Abstract
The effect of human cholesteryl ester transfer protein (CETP) expression on atherogenesis is still under debate. The rs5882 (I405V) polymorphism affect CETP function. We aimed to examine the relationship between the rs5882 polymorphism and the risk of angiographically determined coronary artery disease (CAD). To define premature CAD (PCAD), an age cutoff of 55 years for women and 45 years for men was used. An age- and sex-matched case-control study was conducted in 560 patients with newly diagnosed angiographically documented PCAD (≥50% luminal stenosis of any coronary vessel) and an equal number of control patients with normal coronary arteries (no luminal stenosis at coronary arteries). The severity of CAD was determined by vessel score and Gensini score. A real-time polymerase chain reaction (PCR) and high resolution melting analysis were used to distinguish between genotypes. The I405V genotype distributions were not statistically different in CAD and non-CAD groups in univariate and multivariable-adjusted logistic regression analyzes. The median and inter-quartile range for Gensini score was not significantly different among the AA (43, 24 to 73), AG (40, 20 to 66), and GG (45, 25 to 72) genotypes (p = 0.097). Furthermore, the distribution of vessel score did not statistically differ between these genotypes (p = 0.691). Our results suggest that there is no significant association between CETP I405V polymorphism and the risk of PCAD presence and severity. Larger prospective studies are needed to investigate such associations in different populations.
KEY WORDS: Coronary artery disease, premature atherosclerosis, coronary angiography, cholesteryl ester transfer protein, I405V polymorphism
INTRODUCTION
Coronary artery disease (CAD) is a complex disease, caused by a combination of multiple environmental and genetic risk factors [1]. Recent findings have suggested that CAD has a significant genetic component and that different types of genetic polymorphisms contribute its risk [2]. In young people, the role of genetic risk factors is expected to be even more important than that of environmental factors [3,4]. Although CAD usually occurs at an older age (age >55 years), premature CAD (PCAD) is a growing entity that imposes physical, psychological, and financial constraints on both patients and their families and above all it can influence the quality of life and result in premature death [4]. The early detection of youth at risk for PCAD on the basis of genetic information provides earlier preventive strategies to improve cardiovascular outcomes [5]; however, the predictors and mechanisms behind this condition are yet to be determined [6].
An isolated reduced high-density lipoprotein cholesterol (HDL-C) is reported to be as a risk factor for premature atherosclerosis [7]. The activity of the human cholesteryl ester transfer protein (CETP), a glycoprotein which mediates the transfer of cholesteryl ester from HDL-C to triglyceride (TG)-rich lipoproteins, results in a decrease in HDL-C levels [8]. However, CETP may also promote reverse cholesterol transport pathway, an anti-atherogenic process, wherein CETP facilitates the uptake of cholesterol from peripheral tissues to the liver. Therefore, the net effect of CETP expression on atherogenesis is still controversial [9].
The human CETP gene is located on chromosome 6q21 encompassing 16 exons and 15 introns and spans approximately 25 kb. CETP rs5882 (A>G) is a missense mutation in which an A-to-G substitution in Exon 14 leads to a change in the primary structure of CETP protein isoleucine to valine variation at codon 405 (I405V). It is a common polymorphism that seems to have only slight effects on CETP function [8,10]. The rs5882 (I405V) polymorphism included in the current study was selected based on its potential functionality, its relatively high frequency, and its impact on CETP activity and HDL-C [8].
Several studies on rs5882 polymorphism and the presence of CAD have been previously published, but this variant is exclusively evaluated among general CAD populations [11-15]; moreover, to the best of our knowledge, there is no study that investigated this association in PCAD patients. Thus, the aim of this study was to investigate the relationship between the rs5882 polymorphism and the risk of angiographically determined PCAD in the Iranian sample.
MATERIALS AND METHODS
Study participants, coronary angiograms
We conducted a case-control study of 560 patients with newly diagnosed angiographically documented PCAD and an equal number of control patients with normal coronary arteries (no luminal stenosis at coronary angiography; Gensini score = 0). The case group consisted of young patients (age ≤45 years for men, and age ≤55 years for women at the disease onset) who, from May 2009 to July 2011, consecutively underwent coronary angiography at the cardiac catheterization laboratory of our center because of suspicious CAD due to symptoms related to CAD or results of non-invasive tests. All cases showed the evidence of atherosclerosis, i.e., ≥50% luminal stenosis in at least one coronary artery or major branch segment in their epicardial coronary tree. A sex-matched sample of 560 unrelated controls (274 men, 45.7%) was randomly selected out of a possible 1547 young control subjects (419 men and 1128 women) who were consecutively admitted for elective coronary angiography at our hospital, between June 2004 and July 2011 (Figure 1). Patients with previous history of acute myocardial infarction (MI), stent implantation, cardiopulmonary resuscitation, and coronary artery bypass graft surgery were excluded. We also excluded those with minimal CAD (coronary lesions with <50% luminal stenosis). Patients were matched with controls at the group level (Table 1). The study protocol was approved by the Local Ethical Committee. Written informed consent was obtained from all patients explicitly provided permission for DNA analyzes and gathering the relevant clinical data.
FIGURE 1.
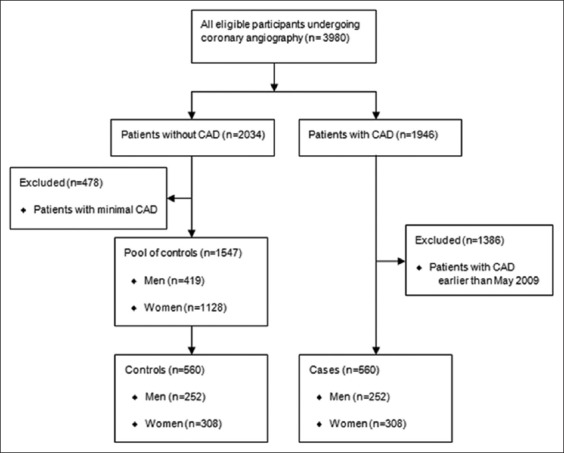
Flowchart of the study population.
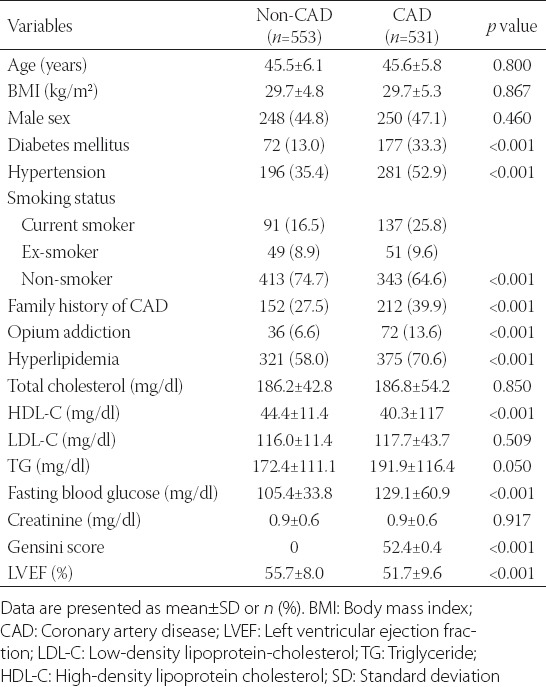
TABLE 1.
Baseline characteristics in the study population based on coronary artery disease presence
Coronary angiographies were done by the percutaneous femoral approach using standard techniques. The presence and severity of CAD were determined by clinical vessel score. The severity of CAD was also determined by a semi-quantitative scoring system (Gensini score) which has been previously described [16]. One single cardiologist, blind to the study results, interpreted all angiograms. Definitions for analyzed risk factors of CAD including age, male sex, hyperlipidemia, opium addiction, hypertension, cigarette smoking, and diabetes have been reported elsewhere [17,18].
Collection of clinical data
For all patients undergoing coronary angiography at our institution, Tehran Heart Center (THC), we prospectively record information about demographic characteristics, cardiovascular risk factors, electrocardiographic interpretations, and echocardiographic findings as well as drug history, history of cardiac events, past history of coronary or any other type of intervention and open heart surgery. These data are gathered on a structured paper form during the patient’s admission (D1-Coronary Angiography Form V2.2). Procedural data detailed by the cardiologists or interventionists are recorded on a separate paper form (D2-Coronary Angiography Report Form V5.2). Then, an operator enters the collected data into a computerized database (THC-Coronary Angiography Data Bank). From this database, the records of all patients included in this study were retrieved for analysis.
Sample collection and DNA extraction
For the purpose of creating a “DNA-bank of patients with PCAD and controls” at THC, from June 2004 to July 2011, all male patients aged 45 years old or younger and all female patients 55 years old or younger were asked to provide a sample of whole blood for DNA extraction. Peripheral venous blood samples were collected from an antecubital vein after 12 hours overnight fasting of participants. Upon arrival at the laboratory, each participant provided about 15 mL venous blood sample. 5 mL blood was collected in plain tubes and used for biochemical assays immediately; the rest of the sample (10 mL) placed in tubes containing ethylenediaminetetraacetic acid (EDTA) and stored deep-frozen until later use. Genomic DNA was extracted from leukocytes using the buffy coat of the stored EDTA whole blood samples. DNA extraction carried out using the standard “salting out” method. DNA quantity was evaluated by calculating absorbance at λ = 260 nm, and the quality were assessed by a ratio of λ = 260/280 nm being close to 1.8. The purified DNA was stored in Tris-EDTA buffer (pH 8.0) at −70°C until further analysis.
Polymerase chain reaction (PCR) amplification and genotype analysis
Real-time PCR and high-resolution melting (HRM) analysis were applied for the genotype analysis using Corbett Rotor-Gene 6000 real-time rotary analyzer (Corbett Life Science Pty. Ltd., Mortlake, NSW, Australia). One set of primers based on common HRM specifications was designed using Beacon Designer software version 7.0 (Premier Biosoft International, Palo Alto, CA, USA) and synthesized by Bioneer Inc. (Daejeon, Korea). The forward primer was 5’- CATTTGATTGGCAGAGCAG -3’ and the reverse was 5’- ACTTACGAGACATGACCTCAG -3’. The final reaction mixture contained Type-it HRM Master Mix (QIAGEN NV, Venlo, Netherlands) with intercalating DNA-binding Evagreen dye (Biotium, Inc. Hayward, CA, USA), DNA polymerase (5 U/mL), primer mix (0.4 µM of each primer), genomic DNA (50 ng), and RNase-Free Water (QIAGEN NV) in a total volume of 20 µL. The PCR cycling conditions consisted of an initial denaturation at 97°C for 15 minutes, followed by 37 cycles of denaturation (97°C for 20 seconds), primer annealing (59°C for 20 seconds), and extension (72°C for 10 seconds). Following the PCR amplification steps, melt curves for the products were generated by heating in 0.1°C increments at a rate of 2 seconds per each step over the temperature range from 65 to 95°C. The HRM data were analyzed using the Rotor-Gene Q software package supplied with the instrument. Sequence variations were distinguished from wild-type samples by the different shapes of normalized and temperature-shifted melting curves. Finally, the results of HRM were reconfirmed by the PCR with restriction fragment length polymorphism technique in 10% of random samples and by direct sequencing in six samples.
Statistical analysis
Statistical calculations were performed by PASW Statistics for Windows, Version 18.0. (SPSS Inc., Chicago, USA). On the basis of a previous study on Iranian patients [12], we calculated a minimum sample size of 509 patients for each group, to achieve a power of 90% at a 0.05 level of significance; sample size was determined at 95% confidence interval (CI), 10% precision with considering 0.366 and 0.348 expected prevalence of G (405V) allele in cases and controls, respectively. We increased the sample size to 560 patients in each group to account for possible genotyping failure and missing data.
The results are reported as mean ± standard deviation for quantitative variables and as numbers and percentages for categorical variables. Gensini score, which was skewed, is presented as a median and inter-quartile range (25-75th percentiles). The case and control groups were compared using the independent two-sample Student’s t-test (or Mann–Whitney U-test if data were not normally distributed) for the continuous variables and the Chi-square test (or the Fisher exact test, as appropriate) for the categorical variables. Since the distributions of Gensini scores were skewed and could not be normalized by several transformation calculations, non-parametric Kruskal–Wallis analysis was chosen to compare Gensini score among I405V genotypes. We used a multivariable logistic regression model to evaluate the independent relationship between the I405V variants and the presence of CAD adjusting for other covariates including, hypertension, diabetes, smoking status, family history of CAD, and opium addiction as well as HDL-C, TG, and left ventricular ejection fraction (LVEF). Variables were incorporated into the regression model if the p values were found to be ≤0.20 in univariable analyzes or based on literature. Odds ratio and 95% CI were calculated. A p ≤ 0.05 was considered statistically significant.
RESULTS
After excluding patients with missing data, a total of 1084 individuals (96.8%) were entered in the analysis including 531 cases and 553 controls. The mean age of the analyzed population was 45.5 ± 6.0 years and 45.9% were men. Clinical and demographic characteristics of the study sample are presented in Table 1. The prevalence of diabetes, family history of CAD, hyperlipidemia, and cigarette smoking, as well as hypertension and opium addiction, were significantly higher in PCAD group while there was no statistically significant difference in age, sex, and body mass index between two groups.
The genotype frequencies observed for CETP I405V among the study population was compatible with the Hardy-Weinberg equilibrium (HWE) expectations (HWE χ2 = 1.72, p = 0.189). Allele and genotype frequencies for the I405V (rs5882) polymorphism in the non-CAD and CAD patients, separated by gender, are demonstrated in Table 2. The G allele distribution was not statistically different in non-CAD and CAD groups (36.2% vs. 36.0%; p = 0.924) and in men (36.5% vs. 34.4%; p = 0.490) and women (35.9% vs. 37.4%; p = 0.603) subgroups after stratification of the study groups by gender. Table 3 shows that I405V polymorphism was not associated with the presence of CAD in the multivariable logistic regression model, after adjustment for diabetes, hypertension, cigarette smoking, opium addiction, and family history of CAD as well as TG, HDL-C, and LVEF (p = 0.933).
TABLE 2.
Genotype and allele frequencies of I405V (rs5882) polymorphism in the CETP gene and its relationship with CAD in whole study group and subgroups separated by gender
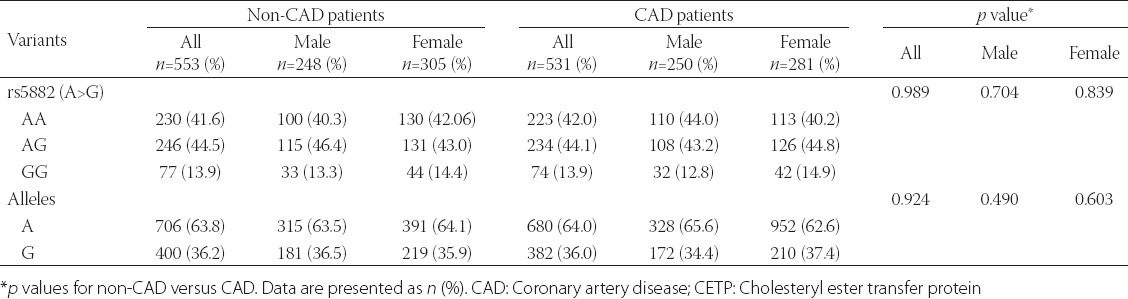
TABLE 3.
Multivariable logistic regression model for determination of independent effect of I405V (rs5882) genotypes on CAD
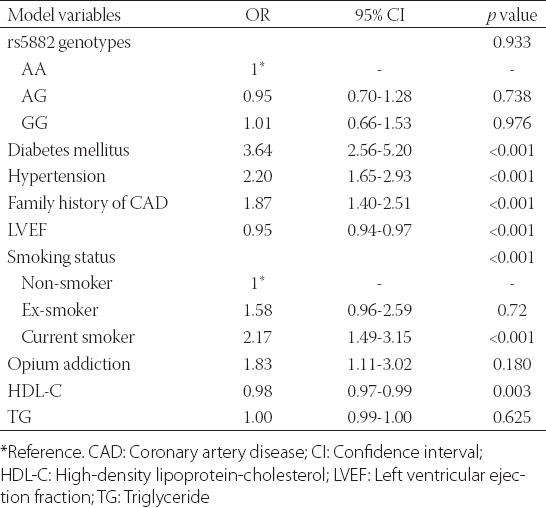
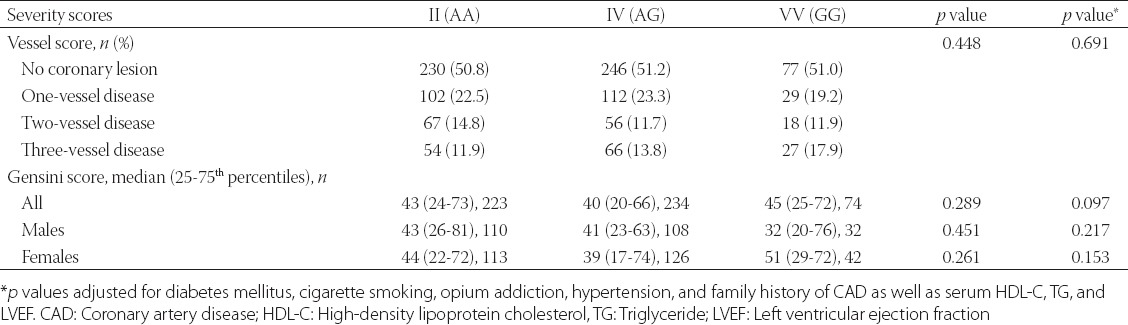
Table 4 shows the association between I405V polymorphism and the severity of CAD with respect to vessel score and Gensini score. The distribution of vessel score was statistically similar among different genotypes of I405V polymorphism (adjusted p = 0.691). In addition, the median and inter-quartile range for Gensini score were not significantly different among the AA (43, 24 to 73), AG (40, 20 to 66), and GG (45, 25 to 72) genotypes (adjusted p = 0.097). The potential associations of examined polymorphism, and the Gensini score were tested within subgroups created by subdividing the population based on total cholesterol (cut-off: 200 mg), low-density lipoprotein-cholesterol (cut-off: 130 mg/), HDL-C (cut-off: 40 mg/dl), and TG (cut-off: 150 mg/dl) levels as well as dyslipidemia and sex. As shown in Table 4, no significant association between the rs5882 polymorphism and the Gensini score was found in men and women when the population was stratified by sex. There were also no significant associations between the examined polymorphism and the severity of CAD in any of the aforementioned subgroups (data not presented).
TABLE 4.
Genotypes of I405V (rs5882) in association with CAD severity
DISCUSSION
An inverse association between serum HDL-C and CAD has been well-established [19], but a positive association of HDL-C levels with elevated CAD has been also reported in patients with CETP deficiency [20]. The CETP has a key role in the metabolism of HDL and several genetic variants in the CETP gene have been described that are generally linked with low CETP activity and accordingly with high HDL concentrations [13,21]. However, the protective role of this CETP-related increase in HDL on the risk of atherosclerosis is still under debate [22]. Moreover, the expression of human CETP in rats, a gene generally not expressed in this species, results in combined hyperlipidemia and CAD, making CETP as a strong candidate locus for CAD [23]. TaqIB polymorphism is the most studied CETP gene variant, and the B2 allele is shown to be associated with the lowest risk of CAD [24]. We investigated the effect of CETP gene I405V variant on the risk of PCAD. The present study focused on relatively young patients with PCAD not only due to the importance of the condition but also because this strategy is less likely to be confounded by the effect of age, and more likely to detect genetic effects. To the best of our knowledge, this is the first study, suggesting that rs5882 genotypes and alleles are not linked with the risk of PCAD and its severity in the Iranian sample.
The frequency of rs5882 mutation is different in various surveys with various sample sizes, in various geographical regions, and in various ethnic populations [25]. The minor allele for rs5882 variant (G allele) exists in all studied populations and generally occurs at a frequency of more than 25% [8]. The G allele frequency, calculated from data on up to 68,134 apparently healthy individuals included in a meta-analysis, in Caucasians and in East Asians was 0.350 and 0.420, respectively [13]. Among our relatively large study population, the prevalence of G allele was 0.361 in total; 0.360 in the case group, and 0.362 in the control group. In line with our results, a study in the northwest of Iran showed the prevalence of G allele to be 0.376 among CAD patients and 0.356 in controls (p > 0.05) [12]. Moreover, in an Asian study, the frequency distribution of 405V allele in control (0.390) and CAD patients (0.385) yielded no significant difference [26].
Previous studies of the influence of CETP I405V polymorphism on CAD have produced discordant results [8,14,25-36]. One large meta-analysis using 10,313 cases with coronary outcomes and 32 244 controls from 18 published studies reported rs5882 genotypes to be linked with moderate inhibition of CETP activity (accordingly with modestly increased HDL-C) and inversely with CAD [13]. However, several studies have found higher CAD risk associated with low CETP activity [27,31]. Bruce et al. [27] failed to show any overall relationship between this polymorphism and CAD status; however, among a subgroup of patients with hypertriglyceridemia, CAD prevalence was higher in those with VV than IV or II subjects. In a sample of 822 male offsprings of early MI victims (age <55 years) and age-matched control subjects, there was a statistically significant association of V allele for I405V variant with 7.0% lower activity of CETP, but no effect on CAD was observed [29]. In line with our results, several more recent studies showed no association between the rs5882 polymorphism and the risk of coronary atherosclerosis [14,32-36].
Even among large studies, the findings are conflicting. In a study embedded in the Rotterdam study cohort, a prospective study of 7983 individuals aged ≥55 years, 6421 (2607 men and 3814 women) were typed for the CETP I405V variant; although elevated HDL-C was found in both sex, the V allele was related to reduced MI risk only in men [28]. Furthermore, there was no relationship between I405V and ischemic heart disease among 4006 healthy Danish men; in 4983 women, the association was not significant until after adjustment for HDL-C levels [31]. Thompson et al. [25] genotyped 9 polymorphisms throughout the gene in 2553 individuals examining potential associations between CETP genotype and history of MI. A highly significant relationship was found between the intron 12 SNP, rs1800774, and MI history in white women but no association with any change in HDL-C. The investigators showed that I405V was also associated with MI history only among Caucasian women. Because rs5882 is in high linkage disequilibrium with rs1800774, the authors proposed this effect to be driven by the much more significant intron 12 association.
A significant part of the discrepancies in the findings from various studies may be attributed to differences in study design, inadequate sample sizes, genetic background, age, gender, and differences in the nature of various populations. For instance, the majority of published association articles in this regards have total populations or subgroups of <300 individuals with only a small number of studies examining >1000 individuals [8,13]. With such small sample sizes, the statistically significant effect is less likely to be detected. In addition, precise definition of the phenotype is an important issue in designing genetic studies. This is especially essential in studies on CAD association because individuals with significant CAD, who are clinically silent may be classified as controls leading to a higher likelihood of null results. To avoid this potential bias in the current study, we defined phenotype based on objective angiographic documentation of coronary artery status. Finally, as Rissanen [37] showed more than 35 years ago, the younger the patient at the diagnosis of a first MI, the more frequent was CAD in his parents and siblings. In families with the onset of CAD after the 45 years of age, heritability ranged from 15% to 30% while in families of PCAD heritability was estimated to be 92-100% [37]. Hence, genetic screening may prove beneficial in cases with premature atherosclerosis.
There are several limitations in the current study that should be addressed. First, a selection bias may have been introduced due to case-control design. Second, the control group was selected from individuals with suspected CAD at clinical assessment who subsequently underwent coronary angiography; this sample may not represent the healthy general population. Third, although we tested a relatively large number of patients, our study was not sufficiently powered to detect small to modest effect sizes. Finally, data come from a single center, and the findings cannot necessarily be generalized to other ethnic groups.
CONCLUSION
In this case-control study, CETP I405V polymorphism was not associated with the risk of CAD among a relatively large number of young Iranian patients undergoing coronary angiography. Larger prospective studies are needed to investigate such association in different populations particularly in those with PCAD.
ACKNOWLEDGMENTS
This work was part of the Ph.D. thesis of the first author at THC with the support of Tehran University of Medical Sciences.
DECLARATION OF INTERESTS
The authors declare no conflict of interest.
REFERENCES
- [1].Hamsten A, Eriksson P. Identifying the susceptibility genes for coronary artery disease:from hyperbole through doubt to cautious optimism. J Intern Med. 2008;263(5):538–52. doi: 10.1111/j.1365-2796.2008.01958.x. http://dx.doi.org/10.1111/j.1365-2796.2008.01958.x . [DOI] [PubMed] [Google Scholar]
- [2].Libby P. Inflammation in atherosclerosis. Nature. 2002;420(6917):868–74. doi: 10.1038/nature01323. http://dx.doi.org/10.1038/nature01323 . [DOI] [PubMed] [Google Scholar]
- [3].Cengel A, Tanindi A. Myocardial infarction in the young. J Postgrad Med. 2009;55(4):305–13. doi: 10.4103/0022-3859.58944. http://dx.doi.org/10.4103/0022-3859.58944 . [DOI] [PubMed] [Google Scholar]
- [4].Celik T, Iyisoy A. Premature coronary artery disease in young patients:an uncommon but growing entity. Int J Cardiol. 2010;144(1):131–2. doi: 10.1016/j.ijcard.2008.12.150. http://dx.doi.org/10.1016/j.ijcard.2008.12.150 . [DOI] [PubMed] [Google Scholar]
- [5].Couch SC, Cross AT, Kida K, Ros E, Plaza I, Shea S, et al. Rapid westernization of children's blood cholesterol in 3 countries:evidence for nutrient-gene interactions? Am J Clin Nutr. 2000;72(5 Suppl):1266S–74. doi: 10.1093/ajcn/72.5.1266s. [DOI] [PubMed] [Google Scholar]
- [6].Vaisi-Raygani A, Ghaneialvar H, Rahimi Z, Nomani H, Saidi M, Bahrehmand F, et al. The angiotensin converting enzyme D allele is an independent risk factor for early onset coronary artery disease. Clin Biochem. 2010;43(15):1189–94. doi: 10.1016/j.clinbiochem.2010.07.010. http://dx.doi.org/10.1016/j.clinbiochem.2010.07.010 . [DOI] [PubMed] [Google Scholar]
- [7].Goldbourt U, Yaari S, Medalie JH. Isolated low HDL cholesterol as a risk factor for coronary heart disease mortality. A 21-year follow-up of 8000 men. Arterioscler Thromb Vasc Biol. 1997;17(1):107–13. doi: 10.1161/01.atv.17.1.107. http://dx.doi.org/10.1161/01.ATV.17.1.107 . [DOI] [PubMed] [Google Scholar]
- [8].Boekholdt SM, Thompson JF. Natural genetic variation as a tool in understanding the role of CETP in lipid levels and disease. J Lipid Res. 2003;44(6):1080–93. doi: 10.1194/jlr.R200018-JLR200. http://dx.doi.org/10.1194/jlr.R200018-JLR200 . [DOI] [PubMed] [Google Scholar]
- [9].Inazu A, Koizumi J, Mabuchi H. Cholesteryl ester transfer protein and atherosclerosis. Curr Opin Lipidol. 2000;11(4):389–96. doi: 10.1097/00041433-200008000-00008. http://dx.doi.org/10.1097/00041433-200008000-00008 . [DOI] [PubMed] [Google Scholar]
- [10].Dachet C, Poirier O, Cambien F, Chapman J, Rouis M. New functional promoter polymorphism, CETP/-629, in cholesteryl ester transfer protein (CETP) gene related to CETP mass and high density lipoprotein cholesterol levels:role of Sp1/Sp3 in transcriptional regulation. Arterioscler Thromb Vasc Biol. 2000;20(2):507–15. doi: 10.1161/01.atv.20.2.507. http://dx.doi.org/10.1161/01.ATV.20.2.507 . [DOI] [PubMed] [Google Scholar]
- [11].Ghatreh Samani K, Noori M, Rohbani Nobar M, Hashemzadeh Chaleshtori M, Farrokhi E, Darabi Amin M. I405V and -629C/A polymorphisms of the cholesteryl ester transfer protein gene in patients with coronary artery disease. Iran Biomed J. 2009;13(2):103–8. [PubMed] [Google Scholar]
- [12].Ghatrehsamani K, Darabi M, Rahbani M, Hashemzadeh Chaleshtory M, Farrokhi E, Noori M. Combined hepatic lipase -514C/T and cholesteryl ester transfer protein I405V polymorphisms are associated with the risk of coronary artery disease. Genet Test Mol Biomarkers. 2009;13(6):809–15. doi: 10.1089/gtmb.2009.0080. http://dx.doi.org/10.1089/gtmb.2009.0080 . [DOI] [PubMed] [Google Scholar]
- [13].Thompson A, Di Angelantonio E, Sarwar N, Erqou S, Saleheen D, Dullaart RP, et al. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299(23):2777–88. doi: 10.1001/jama.299.23.2777. http://dx.doi.org/10.1001/jama.299.23.2777 . [DOI] [PubMed] [Google Scholar]
- [14].Wang J, Wang LJ, Zhong Y, Gu P, Shao JQ, Jiang SS, et al. CETP gene polymorphisms and risk of coronary atherosclerosis in a Chinese population. Lipids Health Dis. 2013;12:176. doi: 10.1186/1476-511X-12-176. http://dx.doi.org/10.1186/1476-511X-12-176 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang Q, Zhou SB, Wang LJ, Lei MM, Wang Y, Miao C, et al. Seven functional polymorphisms in the CETP gene and myocardial infarction risk:A meta-analysis and meta-regression. PLoS One. 2014;9(2):e88118. doi: 10.1371/journal.pone.0088118. http://dx.doi.org/10.1371/journal.pone.0088118 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gensini GG. A more meaningful scoring system for determining the severity of coronary heart disease. Am J Cardiol. 1983;51(3):606. doi: 10.1016/s0002-9149(83)80105-2. http://dx.doi.org/10.1016/S0002-9149(83)80105-2 . [DOI] [PubMed] [Google Scholar]
- [17].Boroumand M, Ghaedi M, Mohammadtaghvaei N, Pourgholi L, Anvari MS, Davoodi G, et al. Association of estrogen receptor alpha gene polymorphism with the presence of coronary artery disease documented by coronary angiography. Clin Biochem. 2009;42(9):835–9. doi: 10.1016/j.clinbiochem.2009.01.005. http://dx.doi.org/10.1016/j.clinbiochem.2009.01.005 . [DOI] [PubMed] [Google Scholar]
- [18].Boroumand M, Pourgholi L, Ziaee S, Anvari MS, Jalali A, Goodarzynejad H. The association between Factor V Leiden with the presence and severity of coronary artery disease. Clin Biochem. 2014;47(6):356–60. doi: 10.1016/j.clinbiochem.2013.12.006. http://dx.doi.org/10.1016/j.clinbiochem.2013.12.006 . [DOI] [PubMed] [Google Scholar]
- [19].Gordon T, Castelli WP, Hjortland MC, Kannel WB, Dawber TR. High density lipoprotein as a protective factor against coronary heart disease. The Framingham Study. Am J Med. 1977;62(5):707–14. doi: 10.1016/0002-9343(77)90874-9. http://dx.doi.org/10.1016/0002-9343(77)90874-9 . [DOI] [PubMed] [Google Scholar]
- [20].Nagano M, Yamashita S, Hirano K, Takano M, Maruyama T, Ishihara M, et al. Molecular mechanisms of cholesteryl ester transfer protein deficiency in Japanese. J Atheroscler Thromb. 2004;11(3):110–21. doi: 10.5551/jat.11.110. http://dx.doi.org/10.5551/jat.11.110 . [DOI] [PubMed] [Google Scholar]
- [21].Ridker PM, Pare G, Parker AN, Zee RY, Miletich JP, Chasman DI. Polymorphism in the CETP gene region, HDL cholesterol, and risk of future myocardial infarction:Genomewide analysis among 18 245 initially healthy women from the Women's genome health study. Circ Cardiovasc Genet. 2009;2(1):26–33. doi: 10.1161/CIRCGENETICS.108.817304. http://dx.doi.org/10.1161/CIRCGENETICS.108.817304 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Boekholdt SM, Sacks FM, Jukema JW, Shepherd J, Freeman DJ, McMahon AD, et al. Cholesteryl ester transfer protein TaqIB variant, high-density lipoprotein cholesterol levels, cardiovascular risk, and efficacy of pravastatin treatment:Individual patient meta-analysis of 13,677 subjects. Circulation. 2005;111(3):278–87. doi: 10.1161/01.CIR.0000153341.46271.40. http://dx.doi.org/10.1161/01.CIR.0000153341.46271.40 . [DOI] [PubMed] [Google Scholar]
- [23].Herrera VL, Makrides SC, Xie HX, Adari H, Krauss RM, Ryan US, et al. Spontaneous combined hyperlipidemia, coronary heart disease and decreased survival in Dahl salt-sensitive hypertensive rats transgenic for human cholesteryl ester transfer protein. Nat Med. 1999;5(12):1383–9. doi: 10.1038/70956. http://dx.doi.org/10.1038/70956 . [DOI] [PubMed] [Google Scholar]
- [24].Niu W, Qi Y. Circulating cholesteryl ester transfer protein and coronary heart disease:Mendelian randomization meta-analysis. Circ Cardiovasc Genet. 2015;8(1):114–21. doi: 10.1161/CIRCGENETICS.114.000748. http://dx.doi.org/10.1161/CIRCGENETICS.114.000748 . [DOI] [PubMed] [Google Scholar]
- [25].Thompson JF, Durham LK, Lira ME, Shear C, Milos PM. CETP polymorphisms associated with HDL cholesterol may differ from those associated with cardiovascular disease. Atherosclerosis. 2005;181(1):45–53. doi: 10.1016/j.atherosclerosis.2005.01.015. http://dx.doi.org/10.1016/j.atherosclerosis.2005.01.015 . [DOI] [PubMed] [Google Scholar]
- [26].Wu JH, Lee YT, Hsu HC, Hsieh LL. Influence of CETP gene variation on plasma lipid levels and coronary heart disease:A survey in Taiwan. Atherosclerosis. 2001;159(2):451–8. doi: 10.1016/s0021-9150(01)00524-x. http://dx.doi.org/10.1016/S0021-9150(01)00524-X . [DOI] [PubMed] [Google Scholar]
- [27].Bruce C, Sharp DS, Tall AR. Relationship of HDL and coronary heart disease to a common amino acid polymorphism in the cholesteryl ester transfer protein in men with and without hypertriglyceridemia. J Lipid Res. 1998;39(5):1071–8. [PubMed] [Google Scholar]
- [28].Isaacs A, Sayed-Tabatabaei FA, Hofman A, Oostra BA, Klungel OH, Maitland-Vander Zee AH, et al. The cholesteryl ester transfer protein I405V polymorphism is associated with increased high-density lipoprotein levels and decreased risk of myocardial infarction:The Rotterdam Study. Eur J Cardiovasc Prev Rehabil. 2007;14(3):419–21. doi: 10.1097/HJR.0b013e32801101aa. http://dx.doi.org/10.1097/HJR.0b013e32801101aa . [DOI] [PubMed] [Google Scholar]
- [29].Gudnason V, Kakko S, Nicaud V, Savolainen MJ, Kesäniemi YA, Tahvanainen E, et al. Cholesteryl ester transfer protein gene effect on CETP activity and plasma high-density lipoprotein in European populations. The EARS Group. Eur J Clin Invest. 1999;29(2):116–28. doi: 10.1046/j.1365-2362.1999.00412.x. http://dx.doi.org/10.1046/j.1365-2362.1999.00412.x . [DOI] [PubMed] [Google Scholar]
- [30].Blankenberg S, Rupprecht HJ, Bickel C, Jiang XC, Poirier O, Lackner KJ, et al. Common genetic variation of the cholesteryl ester transfer protein gene strongly predicts future cardiovascular death in patients with coronary artery disease. J Am Coll Cardiol. 2003;41(11):1983–9. doi: 10.1016/s0735-1097(03)00408-x. http://dx.doi.org/10.1016/S0735-1097(03)00408-X . [DOI] [PubMed] [Google Scholar]
- [31].Agerholm-Larsen B, Nordestgaard BG, Steffensen R, Jensen G, Tybjaerg-Hansen A. Elevated HDL cholesterol is a risk factor for ischemic heart disease in white women when caused by a common mutation in the cholesteryl ester transfer protein gene. Circulation. 2000;101(16):1907–12. doi: 10.1161/01.cir.101.16.1907. http://dx.doi.org/10.1161/01.CIR.101.16.1907 . [DOI] [PubMed] [Google Scholar]
- [32].Todur SP, Ashavaid TF. Association of CETP and LIPC gene polymorphisms with HDL and LDL sub-fraction levels in a Group of Indian Subjects:A cross-sectional study. Indian J Clin Biochem. 2013;28(2):116–23. doi: 10.1007/s12291-012-0259-y. http://dx.doi.org/10.1007/s12291-012-0259-y . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zende PD, Bankar MP, Momin AR, Kamble PS. Study of Cholesteryl Ester Transfer Protein (CETP) I405v Genotype and its association with lipid fractions in myocardial infarction patients:A case control study. J Clin Diagn Res. 2014;8(6):CC01–4. doi: 10.7860/JCDR/2014/7818.4441. http://dx.doi.org/10.7860/jcdr/2014/7818.4441 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Rejeb J, Omezzine A, Boumaiza I, Rebhi L, Rejeb NB, Nabli N, et al. Four polymorphisms of cholesteryl ester transfer protein gene and coronary stenosis in a Tunisian population. J Cardiovasc Med (Hagerstown) 2012;13(9):546–53. doi: 10.2459/JCM.0b013e3283569b24. http://dx.doi.org/10.2459/jcm.0b013e3283569b24 . [DOI] [PubMed] [Google Scholar]
- [35].Peloso GM, Demissie S, Collins D, Mirel DB, Gabriel SB, Cupples LA, et al. Common genetic variation in multiple metabolic pathways influences susceptibility to low HDL-cholesterol and coronary heart disease. J Lipid Res. 2010;51(12):3524–32. doi: 10.1194/jlr.P008268. http://dx.doi.org/10.1194/jlr.P008268 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Padmaja N, Kumar RM, Balachander J, Adithan C. Cholesteryl ester transfer protein TaqIB, -629C>A and I405V polymorphisms and risk of coronary heart disease in an Indian population. Clin Chim Acta. 2009;402(1-2):139–45. doi: 10.1016/j.cca.2008.12.041. http://dx.doi.org/10.1016/j.cca.2008.12.041 . [DOI] [PubMed] [Google Scholar]
- [37].Rissanen AM. Familial occurrence of coronary heart disease: Effect of age at diagnosis. Am J Cardiol. 1979;44(1):60–6. doi: 10.1016/0002-9149(79)90251-0. http://dx.doi.org/10.1016/0002-9149(79)90251-0 . [DOI] [PubMed] [Google Scholar]


