cell Proliferation and Cancer Research. Dordrecht; Springer Netherlands: 2013. The duality of stem cell: double edged sword in tumour evolution and treatment; pp. 391–434. [Google Scholar]