Abstract
Background & Aims
Very early onset inflammatory bowel disease (VEO-IBD), IBD diagnosed ≤5 y of age, frequently presents with a different and more severe phenotype than older-onset IBD. We investigated whether patients with VEO-IBD carry rare or novel variants in genes associated with immunodeficiencies that might contribute to disease development.
Methods
Patients with VEO-IBD and parents (when available) were recruited from the Children's Hospital of Philadelphia from March 2013 through July 2014. We analyzed DNA from 125 patients with VEO-IBD (ages 3 weeks to 4 y) and 19 parents, 4 of whom also had IBD. Exome capture was performed by Agilent SureSelect V4, and sequencing was performed using the Illumina HiSeq platform. Alignment to human genome GRCh37 was achieved followed by post-processing and variant calling. Following functional annotation, candidate variants were analyzed for change in protein function, minor allele frequency <0.1%, and scaled combined annotation dependent depletion scores ≤10. We focused on genes associated with primary immunodeficiencies and related pathways. An additional 210 exome samples from patients with pediatric IBD (n=45) or adult-onset Crohn's disease (n=20) and healthy individuals (controls, n=145) were obtained from the University of Kiel, Germany and used as control groups.
Results
Four-hundred genes and regions associated with primary immunodeficiency, covering approximately 6500 coding exons totaling > 1 Mbp of coding sequence, were selected from the whole exome data. Our analysis revealed novel and rare variants within these genes that could contribute to the development of VEO-IBD, including rare heterozygous missense variants in IL10RA and previously unidentified variants in MSH5 and CD19.
Conclusions
In an exome sequence analysis of patients with VEO-IBD and their parents, we identified variants in genes that regulate B- and T-cell functions and could contribute to pathogenesis. Our analysis could lead to the identification of previously unidentified IBD-associated variants.
Keywords: IBD, inherited defects, common variable immune deficiency, CVID, innate and adaptive immunity
Introduction
Inflammatory bowel disease (IBD) is a multigenic and environmentally triggered disease resulting in a dysregulated immune response. The genomic contribution of IBD has been extensively evaluated through genome wide association studies (GWAS), and over 163 IBD-associated risk loci1 have been identified. However, these studies were primarily performed in adult onset IBD and children 10 years of age and greater and thus did not include children with very early-onset IBD (VEO-IBD), diagnosed at less than 5 years. Furthermore, GWAS often do not capture rare variants, specifically those with minor allele frequency (MAF) less than 5%. VEO-IBD is a heterogeneous disease with different degrees of disease severity, including some children who have a relatively mild disease course.2 However, a subset of patients with VEO-IBD present with a distinct phenotype, including extensive colonic involvement and more severe disease than older children and adults. In addition, due to poor response to conventional therapies, severity of inflammation, and greater duration of disease, there are higher rates of morbidity in this population.3-5 There is currently no standard of care in the evaluation and treatment for this population. While IBD is a complex disease involving an environmental trigger, it is thought that the host genetics plays a more prominent role in this young population. In both mice 6-9 and humans, single gene defects are known to be associated with severe IBD phenotypes.5 For example, several IL-1010 and IL-10 receptor5 gene mutations have been associated with a phenotype of severe perianal disease and colitis in patients with VEO-IBD, particularly in infants.11,12 In general, single gene defects are hypothesized to be enriched in the VEO-IBD population.11,13,14
In addition to IL-10 defects, additional underlying immunodeficiency or genetic disorders have been associated with VEO-IBD.14,15 These include common variable immunodeficiency (CVID), Wiskott-Aldrich syndrome (WAS), Immunodysregulation, Polyendocrinopathy and Enteropathy, X-linked (IPEX), and chronic granulomatous disease (CGD) as well as many others.3 Identifying the driving forces in patients with particularly severe early-onset disease may lead to group-specific therapeutic approaches.
We hypothesized that rare or novel variants, including mutations in genes associated with primary immunodeficiencies, are enriched in patients with severe VEO-IBD and may contribute to the development of disease. Due to the lack of ability to detect rare variants using the GWAS approach, we utilized next generation sequencing technology in order to study specific genes or pathways involved in this disease process. Whole exome sequencing (WES) sequences protein encoding component of the genome and has revolutionized our ability to study rare variants and determining genetic basis of disease. Here we report our experience using WES to identify candidate causal variants in 125 patients with VEO-IBD. The focus of our analysis was on the genes associated with primary immunodeficiences and related pathways.
Materials and Methods
The Institutional Review Board at The Children's Hospital of Philadelphia (CHOP) approved the protocol, 2002-07-2805, and all parents of patients provided written informed consent. Patients with onset of IBD at 5 years of age and younger, and when available their parents, were recruited. This was an unselected cohort, with a heterogeneous disease presentation and severity. Patients with a previously identified immunodeficiency were excluded from the study. All probands had a confirmed diagnosis of IBD by standard methods, including endoscopy, radiologic, laboratory and clinical evaluation. Phenotypic classification was based on the Paris Classification (Table 1).16 Disease activity was quantified by the Pediatric Crohn's Disease Activity Index (PCDAI). Due to early age of onset of disease presentation, the majority of patients underwent an immunology evaluation for primary immunodeficiencies with a gastrointestinal presentation. The extent of this work up varied among subjects, and is described below (Table 1). Clinical information was obtained from the electronic medical records and the following information was extracted: date and age of diagnosis, disease classification and phenotype based on the Paris Classification, disease severity, surgical history, medication history and family history. After consent was obtained, blood samples were drawn from study subjects.
Table 1.
Patient Demographics and Baseline Immunophenotyping
| n | %† | |
|---|---|---|
| Demographics | ||
| Age in years (range) | (0.06-5) | - |
| Male | 69 | 55.2 |
| Disease Location | ||
| Perianal | 21 | 16.8 |
| Ileal | 7 | 5.6 |
| Ileocolonic | 37 | 29.6 |
| Colonic | 85 | 68 |
| Upper | 12 | 9.6 |
| Surgery & Therapy | ||
| Surgery | 17 | 13.6 |
| Diverted | 17 | 13.6 |
| TNF | 59 | 47.2 |
| Immunomodulators | 78 | 62.4 |
| Immunophenotyping | ||
| Immunoglobulins | 53 | 42.4 |
| Vaccine Titers | 46 | 36.8 |
| T Lymphocytes | 45 | 36 |
| B Lymphocytes | 41 | 32.8 |
| DHR | 52 | 41.6 |
| NK Cell Counts | 41 | 32.8 |
| TLR | 12 | 9.6 |
| FOXP3 | 6 | 4.8 |
| IL-10 | 6 | 4.8 |
| None | 56 | 44.8 |
Total sample size = 125
Whole exome sequencing
Exome capture was accomplished using the Agilent SureSelect V4, and sequencing was performed using the Illumina HiSeq platform at an average coverage depth of 100X. Whole exome library preparation was based on modification of the protocol using the Agilent SureSelect Whole Exome, version 4 kit (51 MB target size). Sequence read alignments were completed using Novoalign (V2.07.18) against the human reference genome GRCh37.p10 (http://www.novocraft.com). The Broad Institute's GATK17 best practices for variant detection were utilized for SNP and InDel calls. Annotation of the variants was performed with the use of SNPEff,18 using publicly available information from RefSeq (hg19) and Ensembl (GRCh37.66). Variants with sequence coverage of greater than or equal to 5 and variant quality score of greater than or equal to 10 were then loaded into CBMI's ‘Varify’ database (manuscript in preparation). Additional annotations at the variant, exon, and gene level were obtained using information from the 1000 Genomes Project (www.1000genomes.org/), NHLBI GO Exome Sequencing Project Exome Variant Server (EVS) (http://evs.gs.washington.edu/EVS/), Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org; release 0.3), SIFT,19 PolyPhen2,20 dbSNP,21 HGNC,22 HGMD ,23 CADD,24 and HPO . 25
Following functional annotation, only variants likely to alter protein function, such as missense and loss of function mutations, were kept for subsequent analysis. Variants were further filtered to include only those with a minor allele frequency less than 0.1% in data from the 1000 Genomes Project, EVS and ExAC. Variant frequencies were also checked against those reported in the Children's Hospital of Philadelphia's internal whole exome cohort (containing >400 exomes). We prioritized our focus on a panel of known genes associated with primary immunodeficiencies and related pathways. The list of primary immunodeficiencies associated genes and regions was generated from the NCBI gene database (http://www.mcbi.nlm.nih.gov/gene/; accessed on 2/23/14) using the terms “homo sapiens” and “primary immunodeficiency”, resulting in 400 genes and genomic regions. For each of these, we evaluated the number of probands that carry one or more variants and examined the parents, when available, for presence of the variant.
To identify candidate causative alleles, we utilized as a further filter criterion the Combined Annotation Dependent Depletion (CADD) score,24 and only included variants with scaled CADD of at least 10. The CADD system, a recently described annotation score, is unique in that it scores the deleteriousness of single nucleotide variants as well as insertions/deletions variants through integration of multiple networks and functions.24
Control Whole Exome Sequencing Data
An additional 210 exome samples from pediatric IBD (n=45), adult-onset Crohn's disease (n=20) and healthy controls (n=145) were obtained from the Institute of Clinical Molecular Biology, Christian-Albrechts-University of Kiel, Germany, and were used as control groups. Age of onset for pediatric IBD ranged from 6-18 years. The Illumina TruSeq exome kit was used for library preparation and sequencing was performed using the Illumina HiSeq2000. The same filtering criteria for identifying variants in primary immunodeficiency genes in our VEO-IBD data were applied to the different controls groups. Variants were limited to those with 1) a minor allele frequency less than 0.1% in the 1000 Genomes Project, EVS and ExAC, 2) a minimum CADD score of 10, and 3) a high or moderate effect determined using SNPEff. The total number of variants in the primary immunodeficiency genes satisfying these conditions was calculated for each of the three groups. The average sample variant rate was computed by dividing the total number of variants in a group by the number of samples in that group.
Permutation Bioinformatics Analysis
To assess the enrichment of primary immunodeficiency genes in our VEO-IBD data, we simulated the variant data in the healthy control group, where the sample size was larger than 125. A hundred variant matrices were simulated by randomly assigning the total number of variants observed across the samples in that group. We then randomly selected 125 samples from each of the generated matrixes, equal to the number of VEO-IBD patients in our study, repeating the sampling process 100 times for a total of 10,000 permutations for each dataset. The total number of variants observed in the 125 selected samples was calculated for every iteration and a normal distribution was fit into each dataset to calculate the significance of the number of variants in our cohort compared to the healthy control group.
Sanger sequencing
Sanger sequencing of purified PCR products was performed in the Applied Biosystems 3730 DNA Analyzer and analyzed using Sequencher (GeneCodes, Ann Arbor, MI). Potential novel variants and rare variants identified in this cohort were validated by resequencing in the forward and reverse directions.
IL10RA analysis
Biopsy samples were obtained from the terminal ileum, transverse colon and rectum during colonoscopy from the patient. Two punch biopsies were obtained and the lamina propria cells were isolated following a digestion protocol.26, 27 Peripheral blood was also obtained and peripheral blood mono-nuclear cells (PBMCs) were isolated on a Ficoll gradient (Ficoll-Paque Plus, GE Healthcare). Single cell suspensions were then viably cryopreserved until the day of the experiment. On the day of the experiment, cells were thawed and restimulated with 50 ng/ml PMA and 750 ng/ml Ionomycin in the presence of 10ug/ml of Brefeldin A (all obtained from Sigma-Aldrich) for 4 hours. Cells were then stained intracellularly for cytokines and transcription factors and analyzed on a flow cytometer. Cells were stained with antibodies to the following markers: anti-CD3 PE-Cy7 (clone UCHT1, eBioscience), and anti-CD4 PE-texas red (clone S3.5, Life Technologies). For intracellular staining, cells were fixed and permeabilized utilizing a commercially available kit (eBioscience) and stained with anti-FoxP3 eFluor 450 (clone 236A/E7, eBioscience), anti-TNFa Alexa 700 (clone Mab11, eBioscience) and anti-IFNg FITC (clone GZ-4, eBioscience).
For pSTAT3 analyses, T cells were expanded from PBMCs using anti-CD3 and anti-CD28 coated Dynabeads and recombinant IL-2, according to the manufacturers protocol for 7-10 days (Invitrogen). T cells were then rested for approximately 16 hours and stimulated with 200 ng/mL recombinant (r)IL-10 (eBioscience) for 15 minutes. Cells were then stained for pSTAT3 Y705 Alexa Fluor 647 (clone 4/P-STAT3) using BD Phosphoflow buffers.
For macrophage experiments, macrophages were isolated from PBMCs and stimulated with LPS as previously described28 in the presence or absence of rIL-10 200 ng/mL. Cells were analyzed by intracellular cytokine staining as described above.
Immunologic assessments
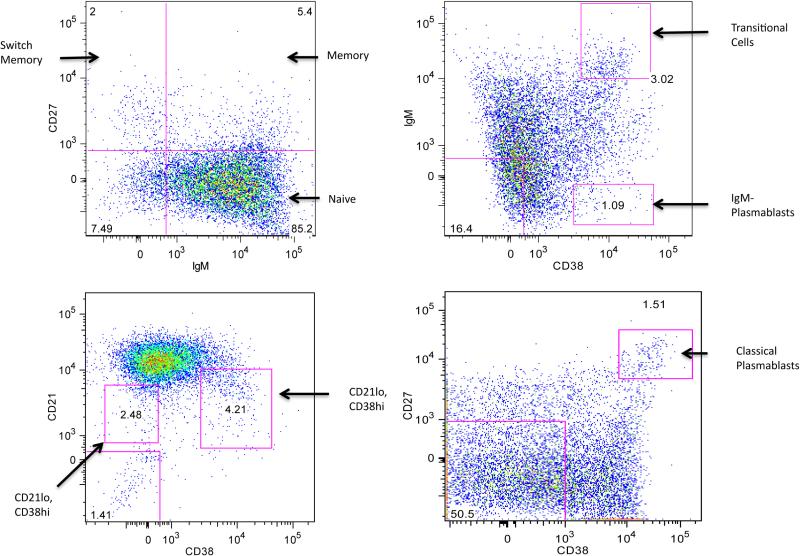
The extent of the immunonology evaluation at baseline was variable among the subjects as depicted in Table 1. Upon evaluation of identified variants, it was evident that many patients had variants related to lymphocyte function and we developed a screen to evaluate for evidence of lymphocyte dysfunction. We focused on B cells where low switched memory B cells are seen in many T cell defects, B cell defects and myeloid defects. A research protocol was developed to analyze these subsets. In addition, many patients had clinical testing of T cell subsets. B cells were identified by physical characteristics and CD19. B cell subsets were identified by additional antibodies: IgM, IgD, CD27, CD38, CD21. Cells were analyzed on a LSR Fortessa (BD Biosciences) in the CHOP Flow Cytometry Core. We defined transitional B cells as IgM+CD27−CD38++, naïve mature B cells as CD27−CD38+, non-switched memory B cells as CD27+IgM+, switched memory B cells as CD27+IgM− and naïve B cells as CD27−IgM+. Plasmablasts were defined as IgM− CD38++ or CD27++.
Results
A total of 125 patients with VEO-IBD were enrolled, including two sets of siblings. We also had access for this analysis, to DNA from 19 parents (both parents for 8 children). The range of age of onset of the affected children was from 3 weeks to 4 years, with the majority (21 or 84%) under 2 years of age. Briefly, 37 (29.6%) of patients had ileocolonic disease, and the remainder had primarily colonic distribution of disease. Disease activity was severe as measured by PCDAI or PUCAI in 76 patients (61%). Forty-six (37%) patients had been treated with anti-TNF Therapy, 76 with immunomodulator (61%). 14 patients had undergone colectomy 15 had diverting ileostomies and 2 had perianal disease requiring surgical intervention (Table 1). There was a positive family history in first-degree relatives in 13 patients (10%), and in second-degree relatives in 23 patients (18%).
Approximately 40% of patients had testing for chronic granulomatous disease with a DHR. A subset of patients had quantitative immunoglobulins (42%) and/or vaccine titers (37%), to identify CVID and immunoglobulin defects. In addition, B and T cell lymphocyte analysis was performed in 33% and 36% of the cohort, respectively, for the evaluation of SCID, WAS or specific B and T cell defects. Specific genetic defect analysis was performed on 6%. (Table 1). There were no identified genetic defects in any subject prior to enrollment in this study.
Analysis of variants in primary immunodeficiency pathways
Four hundred genes (Supplementary Table 2) and genomic regions associated with primary immunodeficiency covering approximately 6,500 coding exons totaling more than 1 Mbp of coding sequence. In these regions, 86.9% of coding exons were fully covered at more than 20x. Percentages of partially covered (between 40-99%) and not covered exons (<40%) were 7.4% and 5.7% respectively. Analysis revealed novel and rare putative causative variants within these genes in multiple patients as described below. Variants remained in this analysis if they were rare (MAF <0.1%) or novel in EVS, 1000 Genomes and ExAC. To meet criteria for potential pathogenicity, we only included variants that were predicted to be deleterious, by altering amino acids or splicing patterns, and had CADD scaled scores of at least 10. In addition, variants in HLA genes were removed due to the inherent variation in their sequence. After applying these filters to the initial 400 genes and regions associated with primary immunodeficiencies, 473 missense and 12 nonsense variants in 267 different genes fulfilled our filter criteria (Supplementary Table 1). There were 204 novel variants identified. All variants occurred in heterozygosity except for 4 homozygous variants found in three different patients. Of the 485 variants, 464 were detected only once each. Four variants, TINF2 Ala436Val, HIVEP3 Glu813Gln, ACOT8 Glu182Lys and RNASEH2A Phe139Leu, were each present in two affected siblings. Of the remaining variants, 13 were found in 2 unrelated patients and 2 in 3 unrelated patients. In addition, the MUC21 Gly386Arg and the ANKS1A Gly49Ser variants were seen in 5 and 8 unrelated patients, respectively. The number of variants per patient ranged from 1 to 17. In the 8 cases in which both parental samples were available, the variants were also detected in one of the parents for 7 out of the 8 children (all heterozygous except two). In the remaining case, all except for one variant were detected in one of the parents. However, reads containing the variant represented only 9% of all reads at that position and therefore the variant is likely to be a sequencing error. Of the 43 variants collectively found in these 8 patients, 5 variants in 3 children came from affected parents.
We also considered gene combinations, where the same two or more genes contained variants in multiple unrelated probands. We found 11 gene combinations (Supplementary Table 3), each harboring different variants found in 2 unrelated probands. Of the 11 combinations, 10 were gene pairs comprised of variants in two genes and one was a three-gene combination, with variants found in HIVEP3, TNXB and GNAI1. However, the significance of these gene combinations cannot be determined without further insight into the inheritance mode of the different variants.
Enrichment of Variants in Primary Immunodeficiency Genes in VEO-IBD
The same filtering criteria were applied to three different control groups: pediatric IBD (N=45), adult IBD (N=20), and healthy controls (N=145), to obtain a total count of variants in primary immunodeficiency genes in each of these cohorts (Table 2). The variant rate in our VEO-IBD cohort was 4.14 variants per child, compared to 3.69, 3.45 and 3.44 variants per sample in pediatric IBD, adult-onset IBD, and healthy controls, respectively. These results support the enrichment of variants in primary immunodeficiency genes in VEO-IBD, as there were 1.12-1.20 times more variants on average per child in our cohort, compared to the older IBD onset and healthy controls.
Table 2.
Summary of the primary immunodeficiency variants found in VEO-IBD and the three control groups.
| Cohort | n | Total # of Variants | Variant Rate | VEO-IBD/Control† | P-Value |
|---|---|---|---|---|---|
| VEO-IBD | 125 | 518 | 4.14 | n/a | n/a |
| Pediatric IBD | 45 | 166 | 3.69 | 1.12 | n/a |
| Adult-Onset IBD | 20 | 69 | 3.45 | 1.20 | n/a |
| Healthy Controls | 145 | 499 | 3.44 | 1.20 | < 1×10−4 |
Ratio of variant rate in VEO-IBD to controls
To further test our hypothesis, we randomly generated 100 datasets from the observed data in the healthy controls. For each iteration, we performed 100 random selections of 125 samples to calculate the expected total number of variants observed in a sample size equivalent to that of our VEO-IBD cohort. The resulting data followed a normal distribution with a mean (standard deviation) of 430.22(±7.74) in the healthy controls data. Comparing the 518 observed variants in our cohort with the expected number of variants in the healthy control groups resulted in a p-value less than 1×10−4, thus confirming the over-representation of variants in primary immunodeficiency genes in our cohort.
Functional analysis of IL10RA variant in terminal ileum biopsy
A known rare IL10RA missense variant (rs143538561; Arg412Trp) was detected in heterozygosity in a male patient born to non-consanguineous parents, who presented during the first few months of life with diarrhea. At 15 months of life he was diagnosed with CD after undergoing endoscopy and colonoscopy. His disease was classified as ileocolonic, characterized as inflammatory with multiple granulomas present throughout the gastrointestinal tract. By 2 years of age, he developed atopic dermatitis and keratosis that was difficult to control. This patient had a normal immunology evaluation, including dihydrorhodamine (DHR) study, T-cell function analysis, immunoglobulin analysis and appropriate vaccination titers. His intestinal disease course progressed and he proved to be refractory to multiple medical therapies, including 5-ASA, antibiotics, immunomodulators and infliximab, to which he developed an anaphylactic reaction at the second dose. This patient had a second variant in a gene of interest, IL21R. This was a novel non-synonymous coding variant (Val341Met).
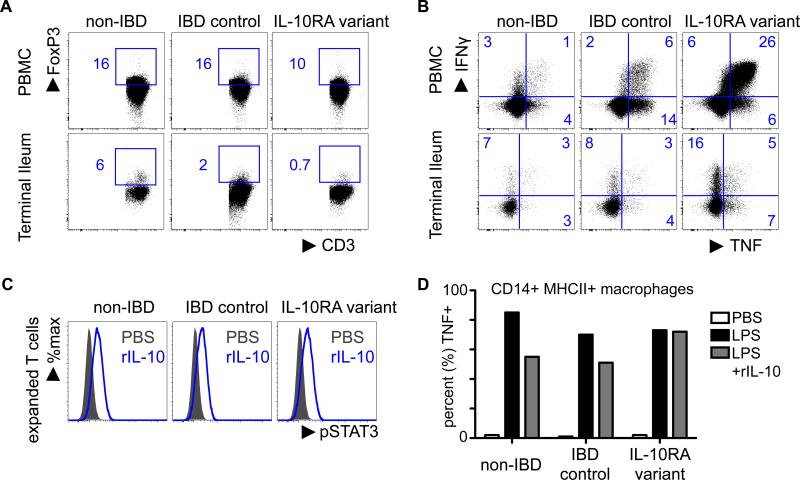
After Sanger sequencing was used to verify variant genotypes in the proband, we sought to determine the functional effect of the IL10RA variant. Analysis of immune cells in the PBMCs and terminal ileum biopsies through gating live, CD4+ CD3+ T-cells revealed a trend towards a reduction in FoxP3+ regulatory T cell frequency in both compartments of the patient as compared to a non-IBD healthy control and a patient with IBD (who lacked the variant) (Fig 1A).
Figure 1.
Preliminary functional analysis of immune cells in the IL-10RA variant patient via punch biopsies from terminal ileum and perhiperal blood. Cells were analyzed intracellularly for cytokines and transcription factors and analyzed on a flow cytometer. (A) Suggests a trend towards decreased expression of FoxP3 in CD4+ T cells and (B) suggest a trend toward an increase in pro-inflammatory cytokine expression in gated (live, CD3+, CD4+ T cells) in both compartments of the IL-10RA variant relative to a non-IBD control and a patient with IBD, but without the variant. (C) CD4 T cells were expanded and rested overnight. Cells were then stimulated with recombinant IL-10, and pSTAT3 (Y705) was analyzed by flow cytometry. (D) Macrophages-derived from blood were rested overnight, stimulated with LPS in the presence of absence of rIL-10, and analyzed for TNF production by flow cytometry.
Consistent with a potential reduction in FoxP3+ regulatory T cells, these analyses also revealed that there was an increase in pro-inflammatory cytokine production from CD4+ T cells as measured by an increased frequency of TNFα+ and IFNγ+ CD4+ T cells in the patient with the IL10RA variant as compared to a non-IBD healthy control and a patient with IBD, but without the variant (Fig 1B). Further analysis showed expanded T cells from all patients can phosphorylate STAT3 in response to IL-10 stimulation in vitro (Fig. 1C), suggesting that it is not a complete loss of function of the IL-10R. However, IL-10 was less efficient at reducing TNF production from LPS-stimulated, blood-derived macrophages in the patient with the IL-10R variant relative to control patients (Fig 1. D). These findings suggest that in the heterozygous state, there may not be complete loss of function, however there are still potential consequences on IL-10-IL-10R function, which is associated with reduced regulatory T cells, increased pro-inflammatory cytokine response and impaired regulation of macrophages. These studies are consistent with the recently identified role for macrophage-intrinsic IL-10R in regulating intestinal homeostasis. 29,30 Additional analyses will be required to determine the exact effects of the variant on immune cell function.
Analysis of variants in CVID associated genes MSH5 and CD19
The genes CD19 and MSH5 were previously identified to be involved in the common variable immunodeficiency (CVID) pathway, a disease whose main phenotype is loss of B cell function and humoral immunity.31
A rare non-synonymous variant in the MSH5 gene (rs370037482; Ser537Thr) was detected in heterozygosity in a 3 year old male subject with severe ileocolonic IBD and fistulizing perianal disease. Laboratory evaluation demonstrated low IgG and IgM, and low end of normal IgA and IgE.
A non-synonymous novel variant (Gln345His) in CD19 was detected in 1 female patient with VEO-IBD diagnosed at the age of 2. This variant is inherited from her unaffected father. This subject had low IgG, IgM and IgE levels.
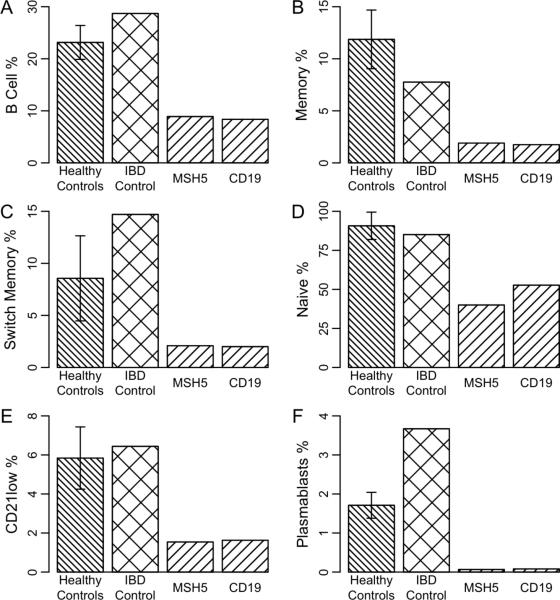
Following Sanger sequencing confirmation of the variants, we sought to investigate the B cell function in the patients with these variants utilizing the B cell assay described in Fig 2. Fig 3 demonstrates stages of B cell development of the patients with variants in MSH5 and CD19, also associated with CVID. There is a decrease in memory B cells (CD27+ / IgM+), switch memory cells (CD27+/IgM−) and plasmablasts (CD38hi/IgM−) cells. In addition, there is an increase in CD21lo/CD38lo, indicative of naïve transition cells, seen often in a stressed state. Together, these findings suggest that in a subset of patients with VEO-IBD, there may be potential effects on the B cell pathway, associated with B cell differentiation functional deficits. Additional analyses will be required to determine the exact effects of these variants in the CVID pathway on immune cell function.
Figure 2.
B Cells were identified by physical characteristics and CD19+. Subsets were identified by additional antibodies: IgM, IgD, CD27, CD38, CD21
Figure 3.
Analysis of (A) B cell function (CD19%), (B) memory B cells (CD27+ / IgM+), (C) switch memory cells (CD27+/IgM−), (D) naïve ransition cells (CD27−IgM+), (E) CD21lo/CD38lo, and (F) Plasmablasts (CD38hi/IgM) in subjects with variants in MSH5, CD19 as well as variant in LRBA, also in CVID pathway. Error bars represent the standard error.
Discussion
VEO-IBD in young children is challenging to both evaluate and treat. This is likely due to the unique genomic and immune triggers of the disease that do not respond to conventional approaches utilized in older-onset IBD. WES provides a method to identify some of the underlying genetic defects and may allow for targeted therapy in these children. We performed a candidate analysis using WES on 125 patients with VEO-IBD to detect causative variants, and subsequent functional analysis on 2 primary immunodeficiency pathways in 3 subjects. This was a heterogeneous unselected cohort, included in this analysis based on age of diagnosis alone. The variants we identified highlight the spectrum of immunological defects potentially relevant in this population, including T cell and regulatory pathways, as well as pathways involving B cell differentiation. Of interest, the relationship between IL-10R and CVID has been explored in several studies, specifically regarding the link between T cell deficiency and subsequent B cell function via impaired IL-10. 32
Prior studies have demonstrated monogenic defects in patients with VEO-IBD, including mutations identified in IL105, XIAP,33 NCF234, MEFV35 and LRBA.36 IL10RA (encoding IL10R1) homozygous mutations have been associated with VEO-IBD, specifically in neonatal onset disease with a phenotype of severe enterocolitis and perianal disease.5 In addition, compound heterozygote mutations of IL10RA have been reported with neonatal CD and enterocolitis.37 IL-10 is an anti-inflammatory cytokine secreted by a variety of cells, including dendritic cells, natural killer cells, eosinophils, mast cells, macrophages, B cells and CD4+ T cell subsets (including Th2, Th1 Th17 and Treg).38 IL-10 maintains homeostasis through suppression of an excessive pro-inflammatory response.39 IL-10 exerts its effect through binding to the tetrametric complex IL-10 receptor. It is composed of 2 distinct chains, 2 molecules of IL10R1 (α chain) and 2 molecules of IL10R2 (β chain). IL-10 binding to IL-10R activates the IL-10/JAK1/STAT3 cascade.40 STAT3 activates effector genes, which subsequently suppresses the pro-inflammatory genes. Our patient presented here was found to have a rare variant of IL10RA. Although he had similar phenotypic findings to IL10R defects depicted in the literature, such as disease onset in infancy, severe disease, and skin rashes, notable distinctions were found, particularly the absence of fistuslae and perianal disease. This novel patient illustrates that varying phenotypic expression may occur in children with very early-onset IBD who have heterozygous IL10R mutations.
Common variable immune deficiency (CVID), a primary immunodeficiency, is a complex and heterogeneous disease, with the responsible mutations known for only a minority of cases. The association between IBD and CVID has been well recognized. In a large cohort study of 473 patients with CVID, 20 developed IBD.41 A recent study demonstrated the importance of the CVID pathway in VEO-IBD, with the identification of IL-21 defect in 3 children in a consanguineous family. These patients had defects in B cell development.42 The variants detected in our subjects in genes associated with CVID included variants in CD19 and MSH5. The B cell analysis performed in our study provides further evidence that these CVID associated genes may be functionally relevant in VEO-IBD.
MSH5 belongs to the MutS family of proteins that are part of DNA mismatch repair and meiotic homologous recombination. There are seven known eukaryote MutS homologues, and MSH5 and MSH4 form an exclusive heterocomplex. This complex has been found to play a role in DNA damage response and double strand base repair, immunoglobulin diversity, and has been linked to neoplasia (including colorectal cancer), immune disease and reproductive disorders.43 MSH5 has also been associated with systemic lupus erythematosus, 44, Kawasaki disease, type 1 diabetes, IgA deficiency, and CVID.43,45,46 Its role in immunoglobulin diversity may be associated with the phenotype of VEO-IBD seen in our subject with the S554T variant, as it relates to B cell function.
The CD19 gene is located on the short arm of chromosome 16 (16p11.2).47 Like CD21, CD19 is a B-cell specific antigen. It is a member of B-cell receptor complex (BCR) together with CD21 and CD81.48,49 B cell development is dependent upon signal transduction through this complex. The complex also acts as a link between the innate and adaptive immune systems.48,49 Defects in CD19 demonstrate decrease in serum Ig secretion and defective response to T-cell-dependent antigens.50,51 Patients with CD19 mutations have been shown to have normal B cell development; however, with the loss of CD19 signal transduction, they have a poor response to antigenic stimuli and thus are unable to have effective humoral response.50 The CD19 variant in our patient may be important in the VEO-IBD phenotype due to B cell functional deficits.
These studies and our findings outlined above, demonstrate the importance of genetic overlap between disease processes. In fact, GWA studies that included multiple disease processes have identified risk variants that demonstrate significant overlap among different immune-related diseases.52 In addition, most complex disorders show a high degree of genetic heterogeneity, and this also seems to be the case in VEO-IBD. It is therefore likely that there are more pathways involved in VEO-IBD, and the outcome of these children can be improved through further study and identification of the associated variants. We identified 518 rare or novel non-synonymous variants in primary immunodeficiency pathways in our cohort of VEO-IBD. Patient presentation varied, as did severity of disease. The phenotypic variation in this population is likely secondary not only to the great genetic heterogeneity of disease causing variants but also to genetic modifiers of disease as well.
We recognize several limitations with this study. Because of the limited sample size, we focused on the known primary immunodeficiency pathways as they represent strong candidate genes for VEO-IBD. However, as IBD is a complex genetic disease, it is very likely that there are variants that affect genes and pathways that are equally important in this cohort, but have not yet been evaluated. Conversely, while we attempted to use strict filter criteria, we may determine as we proceed with further functional analyses that some of the variants we identified may indeed have no effect on gene function.
Despite these limitations, this mode of analysis allows us to begin to understand the complex genomics of VEO-IBD, and may help to identify the pathways that contribute to the disease process in this population. While it is now understood that an immunologic evaluation in children who present with IBD less than 5 years of age is critical, many of the patients in this study had limited or no immune work up prior to WES. As more is understood the about the etiology of disease in this population a systematic approach to evaluating the disease pathogenesis has been suggested.53 As we, and others, have shown, defects in both B and T cell development, migration and proliferation can drive disease in this population. Therefore, a framework to identify potential immune defects is an important part of the analysis and clinical care in these children. We therefore recommend a more complete immunologic evaluation be performed in patients with VEO-IBD, including studies depicted in Supplementary Table 3. As we begin to understand the different components of immune system involved, including B and T cell pathways, we can begin to individualize our therapy to the specific patient.
Supplementary Material
Abbreviations
- VEO-IBD
very early onset IBD
- WES
whole exome sequencing
- MAF
minor allele frequency
- CVID
common variable immunodeficiency
- EVS
Exome Variant Server
- CADD
Combined Annotation Score
- DHR
dihydrorhodamine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: None of the authors have disclosures related to this manuscript.
Author Contributions: All authors contributed to this study, in particular:
Study concept and design: Kelsen, Dawany, Piccoli, Mamula, Artis, Sonnenberg, Baldassano, Sullivan, Devoto Acquisition of data: Kelsen, Pauly-Hubbard, Martinez, Rappaport
Analysis and interpretation of data: Kelsen, Dawany, Petersen, Sarmady, Sasson, Rappaport, Artis, Sonnenberg, Sullivan, Baldassano, Devoto
Drafting of the manuscript: Kelsen, Dawany, Baldassano, Artis, Sonnenberg, Sullivan, Devoto
Critical Revision of Analysis of the manuscript for important intellectual content: Kelsen, Dawany, Moran, Sasson, Piccoli, Mamula, Artis, Sonnenberg, Daly, Sullivan, Baldassano, Devoto
Statistical analysis: Kelsen, Dawany, Sasson, Sarmady, Devoto
Technical or material support: Dawany, Maurer, Soong Sasson, Sarmady, Rappaport
Study Supervision: Kelsen, Baldassano, Devoto
References
- 1.Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–24. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benchimol EI, Mack DR, Nguyen GC, et al. Incidence, Outcomes, and Health Services Burden of Very Early Onset Inflammatory Bowel Disease. Gastroenterology. 2014 doi: 10.1053/j.gastro.2014.06.023. [DOI] [PubMed] [Google Scholar]
- 3.Glocker E, Grimbacher B. Inflammatory bowel disease: is it a primary immunodeficiency? Cell Mol Life Sci. 2012;69:41–8. doi: 10.1007/s00018-011-0837-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannioto Z, Berti I, Martelossi S, et al. IBD and IBD mimicking enterocolitis in children younger than 2 years of age. Eur J Pediatr. 2009;168:149–55. doi: 10.1007/s00431-008-0721-2. [DOI] [PubMed] [Google Scholar]
- 5.Glocker EO, Kotlarz D, Boztug K, et al. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–45. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bristol IJ, Farmer MA, Cong Y, et al. Heritable susceptibility for colitis in mice induced by IL-10 deficiency. Inflamm Bowel Dis. 2000;6:290–302. doi: 10.1002/ibd.3780060407. [DOI] [PubMed] [Google Scholar]
- 7.Mahler M, Leiter EH. Genetic and environmental context determines the course of colitis developing in IL-10-deficient mice. Inflamm Bowel Dis. 2002;8:347–55. doi: 10.1097/00054725-200209000-00006. [DOI] [PubMed] [Google Scholar]
- 8.Mahler M, Most C, Schmidtke S, et al. Genetics of colitis susceptibility in IL-10-deficient mice: backcross versus F2 results contrasted by principal component analysis. Genomics. 2002;80:274–82. doi: 10.1006/geno.2002.6840. [DOI] [PubMed] [Google Scholar]
- 9.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–74. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 10.Glocker EO, Frede N, Perro M, et al. Infant colitis--it's in the genes. Lancet. 2010;376:1272. doi: 10.1016/S0140-6736(10)61008-2. [DOI] [PubMed] [Google Scholar]
- 11.Begue B, Verdier J, Rieux-Laucat F, et al. Defective IL10 signaling defining a subgroup of patients with inflammatory bowel disease. Am J Gastroenterol. 2011;106:1544–55. doi: 10.1038/ajg.2011.112. [DOI] [PubMed] [Google Scholar]
- 12.Moran CJ, Walters TD, Guo CH, et al. IL-10R polymorphisms are associated with very-early-onset ulcerative colitis. Inflamm Bowel Dis. 2013;19:115–23. doi: 10.1002/ibd.22974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Ridder L, Weersma RK, Dijkstra G, et al. Genetic susceptibility has a more important role in pediatric-onset Crohn's disease than in adult-onset Crohn's disease. Inflammatory bowel diseases. 2007;13:1083–92. doi: 10.1002/ibd.20171. [DOI] [PubMed] [Google Scholar]
- 14.Biank V, Broeckel U, Kugathasan S. Pediatric inflammatory bowel disease: clinical and molecular genetics. Inflamm Bowel Dis. 2007;13:1430–8. doi: 10.1002/ibd.20213. [DOI] [PubMed] [Google Scholar]
- 15.Glocker EO, Kotlarz D, Klein C, Shah N, Grimbacher B. IL-10 and IL-10 receptor defects in humans. Ann N Y Acad Sci. 2011;1246:102–7. doi: 10.1111/j.1749-6632.2011.06339.x. [DOI] [PubMed] [Google Scholar]
- 16.Levine A, Griffiths A, Markowitz J, et al. Pediatric modification of the Montreal classification for inflammatory bowel disease: the Paris classification. Inflamm Bowel Dis. 2011;17:1314–21. doi: 10.1002/ibd.21493. [DOI] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cingolani P, Platts A, Wang le L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11:863–74. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0720s76. Chapter 7:Unit7 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–9. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenson PD, Ball EV, Mort M, et al. Human Gene Mutation Database (HGMD): 2003 update. Hum Mutat. 2003;21:577–81. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 24.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson PN, Mundlos S. The human phenotype ontology. Clin Genet. 2010;77:525–34. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 26.Hepworth MR, Fung TC, Masur SH, et al. Group 3 innate lymphoid cells mediate intestinal selection of commensal bacteria-specific CD4+ T cells. Science. 2015 doi: 10.1126/science.aaa4812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sonnenberg GF, Monticelli LA, Alenghat T, et al. Innate lymphoid cells promote anatomical containment of lymphoid-resident commensal bacteria. Science. 2012;336:1321–5. doi: 10.1126/science.1222551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ammann S, Elling R, Gyrd-Hansen M, et al. A new functional assay for the diagnosis of X-linked inhibitor of apoptosis (XIAP) deficiency. Clin Exp Immunol. 2014;176:394–400. doi: 10.1111/cei.12306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shouval DS, Biswas A, Goettel JA, et al. Interleukin-10 receptor signaling in innate immune cells regulates mucosal immune tolerance and anti-inflammatory macrophage function. Immunity. 2014;40:706–19. doi: 10.1016/j.immuni.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zigmond E, Bernshtein B, Friedlander G, et al. Macrophage-restricted interleukin-10 receptor deficiency, but not IL-10 deficiency, causes severe spontaneous colitis. Immunity. 2014;40:720–33. doi: 10.1016/j.immuni.2014.03.012. [DOI] [PubMed] [Google Scholar]
- 31.Park JH, Resnick ES, Cunningham-Rundles C. Perspectives on common variable immune deficiency. Ann N Y Acad Sci. 2011;1246:41–9. doi: 10.1111/j.1749-6632.2011.06338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm AM, Aukrust P, Aandahl EM, Muller F, Tasken K, Froland SS. Impaired secretion of IL-10 by T cells from patients with common variable immunodeficiency--involvement of protein kinase A type I. J Immunol. 2003;170:5772–7. doi: 10.4049/jimmunol.170.11.5772. [DOI] [PubMed] [Google Scholar]
- 33.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–62. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 34.Muise AM, Xu W, Guo CH, et al. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut. 2012;61:1028–35. doi: 10.1136/gutjnl-2011-300078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuloglu Z, Kansu A, Ustundag G, Birsin Ozcakar Z, Ensari A, Ekim M. An infant with severe refractory Crohn's disease and homozygous MEFV mutation who dramatically responded to colchicine. Rheumatol Int. 2012;32:783–5. doi: 10.1007/s00296-009-1326-4. [DOI] [PubMed] [Google Scholar]
- 36.Alangari A, Alsultan A, Adly N, et al. LPS-responsive beige-like anchor (LRBA) gene mutation in a family with inflammatory bowel disease and combined immunodeficiency. J Allergy Clin Immunol. 2012;130:481–8. e2. doi: 10.1016/j.jaci.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shim JO, Hwang S, Yang HR, et al. Interleukin-10 receptor mutations in children with neonatal-onset Crohn's disease and intractable ulcerating enterocolitis. Eur J Gastroenterol Hepatol. 2013;25:1235–40. doi: 10.1097/MEG.0b013e328361a4f9. [DOI] [PubMed] [Google Scholar]
- 38.Moore KW, de Waal Malefyt R, Coffman RL, O'Garra A. Interleukin-10 and the interleukin-10 receptor. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 39.Hutchins AP, Diez D, Miranda-Saavedra D. The IL-10/STAT3-mediated anti-inflammatory response: recent developments and future challenges. Brief Funct Genomics. 2013 doi: 10.1093/bfgp/elt028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murray PJ. The primary mechanism of the IL-10-regulated antiinflammatory response is to selectively inhibit transcription. Proc Natl Acad Sci U S A. 2005;102:8686–91. doi: 10.1073/pnas.0500419102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conley ME, Notarangelo LD, Etzioni A. Diagnostic criteria for primary immunodeficiencies. Representing PAGID (Pan-American Group for Immunodeficiency) and ESID (European Society for Immunodeficiencies). Clin Immunol. 1999;93:190–7. doi: 10.1006/clim.1999.4799. [DOI] [PubMed] [Google Scholar]
- 42.Salzer E, Kansu A, Sic H, et al. Early-onset inflammatory bowel disease and common variable immunodeficiency-like disease caused by IL-21 deficiency. J Allergy Clin Immunol. 2014;133:1651–9. e12. doi: 10.1016/j.jaci.2014.02.034. [DOI] [PubMed] [Google Scholar]
- 43.Clark N, Wu X, Her C. MutS Homologues hMSH4 and hMSH5: Genetic Variations, Functions, and Implications in Human Diseases. Curr Genomics. 2013;14:81–90. doi: 10.2174/1389202911314020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fernando MM, Freudenberg J, Lee A, et al. Transancestral mapping of the MHC region in systemic lupus erythematosus identifies new independent and interacting loci at MSH5, HLA-DPB1 and HLA-G. Ann Rheum Dis. 2012;71:777–84. doi: 10.1136/annrheumdis-2011-200808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lougaris V, Gallizzi R, Vitali M, et al. A novel compound heterozygous TACI mutation in an autosomal recessive common variable immunodeficiency (CVID) family. Hum Immunol. 2012;73:836–9. doi: 10.1016/j.humimm.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 46.Sekine H, Ferreira RC, Pan-Hammarstrom Q, et al. Role for Msh5 in the regulation of Ig class switch recombination. Proc Natl Acad Sci U S A. 2007;104:7193–8. doi: 10.1073/pnas.0700815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Zelm MC, Reisli I, van der Burg M, et al. An antibody-deficiency syndrome due to mutations in the CD19 gene. N Engl J Med. 2006;354:1901–12. doi: 10.1056/NEJMoa051568. [DOI] [PubMed] [Google Scholar]
- 48.Carter RH, Fearon DT. CD19: lowering the threshold for antigen receptor stimulation of B lymphocytes. Science. 1992;256:105–7. doi: 10.1126/science.1373518. [DOI] [PubMed] [Google Scholar]
- 49.Fearon DT, Carroll MC. Regulation of B lymphocyte responses to foreign and self-antigens by the CD19/CD21 complex. Annu Rev Immunol. 2000;18:393–422. doi: 10.1146/annurev.immunol.18.1.393. [DOI] [PubMed] [Google Scholar]
- 50.Bacchelli C, Buckridge S, Thrasher AJ, Gaspar HB. Translational mini-review series on immunodeficiency: molecular defects in common variable immunodeficiency. Clin Exp Immunol. 2007;149:401–9. doi: 10.1111/j.1365-2249.2007.03461.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yazdani R, Hakemi MG, Sherkat R, Homayouni V, Farahani R. Genetic defects and the role of helper T-cells in the pathogenesis of common variable immunodeficiency. Adv Biomed Res. 2014;3:2. doi: 10.4103/2277-9175.124627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Virgin HW, Todd JA. Metagenomics and personalized medicine. Cell. 2011;147:44–56. doi: 10.1016/j.cell.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Uhlig HH, Schwerd T, Koletzko S, et al. The diagnostic approach to monogenic very early onset inflammatory bowel disease. Gastroenterology. 2014;147:990–1007. e3. doi: 10.1053/j.gastro.2014.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.