Abstract
Francisella tularensis is the cause of the zoonotic disease tularemia and is classified among highly pathogenic bacteria (HPB) due to its low infection dose and potential for airborne transmission. In the case of HBP, there is a pressing need for rapid, accurate and reliable identification. Phenotypic identification of Francisella species is inappropriate for clinical microbiology laboratories because it is time-consuming, hazardous and subject to variable interpretation. Matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) was recently evaluated as a useful tool for the rapid identification of a variety of microorganisms. In this study, we evaluated the use of MALDI-TOF MS for the rapid identification of Francisella tularensis and differentiation of its subspecies. Using national collection of Francisella isolates from the National Tularemia Reference Laboratory (Public Health Institution of Turkey, Ankara), a total of 75 clinical isolates were investigated by species and subspecies-specific polymerase chain reaction (PCR) test and MALDI-TOF MS. All isolates were originally identified as F. tularensis subsp. holarctica according to region of difference 1 (RD1) subspecies-specific PCR results. For all isolates MALDI-TOF MS provided results in concordance with subspecies-specific PCR analysis. Although PCR-based methods are effective in identifying Francisella species, they are labor-intensive and take longer periods of time to obtain the results when compared with MALDI-TOF MS. MALDI-TOF MS appeared to be a rapid, reliable and cost-effective identification technique for Francisella spp. Shorter analysis time and low cost make this an appealing new option in microbiology laboratories.
KEY WORDS: Francisella tularensis, tularemia, identification, MALDI-TOF MS, PCR
INTRODUCTION
Tularemia caused by Francisella tularensis, an intracellular obligate aerobic gram-negative coccobacillus, is a zoonosis with natural focality distributed in geographical regions of the Northern hemisphere. Its actual medical and veterinarian importance has been stressed over the last decade by a pronounced activation of natural foci, accompanied by epidemic occurrence of the disease in humans, as well as by fear of bioterrorism and misuse of F. tularensis as a potential biological weapon [1,2].
Tularemia in humans manifests in different ways depending to a large extent on the mode of transmission: arthropod bite, direct contact, ingestion or inhalation of the infectious agent. The clinical presentations of tularemia have been classically divided into six classic forms: ulceroglandular, glandular, oculoglandular, oropharyngeal, respiratory, and typhoidal tularemia [2-4]. However, overlapping of the different symptoms is frequently observed. The occurrence of several different clinical forms of tularemia that often initially presents with non-specific symptoms resembling influenza or other respiratory tract infections makes clinical diagnosis very difficult. Laboratory diagnosis is essential for optimum treatment. Since the mortality rate is high in most severe cases, early diagnosis of tularemia is crucial [3-6].
In Turkey, tularemia outbreaks have been described as early as 1936–1938, but tularemia was not reportable until 2004. Recently, multiple tularemia outbreaks in Turkey have been reported as it is now considered a re-emerging zoonotic disease in Turkey [7-9]. The only F. tularensis subspecies found in most of Eurasia, including Turkey, is holarctica. Genetic diversity is low, probably because it’s a recently emerging pathogen [10]. Laboratory diagnosis is usually based on the detection of bacteria either by culture or by nucleic acid amplification techniques and serology. Despite the fastidious characteristics of F. tularensis which requires sulfhydral compounds (cysteine or cystine) for optimal growth and its highly infectious nature causing laboratory-acquired infections, culture recovery and characterization remains the “gold standard” for laboratory confirmation of tularemia infections [4,5,11]. Conventional culture and biochemical tests, as well as molecular methods, have been used in clinical microbiology laboratories for the identification of Francisella spp. [2,4-6]. Species and subspecies are subsequently determined by testing biochemical reactions/tests such as production of acid from carbohydrates (maltose, d-glucose, lactose, sucrose, and glycerol), citrulline ureidase activity, H2S production in cysteine-supplemented medium, and β-lactamase production. These procedures are inappropriate for clinical microbiology laboratories because they are time-consuming, difficult to perform, and hazardous [11,12]. In the last decade, to decrease the possibility of misidentification and obtain more rapid identification and confirmation, molecular methods such as polymerase chain reaction (PCR)-based diagnostics and 16/23S ribosomal RNA sequencing have been considered as alternative approaches to the phenotypic methods [2,11].
Over the last few years, matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) which enables identification of bacteria by comparing the mass spectra that correspond to protein profiles of bacterial cells to reference spectra stored in the database has been introduced as a fast, reliable and cost-effective technique for routine application in clinical microbiology laboratories [13-15]. Even though the performance of MALDI-TOF MS and phenotypic systems in identifying Francisella isolates have previously been reported [11], information on the performance of the modern methods is scarce in identifying clinical isolates of Francisella species. Furthermore, challenging the MALDI-TOF MS with isolates from different geographical origins is needed to assess its usefulness as a universal method.
The aim of this study was to compare the performance of MALDI-TOF MS with subspecies-specific PCR method for the identification of Francisella species from Turkey.
MATERIALS AND METHODS
Bacterial isolates and reference strains
A total of 75 clinical Francisella strains isolated from humans were enrolled into the study. F. tularensis subsp. holarctica LVS (NCTC 10857) and F. tularensis subsp. tularensis SCHU S4 (FSC237) were included as reference strains. The isolates were carefully selected to ensure geographical diversity among the isolates and collected from different regions of Turkey during the period October 2009 and March 2012. All isolates were stored at −86°C and were subcultured on cysteine heart agar supplemented with 9% heated (chocolatised) sheep blood (CHAB) plates.
PCR method
DNA was extracted from pure cultures of F. tularensis strains grown on CHAB plates by using a commercial kit based on silica-gel-membrane technology (QIAamp DNA mini kit; QIAGEN GmbH, Hilden, Germany). Briefly, a loopful of bacteria were harvested and transferred to a 1.5 mL microcentrifuge tube, bacterial suspension was prepared by adding phosphate-buffered saline to a final volume of 200 μL and 20 μL QIAGEN protease was added to the suspension. The sample was mixed with 200 μL of buffer solution, pulse-vortexed for 15 s and left for incubation in water bath at 56°C for 10 min. After incubation 200 μL ethanol was added and the tube was pulse-vortexed for another 15 s. The mixture was carefully transferred to the QIAamp Mini spin column and centrifuged at 6000 × g for one min. The column was then washed two times with buffers to remove the filtrates. After addition of 200 μL of the elution buffer, the column was incubated at room temperature for five min and then centrifuged at 6000 × g for one min. The eluate obtained in this last step was stored at ‒20°C until PCR analysis. Affiliation to the species Francisella tularensis was confirmed by amplification of the 17 kDa outer membrane lipoprotein gene fragment (species-specific tul4 gene) as described previously [16]. For amplification of the tul4 gene the reaction mixture consisted of 5 μL of 10× PCR buffer, 5 μL MgCL2, 1 μL of dNTP mix (2 mM of each dNTP), 0.1 μL of each of forward and reverse primers (100 pmol/μL), 0.25 μL of Taq DNA polymerase, 1 μL BSA (1 mg/μL), 5 μL bacterial DNA template, and double-distilled water (32.55 μL) to a total reaction mixture volume of 50 μL. The primers used are shown in Table 1. After denaturation at 94°C for 4 min, 30 cycles of amplification were performed according to the following programme: denaturation at 94°C for 40 s, primer annealing at 64°C for 30 s, and primer extension at 72°C for 45 s. After the final extension step at 72°C for 5 min, each reaction mixture was subjected to gel electrophoresis in a 2% agarose gel for 50 min at 110 V, stained with ethidium bromide and the 428 bp fragment corresponding to tul4 gene was visualised by UV light. After confirmation of the isolates as F. tularensis by PCR with tul4 primers, another conventional PCR assay targeting the region of differentiation 1 (RD1) was performed in order to determine subspecies identification [17,18]. For amplification of the RD1 target, the same composition of the reaction mixture was prepared, except RD1 primers were included instead of tul4 primers. The primers used for the amplification of RD1 are shown in Table 1. The PCR steps included denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 50 s, primer annealing at 60°C for 40 s, and primer extension at 72°C for 80 s. After the final extension step at 72°C for 7 min, each reaction mixture was subjected to gel electrophoresis in a 1.5% agarose gel for 1 h at 80 V, and the DNA fragments were detected with ethidium bromide staining. In this PCR assay F. tularensis subsp. holarctica strains yield fragments of 924 bp, whereas F. tularensis subsp. tularensis strains yield fragments of 1522 bp which allowed differentiation at the subspecies level. In each PCR assay F. tularensis subsp. holarctica LVS (NCTC 10857) and F. tularensis subsp. tularensis SCHU S4 (FSC237) reference strains were used as positive controls and a mixture containing water instead of bacterial DNA was used as negative control.
TABLE 1.
Oligunucleotide sequences used for the PCR amplification of tul4 and RD1 targets

MALDI-TOF MS analysis
MALDI-TOF MS analyses in this study followed the protocols described previously [19]. The strains which were incubated for 48 hours at 35°C on CHAB were transferred into 1.5 mL screw cap tubes, suspended with 75% ethanol and incubated for 30 min at room temperature. After vortexing they were centrifuged at 13,000 × g for 5 min. The supernatant was discarded and the pellet was left at room temperature for drying. Afterwards, the pellet was mixed thoroughly with 50 µL of 70% aqueous formic acid. After addition of 50 µL of acetonitrile the mixture was centrifuged at 13,000 × g for 5 min and 1 µL of the microorganism extract supernatant was placed onto the polished steel MALDI target plate (Bruker Daltonics, Bremen, Germany) in duplicate and allowed to dry at room temperature. Each sample was overlaid with 1 µL of matrix solution which consisted of saturated a-cyano-4-hydroxycinnamic acid in 50% acetonitrile-2.5% trifluoroacetic acid and the plate was air dried at room temperature. The plate was then loaded into a Bruker Autoflex III MALDI-TOF mass spectrometer (Bruker Daltonics, Bremen, Germany) and analysis was performed. The spectra were automatically recorded in the linear positive ion mode with delayed extraction at a laser frequency of 20 Hz within a mass range from 2.000 to 20.000 Da. For each spectrum 600 satisfactory shots in 100-shot steps from the sampling area of the target spot were obtained. Spectra were eligible for further analysis when the peaks had a resolution better than 400 intensity a.u. (arbitrary units). Each run included a bacterial test standard with a characteristic peptide and protein profile, provided by Bruker for calibration, a negative extraction control (sterile water) and the reference QC strains (Escherichia coli ATCC 25922 and Staphylococcus aureus ATCC 29213). In case of failures the analyses were repeated using fresh colonies with the same method.
Data analysis
The mass spectra were evaluated with the MALDI Biotyper software version 3.0 (Bruker Daltonik, Bremen, Germany). The list of the best peaks of the spectrum are created automatically by the software after smoothing, normalization and baseline subtraction. The obtained spectra are analyzed by standard pattern matching algorithm, which compares the raw spectra with the spectra of the MALDI Biotyper Library (MBL) by using the standard settings. The results of MALDI Biotyper analysis are listed in a ranking table where the best match gives the identification result depending on its value. The results are expressed as log(score) values, which range from 0 (no spectra) to 3 (perfect match). Log(score) values were interpreted as recommended by the manufacturer: score values of ≥1.7 generally indicate a relationship at the genus level, and values of ≥2.0 generally indicate relationships at the species level. The highest score is used for species identification. However, at the time the study was performed there were no recorded reference F. tularensis strains in the standard MBL but only in a special “SR database” (security relevant database) which was not available in our laboratory. Therefore in-house generated spectral profiles of reference F. tularensis strains, F. tularensis subsp. holarctica LVS (NCTC 10857) and F. tularensis subsp. tularensis SCHU S4 (FSC237), were added to the MALDI Biotyper Support Library (MBSL) and used for the identification of 75 clinical Francisella tularensis isolates.
RESULTS
The PCR test targeting the tul4 gene which is common in Francisella tularensis species was found positive in all isolates investigated in this study (Figure 1). For the determination of the subspecies of Francisella tularensis strains, the region of difference 1 (RD1) subspecies-specific PCR test was employed (Figure 2). All study isolates (n=75) yielded RD1 fragments of 924 bp that corresponds to the RD1 size of F. tularensis subsp. holarctica. The mass spectra obtained by MALDI-TOF MS for the test strains yielded log(score) values ranging between 1.704 and 2.357 (mean 1.897) for matching against the reference strain F. tularensis subsp. holarctica LVS. Whereas the scores obtained for matching against the reference strain F. tularensis subsp. tularensis SCHU S4 were found to be substantially lower, ranging between 1.272 and 1.951 (mean 1.655). For all study isolates the Maldi Biotyper software yielded results for F. tularensis subsp. holarctica LVS as the first match organism which was followed by a second match with F. tularensis subsp. tularensis SCHU S4 with lower scores. Even though the obtained spectra visually looked very similar, the MALDI Biotyper software could correctly identify the study isolates as F. tularensis subsp. holarctica (sample spectral profiles for the study isolate F. tularensis subsp. holarctica TUL S-013 and the two reference strains are shown in Figure 3). Even though the first match organism was F. tularensis subsp. holarctica for all organisms, only 19 (25.3%) isolates received a score of >2.000 indicating reliable identification at the species level, the remaining 56 isolates (74.7%) received scores in the range 1.700 - 2.000 which translates as reliable identification at the genus level. Thus, using the cutoffs established by the manufacturer, MALDI-TOF MS-based identification of clinical F. tularensis isolates exhibited 100% concordance with the species-specific tul4 PCR assay, and 25.3% concordance with the RD1 PCR assay. The general performance of MALDI-TOF MS-based identification of clinical F. tularensis isolates is summarized in Table 2.
FIGURE 1.
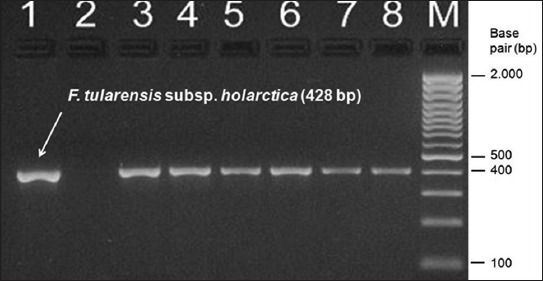
The PCR amplification results for the Francisella species using tul4 primers. Lane 1 is positive control strain F. tularensis subsp. holarctica LVS (NCTC 10857), lane 2 is negative control (water), lanes 3-8 are clinical Francisella strains. Molecular sizes in base pair (bp) are indicated at the right. (M: Molecular mass standard). This PCR assay was used to amplify a 428 bp fragment which is common in Francisella tularensis.
FIGURE 2.
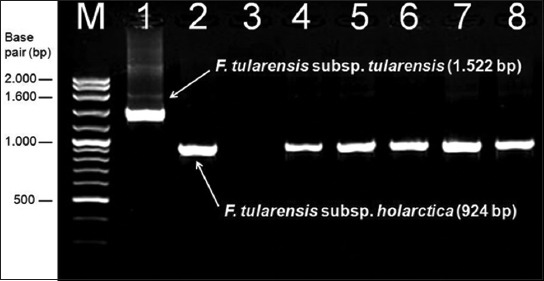
The PCR amplification results for the Francisella species using RD1 primers. Lane 1 and 2 are positive control strains F. tularensis subsp. tularensis SCHU S4 (FSC237) and F. tularensis subsp. holarctica LVS (NCTC 10857), respectively. Lane 3 is negative control (water), lanes 4-8 are clinical Francisella strains. Molecular sizes in base pair (bp) are indicated at the left. (M: Molecular mass standard) In this PCR assay F. tularensis subsp. tularensis yielded a 1.522 bp amplicon, whereas the size of the amplicon for F. tularensis subsp. holarctica was 924 bp which allowed the discrimination of the two subspecies.
FIGURE 3.
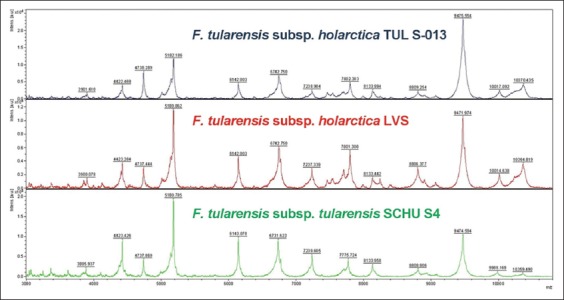
Spectral profiles obtained by Bruker MALDI-TOF MS for the clinical F. tularensis subsp. holarctica isolate TUL S-013 (top) and the reference strains F. tularensis subsp. holarctica LVS (middle) and F. tularensis subsp. tularensis SCHU S4 (bottom). All mass spectra were smoothed, baseline corrected and intensity normalized.
TABLE 2.
Performance of Bruker MALDI Biotyper software for the identification of F. tularensis subsp. holarctica clinical isolates (n=75)
Regarding the duration of each method, the processing of the isolates for MALDI-TOF MS analysis took approximately 1 to 2 h per isolate or per batch but analysis itself took less than 10 min for a single isolate and around 0.5 to 1 h when multiple isolates are processed. For the PCR assay however, it took 4–6 h to complete the runs and analyse the PCR products. When the required hands-on time and the risk of contamination associated with PCR method are considered, MALDI-TOF MS-based identification was found to be easier to perform.
DISCUSSION
The current approach for the identification of Francisella spp. is based mainly on standard slide agglutination and biochemical tests and molecular methods, including PCR, sequencing and hybridization assays using specific probes [4,5,11]. Conventional Francisella identification is both very time-consuming and sometimes inaccurate. Rapid and reliable identification of this agent is of major concern for optimal patient management and especially for the implementation of effective measures for disease control [2,4].
In this study, the most relevant diagnostic methods available in the field of clinical microbiology were compared for the identification of Francisella. All isolates were identified correctly with both PCR and MALDI-TOF MS system. Species-specific PCR and Bruker MALDI-TOF MS identified all isolates belonging to F. tularensis correctly. This result is in accordance with the findings of Seibold et al., who reported a correct identification rate of 100% for Francisella spp. by MALDI-TOF MS [11].
F. tularensis subsp. holarctica is the only Francisella subspecies which has been reported in the Eurasia region, including Turkey [8,9,20,21]. This has been linked to the low genetic diversity and relatively recent emergence of the pathogen in the region [10]. The findings of our study, together with the other reports from Turkey [9,21] have identified F. tularensis subsp. holarctica as the only subspecies present in Turkey.
The ability of MALDI-TOF MS to correctly identify proteomic differences was shown by carrying out comparative proteome analysis of cellular extracts obtained from the Francisella subsp. tularensis, mediaasiatica and holarctica successfully and diagnostic markers and putative factors of virulence were identified [22]. The same concept was further proven by using whole-cell MALDI-TOF MS spectra which identified three specific biomarkers, namely the histone-like protein HU form B, the 10 kDa chaperonin Cpn10, and the 50S ribosomal protein L24 that were found to be relatively conserved amongst the Francisella genus but enabling the distinction of subspecies owing to slight differences in their sequences [23].
Regarding the duration of analysis of both methods, MALDI-TOF MS was found to be more rapid and practical. In our study, analysis procedure of MALDI-TOF MS was completed approximately in one and a half hours with the routinely used extraction method including the preparation step for a single isolate.
On the aspect of costs, the cost of MALDI-TOF MS-based identification is roughly one-eighth of that of phenotypic identification and one-fifth of in-house PCR assay. In our laboratory setting, we calculated the cost of each method as $ 2.0 with MALDI-TOF MS, $ 9.5 with PCR assay including tests for tul4 and RD1, and $ 14-16 when following a conventional identification algorithm. The PCR assay does not require expensive equipment and therefore, in those laboratories where advanced instruments such as MALDI-TOF MS are not yet available and laboratories with small amounts of samples, the PCR assay used in the present study might still be a cost-effective method in the rapid identification of F. tularensis.
The main disadvantage of MALDI-TOF MS-based identification is that the lower limit of detection, when compared to PCR-based identification, is relatively high which requires using pure cultures grown on solid media. The analytical performance of the Bruker system was found to yield a detection limit of 9.0 × 103 to 1.3 × 105 bacteria per μL [24]. This high limit of detection renders direct testing of clinical samples generally not feasible. Among clinical specimens tested for feasibility, promising results were obtained by testing of positive blood cultures [25] and urine [26] owing to the high numbers of bacteria present in these clinical specimens. For monomicrobial urine samples, bacterial counts ≥1 × 107 bacteria/mL yielded successful results to allow direct identification from urine with 87.5% sensitivity [26]. Our study was conducted on isolates that were previously isolated from humans and stored in deep freezers, thus direct testing of clinical specimens by MALDI-TOF MS for the presence of Francisella spp. was not feasible in our set up. The evaluation study performed for the in-house PCR assay used in this study, however, revealed 100% sensitivity and specificity for human clinical specimens at a lower detection limit of 100 genomic equivalent [27].
Even though the studies conducted at single laboratories provide very promising results for the identification of Francisella spp. by MALDI-TOF MS, the results of an interlaboratory ring trial carried out with eleven laboratories in nine countries pointed out the need for a complete and comprehensive database with spectra from a broad strain collection for the accurate microbial identification [28]. In this trial F. tularensis subsp. holarctica along with 15 other bacteria were sent out to laboratories under blinded conditions. The initial MALDI-TOF MS analysis results obtained for the Francisella challenge strain yielded a score of 70% between the participating laboratories but when the mass spectra were collected at the study center (Robert Koch Institute, Berlin, Germany) and subsequently analysed using the own database of the institute, a score of 95% was obtained. The findings of the study indicate that for laboratories which are processing highly pathogenic bacteria on a regular basis, MALDI-TOF MS can serve as a rapid and reliable identification method, however the spectral databases should be supported with additional high-quality spectral entries to enable improved accuracy in identification.
CONCLUSION
In conclusion, identification of F. tularensis by MALDI-TOF MS exhibited good correlation with identification obtained by molecular methods. The PCR methods used in the present study are effective in identifying this fastidous microorganism. However, MALDI-TOF MS represents a rapid and reliable system that allows the identification of bacteria from colonies grown on agar culture plates in just a few minutes, with a very simple methodology and hardly any consumable costs. These advantages make MALDI-TOF MS a good candidate method for the identification of F. tularensis and its subspecies.
ACKNOWLEDGEMENTS
This study was approved by the Acibadem University Medical Research Assessment Committee (ATADEK Decision No. 2012-36).
DECLARATION OF INTERESTS
The authors declare that there are no conflicts of interest.
REFERENCES
- [1].Gurycova D. Epidemiologic characteristics of tularemia in Slovakia. Bratisl Lek Listy. 2006;107(5):224. [PubMed] [Google Scholar]
- [2].World Health Organization (WHO) Epidemic and Pandemic Alert and Response (2007) WHO guidelines on tularaemia. Geneva: WHO; [Accessed 22 November 2015]. http://www.who.int/csr/resources/publications/WHO_CDS_EPR_2007_7.pdf . [Google Scholar]
- [3].Sjöstedt A. Tularemia:history, epidemiology, pathogen physiology, and clinical manifestations. Ann N Y Acad Sci. 2007;1105:1–29. doi: 10.1196/annals.1409.009. http://dx.doi.org/10.1196/annals.1409.009 . [DOI] [PubMed] [Google Scholar]
- [4].Tärnvik A, Chu MC. New approaches to diagnosis and therapy of tularemia. Ann N Y Acad Sci. 2007;1105:378–404. doi: 10.1196/annals.1409.017. http://dx.doi.org/10.1196/annals.1409.017 . [DOI] [PubMed] [Google Scholar]
- [5].Splettstoesser WD, Tomaso H, Al Dahouk S, Neubauer H, Schuff-Werner P. Diagnostic procedures in tularaemia with special focus on molecular and immunological techniques. J Vet Med B Infect Dis Vet Public Health. 2005;52:249–261. doi: 10.1111/j.1439-0450.2005.00863.x. http://dx.doi.org/10.1111/j.1439-0450.2005.00863.x . [DOI] [PubMed] [Google Scholar]
- [6].Splettstoesser W, Guglielmo-Viret V, Seibold E, Thullier P. Evaluation of an immunochromatographic test for rapid and reliable serodiagnosis of human tularemia and detection of Francisella tularensis-specific antibodies in sera from different mammalian species. J Clin Microbiol. 2010;48(5):1629–1634. doi: 10.1128/JCM.01475-09. http://dx.doi.org/10.1128/JCM.01475-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gürcan Ş. Epidemiology of tularemia. Balkan Med J. 2014;31(1):3–10. doi: 10.5152/balkanmedj.2014.13117. http://dx.doi.org/10.5152/balkanmedj.2014.13117 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kılıç S. Tularemia:the pathogen and epidemiology. Turkiye Klinikleri J Inf Dis-Special Topics. 2014;7(2):52–61. [Google Scholar]
- [9].Kılıç S, Birdsell DN, Karagöz A, Çelebi B, Bakkaloğlu Z, Arıkan M, et al. Water as source of Francisella tularensis infection in humans, Turkey. Emerg Infect Dis. 2015;21(12):2213–2216. doi: 10.3201/eid2112.150634. http://dx.doi.org/10.3201/eid2112.150634 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vogler AJ, Birdsell D, Wagner DM, Keim P. An optimized, multiplexed multi-locus variable-number tandem repeat analysis system for genotyping Francisella tularensis. Lett Appl Microbiol. 2009;48(1):140–144. doi: 10.1111/j.1472-765X.2008.02484.x. http://dx.doi.org/10.1111/j.1472-765X.2008.02484.x . [DOI] [PubMed] [Google Scholar]
- [11].Seibold E, Maier T, Kostrzewa M, Zeman E, Splettstoesser W. Identification of Francisella tularensis by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry:fast, reliable, robust, and cost-effective differentiation on species and subspecies levels. J Clin Microbiol. 2010;48(4):1061–1069. doi: 10.1128/JCM.01953-09. http://dx.doi.org/10.1128/JCM.01953-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Sjöstedt A. In: Francisellae. Bergey's Manual of Systematic Bacteriology Volume 2:The Proteobacteria, Part B:The Gammaproteobacteria. 2nd edn. Garrity GB, Don J, Krieg NR, Staley JR, editors. New York: Springer US; 2005. pp. 199–210. [Google Scholar]
- [13].Carbonnelle E, Mesquita C, Bille E, Day N, Dauphin B, Beretti JL, et al. MALDI-TOF mass spectrometry tools for bacterial identification in clinical microbiology laboratory. Clin Biochem. 2011;44:104–109. doi: 10.1016/j.clinbiochem.2010.06.017. http://dx.doi.org/10.1016/j.clinbiochem.2010.06.017 . [DOI] [PubMed] [Google Scholar]
- [14].Croxatto A, Prod'hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012;36:380–407. doi: 10.1111/j.1574-6976.2011.00298.x. http://dx.doi.org/10.1111/j.1574-6976.2011.00298.x . [DOI] [PubMed] [Google Scholar]
- [15].Seng P, Rolain JM, Fournier PE, La Scola B, Drancourt M, Raoult D. MALDI-TOF-mass spectrometry applications in clinical microbiology. Future Microbiol. 2010;5(11):1733–1754. doi: 10.2217/fmb.10.127. http://dx.doi.org/10.2217/fmb.10.127 . [DOI] [PubMed] [Google Scholar]
- [16].Sjöstedt A, Eriksson U, Berglund L, Tärnvik A. Detection of Francisella tularensis in ulcers of patients with tularemia by PCR. J Clin Microbiol. 1997;35(5):1045–1048. doi: 10.1128/jcm.35.5.1045-1048.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Broekhuijsen M, Larsson P, Johansson A, Byström M, Eriksson U, Larsson E, et al. Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. tularensis. J Clin Microbiol. 2003;41(7):2924–2931. doi: 10.1128/JCM.41.7.2924-2931.2003. http://dx.doi.org/10.1128/JCM.41.7.2924-2931.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Huber B, Escudero R, Busse HJ, Seibold E, Scholz HC, Anda P, et al. Description of Francisella hispaniensis sp. nov., isolated from human blood, reclassification of Francisella novicida (Larson et al 1955) Olsufiev et al. 1959 as Francisella tularensis subsp. novicida comb. nov. and emended description of the genus Francisella. Int J Syst Evol Microbiol. 2010;60(Pt 8):1887–1896. doi: 10.1099/ijs.0.015941-0. http://dx.doi.org/10.1099/ijs.0.015941-0 . [DOI] [PubMed] [Google Scholar]
- [19].Mellmann A, Cloud J, Maier T, Keckevoet U, Ramminger I, Iwen P, et al. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry in comparison to 16S rRNA gene sequencing for species identification of nonfermenting bacteria. J Clin Microbiol. 2008;46(6):1946–1954. doi: 10.1128/JCM.00157-08. http://dx.doi.org/10.1128/JCM.00157-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kılıç S, Çelebi B, Acar B, Ataş M. In vitro susceptibility of isolates of Francisella tularensis from Turkey. Scand J Infect Dis. 2013;45(5):337–341. doi: 10.3109/00365548.2012.751125. http://dx.doi.org/10.3109/00365548.2012.751125 . [DOI] [PubMed] [Google Scholar]
- [21].Özsürekci Y, Birdsell DN, Çelik M, Karadağ-Öncel E, Johansson A, Forsman M, et al. Diverse Francisella tularensis strains and oropharyngeal tularemia, Turkey. Emerg Infect Dis. 2015;21(1):173–175. doi: 10.3201/eid2101.141087. http://dx.doi.org/10.3201/eid2101.141087 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Hubálek M, Hernychová L, Brychta M, Lenčo J, Zechovská J, Stulík J. Comparative proteome analysis of cellular proteins extracted from highly virulent Francisella tularensis ssp. tularensis and less virulent F. tularensis ssp. holarctica and F. tularensis ssp. mediaasiatica. Proteomics. 2004;4:3048–3060. doi: 10.1002/pmic.200400939. http://dx.doi.org/10.1002/pmic.200400939 . [DOI] [PubMed] [Google Scholar]
- [23].Durighello E, Bellanger L, Ezan E, Armengaud J. Proteogenomic biomarkers for identification of Francisella species and subspecies by MALDI-TOF mass spectrometry. Anal Chem. 2014;86(19):9394–9398. doi: 10.1021/ac501840g. http://dx.doi.org/10.1021/ac501840g . [DOI] [PubMed] [Google Scholar]
- [24].U.S. Food and Drug Administration. 510(k) Substantial Equivalence Determination Decision Summary for MALDI Biotyper CA System. [Accessed 18 December 2015]. http://www.accessdata.fda.gov/cdrh_docs/reviews/K130831.pdf .
- [25].Nonnemann B, Tvede M, Bjarnsholt T. Identification of pathogenic microorganisms directly from positive blood vials by matrix-assisted laser desorption/ionization time of flight mass spectrometry. APMIS. 2013;121(9):871–877. doi: 10.1111/apm.12050. http://dx.doi.org/10.1111/apm.12050 . [DOI] [PubMed] [Google Scholar]
- [26].March Roselló GA, Gutiérrez Rodríguez MP, de Lejarazu Leonardo RO, Orduña Domingo A, Bratos Pérez MA. Procedure for microbial identification based on matrix-assisted laser desorption/ionization-time of flight mass spectrometry from screening-positive urine samples. APMIS. 2014;122(9):790–795. doi: 10.1111/apm.12208. http://dx.doi.org/10.1111/apm.12208 . [DOI] [PubMed] [Google Scholar]
- [27].Çelebi B, Kılıç S, Yeşilyurt M, Acar B. Evaluation of a newly-developed ready-to-use commercial PCR kit for the molecular diagnosis of Francisella tularensis. Mikrobiyol Bul. 2014;48(1):135–142. [PubMed] [Google Scholar]
- [28].Lasch P, Wahab T, Weil S, PÁlyi B, Tomaso H, Zange S, et al. Identification of highly pathogenic microorganisms by matrix-assisted laser desorption ionization-time of flight mass spectrometry: results of an interlaboratory ring trial. J Clin Microbiol. 2015;53(8):2632–2640. doi: 10.1128/JCM.00813-15. http://dx.doi.org/10.1128/JCM.00813-15 . [DOI] [PMC free article] [PubMed] [Google Scholar]


