Abstract
In 1973, Cohen and coworkers published a foundational paper describing the cloning of DNA fragments into plasmid vectors. In it, they used DNA segments made by digestion with restriction enzymes and joined these in vitro with DNA ligase. These methods established working recombinant DNA technology and enabled the immediate start of the biotechnology industry. Since then, “classical” recombinant DNA technology using restriction enzymes and DNA ligase has matured. At the same time, researchers have developed numerous ways to generate large, complex, multisegment DNA constructions that offer advantages over classical techniques. Here, we provide an overview of “post-Cohen-Boyer” techniques used for cloning single segments into vectors (T/A, Topo cloning, Gateway and Recombineering) and for multisegment DNA assembly (Biobricks, Golden Gate, Gibson, Yeast homologous recombination in vivo, and Ligase Cycling Reaction). We compare and contrast these methods and also discuss issues that researchers should consider before choosing a particular multisegment DNA assembly method.
Keywords: DNA cloning, multisegment DNA assembly, biotechnology, molecular biology, plasmid construction
A) Introduction
Both the first edition of Maniatis et al., Molecular Cloning and Current Protocols in Molecular Biology (CPMB) are consequences of the development of recombinant DNA methods in the 1970s. In 1975, a reasonable list of those methods would have included digestion of DNA with type II restriction endonucleases, purification of at least one DNA fragment after size separation by gel electrophoresis, cloning a copy of the fragment via joining its ends to the ends of a restriction cut plasmid or phage vector in vitro using DNA ligase, transformation of competent Escherichia coli cells with the ligation mix, selection of bacteria that contained plasmids by plating on selective medium, and screening cells in individual bacterial colonies for those that contained the cloned fragment on a self-replicating plasmid. In fact, that is precisely the set of steps in the pioneering paper by Stanley Cohen, Annie Chang, Herbert Boyer, and Robert Helling in 1973 (Cohen et al., 1973) and in the Cohen-Boyer patents (Hughes, 2001). In the vocabulary of the time, a self-replicating plasmid that carried such a fragment was a “chimera.” The double-stranded starting fragment carried on the plasmid that transformed the founding cell in the bacterial colony had been “molecularly cloned”.
The original paper by Cohen et al. described the molecular cloning of single DNA fragments by ligation in vitro. Overhanging or sticky 5′ ends on the DNA fragment (generated by digestion with EcoR1) annealed to complementary 5′ ends on the EcoR1-cut plasmid vector, and E. coli DNA ligase joined the gaps. In the following years this work was extended. E. coli DNA ligase was used to join multiple sticky-ended fragments to a sticky ended cut plasmid, allowing generation of plasmids that carried multiple inserted fragments from a single ligation reaction. Use of other type II restriction enzymes allowed generation of fragments with different 5′ overhangs, 3′ overhangs, and flush ends that could similarly be joined by ligase. Generation of DNA fragments by digestion with multiple enzymes allowed generation of DNA fragments with different ends whose ligation into multisegment stretches could be designed by the investigator. Plasmids that carried desired combinations of DNA fragments could be used as sources of larger DNA fragments for subsequent constructions. By that means, complex stretches of DNA could be assembled from different fragments iteratively, by serial cycles of plasmid construction and isolation. Increasingly, instead of speaking of their plasmids as “clones”, investigators referred to them as “constructs”.
These methods are now more than 40 years old. It’s a testimony to their power that many investigators still make DNA constructions by following the same steps. In fact, classical and improved methods to carry out each of these steps still make up much of the contents of CPMB. Among these, we single out improved means of purifying DNA from gels, which in turn greatly simplified the task of isolating ligatable DNA restriction fragments (unit 2.5A). We will also mention the development of PCR, which provided an alternative route to generating ligatable DNA segments (Unit 15.1 and Unit 3.17). Finally, we should mention the fact that commercially available high transformation efficiency competent E. coli cells have come into wide use (unit 1.8).
Commencing in the 1970s and 1980s, researchers also developed a number of powerful methods for generating recombinant DNA constructions that do not use the same steps as “classical” recombinant methods. Some of these “post-Cohen-Boyer” methods simplify the cloning of individual pieces of DNA, others simplify “assembly” of complex DNA constructs from multiple DNA segments. Many of these are described in individual protocols already included in CPMB. Here, we review key post-Cohen-Boyer methods and give pointers to protocols in CPMB as appropriate.
B) Means to generate single-segment-into-vector constructions
Single segment into vector cloning using prepared vectors
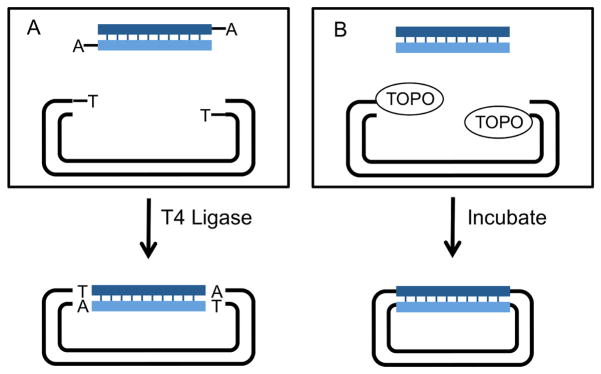
The original Cohen-Boyer methods inserted a single segment into a vector using DNA ligase. During the 1990s, investigators devised, and vendors began to sell, alternative in vitro means to insert single DNA segments into vectors (Figure 1). These include commercially available plasmid preparations whose free DNA ends carry an added 5′ dideoxythymidine added by terminal deoxynucleotydltransferase (TdT), facilitating their ligation in vitro to the 3′ adenines that Thermos aquaticus (Taq) polymerase leaves on PCR products (Holton and Graham, 1991). They also include commercially available cut plasmids with vaccinia virus DNA topoisomerase I immobilized on the ends, permitting the ligase-independent cloning of Taq polymerase PCR products with 3′As, and of blunt-ended PCR products generated by proofreading polymerases (Shuman, 1994).
Figure 1. Means to clone single segments into a plasmid in vitro.
A. TA cloning. A PCR product is generated with Taq polymerase, which adds a non-template-derived 3′ deoxyadenosine to DNA ends. TA cloning vectors are supplied by the manufacturer and have a 3′ deoxythymidine added to their ends by TdT. Annealing of the TA overhangs allows for efficient ligation by T4 DNA ligase. The resulting plasmid carries the PCR segment flanked by the additional A/T residues. Note that blunt-ended PCR products generated by proofreading enzymes can also be cloned by this method after a brief incubation with Taq polymerase
B. Topoisomerase cloning. Vaccinia virus DNA Topoisomerase I can cleave and rejoin ds DNA. Topo cloning vectors are supplied by the manufacturer with Topoisomerase I covalently linked to the linearized vector, allowing ligase free cloning of PCR products.
Single segment recombinational cloning using the Gateway system
While the aforementioned methods simplify the first step, the cloning of single DNA segments into plasmids, downstream steps requiring subcloning of the DNA fragments into expression vectors still requires the use of restriction enzymes and DNA ligases. To circumvent this need, methods now generically called “recombinational cloning” were developed and commercialized (Unit 3.20).
The Gateway system, a collection of plasmids and enzymes sold commercially by Invitrogen, is the prototype of such recombinational systems. While it can be used to clone a single segment into a vector (Figure 2A), its main use is to facilitate single-segment cloning of Open Reading Frames (ORFs) and then to simplify their subcloning into different types of expression vectors (Figure 2B).
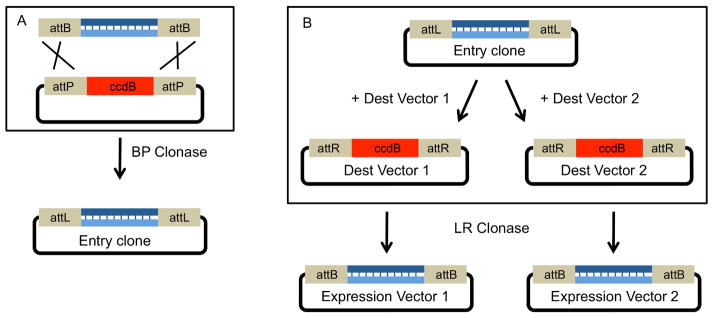
Figure 2. Single segment cloning using Gateway.
A. Single segment Gateway cloning. An attB-flanked, blunt-ended PCR product is recombined with an attP containing plasmid. The acceptor plasmid expresses the positive selection marker ccdB, a bacterial toxin that targets DNA gyrase (Bernard et al., 1994), so that only plasmids that have replaced this gene with the insert can replicate without killing their host. After a successful recombination facilitated by int and IHF (BP Clonase), the ccdB gene is replaced by the insert segment. The final plasmid carries the insert flanked by attL sites and is known as an Entry clone.
B. The insert from an Entry clone can be subcloned into multiple Destination vectors. In this example, the insert from the same Entry clone is subcloned into two distinct Destination vectors via an LR clonase reaction. Again, the ccdB positive selection marker is used. This flexibility allows a researcher to easily create many unique expression vectors from the same Entry clone.
The Gateway system uses the enzymes and DNA sites derived from the site-specific recombination system carried by bacteriophage λ. The phage λ system causes the integration of the phage genome into the E. coli chromosomal DNA during lysogeny and its excision from the genome after phage induction (Echols and Guaneros, 1983). During phage integration, the phage-encoded Int protein and bacterial encoded Integration Host Factor (IHF) bind to attachment sites on the circular phage genome (attP) and the E. coli chromosome (attB) and catalyze strand exchange between a 15bp core region common to the attP and attB sites (Landy, 1989). Because the sequences that flank the attP and attB cores are different, this process results in a genome-integrated prophage with, at its ends, 80–150bp hybrid sites called attL and attR. During phage excision, a second phage protein, Xis, acts with Int and IHF at these hybrid sites to excise the circular phage DNA.
The Gateway system reproduces this enzymology but with mutated attP and attB sequences. In Gateway, an attB1 site can only recombine with an attP1 site, and an attB2 site can only recombine with an attP2 site. The same is true for the sites these recombination events generate, that is, an attL1 site can only recombine with an attR1 and an attL2 site can only recombine with an attR2 site. By flanking cloned DNA segments with unique att sites, a researcher can easily subclone these segments into any compatible expression vector.
Figure 2A shows how the initial cloning works. The DNA fragment of interest is flanked with attB1 and attB2 sequences. Int and IHF proteins (supplied by the “BP Clonase mix”) are added in vitro to recombine the fragments into a Gateway compatible plasmid that contains attP1/attP2 sequences. The resulting plasmid has the DNA segment of interest flanked by attL1 and attL2. In the Gateway lexicon, this plasmid is called an “entry clone”.
The entry clone can be used in subsequent recombination reactions to create different expression vectors (Figure 2B). To do this, the entry clone is mixed with a “destination vector,” which carries attR1 and attR2 sites, and IHF, Int and Xis proteins (LR Clonase mix), which catalyzes the exchange of the cloned segment from the entry clone onto the destination vector. A good example of a destination vector is a plasmid that contains a promoter and a terminator sequence with attR1 and attR2 sites between them. Like in all Gateway assemblies, the final plasmid in this example will contain new att sites flanking the recombined segment, here, new attB1 and attB2 sites flanking the ORF. In the jargon that has emerged for DNA assembly, the attB1 and attB2 sequences between segments are called “scars.”
Researchers can purchase Gateway-compatible expression vectors or generate them in-house by adding compatible att sites to their own constructions (see Unit 3.20, figure 3.20.6). The Gateway system can then be used to subclone ORFs into different plasmids in parallel, for example to evaluate different protein expression constructs, or to generate families of constructs with the same promoter fused to different reporter genes. Single segment cloning using recombineering. Recombineering (unit 1.16) exploits the fact that appropriately engineered E. coli can carry out homologous recombination by double-strand break repair. In wild type E. coli, the RecBCD exonuclease degrades linear DNA. Strains used for recombineering circumvent the action of the exonuclease, allowing homologous recombination between overlapping, complementary single stranded DNA ends.
There are two different versions of recombineering, both based on bacteriophage-encoded recombination proteins. One, the Red system, uses the product of three bacteriophage λ genes: gam, which inhibits RecBCD, exo, a 5′ to 3′ exonuclease that chews back double stranded (ds) DNA to generate free 3′ ends, and beta, which binds to the new single stranded (ss) ends and promotes recombination. The other, the RecET system, uses two proteins encoded by a cryptic prophage, Rac, resident in the E. coli K-12 genome. The phage encoded RecE is a 5′ to 3′ exonuclease analogous to Exo, and RecT is a recombinogenic single stranded (ss) DNA binding protein analogous to Beta. These apparently act on double stranded (ds) DNA ends before RecBCD can degrade them, and convert the ds DNA into RecT coated, recombinogenic ss 3′ ends.
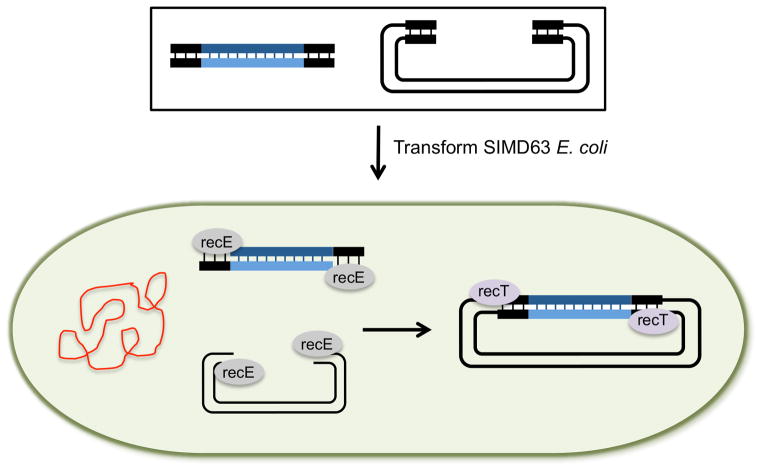
Current recombineering methods are not efficient enough for multisegment assemblies but do allow single segments to be cloned into a vector. For this, the RecET system (Fu et al., 2012) is the most efficient (Unit 1.16.19). Figure 3 shows the steps. Plasmid DNA and the insert segment are amplified as PCR products with 35–50bp of overlapping homologous sequence at each end. The segments are then transformed into an E. coli strain in which the recombinogenic proteins are expressed. This example uses SIMD63 cells, which harbor a defective λ prophage in which transcription of recE and recT is induced. After transformation, RecE chews back the 5′ ends of the insert DNA and the vector allowing RecT to promote annealing of the complementary ss 3′ ends. Recombinant plasmids are recovered from the recombineering strain and transformed into a standard E. coli strain for propagation (sometimes called plasmid rescue).
Figure 3. Single segment cloning into plasmids by recombineering.
Recombineering is a method that is based on homologous recombination of DNA within an E. coli cell. In this example, the starting segments (black box) are a linearized vector and a single PCR product, both generated by PCR with primers that contain 35–50bp of homologous sequence at each end. The starting segments are transformed into a recombineering E. coli strain such as SIMD63, in which the RecE and RecT recombination proteins were induced. recE, a 5′ to 3′ exonuclease chews back the 5′ ends of the plasmid and insert allowing the ss 3′ ends to be bound by recT. RecT, a single strand annealing protein, facilitates the annealing of the homologous ss 3′ ends. After recombination, the newly formed plasmid is purified (rescued) from the recombineering strain and transformed into a standard E. coli cloning strain for propagation.
C) Means to assemble complex, multisegment constructions
Current methods have greatly facilitated construction of complex molecules from multiple DNA segments. These methods either greatly simplify the serial addition of DNA segments into longer constructions, or allow the investigator to make desired constructions from multiple segments in a single reaction. In current usage, such complex multisegment constructs are often called “assemblies” and the process of making them is called “DNA assembly”.
Researchers normally plan the construction of DNA assemblies in two steps. Step one is the design and acquisition of starting single or double stranded DNA segments, and step two is the assembly itself.
Step 1, acquisition of starting segments
Double stranded sequences can come from standard PCR amplifications from plasmids or genomic DNA (Unit 15.1), from overlap extension PCR to generate chimeric segments (Holton and Graham, 1991; unit 3.17) and even from annealing and joining numerous single stranded oligonucleotides (Unit 3.23 and 3.24). They can also come from restriction fragments if those fragments can fulfill the assembly criteria. Finally, starting ds segments can come from commercially synthesized DNA. The most useful synthetic DNAs for assembly projects are short ds fragments or blocks between 100 and 1500bp that can be used as templates for PCR amplification.
In most cases, one of the segments will be a plasmid vector into which the other segments will be cloned. For most DNA assemblies, the vector will be linearized, either by restriction enzyme digestion or by PCR amplification.
Step 2, construction of multisegment assemblies
Here, we present and compare five key classes of methods now in common use. For each, we describe the important features and give a detailed example of at least one common method. Although there are few empirical studies that compare assembly methods, we give such data where it is exists. In these comparisons, we define DNA assembly efficiency as the number of bacterial clones from the final assembly transformation over background (usually, empty vector) and DNA assembly accuracy as the percentage of those clones that have the correct, sequence-confirmed assembly. The five classes are:
-
Assembly in vitro using the λ phage recombination system to carry out exact recombination between defined sites.
We use the Gateway system as an example of this class.
-
Assembly in vitro using restriction enzymes and DNA ligases, also known as “brick” or “parts” methods.
We use the Biobricks and Golden Gate methods as examples of this class.
-
Assembly in vitro of overlapping DNA fragments.
We use Gibson Assembly as an example of this class.
-
Assembly in vivo of overlapping DNA fragments by homologous recombination in yeast.
We use a construction by homologous recombination in S. cerevisiae as an example of this class.
-
Assembly in vitro using a thermostable ligase and bridge oligos.
We use implementations of the Ligase Cycling Reaction (LCR) as an example of this class.
1. Use of Gateway to generate multisegment assemblies
As explained in the previous section, Gateway uses phage proteins to promote recombination between att sites. Researchers can use the Gateway system to assemble two, three or four different DNA inserts on a single plasmid vector in a series of steps. An example of such a construction might be the assembly of a promoter, a gene, and a transcription terminator (called a “transcription unit” or “expression cassette”) into a compatible expression vector. The process for such serial constructions is similar to that described in the previous section for single segments, with the major differences being that each step requires the use of additional sequence-variant att sites as well as a second round of recombination reactions to generate the final multisegment construction.
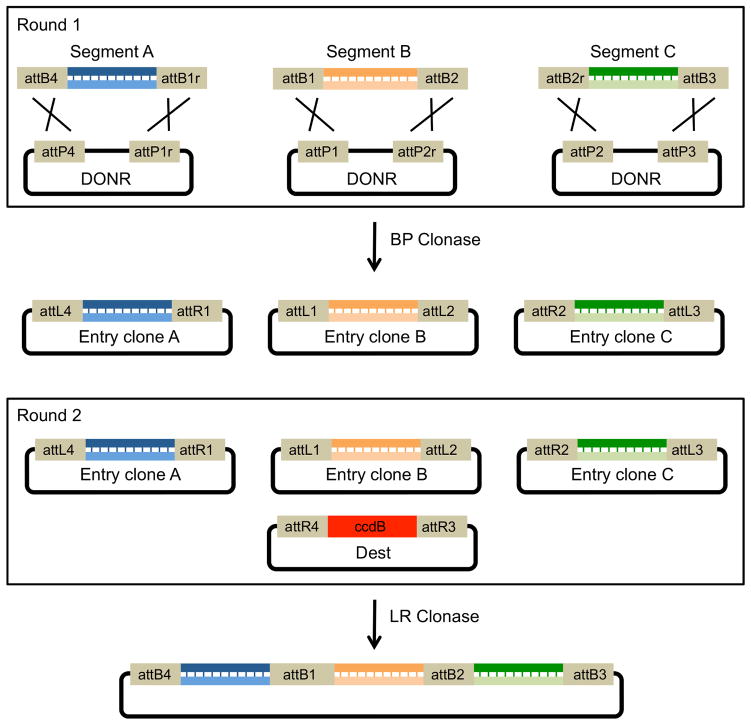
Figure 4 shows an example three-insert assembly using the Gateway system. In round 1, the investigator generates entry clones from att-sequence-flanked PCR products. In this example, segment A is flanked by attB4 and attB1r, segment B by attB1 and attB2 and segment C by attB2r and attB3. The investigator then mixes each PCR product with a “DONR” plasmid carrying corresponding attP-variant sequences along with Int and IHF (BP Clonase) to generate attL and attR entry clones. Once the entry clones are established, they, or a library of them, can be stored and used in subsequent LR recombination reactions to generate expression clones. In round 2, all three entry clones are mixed with a destination vector plus Int, IHF and Xis (LR Clonase), resulting in a final construction which contains all three DNA inserts separated by new attB scar sequences (Figure 5D).
Figure 4. Multiple segment assembly using Gateway.
Multisegment Gateway cloning works by the same means as single segment Gateway (see Figure 2) except that it requires additional sets of att sites of slightly different sequence. The att sites flanking each segment determine the order of the segments in the final recombined vector. In round 1, Entry clones are generated by flanking PCR products with unique attB sites and recombining them with vectors containing unique attP sites. The resulting entry clones can then be used to generate multisegment inserts. In round 2, Entry clones containing the different DNA segments flanked by attL and attR sites are recombined with an attR containing destination vector. The destination vector (Dest) also contains the ccdB positive selection marker (Bernard et al., 1994). After a successful LR recombination, the ccdB cassette is replaced by the full insert, which contains all three segments separated by attB scar sites (Figure 5D).
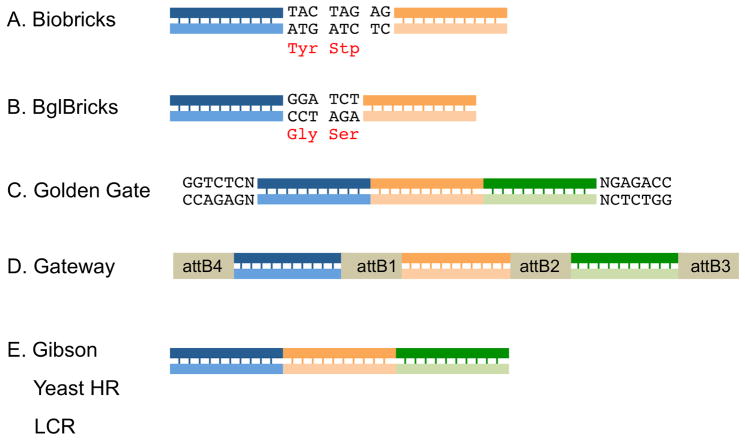
Figure 5. Scar sequences left by different multisegment assembly methods.
A) Biobricks assembly leaves an 8 nucleotide scar sequence between insert fragments. This scar codes for a tyrosine residue and a stop codon, limiting the utility of a Biobricks assembly for protein engineering. B) The BglBricks standard alleviates this constraint by replacing the isocaudomer pairs of SpeI and XbaI with BglII and BamH1. The 6 nucleotide scar sequence between inserts codes for Gly and Ser and keeps both inserts in-frame. C) “Quasiscarless” assembly of three segments by Golden Gate. All segments are seamlessly joined but the original BsaI recognition sequences flank the new multisegment insert. D) In Multisite Gateway assembly, the resulting expression clone has each insert flanked by att site scar sequences. E) Gibson assembly, yeast HR and LCR are all scarless assembly methods; all segments are joined seamlessly and there are no new sequences flanking the inserts.
2. Assembly in vitro from parts or bricks using restriction enzymes and ligases
During the 1990s and 2000s, engineers interested in development of “synthetic biology” devised schemes that used specific combinations of restriction enzymes in conjunction with DNA ligases, sometimes within the same reaction mix, to assemble DNA fragments in a desired order. One of the drivers for this development was a desire among engineers to bring engineering principles to construction of DNA molecules. One tenet of engineering embraced by these groups was to articulate standards for building blocks and to suggest as common practice that constructions be made from such collections of standardized parts (Canton et al., 2008; Endy, 2005). Tom Knight articulated one such approach, called “Biobricks”, in 2003 (Knight, 2003). This and other brick methods are most useful when all of the parts are preassembled into compatible starting or entry plasmids.
The Biobricks approach, and many subsequent approaches based on DNA “bricks” or “parts,” uses restriction enzymes that are “isocaudomers,” (from Greek roots meaning “same tails”): restriction enzymes that generate complementary overhangs but which have different recognition sequences. When isocaudomer ends are ligated, the starting restriction enzyme recognition sequences are destroyed. An example isocaudomer pair, and the one described in the first Biobrick standard, is Xba1 (T^CTAGA) and Spe1 (A^CTAGT) which both leave CTAG tails after digestion.
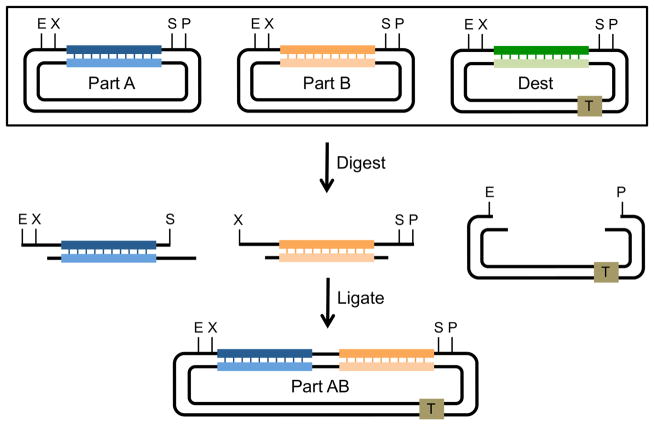
Figure 6 illustrates how Biobricks can be used to assemble two different DNA segments carried on separate starting plasmids into a destination plasmid. Part A, the upstream segment, is generated by cutting one starting plasmid with EcoR1 and Spe1. Part B, the downstream segment, is generated by cutting another starting plasmid with Xba1 and Pst1. The destination plasmid (Dest) is digested with EcoR1 and Pst1. The digestion reactions are then mixed with T4 DNA ligase and then transformed into competent E. coli. If fragments A and B and the destination vector are gel purified before ligation, then only one product will be formed, a correctly assembled plasmid with parts A and B, separated by an 8nucleotide scar sequence, ligated between the EcoR1 and Pst1 sites of the destination vector. Because the same original restriction sites are flanking the new fused part AB, the new segment can be used in subsequent Biobrick assemblies.
Figure 6. Multiple segment assembly using Biobricks.
Each segment or “part” for a Biobrick assembly is stored on a separate plasmid with each one flanked by restriction enzyme recognition sites EcoRI (E), XbaI (X), SpeI (S) and PstI (P). The destination vector contains the same restriction enzyme recognition sites as well as any additional elements desired for the construction (in this case, a transcription terminator, square T). Part A, the upstream segment, is digested with EcoR1 and SpeI while Part B, the downstream segment, is digested with XbaI and PstI. The destination vector (Dest) is digested with EcoRI and PstI. Insert segments and the vector are ligated by T4 ligase in a three-way ligation reaction. The resulting plasmid contains both segments in the predetermined order and separated by a scar sequence (Figure 5A). Because SpeI and XbaI are isocaudomers, the sites for both of these are destroyed when the two insert segments are ligated, whereas the sites flanking the full insert are regenerated. The new Part AB insert can then be used in future Biobrick constructions.
To save time, an investigator can mix unpurified digestion reactions with T4 ligase and then use the resulting mix of ligated DNAs to transform competent E. coli. By making sure that all starting plasmids have different bacterial selection markers, and plating the final assembly-transformed bacteria onto the selective media that corresponds to the destination vector marker, only a properly assembled plasmid will be recovered. This approach is now known as three antibiotic or “3A” assembly (Shetty et al., 2011).
As mentioned above, fragments joined at SpeI/XbaI sticky ends are separated by 8 nt scars (Figure 5A). Their sequence means that protein coding sequences on joined segments will be out of frame. In order to address this limitation, numerous additional Biobrick standards have been proposed. One standard, proposed by Anderson et al. (BBF_RFC21, known as “Bglbricks”) addresses the coding sequence issue by changing the isocaudomers Spe1 and Xba1 to BglII and BamH1 (Anderson et al., 2010). As shown in Figure 5B, ligation of BglII and BamHI ends leaves a GGATCT scar between parts A and B. This leaves the fragments in-frame, with the amino acids in the resulting protein separated by Gly and Ser, amino acids frequently used in flexible linkers for protein engineering.
A second brick-based method called Golden Gate also addresses some of the limitations of the other Biobrick standards (Engler et al., 2008). It also requires the use of restriction enzymes to generate complementary or sticky ends. However, it utilizes type IIs restriction enzymes which cut at sites outside their recognition sequences. Examples of frequently used type IIs enzymes are BsaI (GGTCTCN^NNNN) and BpiI (GAAGACNN^NNNN).
The advantage of using type IIs enzymes is that the same enzyme can be used to generate hundreds of investigator-designed unique overhangs (in the case of BsaI, 256 unique 4bp overhangs). With careful attention to design of the overhangs, DNA assembly can be quasi-seamless, that is, ligation can take place without scar sequences between ligated segments but the restriction enzyme sites will remain flanking the full insert (Figure 5C).
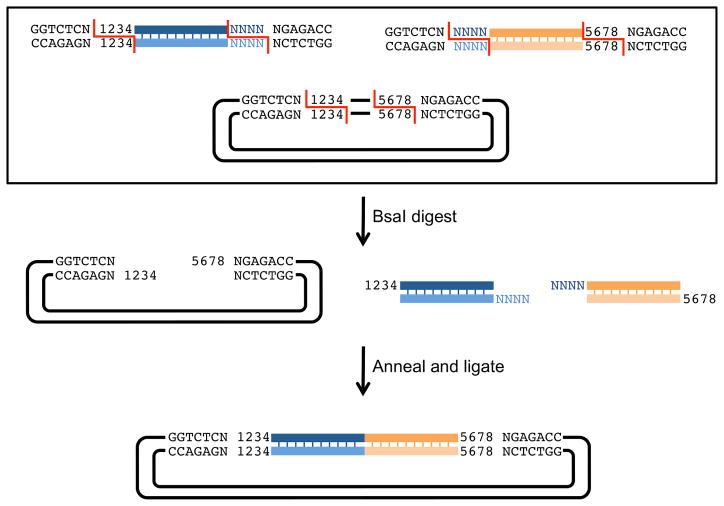
Figure 7 shows a quasi-seamless assembly of 2 PCR segments into a vector by Golden Gate. Segments are generated by PCR using primers that add specific sequences at 5′ and 3′ ends including BsaI recognition sequences. Part A has at its 5′ end a BsaI recognition site followed by any nucleotide (N) and the complement of the 4bp 3′ overhang of the vector (1234), and at its 3′ end, any nucleotide (N) followed by an inverted BsaI site. The last 4 nucleotides of part A are indicated (NNNN). Part B has at its 5′ end a BsaI recognition site followed by any nucleotide (N) and the complement of the 4bp 3′ overhang of segment 1 (NNNN), and at its 3′ end, the complement of the 4bp 3′ overhang of the vector (black 5678) followed by any nucleotide (N) and an inverted BsaI recognition site. The destination vector contains inverted BsaI recognition sequences separated by any number of nucleotides to be removed after BsaI digestion. After BsaI digestion, the 4bp sticky ends anneal and are ligated by T4 DNA ligase. The final construction contains parts A and B seamlessly ligated together between the predetermined nucleotides of the vector and flanked by inverted BsaI recognition sites.
Figure 7. Multiple segment assembly using Golden Gate.
Golden Gate starting segments can be carried on separate plasmids or generated by PCR. In this example, PCR generated inserts contain flanking sequences that allow for quasi-seamless cloning of two inserts into a vector. The upstream insert has, at its 5′ end, a BsaI recognition sequence (GGTCTCN) followed by 4 nucleotides homologous to the vector (1234), and at its 3′ end, the last 4 nucleotides of the upstream insert (shown as NNNN) followed by an inverted BsaI recognition site (NGAGACC). The downstream insert has, at its 5′ end, a BsaI recognition sequence (GGTCTCN) followed by 4 nucleotides homologous to the upstream insert (NNNN), and at its 3′ end, the last 4 nucleotides of the vector (5678) followed by an inverted BsaI recognition site (NGAGACC). The vector is linearized by digestion with BsaI. After the insert segments and vector are digested, the unique 4 nucleotide overhangs anneal and are ligated by T4 ligase. The resulting plasmid has both inserts ligated without scars and flanked by BsaI recognition sites. This new assembly can be used in future constructions by the same means.
3. Assembly in vitro by annealing of complementary single stranded ends
In 1990, Aslanidis and De Jong demonstrated it was possible to insert PCR products into a plasmid vector without using restriction enzymes or DNA ligase (Aslanidis and de Jong, 1990). The key to what the authors called “ligation independent cloning” or LIC was to generate complementary ss ends in a PCR product and in a plasmid vector by treatment with T4 DNA polymerase and allow the complementary ends to anneal before transformation into E. coli. In the absence of dNTPs, T4 DNA polymerase has a 3′ to 5′ exonuclease activity. In the presence of a single dNTP (for example, dGTP) the enzyme will chew back the 3′ end until it reaches the first G residue.
In their original paper, the authors generated both the insert and plasmid vector segments by PCR, using primers whose 5′ ends lacked Cs. Next, they digested each segment with T4 polymerase in the presence of a single dNTP, the insert with dGTP and the vector with dCTP, resulting in 12 bp complementary 5′ tails which could anneal. They introduced the annealing mix into competent E. coli, where the nicked circular molecule was repaired and replicated.
Since 1990, many similar methods have been developed. All generate complementary ss ends and all anneal the tails. They differ in the means by which they generate ss overhangs and in the extent to which they repair the circular molecule before transformation into E. coli. For example, Li and Elledge demonstrated a method, sequence and ligation independent cloning (SLIC), that generates overhangs with T4 DNA polymerase but without the control of the digestion provided by the addition of the single dNTP to the reaction mix (Li and Elledge, 2007). Instead, a short T4 polymerase digestion of the 20 to 40 bp of overlapping sequence between DNA segments generates sufficient free 5′ tails for annealing to occur. The investigators showed SLIC could be used to assemble five-segment constructions (4 inserts of 4kbp total insert size into a vector) with high efficiency and 100% accuracy. However, for ten-segment assemblies (nine inserts of 4.8kbp total insert size into a vector), the accuracy dropped to ~20% (Li and Elledge, 2007). This is consistent with (Hill and Eaton-Rye, 2014) where the practical limit of SLIC has been described as a six-segment assembly.
The most commonly used method of this type is Gibson assembly (unit 3.11). Gibson assembly was used to construct 144kbp fragments of the Mycoplasma genitalium genome (Gibson et al., 2008), which were in turn assembled in yeast into larger constructions that contained the entire bacterial genome. This method gained some prominence from the press coverage it received because prior to its publication, the largest DNA ever assembled was 32kbp (Kodumal et al., 2004). The current, optimized method consisting of a single isothermal reaction step has been used to generate synthetic DNAs of 500kbp and to clone 300kpb constructions in E. coli (Gibson, 2009).
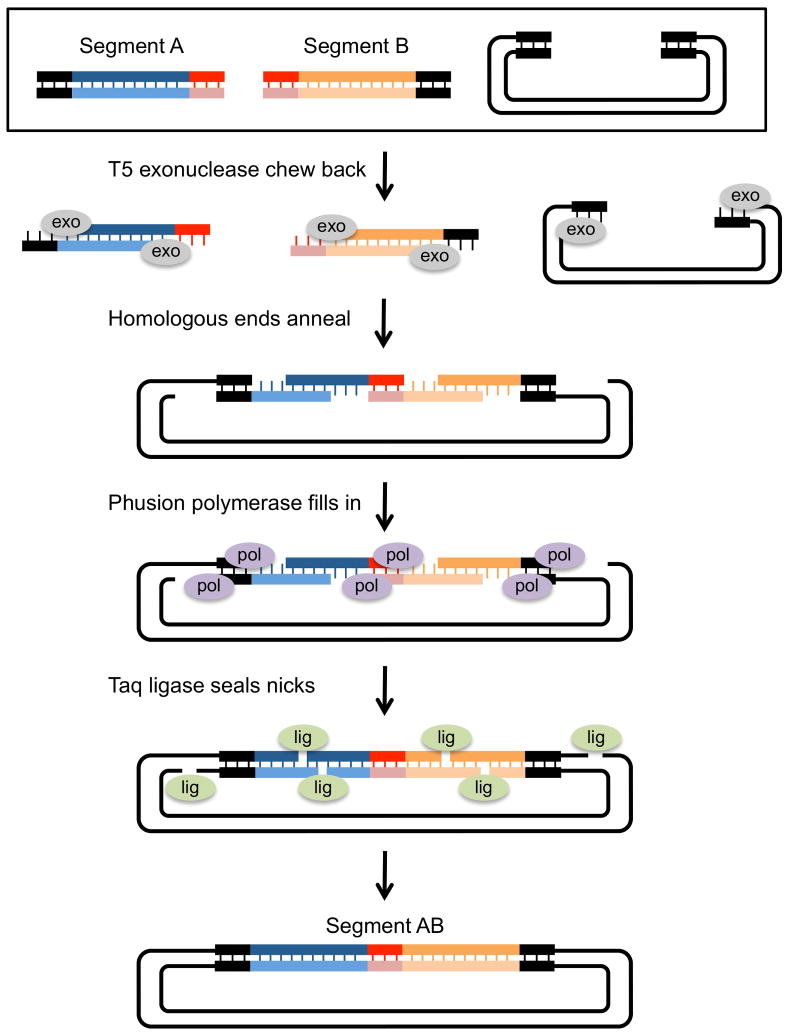
Figure 8 shows the steps of isothermal Gibson assembly. In this example, insert segments are PCR amplified such that there is 15–40bp of overlapping homologous sequence at each end. Segment A has at its 5′ end sequence homology to the linearized vector, and at its 3′ end, sequence homology to segment B. Segment B has at its 5′ end sequence homology to segment A, and at its 3′ end, sequence homology to the vector.
Figure 8. Multiple segment assembly using Gibson assembly.
Gibson assembly allows the seamless cloning of multiple segments into a plasmid vector. In this example, two PCR generated inserts are assembled into a linearized plasmid. Each insert segment is amplified with primers that contain sequence homology to the plasmid ends (dark ends) and to each other (light ends). The starting segments (black box) are mixed with T5 exonuclease, Phusion DNA polymerase and Taq ligase and incubated at 50 degrees for 15min to 1 hour. T5 exo chews back the 5′ ends of each segment, exposing ss 3′ homologous ends. After the 3′ ends anneal, Phusion polymerase fills in the gaps between annealed segments. In the final step, Taq ligase seals the nicks. The resulting plasmid has both inserts (Segment AB) seamlessly joined and it can be used directly as a template in PCR reactions or transformed into E coli for propagation.
Assembly of those overlapping DNA segments proceeds in four steps. First, bacteriophage T5 5′-> 3′ exonuclease generates free 3′ ends. Second, complementary DNA sequences at the ends of the free 3′ overhangs anneal, leaving gaps between annealed fragments. Third, a proofreading DNA polymerase binds the 3′ ends of the annealed stretches and initiates polymerization. Polymerization proceeds until the polymerase reaches the end of the single-stranded stretch. Finally, Taq DNA ligase seals the nicks and the resulting circular, sealed plasmids are transformed into competent E. coli. Commercial kits containing mixtures of these enzymes and needed cofactors (dNTPs for the polymerases, NADPH for E. coli DNA ligase) can carry out the entire procedure in 15 mins at 50°.
Gibson assembly differs from other members of this class of assembly methods in three ways, all of which increase its utility. First, instead of using T4 DNA polymerase to generate 5′ ss tails, it uses T5 exonuclease to generate 3′ ss tails. By chewing back the 5′ ends instead of the 3′ ends, T5 exonuclease does not compete with the action of the DNA polymerase which is used in the next step of assembly.
Second, it uses a highly processive, proofreading DNA polymerase (e.g., Phusion) to fill in the gaps between annealed fragments, leaving only single stranded nicks. This decreases the chance for errors compared to other methods, including SLIC, which use Taq polymerase for the extension step
Third, and most importantly, it uses Taq Ligase to seal the nicks in vitro before transformation into E. coli. The end product of isothermal Gibson is a covalently closed circular DNA. This is likely the reason that Gibson assembly end products can transform E. coli at high efficiency compared to other methods and why the transformation of very large DNAs (such as BACs) is possible. Moreover, because Gibson assembly generates nick-free molecules, its products can be used directly as templates for PCR, allowing a multisegment assembly to be amplified without the intermediate steps of E. coli transformation and plasmid isolation.
4. Assembly of overlapping DNA segments in vivo using homologous recombination in yeast
For large (eg >~30–100kbp and complex, eg > ~6 segment) assemblies, yeast homologous recombination (HR) has become the assembly method of choice (Unit 3.22). The double-strand break repair mechanism that Saccharomyces cerevisiae uses to perform homologous recombination was described in the early 1980s (Orr-Weaver et al., 1981). Researchers have used it, recombining linear fragments with cut plasmid vectors, to carry out single-segment-into-vector constructions since 1987 (Ma et al., 1987). Some years ago, Gibson et al. described the construction of a 583kbp synthetic genome from 25 overlapping > 10kbp fragments by homologous recombination in yeast (Gibson, 2008). In 2014, Annaluru et al. used two rounds of yeast homologous recombination to assemble 750 kbp PCR products (made from overlapping oligonucleotides) into a re-engineered, wholly functional 273kbp version of yeast Chromosome III (Annaluru et al., 2014).
Figure 9 illustrates assembly via yeast homologous recombination of two overlapping ds inserts and a plasmid vector. Segments to be joined, designed to overlap by 20–40bp, are transformed into competent yeast cells along with a linearized “shuttle” vector. A yeast shuttle vector contains sequences needed for replication and selection in both E. coli and S. cerevisiae. 48 hours after transformation, yeast plasmid DNA is recovered and used directly (e.g., as a template for PCR) or “rescued” into E. coli to allow large-scale preparations of plasmid DNA. Presumably, constructs as large as a S. cerevisiae chromosome (ch IV is 1.5Mbp) or larger could be constructed by homologous recombination, so long as all of the transformed fragments can get into the nucleus and the assembled molecule can stably replicate. However, for the general DNA assembly projects that most biology labs will undertake, one can assume near 100% accuracy of assemblies under 30kbp and good assembly efficiency of larger constructions (e.g., 5–10 fragments and a total size up to100kbp, see unit 3.22 for more details).
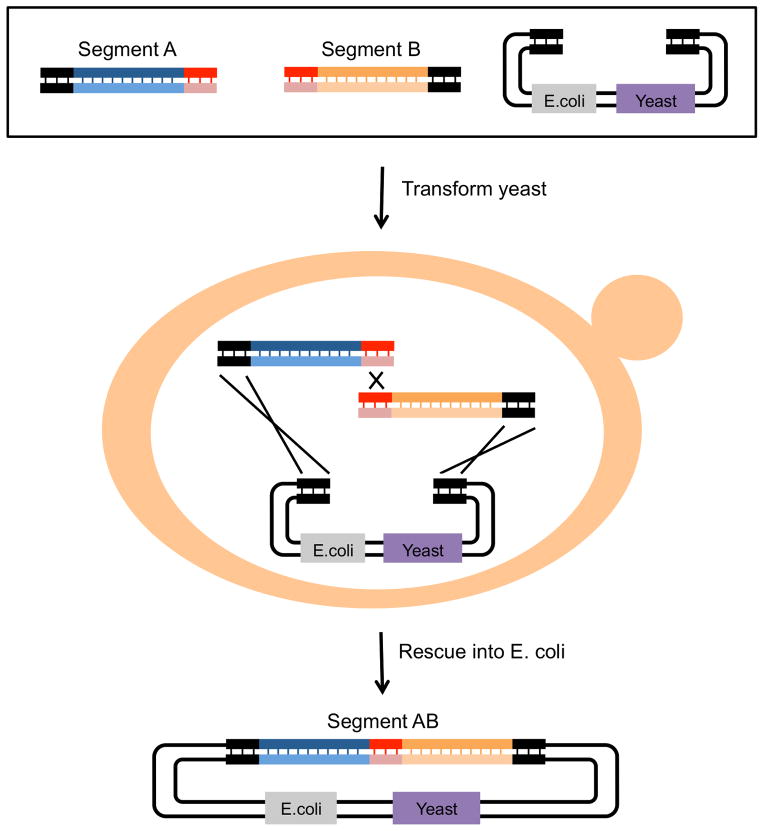
Figure 9. Multiple segment assembly in vivo by yeast homologous recombination.
In this example, the highly efficient HR system of the yeast Saccharomyces cerevisiae is used to assemble two insert segments into a linearized plasmid vector. The “shuttle vector” contains all the necessary elements for selection and propagation in both S. cerevisiae and E. coli. As for Gibson assembly, each starting insert segment is generated by PCR with primers that contain 35bp of sequence overlap, here, homology with the vector (dark ends) and with one another (light ends). The starting segments are transformed into competent S. cerevisiae and plated on selective medium. After 48 hours, the assembled plasmid is purified from yeast and transformed into competent E. coli for propagation. The new insert segment AB consists of both starting segments seamlessly joined together.
5. Assembly of multiple adjacent DNA sequences using the Ligase Cycling Reaction
The Ligase Cycling Reaction (LCR) was initially developed as a non PCR-based diagnostic means to detect the major sickle cell anemia allelic form of the human beta-globin gene (Barany, 1991). It was subsequently used to detect other human single nucleotide polymorphisms and to distinguish between closely related bacteria and viruses reviewed in (Wiedmann et al., 1994). Later, Pachuk et al. used LCR to join 6 blunt end restriction fragments into a functional plasmid (Pachuk et al., 2000). Recently, de Kok et al. used it to assemble up to 12 DNA segments into a ~20kbp plasmid (de Kok et al., 2014).
Unlike the methods described above, LCR uses bridge oligonucleotides to join non-sequence-overlapping segments. Bridge oligos bring together the 3′ end of an upstream segment with the 5′ end of its neighboring segment, so that the two neighboring segments can be ligated. Figure 10 shows the steps of a LCR reaction in which two DNA segments are assembled into a single linear fragment. The assembly mix contains the ds DNA fragments to be assembled along with the bridge oligos and a thermostable ligase (e.g., Ampligase). Each bridge oligo contains sequence complementing the ends of the DNA segments to be joined. The reaction mix is heated to denature ds DNA, then cooled to allow bridge oligos to anneal and bring the ss ends together. DNA ligase then seals the nick between neighboring fragments. Over multiple temperature cycles of denaturing and annealing, newly assembled multimers act as bridges to bring together even more single stranded DNA segments. If the goal is to clone the segments onto a plasmid, additional bridge oligos that overlap the vector and insert ends can be added to the reaction mix along with the linearized vector.
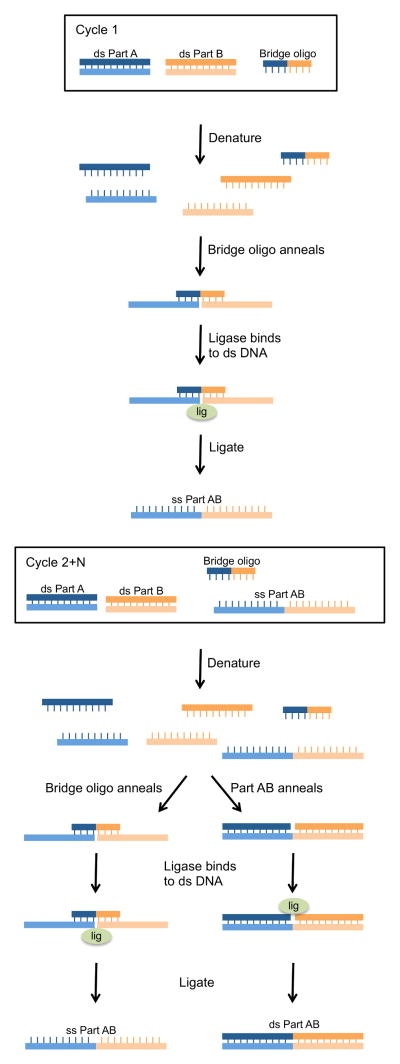
Figure 10. Multiple segment assembly using the Ligase Cycling Reaction (LCR).
LCR is a method that utilizes bridge oligonucleotides to align adjoining segments so that a thermostable ligase can join the segments together. In this example, the upstream segment (ds Part A) and downstream segment (ds Part B) are mixed with a bridge oligonucleotide and thermostable ligase and then temperature cycled. During the first cycle, the ds insert segments are denatured so that the bridge oligo can anneal to complementary nucleotides at the 3′ end of part A and the 5′ end of part B. A thermostable ligase then joins the termini of the two segments, creating a new ss DNA with both segments seamlessly ligated together (ss Part AB). In the subsequent cycles, the bridge oligo continues to bring new ss parts together. In addition, the newly formed ss part AB will also act as a “bridge” by annealing to both ss Part A and ss Part B. The ligase will then seal the nicks to generate a new ss Part AB as well as a new ds Part AB. ss Part AB and both strands of ds Part AB will act as “bridges” in subsequent rounds of thermocycling.
de Kok et al. have tested many LCR parameters in order to develop an optimized assembly method (de Kok et al., 2014). They found that the parameters that had the most effect on assembly were use of short denaturation times (10 sec), addition of DMSO and betaine in order to facilitate strand separation, use of low DNA concentrations (3nM each segment), and use of high bridge oligo conentrations (30nM) with the Tm of each half of the bridge oligo of 70°. They used this optimized protocol to assemble 20kbp plasmids from 12 starting segments (plus a vector) with 80–100% of resulting constructions properly assembled.
Conclusion
DNA construction methods have come a long way since 1973. It has never been easier to construct desired DNA molecules.
The ubiquity of PCR, the availability of high fidelity polymerases, and availability of commercially synthesized DNA, have made it very simple for investigators to obtain the starting DNA segments for their constructions.
And certainly constant development has ensured that there are now numerous good means to assemble single-segment-into-vector constructions.
By contrast, the perfect multisegment assembly method: scarless, sequence-independent, adaptable to robotic workflows, and perfectly accurate, does not now exist. As a consequence, an investigator must weigh the strengths and weaknesses of different methods and choose the approach that best aligns with the project goals and the lab’s expertise.
Although there are no defined rules to dictate the choice of assembly method, there are some considerations we believe helpful to keep in mind. We show these in table 1 and discuss them in more detail here.
Table 1.
Comparison of Multisegment DNA Assembly Methods
| Gateway | Biobricks | Gibson | Yeast HR | LCR | |
|---|---|---|---|---|---|
| Days to Completiona | 4 | 3 | 3 | 5 | 3 |
| Hands-0n Time (hrs)b | 1–2 | 1–2 | 1–2 | 1–2 | 1–2 |
| Premade Partsc | yes | yes | no | no | no |
| Scarless | no | noi | yes | yes | yes |
| Restriction Site Dependentd | no | yes | no | no | no |
| Construction Over 30kbpe | no | no | yes | yes | yes |
| Edit Existing Assemblyf | no | no | no | yes | no |
| Additional Ordered Assemblyg | no | yes | no | yes | yes |
| Costh | $$$ | $ | $ | $ | $ |
| Commercial Kits Available | yes | yes | yes | no | no |
Time in days for a 3-segment assembly (two inserts plus a vector) starting with PCR segments or restriction fragments and ending with an assembled plasmid purified from E. coli.
Total bench time. Total time pipetting at the bench, including reaction set up and transformation into yeast or E. coli. Does not include overnight growth of yeast and E. coli, incubation times or sequencing. Assumes some expertise in conventional cloning.
Biobricks and Gateway require premade parts or entry plasmids that significantly increase the time to final assembly. Since the parts or entry plasmids can be reused, this extra time is only required for the first unique assembly.
Limited by restriction enzyme recognition sites. Sites found within segments need to be mutated before assembly.
Constructions over 30kpb will result in adequate efficiency and accuracy.
Possible to make targeted changes to a single element of a construction. This is most easily accomplished with yeast HR though a unique restriction site somewhere within the element is necessary.
Each assembly can be used to generate additional ordered assemblies by the same method. One example would be combining multiple expression cassettes (transcription units) in order to generate a synthetic metabolic pathway.
Up front and consumable supply cost.
Golden Gate can be quasi-scarless, see text
Time to completion
All multisegment assembly methods take between 3 and 5 days to complete, measured from the generation of starting segments by PCR or other means and ending with a purified, assembled plasmid ready for sequencing. However, this does not take into consideration the time needed to generate the premade parts or entry plasmids or to remove restriction enzyme recognition sequences from segments. For Gateway or brick methods, generating such constructions essentially doubles the time to completion, but, because the parts are reusable, this extra time can speed future construction efforts.
For yeast based methods, the fact that S. cerevisiae doubles more slowly than E. coli and the need to rescue the yeast-assembled plasmid into E. coli for plasmid preparation are the reasons that yeast HR assembly takes two more days to complete than the other methods. However, at day 3, a small amount of plasmid can be purified from yeast and used as a template for PCR. Moreover, yeast clones can be grown overnight in liquid culture for larger plasmid DNA preparations on day 4. However, if the project requires large amounts of plasmid DNA, rescue of the plasmid into E. coli is recommended.
Yeast HR assembly relies on shuttle vectors, plasmids that have replication origins and selectable markers that function in S. cerevisiae and in E. coli (Sikorski and Hieter, 1989 and Unit 13.4.1). Since it is unlikely that a preferred expression vector will be a yeast shuttle vector, the investigator needs to add these elements to their preferred plasmid backbone, either before assembly or as additional elements in a single multisegment assembly.
Bench time
Despite each method having different components and steps per reaction, hands-on bench time between the methods is very similar.
Planning time
An investigator experienced in PCR and Cohen-Boyer construction should have little problem learning to use any of these methods. Where a problem might arise is in the planning stages, as all multisegment assembly methods are more complex than Cohen-Boyer cloning. Sequence design software is invaluable here, especially when one wants to plan multiplexed assemblies and/or generate very large constructions.
Scarlessness
The ability to seamlessly join multiple DNA segments is important for many constructions (e.g., protein engineering). For that reason, Gibson, yeast HR and LCR are better choices than Gateway and most brick methods for these constructions (see Figure 5). Of the brick methods, Golden Gate is the most useful for protein engineering because the DNA segments can be joined seamlessly within an open reading frame.
Restriction site dependence
As multisegment assemblies get larger, the probability that a particular restriction enzyme site exists within a DNA segment increases. For brick methods, reliance on restriction enzymes means that for some constructions, there will be a need to mutate recognition sites within assembly segments. This can be done after the segment is cloned or during the PCR step. In both cases, the need to mutate the restriction site increases the workload to final assembly. And of course there are instances (for example, promoter or other regulatory sequences) when it might be unacceptable to mutate the site. For these reasons, we find that for single multisegment constructions over 30kpb, brick methods require too much editing to be feasible. That said, we realize that this constraint might eventually be overcome by engineering novel type IIs restriction endonucleases with longer recognition sequences, resulting in far fewer recognition sites within long DNA assemblies. In fact, brick methods that utilize homing endoucleases have been reported (Liu et al., 2014) and we note that a chimeric protein that binds to the 18bp recognition sequence of the homing endonuclease I-SceI but leaves FokI overhangs has been developed (Lippow et al., 2009).
Constructions over 30kbp
When the goal is to generate very large constructions from many segments in a single assembly, yeast HR is the most widely used method (see for example Annaluru et al., 2014). Despite the remarkable success Gibson et al. demonstrated in generating very large constructions, including cloned DNAs over 300kpb, we, as well as others, have not been able to reproduce those results. In one of the only side-by-side comparisons, Gibson assembly significantly underperformed in multisegment assembles compared to yeast HR and LCR (de Kok et al., 2014). Consistent with this, NEB, the supplier of commercial Gibson assembly kits states in its product information sheet that “frequencies of recovery of correct assemblies into plasmid vectors drop 5–10 fold when more than 2 fragments are assembled.” Generally speaking, for large, complex (more than 5 segments) constructions over 30kbp, one can expect a decrease in the overall number of colonies from an assembly reaction (efficiency) as well as a decrease in the number of colonies that harbor a correctly assembled plasmid (accuracy). Thus, for large-plasmid assemblies, one should expect to spend more time and effort screening for correctly assembled plasmids.
Construction efficiency, ease and versatility
While the paper from de Kok et al. did not compare constructions over 30kbp, they did show that in 12-segment assembles up to 17kbp in length, LCR and yeast HR show nearly identical assembly efficiency and accuracy rates (de Kok et al., 2014). The ability to use LCR instead of yeast HR for large and complex constructions offers two particular advantages. First, LCR assembly is faster than yeast HR, taking 3 days to complete instead of 5. Second, because LCR uses bridge oligos to bring together DNA segments, the segments themselves can be blunt-ended “bricks,” which can be reused and, more importantly, assembled in a different order in future constructions simply by changing the bridge oligos. LCR then maximizes the future usefulness of generated segments which can be stored as fragment libraries.
Because in yeast HR the DNA segments have overlapping homologous sequences, assembly of existing segments in a different order is not possible. Use of standardized linkers between segments can overcome this deficiency (Serber et al., 2012), but of course, constructions generated in this fashion are no longer seamless (the linkers become scars between segments).
Yeast HR does have one significant advantage over other methods, including LCR, which is the ability to make changes to the DNA of existing plasmid assemblies without having to re-assemble the entire construction. This can be done as long as a unique restriction site exists within the segment to be changed. It is accomplished by opening the existing construction at a unique restriction site within the element to be changed and repairing that site with a linear segment bearing the desired change plus 25–35nt of flanking sequence homology. The ability to make such changes quickly can be useful, for example, when an investigator wants to replace the sequence encoding one protein moiety for another (e.g., one fluorescent reporter tag for another). Moreover, because the majority of the existing construction remains unchanged, the time and costs associated with sequencing of the final construction are minimal.
Brick or parts assembly methods are most useful when an investigator has access to a large library of defined DNAs. This advantage is most obvious in projects requiring gene shuffling (Engler et al., 2009) or metabolic pathway engineering (Werner et al., 2012), where the same parts are being repeatedly used or mixed. In such cases, brick methods can be very useful and the time necessary to generate the parts and remove restriction sites is worth the extra effort.
Cost
When comparing costs between assembly strategies, one should consider both up-front costs such as purchasing compatible vectors and the time to generate entry plasmids, as well as the consumable supply costs associated with DNA assembly (which may change depending on scale). Of the methods mentioned here, Gateway has the largest up front cost, as well as high consumable reagent costs due to the need for proprietary enzyme mixes. At small scale (just a few assemblies), bricks, Gibson, LCR and yeast HR have similar per reaction consumable supply costs though the monies are spent on different classes of supplies; enzymes, oligobucleotides or a combination of the two. Yeast HR requires longer, more expensive oligos than those required in conventional cloning because of the added 25–35nts of flanking sequence on the end of each oligo. LCR requires both PCR and bridge oligos. Moreover, for the ligase to join neighboring segments, the 5′ ends of the strands need to be phorphorylated. If the segments to be joined are PCR products, those can be made using custom primers that contain 5′ phosphates, significantly increasing the cost of primer synthesis but keeping hands-on time to a minimum. Alternatively, investigators can phosphorylate PCR primers with T4 polynucleotide kinase (PNK), decreasing the cost of phosphorylation, but adding more hands-on time.
For Gibson and brick methods, the main supply costs are enzyme mixes, though, longer oligos also contribute to the cost of Gibson assembly. The cost of sequencing the final construction should also be considered. For methods where each segment is a new PCR product (e.g., Yeast HR and LCR), the entire final assembly might need to be sequenced, whereas with brick and Gateway assemblies, one might only need to sequence the segment junctions in the final construction.
Future outlook
The use of synthetic DNA in plasmid constructions continues to increase. As we stated above, the most useful synthetic DNAs for assembly are short blocks that can be used as template for PCR. Depending on the synthesis company, these can cost between $85.00 and $350.00 and can be delivered in about one week. For larger synthetic DNAs cloned into plasmids, the price is higher and the time to delivery is longer. For example, a 5000bp synthetic gene could cost over $3000.00 and take as long as a month to synthesize. Thus, it is currently more cost effective and more efficient to assemble larger stretches of DNA from smaller synthetic blocks. As the cost of DNA synthesis continues to drop, we imagine that the use of de novo synthesized segments will continue to increase, reducing the time and effort needed to assemble large constructions and increasing accuracy of the assemblies themselves.
We also imagine that future DNA assembly will soon be directed by software that systematizes design and controls the operation of automated platforms that generate the starting segments, assemble them, purify the final products and then confirm them by DNA sequencing. Until then, investigators have numerous good options for generating large DNA constructions at the bench. These “post-Cohen-Boyer” methods allow the construction of large, complex, multisegment DNA assemblies that were considered impossible just a short time ago.
Acknowledgments
BS and RB were supported by NIH grants R01GM97479 to RB and U54CA132381 between FHCRC and NMSU
References
- Anderson JC, Dueber JE, Leguia M, Wu GC, Goler JA, Arkin AP, Keasling JD. BglBricks: A flexible standard for biological part assembly. Journal of biological engineering. 2010;4:1. doi: 10.1186/1754-1611-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annaluru N, Muller H, Mitchell LA, Ramalingam S, Stracquadanio G, Richardson SM, Dymond JS, Kuang Z, Scheifele LZ, Cooper EM, Cai Y, Zeller K, Agmon N, Han JS, Hadjithomas M, Tullman J, Caravelli K, Cirelli K, Guo Z, London V, Yeluru A, Murugan S, Kandavelou K, Agier N, Fischer G, Yang K, Martin JA, Bilgel M, Bohutski P, Boulier KM, Capaldo BJ, Chang J, Charoen K, Choi WJ, Deng P, DiCarlo JE, Doong J, Dunn J, Feinberg JI, Fernandez C, Floria CE, Gladowski D, Hadidi P, Ishizuka I, Jabbari J, Lau CY, Lee PA, Li S, Lin D, Linder ME, Ling J, Liu J, Liu J, London M, Ma H, Mao J, McDade JE, McMillan A, Moore AM, Oh WC, Ouyang Y, Patel R, Paul M, Paulsen LC, Qiu J, Rhee A, Rubashkin MG, Soh IY, Sotuyo NE, Srinivas V, Suarez A, Wong A, Wong R, Xie WR, Xu Y, Yu AT, Koszul R, Bader JS, Boeke JD, Chandrasegaran S. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–58. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslanidis C, de Jong PJ. Ligation-independent cloning of PCR products (LIC-PCR) Nucleic acids research. 1990;18:6069–6074. doi: 10.1093/nar/18.20.6069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barany F. Genetic disease detection and DNA amplification using cloned thermostable ligase. Proceedings of the National Academy of Sciences of the United States of America. 1991;88:189–193. doi: 10.1073/pnas.88.1.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Gabant P, Bahassi EM, Couturier M. Positive-selection vectors using the F plasmid ccdB killer gene. Gene. 1994;148:71–74. doi: 10.1016/0378-1119(94)90235-6. [DOI] [PubMed] [Google Scholar]
- Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nature biotechnology. 2008;26:787–793. doi: 10.1038/nbt1413. [DOI] [PubMed] [Google Scholar]
- Cohen SN, Chang AC, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro. Proceedings of the National Academy of Sciences of the United States of America. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kok S, Stanton LH, Slaby T, Durot M, Holmes VF, Patel KG, Platt D, Shapland EB, Serber Z, Dean J, Newman JD, Chandran SS. Rapid and reliable DNA assembly via ligase cycling reaction. ACS synthetic biology. 2014;3:97–106. doi: 10.1021/sb4001992. [DOI] [PubMed] [Google Scholar]
- Echols H, Guaneros G. Control of integration and excision. Cold Spring Harbor Laboratory Press; New York: 1983. [Google Scholar]
- Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. [DOI] [PubMed] [Google Scholar]
- Engler C, Gruetzner R, Kandzia R, Marillonnet S. Golden gate shuffling: a one-pot DNA shuffling method based on type IIs restriction enzymes. PloS one. 2009;4:e5553. doi: 10.1371/journal.pone.0005553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. A one pot, one step, precision cloning method with high throughput capability. PloS one. 2008;3:e3647. doi: 10.1371/journal.pone.0003647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Muller R, Stewart AF, Zhang Y. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nature biotechnology. 2012;30:440–446. doi: 10.1038/nbt.2183. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA, Merryman C, Young L, Noskov VN, Glass JI, Venter JC, Hutchison CA, 3rd, Smith HO. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–1220. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Hill RE, Eaton-Rye JJ. Plasmid construction by SLIC or sequence and ligation-independent cloning. Methods in molecular biology. 2014;1116:25–36. doi: 10.1007/978-1-62703-764-8_2. [DOI] [PubMed] [Google Scholar]
- Holton TA, Graham MW. A simple and efficient method for direct cloning of PCR products using ddT-tailed vectors. Nucleic acids research. 1991;19:1156. doi: 10.1093/nar/19.5.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes SS. Making dollars out of DNA. The first major patent in biotechnology and the commercialization of molecular biology, 1974–1980. Isis; an international review devoted to the history of science and its cultural influences. 2001;92:541–575. doi: 10.1086/385281. [DOI] [PubMed] [Google Scholar]
- Knight T. [Accessed 8-30-2015];Idempotent Vector Design for Standard Assembly of Biobricks. 2003 http://hdl.handle.net/1721.1/21168.
- Kodumal SJ, Patel KG, Reid R, Menzella HG, Welch M, Santi DV. Total synthesis of long DNA sequences: synthesis of a contiguous 32-kb polyketide synthase gene cluster. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15573–15578. doi: 10.1073/pnas.0406911101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li MZ, Elledge SJ. Harnessing homologous recombination in vitro to generate recombinant DNA via SLIC. Nature methods. 2007;4:251–256. doi: 10.1038/nmeth1010. [DOI] [PubMed] [Google Scholar]
- Lippow SM, Aha PM, Parker MH, Blake WJ, Baynes BM, Lipovsek D. Creation of a type IIS restriction endonuclease with a long recognition sequence. Nucleic acids research. 2009;37:3061–3073. doi: 10.1093/nar/gkp182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Chen WH, Ren SX, Zhao GP, Wang J. iBrick: a new standard for iterative assembly of biological parts with homing endonucleases. PloS one. 2014;9:e110852. doi: 10.1371/journal.pone.0110852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Kunes S, Schatz PJ, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Yeast transformation: a model system for the study of recombination. Proceedings of the National Academy of Sciences of the United States of America. 1981;78:6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pachuk CJ, Samuel M, Zurawski JA, Snyder L, Phillips P, Satishchandran C. Chain reaction cloning: a one-step method for directional ligation of multiple DNA fragments. Gene. 2000;243:19–25. doi: 10.1016/s0378-1119(99)00508-9. [DOI] [PubMed] [Google Scholar]
- Roth TL, Milenkovic L, Scott MP. A rapid and simple method for DNA engineering using cycled ligation assembly. PloS one. 2014;9:e107329. doi: 10.1371/journal.pone.0107329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serber Z, Lowe R, Ubersax JA, Chandran SS. Compositions and methods for the assembly of polynucleotides. 8110360B2 US Patent No. 2012
- Shetty R, Lizarazo M, Rettberg R, Knight TF. Assembly of BioBrick standard biological parts using three antibiotic assembly. Methods in enzymology. 2011;498:311–326. doi: 10.1016/B978-0-12-385120-8.00013-9. [DOI] [PubMed] [Google Scholar]
- Shuman S. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. The Journal of biological chemistry. 1994;269:32678–32684. [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S, Engler C, Weber E, Gruetzner R, Marillonnet S. Fast track assembly of multigene constructs using Golden Gate cloning and the MoClo system. Bioengineered bugs. 2012;3:38–43. doi: 10.4161/bbug.3.1.18223. [DOI] [PubMed] [Google Scholar]
- Wiedmann M, Wilson WJ, Czajka J, Luo J, Barany F, Batt CA. Ligase chain reaction (LCR)--overview and applications. PCR methods and applications. 1994;3:S51–64. doi: 10.1101/gr.3.4.s51. [DOI] [PubMed] [Google Scholar]