ABSTRACT
When trophoblasts migrate and invade in vivo, they do so by interacting with a range of other cell types, extracellular matrix proteins, chemotactic factors and physical forces such as fluid shear stress. These factors combine to influence overall trophoblast migration and invasion into the decidua, which in turn determines the success of spiral artery remodelling, and pregnancy itself. Our understanding of these important but complex processes is limited by the simplified conditions in which we often study cell migration in vitro, and many discrepancies are observed between different in vitro models in the literature. On top of these experimental considerations, the migration of individual trophoblasts can vary widely. While time-lapse microscopy provides a wealth of information on trophoblast migration, manual tracking of individual cell migration is a time consuming task that ultimately restricts the numbers of cells quantified, and thus the ability to extract meaningful information from the data. However, the development of automated imaging algorithms is likely to aid our ability to accurately interpret trophoblast migration in vitro, and better allow us to relate these observations to in vivo scenarios. This commentary discusses the advantages and disadvantages of techniques commonly used to quantify trophoblast migration and invasion, both from a cell biology and a mathematical perspective, and examines how such techniques could be improved to help us relate trophoblast migration more accurately to in vivo function in the future.
KEYWORDS: cell tracking algorithms, Placenta, trophoblast migration, time-lapse microscopy
Introduction
The placenta is a fetal organ responsible for all nutrient, gas and waste exchange between mother and baby during pregnancy. As such, the function of the placenta is vital for fetal growth and pregnancy success. The human placenta contains specialized populations of epithelial cells called trophoblasts. One such population–called extravillous trophoblasts (EVTs)–grow out from the placental villi and invade into the maternal decidua where they breach the maternal spiral arteries that supply blood to the placental surface and remodel these vessels into wide non-vasoactive structures.1,2 For this process of spiral artery remodelling to be successful, 2 key trophoblast migration events are required 1) the migration of EVTs within the decidual stroma prior to arterial colonisation, and 2) the migration of a subset of EVTs (termed endovascular trophoblasts) along the arterial lumen down to the inner third of the myometrium. Trophoblast-induced remodelling of the spiral arteries results in an increased and constant supply of blood to the placenta as pregnancy progresses, and the importance of this process is highlighted in pregnancy disorders such as pre-eclampsia that are associated with inadequate spiral artery remodelling.3
As EVTs grow out from the placenta they differentiate from a proliferative phenotype in those cells proximal to the placental villus, to an invasive phenotype capable of migrating into the decidua. Understanding the factors that underlie successful trophoblast invasion and migration is key to understanding how they contribute to normal and abnormal placentation. As the extent of trophoblast migration and invasion in humans is relatively unique within the animal kingdom, findings from animal models cannot necessarily be directly extrapolated to the human. However, as access to human implantation sites is extremely limited for obvious ethical reasons, animal models of trophoblast invasion do provide one of the only ways that trophoblast invasion can be studied in vivo, and have the advantage of allowing us to manipulate the physical and genetic environment in ways not possible in human studies. For example, the ability to generate murine gene knockout or overexpression models allows us to distinguish how specific genes may contribute to trophoblast invasion.4,5 More recently the rat has gained favor as a model of trophoblast invasion as it confers several advantages over the mouse in that the depth of trophoblast invasion follows a similar pattern to that observed in the human and trophoblasts also colonize the spiral arteries and contribute to their remodeling.6-8 Finally, animal models allow the complex in vivo interactions between the trophoblast and endometrium to be examined, an aspect of trophoblast invasion that is particularly difficult to accurately replicate in vitro. Despite these advantages, it is important to remember that rodent placentation is anatomically distinct from human placentation, and thus it is important to relate findings from such animal models back to human trophoblast. To do this, researchers have used a range of in vitro tools from cell lines to explant models to try to better understand the factors that regulate trophoblast migration. Our ability to obtain meaningful information from these models is dependent both on understanding the limitations of each model, and developing accurate methods to quantify cell migration in order to relate it back to the complex in vivo situation. This commentary aims to discuss the advantages and disadvantages of techniques commonly used to quantify human trophoblast migration and invasion in vitro, both from a cell biology and a mathematical perspective, and examines how such techniques could be improved to help us understand trophoblast migration more accurately.
Studying trophoblast migration in vitro
The acquisition of a migratory EVT phenotype, and the subsequent ability of these cells to invade the decidua does not result from one discrete differentiation event that can be directly measured, but rather is a continuous and complex process involving multiple changes in adhesion molecules and secreted proteases that is regulated by a large number of interacting cytokines arising from multiple cell types at the materno-fetal interface.9 In order to better understand and quantify the complex factors regulating trophoblast invasion and migration, a wide range of in vitro models of trophoblast migration have been employed.
Cell lines
The cell lines used to study trophoblast migration/invasion can be broadly grouped into 2 categories; choriocarcinoma cell lines (including Jeg3, Jars, BeWos),10,11 and those derived from immortalized isolated primary trophoblast populations (including HTR-8/SVneo, TEV-1 and SGHPL-4).12-14 While cell lines provide a convenient, high-throughput tool to study trophoblast migration, they exhibit some important differences from primary EVTs, meaning that caution needs to be exercised in interpreting the results of these studies. For example, choriocarcinoma derived cell lines are characterized by their transformation from normal EVTs to highly invasive cancerous cells with extensive abnormalities in chromosome number (>70) and rearrangement.15 The pathological extent of choriocarcinoma invasion in vivo also far exceeds normal EVT invasion, demonstrating that choriocarcinoma invasion is regulated differently from normal EVTs. While cell lines generated from isolated primary cytotrophoblasts or EVTs improve on choriocarcinoma cell lines, by virtue of their immortalisation these cell lines are proliferative, whereas in vivo, differentiated invasive EVTs have exited the cell cycle. Thus, no cell line is truly able to represent invasive primary EVTs.
Isolated primary trophoblasts
The isolation of trophoblasts from first trimester placentae has an advantage over cell lines in that it enables researchers to study the cells of interest directly. The most commonly isolated cell population are cytotrophoblasts, which can be extracted from first trimester or term placentae.16,17 When employing these cells to study trophoblast migration it is crucial to assess purity of cell isolates, as cytotrophoblast isolation procedures are prone to contamination with low levels of fibroblasts that are highly motile and multiply rapidly in culture.18 A number of researchers have reported that first trimester cytotrophoblasts exhibit invasive activity (indicative of EVT differentiation) following culture on Matrigel,19-21 but the resulting cell population is frequently not well characterized, and the proportion of differentiated EVT progeny obtained is unclear. In contrast, cytotrophoblasts isolated from term placentae appear to demonstrate a greater propensity to syncytialize in culture and do not differentiate into an invasive EVT phenotype.22
In order to overcome these difficulties, methods of isolating EVTs directly from first trimester placentae have been developed.23 Isolated EVTs that express cytokeratin, HLA-G and c-erbB2, and do not express vimentin, can be obtained with >95% purity.24 The use of such EVTs in in vitro models of migration and invasion is therefore as close as we can get to the in vivo situation. However, when we consider the practical drawbacks of working with isolated primary EVTs it becomes clear why they are not more widely used even in laboratories with access to this tissue. First, only moderate numbers of EVTs (ranging from 100,000 to 1.2 million cells per gram of tissue) can typically be isolated from the amounts of first trimester villous tissue routinely obtained.23,25 Furthermore, as EVTs do not proliferate in culture they cannot be propagated or expanded in vitro. Finally, isolated cells only survive for approximately one week in culture, meaning a significant ongoing time commitment is required to prepare fresh isolates.
Explant culture models
Primary EVTs can also be generated in vitro from explant cultures. Such villous explant models involve the culture of small pieces of first trimester placental tissue on type I collagen or Matrigel, resulting in the formation of EVT outgrowths from the tips of anchoring villi in contact with the extracellular matrix substrate.25-29 In any culture only a proportion of explants produce trophoblast outgrowths, dependent on the culture conditions employed and the tissue gestation.27,29,30 The formation of these EVT outgrowths represents the first stages of villous cytotrophoblast differentiation into the EVT lineage, and further expansion of the outgrowth can then be used to study EVT migration and/or invasion. Explant models allow some of the complex multi-cellular interactions driving EVT migration/invasion in vivo to be replicated in vitro by ensuring structural and paracrine relationships between cells in the villi and those in the outgrowths are maintained. However, like primary EVTs, the villous explant tissue itself has a limited period of viability in culture, meaning that outgrowth expansion can only be tracked for a few days.31
Functional assays to study trophoblast migration
The above cell models can be used in a number of functional assays to study trophoblast migration in vitro. The migratory or invasive capacity of a cell can be correlated with changes in expression of well characterized adhesion molecules and matrix digesting enzymes, which in EVTs include α5β1 and α1β1 integrins, matrix metalloproteinases 2 and 9, and urokinase plasminogen activator. Here however we will focus on the use of cell-based assays to directly visualize trophoblast migration and invasion. The simplest of these is wound healing assays,32-34 where cell lines are grown to near-confluence, and then a ‘wound’ in the monolayer is created by mechanical or enzymatic means.35 The migration of cells into this cell-free region is then monitored either by time-lapse microscopy, or over a set time-course, and the proportion of the wound filled with cells quantified by digital image analysis. Thus, the longer a wound takes to fill, the less migratory the cells in those culture conditions are considered to be. The major limitation to this approach is that cell proliferation is not frequently quantified alongside wound healing, and thus the relative effects of proliferation and migration in closing the wound can become difficult to determine by simple endpoint analysis. Furthermore, the most commonly used method of mechanical scraping to create the cell-free region often results in non-uniform wounds and variable levels of cell injury, meaning experiments can be difficult to replicate.35 Finally, wound healing assays tell us only about trophoblast migration, and not about the capacity of trophoblasts to invade through extracellular matrices or into the decidua. Thus, to relate these findings to the in vivo situation such experiments should be backed up by additional studies of trophoblast invasion.
Matrigel invasion assays provide a more direct measure of the invasive capabilities of trophoblasts. In these experiments, trophoblasts (cell lines, isolated primary cytotrophoblasts or EVTs) are plated into the upper chamber of Matrigel-coated polycarbonate membrane Transwell inserts (usually 8 or 12um pore size), and the invasion of cells through the membrane is quantified over 24-72 hrs of culture.32,36 Trophoblasts are then visualised by immunohistochemistry, histological staining or pre-labeling with fluorescent cell tracker dyes, and the number of cells on the lower portion of the membrane quantified.32,37 To drive trophoblast migration, cells are usually plated in serum-free media, and migrate toward serum-containing medium in the lower chamber, but experiments to test specific chemoattractants can also easily be established. As light cannot be transmitted through the membrane, and cells cannot be imaged by time-lapse microscopy while moving through multiple focal planes, Matrigel invasion assays are limited to endpoint analysis. Thus it is important to optimise the chosen endpoint to the experimental conditions used. An alternative approach to Matrigel invasion assays is to embed trophoblast spheroids in fibrin gel, then quantify the average number and length of invasive processes from each spheroid following a set culture period.38 While this method allows trophoblasts to be visualised in situ, and provides more detailed morphological information, invasive processes can only be imaged in 2-dimensions and the requirement for spheroid generation means that while this method has been undertaken with primary trophoblasts,39 it is more commonly used with cell lines.
In vivo, trophoblasts migrate away from the placental villus and into the maternal decidua, and their stimulus to do this is driven by complex chemotactic signals arising from both of these locations. In vitro, this can in part be replicated by tracking trophoblast migration toward specific chemoattractants,38,40 but to more closely mimic the complex in vivo situation researchers have used explant models of EVT outgrowth that allow structural and paracrine relationships to be maintained. Traditionally, many villous explant models embedded explants in a thick layer of Matrigel.26,28,41 In these systems the EVT outgrowths can expand and migrate in 3-dimensions by actively invading through the Matrigel, but the accurate vizualization and quantification of the resulting outgrowth volume can become problematic,26,28,41 It is also possible to culture villous explants on top of an extremely thin layer of Matrigel (Fig. 1A and B).25,29,38 This model has the advantage that EVT outgrowths grow in a 2-dimensional (2D) layer that spreads across the Matrigel surface, making outgrowths easily detectable and readily quantifiable. In addition, growth factors often found in Matrigel are kept to a minimum and are less likely to confound experimental results. However, while 2D models can accurately portray EVT expansion and migration across the surface of the Matrigel, as EVTs do not penetrate the extracellular matrix to facilitate outgrowth expansion such models do not provide data on EVT invasion.
Figure 1.
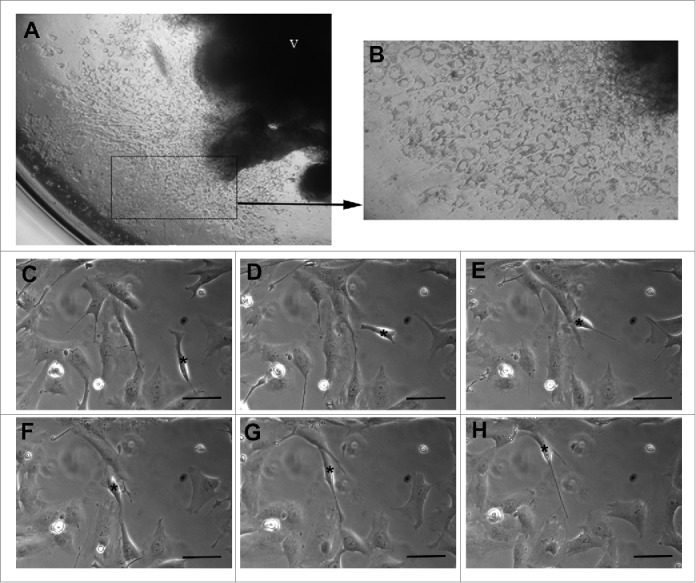
In vitro methods used to analyze trophoblast migration. (A-B) 2D outgrowth of extravillous trophoblasts from a first trimester villous explant (V) across a thin layer of Matrigel at 4x (A) and 10x (B) magnification. (C-H) Time-lapse microscopy images of SGHPL-4 cells cultured under 6dyne/cm of shear stress demonstrating changes in cell morphology that occur with migration. Images presented were captured one hour apart. The asterisk delineates the same cell in each image as it migrates. Scale bars represent 50 µm.
Even in a 2D system, the quantification of EVT outgrowth presents some challenges. Inter-placental and inter-explant data often shows high levels of variance, requiring in a large number of explant replicates to obtain meaningful results.29,41 This need for high-throughput experiments is further confounded by the difficulties in developing automated methods to readily quantify EVT outgrowth expansion and migration in these models as images are often captured in phase contrast, which presents problems for automated image analysis algorithms that need to clearly delineate cell boundaries in a binary manner.
Finally, co-culture models have been used to replicate some of the cell-cell interactions that may influence trophoblast migration in vivo. Such models usually involve the culture of trophoblasts with either endothelial cells to mimic migration down the spiral artery lumen,40,42 or endometrial stromal cells to mimic the interactions that occur during invasion through the decidua.43,44 In such scenarios endothelial or stromal cells are grown to confluence, then trophoblast seeded on top of these monolayers, either in a single cell suspension, or in pre-formed trophoblast spheroids that mimic the first trophoblast migration and invasion events during blastocyst implantation. In some instances, co-culture models have been advanced even further toward the in vivo scenario by culturing trophoblasts with segments of uterine spiral arteries45 or culturing placental villous explants with decidual tissue.46,47 Different cell types can be distinguished either by immunohistochemistry at the end of the culture period, or pre-labeling with fluorescent tracker dyes. In 2D cultures, fluorescent labeling has the advantage of allowing migration to be monitored in real-time by fluorescent time-lapse microscopy, from which the speed and direction of both cell populations, and the influence of co-culture on these parameters, can be quantified.40
Quantifying trophoblast migration
Data describing EVT migration and invasion can thus be divided into 3 broad groups; 1) surrogate markers of invasion such as matrix metalloprotease production and adhesion molecule expression, 2) endpoint assays where the number of invaded cells is quantified after a set culture period, or 3) analysis of time-lapse microscopy data that allows cellular speed and directionality to be tracked in real-time. This final option allows for a more readily quantifiable and accurate understanding of cell migration, but is under-utilised used in the study of trophoblast migration. This perhaps results from the labor intensive frame by frame analysis required to manually track cell migration in time-lapse datasets,38 which restricts the numbers of cells able to be quantified, and thus the ability to extract meaningful information from the data. Here, we present different approaches to the development of automated imaging algorithms that may aid our ability to accurately interpret trophoblast migration in vitro.
Time-lapse imaging provides snapshots of cell locations as they migrate and proliferate in culture (Fig. 1C-H). To manage the size of the data set, images must be acquired at physiologically appropriate time points. In the case of trophoblast migration assays images are typically acquired at 10-15 minute intervals over periods of several hours, with a number of repeat assays,40,48,49 Therefore, tens to hundreds of cells need to be tracked over 50-100 time points, which is a lengthy process if conducted manually. Automatic algorithms to detect cellular regions, and to identify cell types via fluorescent properties, are the ideal way to analyze time-lapse data. Useful summaries of available tracking tools and the current limitations of these tools is given by Meijering et al,50,51 and their differences quantified.52 Automatic algorithms often have to be tested and modified on a case-by-case basis to account for levels of fluorescence or noise in a particular experiment, and often fail to track individual cells through time if cells can ‘climb over each other' in a monolayer or if they change shape and 2D size when they move, both common occurrences in trophoblast cultures (Fig. 1).48 Automatic algorithms can also have difficulty detecting individual cells if numerous cells of the same type are clustered together, or as is the case in many primary cell cultures, if effects on cell viability mean that fluorescent images can only be acquired at a subset of image time-points to maintain the integrity of the culture.40,48 As a result, in many time-lapse assays subsets of cells are identified manually,38,40,49,53 or cells are identified using semi-automated procedures where the user corrects errors in the automated process.48 Alternatively, the movement of individual cells is neglected, doing away with the inherent difficulties in automated tracking of cells, and the cells are treated as a network of brightness patterns that move through an image sequence,40,54 or alternative measures of migration such as cell-free space are considered as proxies for individual cell behavior.34 The final option is by far the simplest way to quantify migration but it makes use of little of the rich data available from time-lapse imaging, and so provides the least mechanistic insight. Automatic cell detection methods are continually improving,52,55 however at this time manual or semi-automatic methods with manual correction are still commonly used in practice and can provide the most accurate methods for analysis.
Time-lapse imaging of cell behavior provides significant amounts of information that can be used to build a picture of the mechanisms of cell migration, proliferation and cell-cell interactions. Many important metrics of cell behavior (such as cell diffusivity, cell velocities under stimuli, and directional bias) can be calculated from cell displacement and/or velocities over time. Both individual cell tracking and network-based approaches can therefore be used to quantify these behaviors, allowing meaningful comparisons between data obtained using the same cell type but under different culture conditions. However, analysis techniques can influence results even when employed in the same dataset. In order to illustrate the importance of a judicious choice of analysis technique, here we consider both individual cell tracking and network analysis of a previously published time-lapse data set of the trophoblast cell line SGHPL-4 cultured alone or on top of a monolayer of human umbilical vein endothelial cells (HUVECs),40 These cultures were then subjected to shear stress (with flow over the surface of the monolayer) of 0.5-6 dyne/cm2 Here only results showing SGHPL-4 migrating in the absence of HUVECs and at 0.5 and 6 dyne/cm.2 are shown, however results hold across experimental conditions. Network analysis is as presented by James et al.40 and individual cell tracking methods are given in Appendix A.
First, let us consider manual tracking of a subset of cells per image sequence/experimental replicate, which is the simplest way to analyze time-lapse imaging. Typically 10-20 cells per image sequence are analyzed,38,40,49,53 compared with 40-80 cells seen in our sample image sequences. Manual tracking of 10 cells in the data presented here, shows an overall trend for right-to-left movement (with the flow) at 6 dyne/cm2, while at 0.5dyne/cm2 a more uniform movement across all angles (a diffusive movement) is evident (Fig. 2). However, this clearly illustrates that there are significant variations in the trajectory of a single cell, as well as variations in the direction and total distance traveled between cells. As a result of this high degree of variation, the number of individual cells analyzed is critical to capture group dynamics and draw meaningful comparisons between experimental conditions. When compared with the very low variation between image sequences when all cells are considered, there is significant variation in velocity and directional bias values and between experimental replicates (image sequences) when low numbers of cells are sampled (Fig. 3). This illustrates how the choice of sampled cells can result in widely varying results even for simple measures of migration. In particular, the variability between image sequences is greatly increased when low numbers of cells are sampled, so although some samples by chance give good estimates of cell migratory properties, sampling 10 cells per image sequence is not sufficient to give a true representation of cell migration in the population (Fig. 3). Rather, the data presented here suggests on the order of 30 cells per image should be analyzed to provide a realistic picture of cell dynamics, or data from multiple sequences pooled when too few cells imaged in a single image sequence.(Fig. 3)
Figure 2.
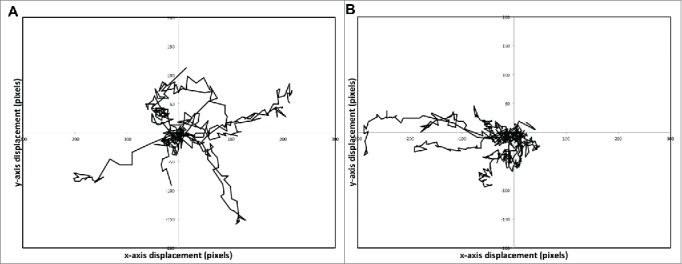
The trajectories of 10 individual SGHPL-4 cells cultured under shear stress conditions of (A) 0.5 dyne/cm2 and (B) 6 dyne/cm2. The origin of each cell is transposed to the start point. Although there is a right-to-left trend in cell movement under 6 dyne/cm2 conditions there is significant variation within an individual cell's trajectory as well as between cells.
Figure 3.
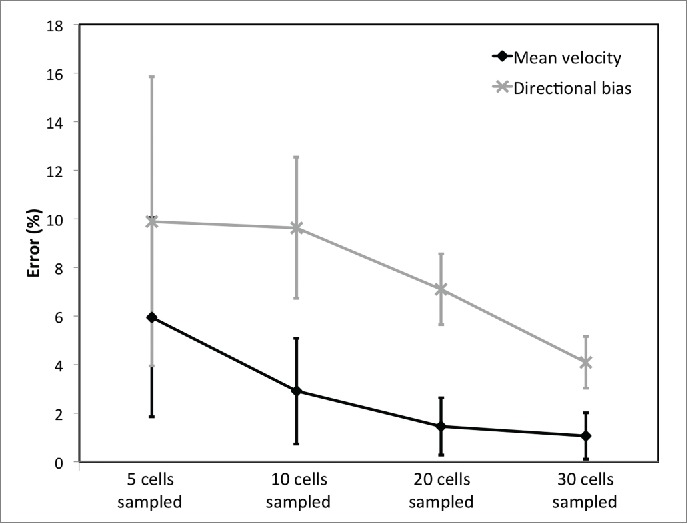
The error in cell migration metrics when subpopulations of cells (5, 10, 20 and 30 per sequence) are sampled compared with an analysis of the trajectories of every imaged cell (40-80 per sequence). To sample cells, they were selected at random from each of 5 image sequences and 1) the mean cell velocity (displacement per time step/time between frames) and 2) directional bias with the flow (velocity of cells moving between 90° and 270° divided by overall cell velocity) were calculated. Subpopulation sampling was repeated 10 times with different randomly selected cells to provide a mean error and standard deviation of error from the ‘gold standard’ case where all cells were analyzed. Error bars on individual data points represent variation in results as a result of the choice of sampled cells. Data are presented for isolated SGHPL-4 cultured in isolation under 6 dyne/cm2 shear but hold for other cultures analyzed.
In contrast to individual cell tracking, the network approach to analyzing time-lapse imaging considers the image sequence as a whole by assessing changes in image pixel intensity over time.40 Phase images at each time point provide a matrix with variable intensities allowing discrimination between cellular and acellular regions. Combined with fluoresence imaging one can distinguish between cell types. As cells move the intensity of image pixels changes from frame to frame in the image sequence and so the velocity of cells and/or cell clusters can be estimated from time-lapse imaging over time without having to identify the location of discrete cells. This analysis technique has been tested against manual tracking of sampled cells within a population and has been shown to perform well in assessing cell velocity or directional bias in cell movement in comparison to cells sampled from time-lapse imaging.40,54 When the dataset presented here was assessed tracking every cell individually, the same trends as network analysis were observed, and thus both techniques provide similar experimental outcomes. However, as network methods do not distinguish between individual cells, they cannot assess cell clustering, which is problematic for the assessment of cells like trophoblasts that change in 2D shape or size as they migrate, thus affecting the algorithms ability to treat them as a single cell, or cells that migrate toward each other to create multi-cellular groups.
Future directions
Despite the wealth of information contained in time-lapse imaging of trophoblast migration, most studies have focused on identifying differences in very simple measures (such as average cell velocity), whereas more subtle and interesting differences may be present when stimuli (chemoattractants or flow) are applied to the system. Across biological fields similar data has been used successfully in conjunction with computational models to gain more in depth understanding of the mechanisms of cell behavior in vitro.56 These models can help to reconcile data between in vitro systems and to hypothesize how the in vitro observations translate to the in vivo system. Computational models can be used to predict how individual contributors to cell migration (e.g. oxygen levels and blood flow) result in whole population dynamics, and can be built upon to include complex cell-cell signaling networks.57,58 The future of trophoblast migration studies may be to apply these tested techniques to build upon our knowledge of the mechanisms controlling trophoblast invasion and spiral artery remodelling. Indeed, our ability to capture data on trophoblast migration data that can be used in this way to understand normal and pathological placentation is of particular importance given the difficulties that studying the process in vivo entails.
Regardless of the experimental technique employed in assessing trophoblast migration a consistent set of metrics can be calculated to allow model development. Quantities of interest include:
The diffusivity of cells, or the random movement of cells in the absence of – or in addition to – movement under mechanical or chemoactive stimuli. A diffusive process has an equal probability of a cell moving in any direction (there is no directional bias) and a ‘diffusion coefficient’ is typically obtained from time-lapse imaging by assessment of the cumulative root mean square displacement of cells over time,50 and from wound healing or cell spreading assays by tracking the front of moving cells.59 It should be noted that diffusivity can be influenced by geometry, and the density of cells in a culture.59
The level of active movement, or the influence of stimuli on cell movement. This can be assessed by calculating the distribution of cell movement (do cells preferentially move toward or away from a stimulus),40,48 whether cells move more rapidly toward or away from stimuli,40 and whether there a persistence in cell movement (once a cell starts moving in a particular direction does it keep moving that way).48
The extent of cell clustering including whether cell types attract/repulse one another or if they behave differently in the vicinity of other cells. This metric is subtler than cell velocities; cell clustering can occur as a result of experimental error, and clustering that appears apparent when a user views time-lapse imaging may be random and so insignificant. However, computational techniques are available to quantify whether cell clustering is significant in an image sequence.60
The relative contribution of each of the above factors, and other metrics of interest in a particular assay such as cell proliferation and apoptosis, to collective motion. This balance cannot always be assessed directly from standard metrics obtained via time-lapse imaging. Therefore, computational models provide an invaluable tool in building a complete picture of cell motility.
Each of these metrics can be extrapolated from time-lapse imaging data sets using the location of cells over time, and do not necessarily rely on knowledge of the trajectory of individual cells.54 While it may not be practical to report these metrics in each study, storage of data in curated repositories, which can be linked to publication data should be encouraged.61 This will allow their future use in parameterising computational models, which may like their counterparts in other biological fields ultimately provide a library of cell behaviors that can be combined to assess the relative influence of stimuli and cell-cell interactions in vivo.
Conclusion
Trophoblast migration plays an important role in human placentation, and as a result many laboratories use a range of in vitro models to assess the how environmental cues such as cytokines, shear stress or oxygen tension affect trophoblast migration. The choice of model used is likely to be restricted by the tissue and microscopy resources available, and while cell lines are attractive options for high throughput experiments or optimisation, where possible primary cells should be used to validate experimental findings. Overall, appropriate numbers of experimental replicates combined with multiple experimental and analytical approaches are required to obtain meaningful conclusions about trophoblast migration/invasion. In comparison to endpoint assays, time-lapse microscopy provides a much richer repository of data about the dynamic process of EVT migration. However, the ability to extract meaningful data from time-lapse experiments is dependent on the ability to analyze a large number of cells individually, or use more sophisticated network analysis methodologies. As a result, future interdisciplinary collaborations between placental biologists and computational modellers may enable a more comprehensive understanding of trophoblast migration datasets, in order to better relate these in vitro findings to the in vivo situation.
Appendix A
To track SGHPL-4 cells through time-lapse image sequences, fluorescent images obtained at a subset of time points were first used to identify the center of mass of each SGHPL-4 cell. To do this images were imported into Matlab (Version R2013a, The Mathworks Inc..) and noise is reduced in the image sequence by first blurring by opening with a 40 pixel radius disk and subtracting the resulting image from the original. A threshold is then applied to the image so high intensity regions (cell interiors) remain in a binary image. Holes in the image are filled using Matlab's “imfill” function. To find cell clusters that potentially contain multiple cells by taking bounding boxes around cell regions with more than double the average area and re-thresholded to find the edges between cells. Cell locations are then manually edited through time to provide a complete description of each cell's location through time.
Abbreviations
- 2D
2-dimensional
- EVT
extravillous trophoblasts
- HUVEC
human umbilical vein endothelial cell
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This research was supported by a Royal Society of New Zealand Marsden FastStart Grant (13-UOA-032). ARC is supported by an Aotearoa Foundation Fellowship.
References
- 1.Pijnenborg R, Bland J, Robertson W, Dixon G, Brosens I. The pattern of interstitial trophoblastic invasion of the myometrium in early human pregnancy. Placenta 1981; 2:303-16; PMID:7301778; http://dx.doi.org/ 10.1016/S0143-4004(81)80027-6 [DOI] [PubMed] [Google Scholar]
- 2.Cartwright JE, Fraser R, Leslie K, Wallace AE, James JL. Remodelling at the maternal-fetal interface: relevance to human pregnancy disorders. Reproduction 2010; 140:803-13; PMID:20837731; http://dx.doi.org/ 10.1530/REP-10-0294 [DOI] [PubMed] [Google Scholar]
- 3.Pijnenborg R, Anthony J, Davey D, Rees A, Tiltman A, Vercruysse L, van Assche A. Placental bed spiral arteries in the hypertensive disorders of pregnancy. British J Obstetrics Gynaecol 1991; 98:648-55; PMID:1883787; http://dx.doi.org/ 10.1111/j.1471-0528.1991.tb13450.x [DOI] [PubMed] [Google Scholar]
- 4.Cui Y, Wang W, Dong N, Lou J, Srinivasan DK, Cheng W, Huang X, Liu M, Fang C, Peng J, et al. . Role of corin in trophoblast invasion and uterine spiral artery remodelling in pregnancy. Nature 2012; 484:246-50; PMID:22437503; http://dx.doi.org/ 10.1038/nature10897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tyberghein K, Goossens S, Haigh JJ, van Roy F, van Hengel J. Tissue-wide overexpression of α-T-catenin results in aberrant trophoblast invasion but does not cause embryonic mortality in mice. Placenta 2012; 33:554-60; PMID:22534068; http://dx.doi.org/ 10.1016/j.placenta.2012.04.002 [DOI] [PubMed] [Google Scholar]
- 6.Hayes EK, Tessier DR, Percival ME, Holloway AC, Petrik JJ, Gruslin A, Raha S. Trophoblast invasion and blood vessel remodeling are altered in a rat model of lifelong maternal obesity. Reprod Sci 2014; 21:648-57; PMID:24155067; http://dx.doi.org/ 10.1177/1933719113508815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fonseca BM, Correia-da-Silva G, Teixeira NA. The rat as an animal model for fetoplacental development: a reappraisal of the post-implantation period. Reprod Biol 2012; 12:97-118; PMID:22850465; http://dx.doi.org/ 10.1016/S1642-431X(12)60080-1 [DOI] [PubMed] [Google Scholar]
- 8.Soares MJ, Chakraborty D, Karim Rumi MA, Konno T, Renaud SJ. Rat placentation: an experimental model for investigating the hemochorial maternal-fetal interface. Placenta 2012; 33:233-43; PMID:22284666; http://dx.doi.org/ 10.1016/j.placenta.2011.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Menkhorst E, Winship A, Van Sinderen M, Dimitriadis E. Human extravillous trophoblast invasion: intrinsic and extrinsic regulation. Reprod Fertil Dev 2014; http://dx.doi.org/ 10.1071/RD14208 [DOI] [PubMed] [Google Scholar]
- 10.Kohler P, Bridson W, Hammond J, Weintraub B, Kirschner M, Van Thiel D. Clonal lines of human choriocarcinoma cells in culture. Acta Endocrinol Supple (Copenhagen) 1971; 153:137-53 [DOI] [PubMed] [Google Scholar]
- 11.Pattillo R, Gey G. The establishment of a cell line of human hormone-synthesizing trophoblastic cells in vitro. Cancer Res 1968; 28:1231-6; PMID:4299001 [PubMed] [Google Scholar]
- 12.Feng H, Choy M, Deng W, Wong H, Lau W, Cheung A, Ngan N, Tsao S. Establishment and characterization of a human first -trimester extravillous trophoblast cell line (TEV-1). J Soc Gynecol Investig 2005; 12:21-32; http://dx.doi.org/ 10.1016/j.jsgi.2005.02.008 [DOI] [PubMed] [Google Scholar]
- 13.Graham C, Hawley T, Hawley R, MacDougall J, Kerbel R, Khoo N, Lala P. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp Cell Res 1993; 206:204-11; PMID:7684692; http://dx.doi.org/ 10.1006/excr.1993.1139 [DOI] [PubMed] [Google Scholar]
- 14.Cartwright JE, Holden DP, Whitley GS. Hepatocyte growth factor regulates human trophoblast motility and invasion: a role for nitric oxide. British J Pharmacol 1999; 128:181-9; http://dx.doi.org/ 10.1038/sj.bjp.0702757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Surti U, Habibian R. Chromosomal rearrangement in choriocarcinoma cell lines. Cancer Genetics Cytogenetics 1989; 38:229-40; PMID:2720636; http://dx.doi.org/ 10.1016/0165-4608(89)90664-X [DOI] [PubMed] [Google Scholar]
- 16.Kliman H, Nestler J, Sermasi E, Sanger J, Strauss J. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinol 1986; 118:1567-82; http://dx.doi.org/ 10.1210/endo-118-4-1567 [DOI] [PubMed] [Google Scholar]
- 17.Fisher SJ, Cui TY, Zhang L, Hartman L, Grahl K, Zhang GY, Tarpey J, Damsky CH. Adhesive and degradative properties of human placental cytotrophoblast cells in vitro. J Cell Biol 1989; 109:891-902; PMID:2474556; http://dx.doi.org/ 10.1083/jcb.109.2.891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frank H, Morrish D, Potgens A, Genbacev O, Kumpel B, Caniggia I. Cell Culture Models of Human Trophoblast: Primary Culture of Trophoblast - A Workshop Report. Placenta 2001; 15:S107-S9; http://dx.doi.org/ 10.1053/plac.2001.0644 [DOI] [PubMed] [Google Scholar]
- 19.Schanz A, Red-Horse K, Hess AP, Baston-Bust DM, Heiss C, Krussel JS. Oxygen regulates human cytotrophoblast migration by controlling chemokine and receptor expression. Placenta 2014; 35:1089-94; PMID:25293376; http://dx.doi.org/ 10.1016/j.placenta.2014.09.012 [DOI] [PubMed] [Google Scholar]
- 20.Librach CL, Werb Z, Fitzgerald ML, Chiu K, Corwin NM, Esteves RA, Grobelny D, Galardy R, Damsky CH, Fisher SJ. 92-kD type IV collagenase mediates invasion of human cytotrophoblasts. J Cell Biol 1991; 113:437-49; PMID:1849141; http://dx.doi.org/ 10.1083/jcb.113.2.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Onogi A, Naruse K, Sado T, Tsunemi T, Shigetomi H, Noguchi T, Yamada Y, Akasaki M, Oi H, Kobayashi H. Hypoxia inhibits invasion of extravillous trophoblast cells through reduction of matrix metalloproteinase (MMP)-2 activation in the early first trimester of human pregnancy. Placenta 2011; 32:665-70; PMID:21764444; http://dx.doi.org/ 10.1016/j.placenta.2011.06.023 [DOI] [PubMed] [Google Scholar]
- 22.Morrish D, Dakour J, Li H, Xiao J, Miller R, Sherburne R, Berdan R, Guilbert L. In vitro cultured human term cytotrophoblast: a model for normal primary epithelial cells demonstrating a spontaneous differentiation programme that requires EGF for extensive development of syncytium. Placenta 1997; 18:577-85; PMID:9290154; http://dx.doi.org/ 10.1016/0143-4004(77)90013-3 [DOI] [PubMed] [Google Scholar]
- 23.Tarrade A, Kuen R, Malassine A, Tricottet V, Blain P, Vidaud M, Evain-Brion D. Characterization of human villous and extravillous trophoblasts isolated from first trimester placenta. Lab Invest 2001; 81:1199-211; PMID:11555668; http://dx.doi.org/ 10.1038/labinvest.3780334 [DOI] [PubMed] [Google Scholar]
- 24.Malassine A, Handschuh K, Tsatsaris V, Gerbaud P, Cheynet V, Oriol G, Mallet F, Evain-Brion D. Expression of HERV-W Env glycoprotein (syncytin) in the extravillous trophoblast of first trimester human placenta. Placenta 2005; 26:556-62; PMID:15993705; http://dx.doi.org/ 10.1016/j.placenta.2004.09.002 [DOI] [PubMed] [Google Scholar]
- 25.Prossler J, Chen Q, Chamley L, James JL. The relationship between TGFbeta, low oxygen and the outgrowth of extravillous trophoblasts from anchoring villi during the first trimester of pregnancy. Cytokine 2014; 68:9-15; PMID:24787051; http://dx.doi.org/ 10.1016/j.cyto.2014.03.001 [DOI] [PubMed] [Google Scholar]
- 26.Aplin J, Haigh T, Jones C, Church H, Vicovac L. Development of cytotrophoblast columns from explanted first-trimester human placental villi: role of fibronectin and integrin alpha5beta1. Biol Reprod 1999; 60:828-38; http://dx.doi.org/ 10.1095/biolreprod60.4.828 [DOI] [PubMed] [Google Scholar]
- 27.Caniggia I, Taylor C, Knox Ritchie J, Lye S, Letarte M. Endoglin regulates trophoblast differentiation along the invasive pathway in human placental villous explants. Endocrinol 1997; 138:4977-88 [DOI] [PubMed] [Google Scholar]
- 28.Genbacev O, Schubach S, Miller R. Villous culture of first trimester human placenta - a model to study extravillous trophoblast (EVT) differentiation. Placenta 1992; 13:439-61; PMID:1470605; http://dx.doi.org/ 10.1016/0143-4004(92)90051-T [DOI] [PubMed] [Google Scholar]
- 29.James J, Stone P, Chamley L. The effects of oxygen concentration and gestational age on extravillous trophoblast outgrowth in a first trimester villous explant model. Hum Reprod 2006; 21:2699-705; PMID:16807282; http://dx.doi.org/ 10.1093/humrep/del212 [DOI] [PubMed] [Google Scholar]
- 30.Tartakover Matalon S, Ornoy A, Fishman A, Drucker L, Lishner M. The effect of 6-mercaptopurine on early human placental explants. Hum Reprod 2005; 20:1390-7; PMID:15760953; http://dx.doi.org/ 10.1093/humrep/deh721 [DOI] [PubMed] [Google Scholar]
- 31.James J, Stone P, Chamley L. Cytotrophoblast differentiation in the first trimester of pregnancy: evidence for separate progenitors of extravillous trophoblasts and syncytiotrophoblast. Reproduction 2005; 130:95-103; PMID:15985635; http://dx.doi.org/ 10.1530/rep.1.00723 [DOI] [PubMed] [Google Scholar]
- 32.Desforges M, Harris LK, Aplin JD. Elastin-derived peptides stimulate trophoblast migration and invasion: a positive feedback loop to enhance spiral artery remodelling. Mol Hum Reprod 2015; 21:95-104; http://dx.doi.org/ 10.1093/molehr/gau089 [DOI] [PubMed] [Google Scholar]
- 33.Wang SC, Yu M, Li YH, Piao HL, Tang CL, Sun C, Zhu R, Li MQ, Jin LP, Li DJ, et al. . Cyclosporin A promotes proliferating cell nuclear antigen expression and migration of human cytotrophoblast cells via the mitogen-activated protein kinase-3/1-mediated nuclear factor-kappaB signaling pathways. Int J Clin Exp Pathol 2013; 6:1999-2010 [PMC free article] [PubMed] [Google Scholar]
- 34.Suga N, Sugimura M, Koshiishi T, Yorifuji T, Makino S, Takeda S. Heparin/heparan sulfate/CD44-v3 enhances cell migration in term placenta-derived immortalized human trophoblast cells. Biol Reprod 2012; 86:134, 1-8; http://dx.doi.org/ 10.1095/biolreprod.111.093690 [DOI] [PubMed] [Google Scholar]
- 35.Riahi R, Yang Y, Zhang DD, Wong PK. Advances in wound-healing assays for probing collective cell migration. J Lab Automation 2012; 17:59-65; http://dx.doi.org/ 10.1177/2211068211426550 [DOI] [PubMed] [Google Scholar]
- 36.Lacroix M, Guibourdenche J, Fournier T, Laurendeau I, Igout A, Goffin F, Pantel J, Tsatsaris V, Evain-Brion D. Stimulation of human trophoblast invasion by placental growth hormone. Endocrinol 2005; 146:2434-44; http://dx.doi.org/ 10.1210/en.2004-1550 [DOI] [PubMed] [Google Scholar]
- 37.Tang C, Mei L, Pan L, Xiong W, Zhu H, Ruan H, Zou C, Tang L, Iguchi T, Wu X. Hedgehog signaling through GLI1 and GLI2 is required for epithelial-mesenchymal transition in human trophoblasts. Biochim Biophys Acta 2015; 1850:1438-48; PMID:25888497; http://dx.doi.org/ 10.1016/j.bbagen.2015.04.005 [DOI] [PubMed] [Google Scholar]
- 38.Wallace AE, Host AJ, Whitley GS, Cartwright JE. Decidual natural killer cell interactions with trophoblasts are impaired in pregnancies at increased risk of preeclampsia. Am J Pathol 2013; 183:1853-61; http://dx.doi.org/ 10.1016/j.ajpath.2013.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korff T, Krauss T, Augustin HG. Three-dimensional spheroidal culture of cytotrophoblast cells mimics the phenotype and differentiation of cytotrophoblasts from normal and preeclamptic pregnancies. Exp Cell Res 2004; 297:415-23; http://dx.doi.org/ 10.1016/j.yexcr.2004.03.043 [DOI] [PubMed] [Google Scholar]
- 40.James JL, Cartwright JE, Whitley GS, Greenhill DR, Hoppe A. The regulation of trophoblast migration across endothelial cells by low shear stress: consequences for vascular remodelling in pregnancy. Cardiovascular Res 2012; 93:152-61; http://dx.doi.org/ 10.1093/cvr/cvr276 [DOI] [PubMed] [Google Scholar]
- 41.Newby D, Marks L, Cousins F, Duffie E, Lyall F. Villous explant culture: characterization and evaluation of a model to study trophoblast invasion. Hypertens Pregnancy 2005; 24:75-91; PMID:16036393; http://dx.doi.org/ 10.1081/PRG-45785 [DOI] [PubMed] [Google Scholar]
- 42.Campbell S, Rowe J, Jackson CJ, Gallery ED. In vitro migration of cytotrophoblasts through a decidual endothelial cell monolayer: the role of matrix metalloproteinases. Placenta 2003; 24:306-15; PMID:12657503; http://dx.doi.org/ 10.1053/plac.2002.0911 [DOI] [PubMed] [Google Scholar]
- 43.Gellersen B, Wolf A, Kruse M, Schwenke M, Bamberger AM. Human endometrial stromal cell-trophoblast interactions: mutual stimulation of chemotactic migration and promigratory roles of cell surface molecules CD82 and CEACAM1. Biol Reprod 2013; 88:80; http://dx.doi.org/ 10.1095/biolreprod.112.106724 [DOI] [PubMed] [Google Scholar]
- 44.Irwin JC, Giudice LC. Insulin-like growth factor binding protein-1 binds to placental cytotrophoblast alpha5beta1 integrin and inhibits cytotrophoblast invasion into decidualized endometrial stromal cultures. Growth Hormone IGF Res 1998; 8:21-31; http://dx.doi.org/ 10.1016/S1096-6374(98)80318-3 [DOI] [PubMed] [Google Scholar]
- 45.Cartwright JE, Wareing M. An in vitro model of trophoblast invasion of spiral arteries. Methods Mol Med 2006; 122:59-74; PMID:16511975 [DOI] [PubMed] [Google Scholar]
- 46.Babawale MO, Van Noorden S, Pignatelli M, Stamp GW, Elder MG, Sullivan MH. Morphological interactions of human first trimester placental villi co-cultured with decidual explants. Hum Reprod 1996; 11:444-50; PMID:8671240; http://dx.doi.org/ 10.1093/HUMREP/11.2.444 [DOI] [PubMed] [Google Scholar]
- 47.Vivovac L, Jones C, Aplin J. Trophoblast differentiation during anchoring villus formation in a coculture model of the human implantation site in vitro. Placenta 1995; 16:41-56; PMID:7716127; http://dx.doi.org/ 10.1016/0143-4004(95)90080-2 [DOI] [PubMed] [Google Scholar]
- 48.Hamzic E, Cartwright J, Keogh R, Whitley G, Greenhill D, Hoppe A. Live cell image analysis of cell-cell interactions reveals the specific targeting of vascular smooth muscle cells by fetal trophoblasts. Exp Cell Res 2008; 314:1455-64; PMID:18314101; http://dx.doi.org/ 10.1016/j.yexcr.2008.01.017 [DOI] [PubMed] [Google Scholar]
- 49.Soghomonians A, Barakat A, Thirkill T, Douglas C. Trophoblast migration under flow is regulated by endothelial cells. Biol Reprod 2005; 73:14-9; http://dx.doi.org/ 10.1095/biolreprod.104.036509 [DOI] [PubMed] [Google Scholar]
- 50.Meijering E, Dzybachyk O, Smal I. Methods for cell and particle tracking. In: Conn PM, ed. Methods in Enzymology: Academic Press, 2012:183-200 [DOI] [PubMed] [Google Scholar]
- 51.Meijering E, Dzybachyk O, Smal I, van Cappellen W. Tracking in cell and developmental biology. Semininars Cell Dev Biol 2009; 20:894-902; http://dx.doi.org/ 10.1016/j.semcdb.2009.07.004 [DOI] [PubMed] [Google Scholar]
- 52.Chenouard N, Smal I, De Chaumont F, Sbalzarini I, Gong Y, Cardinale J, Carthel C, Coraluppi S, Winter M, Cohen A, et al. . Objective comparison of particle tracking methods. Nat Methods 2014; 11:281-9; PMID:24441936; http://dx.doi.org/ 10.1038/nmeth.2808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soghomonians A, Barakat A, Thirkhill T, TN B, Douglas C. Effect of shear stress on migration and integrin expression in macaque trophoblast cells. Biochimica Et Biophysica Acta 2002; 1589:233-46; PMID:12031791; http://dx.doi.org/ 10.1016/S0167-4889(02)00179-9 [DOI] [PubMed] [Google Scholar]
- 54.Seigert F, Weijer C, Nomura A, Heidetoshi M. A gradient method for the quantitative analysis of cell movement and tissue flow and its application to the analysis of multi-cellular Dictyostellium. J Cell Sci 1994; 107:97-104; PMID:8175927 [DOI] [PubMed] [Google Scholar]
- 55.Maška M, Ulman V, Svoboda D, Matula P, Matula P, Ederra C, Urbiola A, España T, Venkatesan S, Balak D, et al. . A benchmark for comparison of tracking algorithms. Bioinformatics 2014; 30:1609-17; http://dx.doi.org/ 10.1093/bioinformatics/btu080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hatzikirou H, Deutsch A. Cellular automata as microscopic models of cell migration in heterogeneous environments. Curr Top Dev Biol 2008; 81:401-34; PMID:18023736; http://dx.doi.org/ 10.1016/S0070-2153(07)81014-3 [DOI] [PubMed] [Google Scholar]
- 57.Weif E, Haugh J. Signaling pathways that control cell migration: Models and analysis. Wiley Interdiscip Rev Syst Biol Med 2011; 3:231-40; http://dx.doi.org/ 10.1002/wsbm.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deisboeck T, Wang Z, Macklin P, Cristini V. Multiscale Cancer Modeling. Annu Rev Biomed Eng 2011; 13:127-55; http://dx.doi.org/ 10.1146/annurev-bioeng-071910-124729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Treloar K, Simpson M, McElwain D, Baker R. Are in vitro estimates of cell diffusivity and cell proliferation rate sensitive to assay geometry. J Theoretical Biol 2014; 356:71-84; http://dx.doi.org/ 10.1016/j.jtbi.2014.04.026 [DOI] [PubMed] [Google Scholar]
- 60.Agnew D, Green J, Brown T, Simpson M, Binder B. Distinguishing between mechanisms of cell aggregation using pair correlation functions. J Theoretical Biol 2014; 352:16-23; http://dx.doi.org/ 10.1016/j.jtbi.2014.02.033 [DOI] [PubMed] [Google Scholar]
- 61.Masuzzo P, Martens L. An open data ecosystem for cell migration. Trends Cell Biol 2015; 25:55-8; PMID:25484346; http://dx.doi.org/ 10.1016/j.tcb.2014.11.005 [DOI] [PubMed] [Google Scholar]


