ABSTRACT
The placental syncytiotrophoblast, which is formed by the fusion of cytotrophoblast cells, is indispensable for the establishment and maintenance of normal pregnancy. The human endogenous retrovirus envelope glycoprotein syncytin-2 is the most important player in mediating trophoblast cell-cell fusion as a fusogen. We constructed expression plasmids of wild-type and 21 single-amino-acid substitution mutants of syncytin-2, including 10 N-glycosylation sites individually silenced by mutagenizing N to Q, 1 naturally occurring single-nucleotide polymorphism (SNP) N118S that introduced an N-glycosylation site, and another 10 non-synonymous SNPs located within important functional domains. We observed that syncytin-2 was highly fusogenic and that the mutants had different capacities in merging 293T cells. Of the 21 mutants, N133Q, N312Q, N443Q, C46R (in the CXXC motif) and R417H (in the heptad repeat region and immunosuppressive domain) lost their fusogenicity, whereas N332Q, N118S, T367M (in the fusion peptide), V483I (in the transmembrane domain) and T522M (in the cytoplasmic domain) enhanced the fusogenic activity. We also proved that N133, N146, N177, N220, N241, N247, N312, N332 and N443 were all glycosylated in 293T cells. A co-immunoprecipitation assay showed compromised interaction between mutants N443Q, C46R, T367M, R417H and the receptor MFSD2A, whereas N118S was associated with more receptors. We also sequenced the coding sequence of syncytin-2 in 125 severe pre-eclamptic patients and 272 normal pregnant Chinese women. Surprisingly, only 1 non-synonymous SNP T522M was found and the frequencies of heterozygous carriers were not significantly different. Taken together, our results suggest that N-glycans at residues 133, 312, 332 and 443 of syncytin-2 are required for optimal fusion induction, and that SNPs C46R, N118S, T367M, R417H, V483I and T522M can alter the fusogenic function of syncytin-2.
Keywords: cell-cell fusion, N-glycosylation, placenta, syncytin-2, single-nucleotide polymorphism
Introduction
The human placenta is a transient organ of embryonic origin whose main function involves the exchange of metabolic and gaseous products between the fetal and maternal circulation. It also acts as an important endocrine secretion organ for normal pregnancy and a barrier between the fetus and its mother for immune tolerance, ensuring the preservation of the fetus. The major constituent of human placental tissue is the trophoblast, which stems from the trophectoderm of the blastocyst and develops before any embryonic tissue arises. Villous cytotrophoblast cells (CTBs) have 2 distinct ways of differentiation: to form highly invasive extravillous trophoblast cells or to fuse and form a multinucleated syncytiotrophoblast (STB) layer, a process designated syncytialization. The STB comes into direct contact with maternal blood, secreting important hormones to maintain pregnancy and mediating gas, nutrient and waste exchange. Inadequate syncytialization is correlated with severe pregnancy disorders such as pre-eclampsia (PE), which is a leading cause of maternal and fetal mortality.1
Our understanding of the mechanisms underlying syncytialization in the placenta came from the identification of the human endogenous retrovirus (HERV) envelope proteins, syncytins, which are fusogens specifically expressed in the placenta.2-6 HERVs are remnants of infections of former exogenous retroviruses, occupying at least 8% of the human genome at various stages of “fossilization.”7 The 2 bona fide envelope glycoproteins, syncytin-1 and syncytin-2, are encoded by HERV-W (the official name is ERVW-1) and HERV-FRD (official name ERVFRD-1), and the retroviruses carrying them entered the primate lineage 25 and 40 million years ago, respectively.4,8,9 Indeed, these 2 HERV envelope genes have undergone purifying selection during primate evolution to play important functions in placental physiology.10,11 The existence of 2 murine endogenous retroviral envelope genes (syncytin-A and -B), homologous but not orthologous to the human syncytin genes and also critical for trophoblast cell fusion, have been reported.12 The syncytin-A-deficient mouse placenta shows specific disruption of the architecture of the syncytiotrophoblast layer I (ST-I).13 The syncytin-B null placenta exhibits impaired formation of the syncytiotrophoblast layer II (ST-II).14 Furthermore, syncytin genes with fusogenic activity and placenta-specific expression have been identified successively in the leporidae, carnivora, ruminant, tenrecidae, rodentia squirrel-related clade and marsupial opossum, namely syncytin-Ory1,15 syncytin-Car1,16 syncytin-Rum1,17 syncytin-Ten1,18 syncytin-Mar119 and syncytin-Opo1,20 respectively. This indicates that different syncytins have been acquired by different genomes and facilitate trophoblast cell-cell fusion in various animals.
Syncytin-1 has been visualized in almost all types of trophoblastic cells, including villous and extravillous trophoblast.3,5,6,21-24 Syncytin-1-induced cell-cell fusion is dependent on both ASCT1 and ASCT2 (alanine/serine/cysteine/threonine transporter type 1 and 2), also called SLC1A4 and SLC1A5, respectively, which are Na+-dependent transporters for polar neutral amino acids and a common cell surface receptor for retroviruses.6,25-27 Systematic ASC activity is present in basal but not in apical membranes of the STB,28,29 and ASCT2 appears to be ubiquitously expressed in most human placental tissues, including the CTBs and STB.28,30-32 By contrast, the expression of syncytin-2 is restricted to some of the CTBs throughout pregnancy, and more often at the interface between the CTBs and STB.33 The receptor for syncytin-2, major facilitator superfamily domain containing 2A (MFSD2A), is a carbohydrate transporter with 10–12 transmembrane domains and is specifically expressed in the STB layer.34 The highly specific localization of syncytin-2 and its receptor MFSD2A might argue for a major role of syncytin-2 in synergy with syncytin-1 during the fusion of the mononucleated CTBs into the multinucleated STB in vivo, therefore favoring the growth and renewal of the STB and precluding the fusion of CTBs between themselves.9
The 1,617 bp sequence of the syncytin-2 envelope (env) gene mapped at 6p24.1 is within the HERV-FRD locus and encodes a ˜73 kDa glycosylated precursor protein.4,33,35,36 Similar to a retroviral envelope protein, syncytin-2 is formed as a homotrimer connected by an intersubunit disulfide bond after posttranslational cleavage into the surface (SU) and transmembrane (TM) subunits by proteolytic furin, with the SU interacting with its receptor on the cell surface and the TM carrying the fusion activity and serving to mediate membrane fusion.36,37 The proper association of SU and TM subunits is necessary for the functional integrity of syncytin-1 and -2 because the chimeras of syncytin-1 and -2 swapping their subunits lose their fusogenicities.36 As shown in Fig. 1A, the syncytin-2 precursor polypeptide contains the following important regions: (a) a signal peptide (SP) in the amino-terminal end (1-15 aa); (b) a disulfide bond isomerase motif (43–46 aa), Cys-X-X-Cys (CXXC); (c) a furin cleavage site (347-350 aa), RVRR, which separates the surface (SU) (16–350 aa) and transmembrane (TM) subunits (351-538 aa); (d) a conserved hydrophobic domain as a putative fusion peptide (FP) (354-374 aa) mediating the fusion of cell membranes; (e) a highly conserved putative immunosuppressive domain (ISD) (414-430 aa); (f) a CX6CC motif (431–439 aa), in which the first 2 cysteines can form an intersubunit disulfide bond and the third cysteine can form a intrasubunit disulfide bridge with the CXXC motif; (g) 2 heptad repeat regions (HR1 and HR2) (384–422 aa and 447–456 aa); (h) a transmembrane domain (TMD) (479–499 aa); and (i) a 39-amino-acid cytoplasmic domain (CTM) (500–538 aa).36,38-40 Functional domain integrality and proper maturation conformation are required for the fusogenic activity of syncytin-2.
Figure 1.
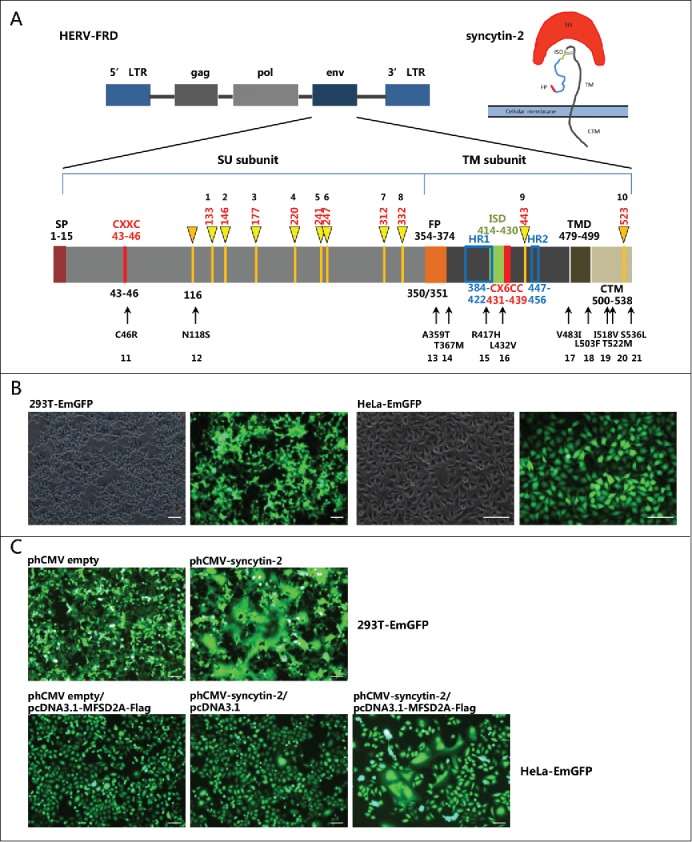
Schematic of the syncytin-2 gene structure indicating the location of candidate N-glycosylation sites and non-synonymous single-nucleotide polymorphic (SNP) sites and the establishment of the fusion assay for functional evaluation of syncytin-2 and its mutants. (A) Upper panel: structure of a human endogenous retrovirus (HERV)-FRD gene locus and its encoded envelope protein syncytin-2.4,9 HERV is typically composed of 5′ and 3′ LTRs (long terminal repeats), gag (group-specific antigen gene), pol (polymerase), and env (envelope). Syncytin-2 encoded by the env gene is composed of SU and TM subunits. Lower panel: schematic diagram of syncytin-2 indicating the positions of 10 potential N-glycosylation sites (numbered 1–10) and 10 naturally occurring SNPs from SNP databases (http://www.ncbi.nlm.nih.gov/snp) in predicted functional domains (numbered 11–21). The positions of amino acids of different domains/motifs are indicated. SU, surface protein; TM, transmembrane protein; SP, signal peptide; FP, fusion peptide; HR, heptad repeat region; ISD, immunosuppressive domain; TMD, transmembrane domain. CTM, cytoplasmic domain. (B) Establishment of 293T and HeLa cell lines that stably express EmGFP. (C) Cell-cell fusion mediated by exogenous syncytin-2. The 293T-EmGFP cells were transfected with phCMV empty vector or phCMV-syncytin-2 for 36 h. HeLa-EmGFP cells were co-transfected with phCMV-syncytin-2 and pcDNA3.1-MFSD2A-FLAG for 36 h. Single-plasmid transfection with either phCMV-syncytin-2 or pcDNA3.1-MFSD2A-FLAG did not cause cell-cell fusion. All experiments were repeated 3 times. Bars = 100 µm.
In the field of cell-cell fusion, interest in syncytins is emerging. Besides syncytins, only one another unrelated family of fusogens has been discovered, the F proteins in Caenorhabditis elegans.41 To date, critical functional characterization of single-amino-acid mutations of syncytins and their relevance to diseases are still lacking. In the present study, we investigated whether disruption of potential N-glycosylation sites and naturally occurring non-synonymous single-nucleotide polymorphisms (SNPs) (http://www.ncbi.nlm.nih.gov/snp) in functional domains affected the fusogenicity of syncytin-2, and whether naturally occurring SNPs that affected the fusogenic function of syncytin-2 were associated with pregnancy-related complications such as PE. We approached these issues by generating 21 site-specific mutants, including 10 potential N-glycosylation site mutants and 11 naturally occurring SNPs, and assessed their fusogenic activities in 293T and HeLa cells, which do not express syncytin-2 and serve as models for the cell-cell fusion assay. To define the receptor and other molecules interacting with syncytin-2, co-immunoprecipitation and protein mass spectrum analysis were utilized. Our results indicate that N-glycan and naturally occurring individual SNPs have different implications in regulating syncytin-2 function and provide a better understanding of the structure-function relationship of syncytin-2.
Results
Searching for single mutation sites that might affect the fusogenicity of syncytin-2 and the establishment of a syncytin-2-mediated fusion assay in 293T and HeLa cells
We first analyzed the putative N-glycosylation sites of syncytin-2 using the tool provided by the UniProt database. In total, there are 10 potential N-glycosylation sites, with the first 8 located in the SU and the last 2 located in the TM (Fig. 1A, numbered as 1–10). We also searched the SNP database and found that 162 naturally occurring non-synonymous SNPs in the open reading frame (ORF) of syncytin-2 had been recorded. We selected 10 sites that were located within important functional domains or motifs. Of these, N118S probably introduced a new N-glycosylation site, C46R was located in the CXXC domain, A359T and T367M were located in the FP, R417H was located in the HR1 and ISD domains, L432V was located in the CX6CC motif, V483I was located in the TMD, and L503F, I518V, T522M and S536L were located in the CTM (Fig. 1A, numbered as 11–21). To characterize the effects of these N-glycosylation sites and non-synonymous SNPs on the fusogenic function of syncytin-2, we established a fusion assay using 293T-EmGFP and HeLa-EmGFP cell lines that stably overexpress green fluorescent protein (Fig. 1B) to monitor cell-cell fusion. The syncytin-2 receptor, MFSD2A, is expressed in 293T cells.4 Therefore, syncytin-2-mediated cell-cell fusion could be observed 24 h after transfection with a syncytin-2 expression plasmid. As fusion proceeded, patches of green fluorescence were seen as the formation of multinucleated syncytia (Fig. 1C). Moreover, as HeLa cells do not express MFSD2A,34 syncytin-2 mediated cell-cell fusion only occurred when co-transfecting syncytin-2 and MFSD2A overexpression plasmids, but not either plasmid alone (Fig. 1C).
Production and analysis of the effects of 21 single-amino-acid-substitution mutants that either disrupt N-glycosylation sites or are encoded by naturally occurring non-synonymous SNPs on the fusogenicity of syncytin-2
We first explored the requirement of each N-glycosylation site for the fusogenic function of recombinant syncytin-2 by N to Q substitution at N133, N146, N177, N220, N241, N247, N312, N332, N443 and N523 (Table 1). Meanwhile, the naturally occurring N118S, which probably introduces an N-glycosylation site, was also investigated. Of these mutants, N146Q, N177Q, N220Q, N241Q and N247Q in the SU subunit maintained their original fusion efficiency compared to wild-type (WT) syncytin-2, whereas N133Q, N312Q and N443Q lost their fusogenicity and N118S and N332Q acquired enhanced fusogenic activity (Fig. S1). Importantly, as shown in Fig. 2, when the cell samples were subjected to Western blot analysis, all N to Q substitutions except for N523Q revealed bands with lower molecular weight (arrowhead) than WT syncytin-2, indicating that N-glycosylation of syncytin-2 was widely distributed at these sites. Moreover, the naturally occurring N118S caused a higher molecular weight (arrow), indicating that the introduction of an extra N-glycosylation site occurred. Furthermore, the expression levels of the WT and mutant syncytins were consistent, showing that the transfection efficiency of the different plasmids and the exogenous expression of syncytin-2 were at a same level. Although the N118S immunostaining was reduced, it still significantly elevated the fusion efficiency. As a control, when all the N-glycans were removed by treatment with PNGase F N-glycanase, only a ˜55 kDa band was seen, whereas the precursor of syncytin-2 protein appeared to be ˜73 kDa, in accordance with previous results.33,35, 36
Table 1.
Nucleotide and amino acid changes introduced to generate syncytin-2 mutants by site-directed mutagenesis.
| Mutant site | Residue number | Forward Primer (5′ - 3′) | Allele change | Fusion |
|---|---|---|---|---|
| 1 | 133 | GTGTTATGGCCAAAAGGAAACAGGGAACAAATGTAGGCACTC | aat→cag N→Q | − |
| 2 | 146 | CTCTTCCAAGTACAGTCTGTCAGGTTACTTTCACTGTAGATTC | aat→cag N→Q | + |
| 3 | 177 | CAAGATTCCCCAAACCTCCACAGATTACTTTTCCTCAGGGAAC | aat→cag N→Q | + |
| 4 | 220 | CCAATGTCTGCAAATTTCCCAGCTCAGCTCTACAGCGGAATGG | aac→cag N→Q | + |
| 5 | 241 | CGAAATTCTCTTTTTTGGGAACAGAAAACCAAGGGAGCTAACCAG | aat→cag N→Q | + |
| 6 | 247 | GGAAAATAAAACCAAGGGAGCTCAACAGAGCCAAACACCCTGCGTCC | aac→caa N→Q | + |
| 7 | 312 | CACCAATGCCTCCCCAGTCAATGGACTGGAACTTGTACC | aac→caa N→Q | − |
| 8 | 332 | CTTCATAGCCCCTGGCCAGCTCTCTCTTCCAATAC | aat→cag N→Q | ++ |
| 9 | 443 | GAAAAATGTTGCTTTTGGGTACAGCAATCAGGAAAAGTACAAGAC | aat→cag N→Q | − |
| 10 | 523 | GGCCATAAAGCTCCAGACGCAACTCAGTGCAGGACGCCATC | aat→caa N→Q | + |
| 11 | 46 | CCACCAATTGCTGGTTACGTACTAGCTCTTCCAC | tgt→cgt N→Q | − |
| 12 | 118 | CCATCTTTACTAATATCAGCCTAATGGGAATAGCCCC | aac→agc N→Q | ++ |
| 13 | 359 | CATTTCATTCCCCTTCTCACGGGACTCGGCATTCTAG | gcg→acg N→Q | + |
| 14 | 367 | CGGCATTCTAGCTGGTATGGGAACCGGAATTGCTG | acg→atg N→Q | ++ |
| 15 | 417 | CAGCCGTAGTCCTTCAAAATCATCGAGGACTAGACATGTTAAC | cgt→cat N→Q | − |
| 16 | 432 | GCACAGGGAGGAATTTGTGTGGCCTTAGATGAAAAATG | ttg→gtg N→Q | + |
| 17 | 483 | GGAAATGGTTCTCTTGGATTCTTCCCCTTACAGGC | gtt→att N→Q | ++ |
| 18 | 503 | GCTCCTTTTTGGTCCATGTTTCCTAAATCTAATAACCC | ctc→ttc N→Q | + |
| 19 | 518 | GTCTCCTCTCGCCTTCAGGCCGTAAAGCTCCAGACGAATCTCA | ata→gta N→Q | + |
| 20 | 522 | GGCCATAAAGCTCCAGATGAATCTCAGTGCAGGAC | acg→atg N→Q | ++ |
| 21 | 536 | CCTCGCAATATTCAAGAGTTACCCTTCTAGTGACTGCAG | tca→tta N→Q | + |
a Cell fusion was monitored 36 h after transfection. −, no fusion; +, moderate fusion; ++, excessive fusion.
Figure 2.
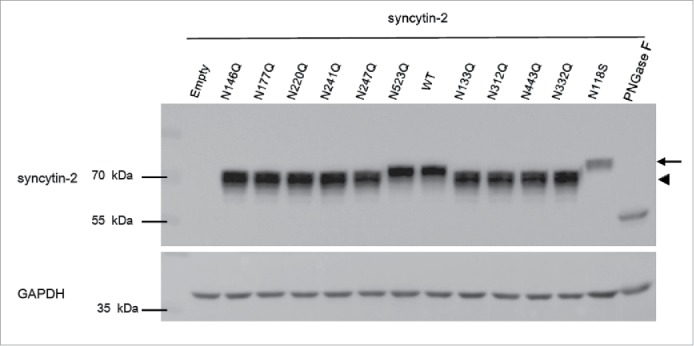
Western blot analysis of the expression of syncytin-2 and its single N-glycosylation site mutants in 293T-EmGFP cells after 36 h of transfection. All N-glycans of wild-type (WT) syncytin-2 were removed by treatment with PNGase F N-glycanase. phCMV empty vector (empty) was used as a control. The arrow and arrowhead represent bands with higher and lower molecular weight than for WT syncytin-2, respectively.
We also investigated the effects of the 10 selected non-synonymous SNPs located within important functional domains/motifs on the fusogenic function of syncytin-2. Of these, A359T, L432V, L503F, I518V and S536L had no impact on the fusion efficiency mediated by syncytin-2 (Fig. S1). Notably, C46R and R417H inhibited normal cell-cell fusion, whereas T367M, V483I and T522M enhanced fusogenic activity compared with the WT (Fig. S1). Again, the expression levels of these mutants were consistent with that of WT (Fig. S2).
Identification of dysfunctional syncytin-2 mutants that stimulate or inhibit syncytin-2-mediated fusion
We next wanted to further characterize the above dysfunctional mutants together with WT syncytin-2, and the effects of the different stimulatory mutants (including N118S, N332Q, T367M, V483I and T522M) and inhibitory mutants (including N133Q, N312Q, N443Q, C46R and R417H) were confirmed (Fig. 3A). N118 and N133 are close to each other and within the N-terminal half of the SU subunit. Gaining an extra N-glycosylation site at N118 and removal of the N-glycan at N133 stimulated and inhibited the fusion efficiency, respectively. However, although both N312 and N332 are located in the C-terminal region of the SU subunit, disruption of both sites individually caused contradictory effects. Furthermore, C46R within the CXXC motif, T367M within the FP, R417H within the HR1 and ISD, V483I within the TMD and T522M within the CTM all caused significantly altered fusogenic activity, indicating that these functional domains were sensitive to single-amino-acid substitution and that these sites were required for optimal fusion induction (Fig. 3A). Importantly, Western blot (Fig. 3B) and quantitative real-time PCR (Fig. 3C) analysis were used to ensure that all the WT and mutant syncytins were expressed at similar levels both at the mRNA and protein levels. Again, N118S stimulated the fusogenic activity of syncytin-2, despite the reduced immune-reactivity (Fig. 3B). Because its mRNA was expressed normally, one explanation for this could be the loss of an epitope due to the amino acid substitution.
Figure 3.
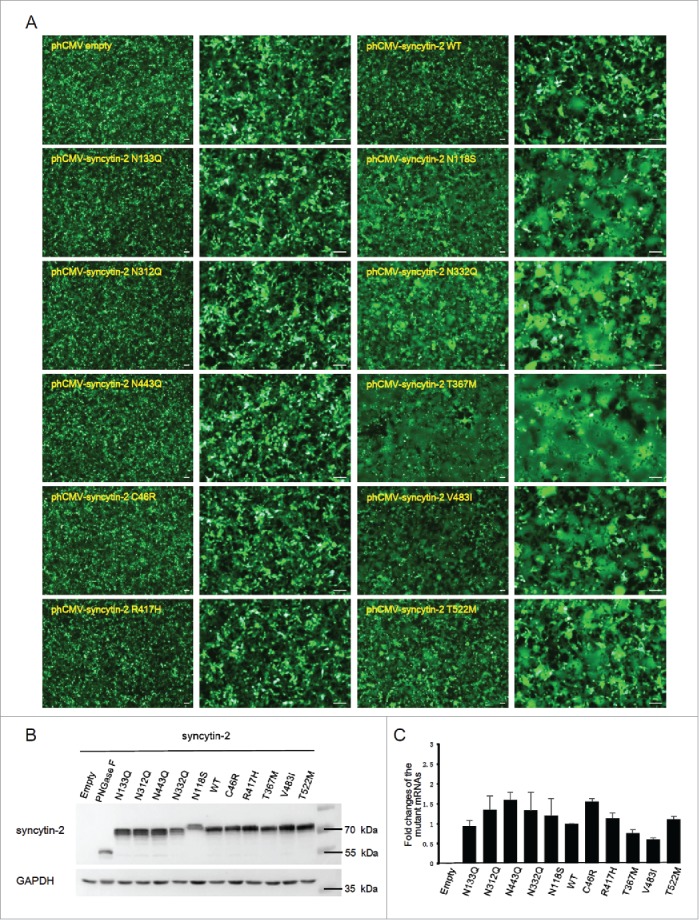
Expression of WT and mutant syncytin-2 in 293T-EmGFP cells and effects on cell-cell fusion. (A) Cell-cell fusion assay mediated by wild-type (WT) or mutant syncytin-2. 293T-EmGFP cells in 35 mm plates were transfected with 1.5 μg phCMV-syncytin-2 WT or mutant expression plasmids for 36 h. Patches of green fluorescence were monitored as the formation of multinucleated syncytia. Compared with WT syncytin-2, more obvious syncytia were observed by transfection with syncytin-2 mutants N118S, N332Q, T367M, V483I and T522M. No syncytia were visible after overexpression of syncytin-2 mutants C46R, N133Q, N312Q, N443Q and R417H followed by microscopic examination at ×40 and ×100 magnification. Bars = 100 µm. (B) Western blot analysis of the expression of syncytin-2 and its mutants in 293T-EmGFP cells after 36 h of transfection. The cells shown in A were harvested for immunoblotting using syncytin-2 and GAPDH antibodies, respectively. (C) The transcription levels of syncytin-2 were simultaneously quantified by real-time PCR using GAPDH as an internal control. Mean values and SDs are obtained from 3 independent experiments. Empty, phCMV empty vector.
We then tested whether the 5 inhibitory mutants (N133Q, N312Q, N443Q, C46R and R417H) blocked syncytin-2-mediated fusion in 293T-EmGFP cells. We found that 293T-EmGFP cells in 6-well plates transfected with 1.5 μg WT syncytin-2 expression vector for 36 h formed obvious green multinucleated syncytia. The induction of cell-cell fusion upon syncytin-2 overexpression was dose-dependent, and higher doses of expression plasmids triggered cell-cell fusion to a larger extent (Fig. 4A-B). We then co-transfected the cells with 1.5 μg phCMV-syncytin-2 WT and 1.5 μg inhibitory mutant plasmids. As a control, the plasmids expressing the mutants were replaced with phCMV empty vector. As shown in Fig. 4C, the C46R and R417H mutants almost completely blocked the fusogenicity of WT syncytin-2. Mutants N133Q, N312Q and N443Q significantly suppressed the fusogenic properties of the native syncytin-2 protein. These results suggest that these mutants cause significant repressive effects on the fusogenic activity of syncytin-2 and therefore may serve as competitive inhibitors in WT syncytin-2-mediated cell-cell fusion.
Figure 4.
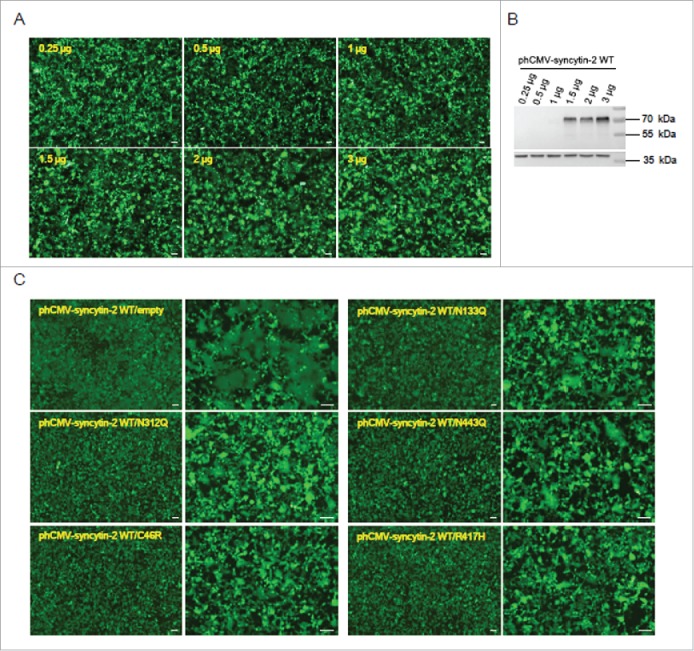
The 5 mutants N133Q, N312Q, N443Q, C46R and R417H exerted inhibitory effects on syncytin-2-mediated cell-cell fusion. (A) Dose-dependent promotion of cell-cell fusion after overexpression of syncytin-2. The 293T-EmGFP cells in 35 mm plates were transfected with 0.25 μg, 0.5 μg, 1 μg, 1.5 μg, 2 μg and 3 μg phCMV-syncytin-2 WT for 36 h. Cell-cell fusion was monitored by microscopy at ×40 magnification. (B) The cells shown in A were harvested for Western blotting analysis with syncytin-2 antibody. (C) The N133Q, N312Q, N443Q, C46R and R417H mutants of syncytin-2 interfere with the fusogenic capacity of WT syncytin-2 proteins. The 293T-EmGFP cells in 35 mm plates were transfected with 1.5 μg WT syncytin-2 and 1.5 μg mutant expression plasmids, followed by microscopic examination at ×40 and ×100 magnification. As a control, the cells were transfected with 1.5 μg WT syncytin-2 expression vector and 1.5 μg phCMV empty vector. All experiments were repeated 3 times. WT, wild-type. Bars = 100 µm.
The abnormal interaction between syncytin-2 mutant and its receptor MFSD2A during cell-cell fusion
To verify the interaction between syncytin-2 and its receptor MFSD2A, they were co-overexpressed in HeLa cells that express neither protein and analyzed by co-immunoprecipitation. We firstly detected the expression of FLAG-tagged MFSD2A in HeLa cells by Western blotting using an anti-FLAG antibody. No immunoreactive bands were detected in control mock-transfected cells and pEF6-MFSD2A-transfected cells (Fig. 5A). Monoclonal anti-FLAG antibody was then used for the co-immunoprecipitation of syncytin-2 in HeLa cells. As shown in Fig. 5B, the lysed cell samples precipitated by the anti-FLAG antibody contained significant amounts of syncytin-2 protein, indicating that full-length MFSD2A associates with syncytin-2. Syncytin-2 was not detected either in the co-transfection of pcDNA3.1-MFSD2A-FLAG and phCMV empty vector or of pcDNA3.1-GFP-FLAG and phCMV-syncytin-2, indicating the adequate protein expression in this system and that the antibodies in this expression system are largely devoid of nonspecific protein binding. To infer the sensitivity and specificity of syncytin-2 with its receptor MFSD2A, we further examined the interaction between different syncytin-2 mutants and FLAG-tagged MFSD2A. The expression levels of WT syncytin-2 and syncytin-2 mutants were adjusted to similar levels. MFSD2A-FLAG strongly co-immunoprecipitated with WT syncytin-2 and mutants N133Q, N312Q, N332Q, V483I and T522M, whereas minimal amounts were detected in N443Q, C46R, T367M and R417H immunoprecipitated complexes. N118S was associated with more receptors than the WT (Fig. 5C). We also employed preliminary mass spectrometry studies to find syncytin-2 interacting proteins during membrane merging (see Supplementary data and Fig. S3).
Figure 5.
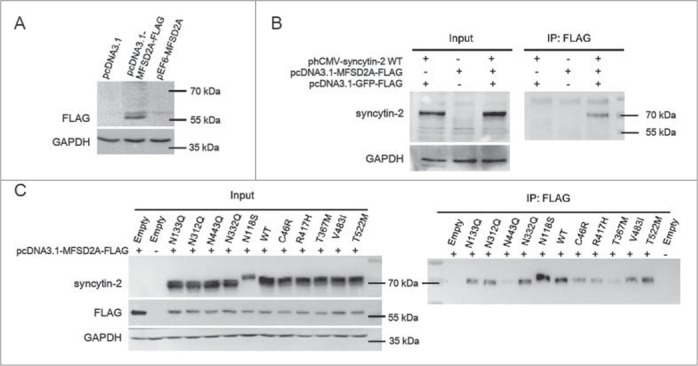
Interaction between syncytin-2/mutants and the MFSD2A receptor. (A) HeLa cells were transiently transfected with expression plasmids encoding either FLAG-tagged MFSD2A (pcDNA3.1-MFSD2A-FLAG) or MFSD2A without a FLAG tag (pEF6-MFSD2A) and subjected to Western blotting assay using an anti-FLAG antibody. The pcDNA3.1 empty vector was used as a control. (B) pcDNA3.1-MFSD2A-FLAG and phCMV-syncytin-2 were co-transfected into HeLa cells to test the sensitivity and specificity of the interaction between syncytin-2 and MFSD2A. Cell lysates were immunoprecipitated with an anti-FLAG antibody and immunoblotted with an anti-syncytin-2 antibody. phCMV empty vector and pcDNA3.1-GFP-FLAG were used as a control. (C) Cells expressing either syncytin-2 WT or the mutants were lysed, immunoprecipitated with the anti-FLAG antibody and immunoblotted with the anti-syncytin-2 antibody. phCMV empty vector and pcDNA3.1-GFP-FLAG were used as a control. WT, wild-type. Empty, phCMV empty vector.
Identification of the T522M SNP in the coding region of syncytin-2 by genomic DNA sequencing in sPE patients and normal pregnant women
We further set out to identify non-synonymous SNPs in the coding region of syncytin-2 in 125 sPE patients and 272 normal pregnant women by genomic DNA sequencing. Surprisingly, only 1 missense mutation (rs138651238; c.1565C>T and p.T522M) was discovered (Fig. 6). In total, 18 heterozygous carriers of T522M out of 272 normal pregnant women were found. However, only 4 heterozygous carriers out of 125 sPE patients were identified. The frequency of T522M heterozygous carriers was 0.0618 in the control group versus 0.0320 in sPE patients. A chi-square test was used to analyze the difference, revealing a chi-square of 0.167 and a P value of 0.124. The heterozygosity and MAF of T522M recorded by the 1000 Genomes Project are 0.009 and 0.0044, respectively. Therefore, the frequency of this SNP appeared to be higher in the Chinese population.
Figure 6.

Identification of heterozygous carriers of syncytin-2 T522M (rs138651238) in normal pregnant women and severe pre-eclamptic (sPE) patients. (A) Frequency of T522M in normal pregnant women (n = 272) and sPE patients (n = 125). (B) Sequencing of syncytin-2 coding region revealed a C > T single-nucleotide polymorphism with amino acid substitution at 522 (T522M). MAF, minor allele frequency.
Discussion
We identified 10 single-amino-acid-substitution human syncytin-2 mutants with abnormalities in fusogenic function, including N133Q, N312Q, N332Q and N443Q, which silenced the N-glycosylation sites individually, and the naturally occurring SNPs C46R, N118S, T367M, R417H, V483I and T522M. N118S introduced a new N-glycosylation site. Of these 10 mutants, N133Q (in the N-terminal half of the SU), N312Q (in the C-terminal region of the SU), N443Q (in the TM, close to the CX6C motif and HR2 domain), C46R (in the CXXC motif) and R417H (in the HR1 and ISD domain) exerted inhibitory effects on the fusogenic activity of syncytin-2, whereas N118S (in the N-terminal half of the SU), N332Q (in the C-terminal region of the SU), T367M (in the FP), V483I (in the TMD) and T522M (in the CTM) were stimulatory.
Several lines of evidence suggest that the sequence polymorphisms of HERV family genes provide susceptibility to human diseases.42,43 For example, ERVW-1 3′-long-terminal repeat (LTR) SNPs (142 T>C and 277 A>G) are associated with syncytin-1 overexpression and may be an indicator of the risk of urothelial cell carcinoma of the bladder.43 However, there is also a report suggesting the conservation of the ERVW-1 locus, including the LTR transcriptional elements and the splice sites involved in env mRNA maturation in 155 individuals.11 The authors also sequenced the envelope ORF that encodes syncytin-1 in 24 individuals and found 4 non-synonymous mutations that occurred at a frequency of 25%, 4%, 2%, and 2%, respectively. However, these polymorphic variants are functionally preserved and do not affect the fusogenic function of WT syncytin-1 in a heterotypic cell-cell fusion assay.11 To our knowledge, this is the only published study related to the functional analysis of the non-synonymous SNPs in the coding region of syncytins. Despite the availability of public SNP databases in which 162 non-synonymous SNPs of syncytin-2 have been recorded, their functional study and relevance to diseases remain surprisingly elusive.
As one of the most common post-translational protein modifications, N-glycosylation is crucial for many elementary biological and pathological processes. Syncytins belong to the class I fusion proteins that include the envelope proteins of many viruses, such as the human immunodeficiency virus (HIV). Typically, class I fusion proteins are synthesized as glycosylated precursors in the endoplasmic reticulum and then modified by N-glycosylation and disulfide bond formation.40 As such, syncytin-2 is also glycosylated.33,36 In the case of the HIV envelope protein, N-glycosylation is critical for viral infectivity, immunogenicity, cytopathicity and transmission.44-46 In an extensive study, 23 N-glycosylation sites in the gp120 subunit of the simian immunodeficiency virus (SIV) envelope protein are individually silenced, and the mutations result in different effects on viral infectivity, replication and recovery.47 Therefore, we set out to screen all the N-glycosylation sites in syncytin-2 and to investigate whether they are necessary for the fusogenic function.
Using individual deglycosylation studies, we verified that except for N523, the other 9 predicted N-glycosylation sites, including N133, N146, N177, N220, N241, N247, N312, N332 and N443, were indeed glycosylated in 293T cells. Moreover, the naturally occurring N118S introduced an extra N-glycosylation site and produced a protein with a higher molecular weight. Of these 10 glycosylated sites, the first 9 were all located within the SU subunit. Interestingly, only the mutations of the first 2 and the last 2 sites (N118S, N133Q, N312Q and N332Q), but not of the middle 5 sites (N146Q, N177Q, N220Q, N241Q and N247Q) altered the fusogenicity of syncytin-2. The binding of the SU to the receptor is the prerequisite for the FP at the N-terminus of the TM to drive membrane fusion.37,40 Therefore, our results indicate that the N-glycans located at the N-terminal half and C-terminal arm of the SU subunit are important for syncytin-2-mediated cell-cell fusion probably by affecting the interaction between the SU and the receptor. The non-functional sites from N146 to N247 indicate that this region is flexible in conformation and tolerates losing the glycosylation. In the case of syncytin-1, the N-terminal 124 amino acids of the SU are the minimal receptor-binding domain containing a conserved 18-residue motif essential for syncytin-1-ASCT2 interaction.48 Moreover, N443Q, which is in proximity to the CX6CC motif and HR2 domain in the central region of the ectodomain TM, also significantly suppressed the fusogenic property of the native syncytin-2 protein. As reported, the 2 heptad repeat regions HR1 and HR2 specifically form intramolecular interaction between each other and are necessary for syncytin-mediated cell-cell fusion.38 Our results add new evidence that the N443Q might destabilize the hairpin conformation of HR1 and HR2, or hinder the formation of the disulfide bond, and discourage cell-cell fusion. Taken together, our results indicate that syncytin-2 has 9 N-linked glycosylation sites, among which glycans at residues 133, 312, 443 and 332 are required for optimal fusion induction, and that appropriate sugar chain density over the specific region is preserved for functional integrity.
We also generated another 10 mutants carrying the naturally occurring SNPs C46R, A359T, T367M, L432V, R417H, V483I, L503F, I518V, T522M and S536L, which are located in important functional domains or motifs. Of these, 2 defective mutants (C46R and R417H) and 3 stimulatory mutants (T367M, V483I and T522M) were identified. C46R is located in the CXXC motif of the SU subunit, which is highly conserved among retroviral envelope proteins and participate in the formation of a labile intersubunit disulfide bond with the CX6CC motif of the TM subunit for isomerization.36,49 Therefore, the silencing effect of C46R on the fusogenic activity confirmed the significance of the CXXC motif for fusogenesis and the necessity of the cysteine for the formation of an intersubunit disulfide bridge. This agrees well with the effect of C46A from a previous extensive study, which shows mutations in the CXXC motif not only inhibit fusion but also can function as dominant-negative mutants.36 However, the naturally occurring SNP L432V located in the CX6CC motif in the TM subunit did not alter the fusion efficiency, indicating the flexibility of the 6 amino acids between the 3 cysteines. R417H is located in 2 domains, the HR1 domain discussed above and the 17-amino-acid ISD present in several retroviral envelope proteins that is highly immunosuppressive for inhibiting immune function.50,51 R417H led to a complete loss of its fusogenic activity, implying the significance of both domains for syncytin-2 fusion ability.
Of the 3 stimulatory mutants that can enhance the fusogenic activity of syncytin-2, the T367M is located within the FP. FP is a conserved hydrophobic domain located at the N-terminus of the TM subunit mediating cell membrane fusion.38 The SNP V483I is located at the hydrophobic stretch TMD domain. This suggests that the conformation of the hydrophobic transmembrane region might be sensitive to single-amino-acid substitution, although both valine and isoleucine are nonpolar amino acids. The CTM of syncytin-1 and syncytin-2 is essential for their fusogenic activities. The comparison of human ERVW-1 elements and the orthologous loci in simians reveals an syncytin-1-specific signature in the intracytoplasmic tail is crucial for the envelope fusogenic activity.10 A systematic study involves a series of C-terminal truncated mutants of syncytin-1 shows that cytoplasmic sequences immediately adjacent to the transmembrane region are necessary for inducing optimal cell-cell fusion, whereas the extreme C-terminus partially inhibits its fusogenic function.39 Similarly, different C-terminally truncated syncytin-2 mutants led to a decrease in the size and number of syncytia at different levels.36 Of the 4 SNPs (L503F, I518V, T522M and S536L) in the CTM tested, only T522M changed the fusogenic activity compared with the WT. This is also consistent with previous findings that some regions in the CTM of syncytin-1 are flexible in the conformation.38 Altogether, we have characterized 5 SNPs (C46R, T367M, R417H, V483I and T522M) in conserved domains as strong candidates that can alter the fusogenic function of syncytin-2.
To further define the functional properties of the 5 defective mutants, WT and mutant syncytin-2 expression plasmids were co-transfected. We found that the C46R and R417H mutants almost completely blocked syncytin-2-mediated cell-cell fusion, and N133Q, N312Q and N443Q mutants significantly suppressed the fusogenic properties of the native syncytin-2 protein. The molecular mechanism of inhibition is unclear, but it is likely that the mutants with single-amino-acid substitutions in important functional domains can efficiently compete with native syncytin-2 to bind the receptor or to trigger signaling pathways for merging membranes. We therefore employed co-immunoprecipitation experiments to study the sensitivity and specificity of the interaction between syncytin-2 and its receptor MFSD2A. Of the 5 defective mutants, N133Q and N312Q could be pulled down normally by MFSD2A, whereas N443Q, C46R and R417H showed compromised interaction with the receptor. The results with R417H and N443Q are quite unexpected because they are located within the TM subunit. This finding probably provides insight into the significance of the TM subunit serving as an indispensable component of the SU-TM homotrimer during the interaction between the SU and the receptor. Notably, the significantly induced fusion with the N118S mutant correlated well with its higher ability to bind with the receptor. However, whether the T367M mutant that showed reduced interaction with the receptor while promoting fusion is related to the activation of other signaling pathways remains to be studied.
PE is a common, multi-system and serious complication of human pregnancy affecting 3–5% of all women. Characterized by new-onset hypertension and proteinuria after 20 weeks of gestation, PE is a major contributor to maternal morbidity and mortality worldwide, and unbalanced syncytialization has been implicated in its pathology.52 Expression of both syncytin-1 and syncytin-2 are significantly decreased in sPE placentas vs. normal control.23,36,53-61 In our attempts to identify polymorphic sites within the coding sequence of syncytin-2 in 125 sPE patients and 272 normal pregnant women, only 1 missense mutation [rs138651238 (T522M)] was found. This mutation enhanced syncytin-2-mediated cell-cell fusion in vitro and occurred at a frequency of 0.0320 in sPE patients and 0.0618 in the control group. However, the statistical analysis revealed no significant difference. Considering the large number of non-synonymous SNPs in the coding region of syncytin-2 in the SNP database, further studies in a larger sample size are warranted.
Taken together, we provided novel in vitro experimental evidence for the structure-function relationship of syncytin-2 through investigating 21 single-amino-acid mutants. We proved that N133, N146, N177, N220, N241, N247, N312, N332 and N443 were glycosylation sites and that N133, N312, N332 and N443 played important roles in the fusogenic function of syncytin-2. The naturally occurring polymorphisms C46R, R417H, N118S, T367M, V483I and T522M led to abnormal fusogenicity. The binding between syncytin-2 and its receptor MFSD2A was compromised by the N443Q, C46R, T367M and R417H mutations, whereas N118S was associated with more receptors. We sequenced the syncytin-2 coding sequences in sPE patients and normal pregnant women and found 1 missense mutation T522M. The above results suggest some key residues in determining the fusogenic function of syncytin-2, and investigation of the mechanisms will help to better understand how this molecule merges the membranes of CTBs.
Materials and methods
Blood and DNA samples
A total of 397 blood samples were collected from 272 normal pregnant Han Chinese women and 125 sPE patients undergoing legal procedures in Beijing Obstetrics and Gynecology Hospital and Peking University Third Hospital. A woman was determined to have sPE when either severe hypertension (a systolic blood pressure ≥ 160 mmHg or a diastolic blood pressure ≥ 110 mmHg), severe proteinuria (more than 5 g of protein was collected in a 24 h urine specimen), or both, were present after 20 weeks of gestation.62 None of the sPE patients involved in this study had other maternal complications. All the sPE patients were aged 23–35 and delivered at 25 to 40 weeks. In all cases, blood samples were anonymized after routine analysis was completed. Ethical approval and utilization of samples under standard experimental protocols was granted by the Ethics Committee of Beijing Obstetrics and Gynecology Hospital, Peking University Third Hospital and the Institute of Zoology, Chinese Academy of Sciences. Genomic DNA from blood lymphocytes was extracted using a genomic DNA extraction kit (DN0113, Biomed Biotechnology Corp., Beijing, China).
Plasmid construction and site-directed mutagenesis
The human syncytin-1 expression construct phCMV-syncytin-111 is a kind gift from Dr. Francois Mallet, Laboratoire Commun de Recherche Hospices Civils de Lyon-bioMérieux, Cancer Biomarkers Research Group, Ecole Normale Supérieure, Lyon, France. The phCMV empty vector was generated by removing the syncytin-1 cDNA. The syncytin-2 cDNA fragment was amplified from pTM1-syncytin-236 (a kind gift from Dr. Hungwen Chen, Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan) and cloned into phCMV. A full-length cDNA encoding the human MFSD2A with FLAG was amplified from pEF6-MFSD2A63 (a kind gift from Dr. Tommaso Dragani and Dr. Francesca Colombo, Department of Predictive and for Prevention Medicine, Fondazione IRCCS Istituto Nazionale Tumori, Milan, Italy) and cloned into an XhoI/XbaI-digested pcDNA3.1 to generate pcDNA3.1-MFSD2A-FLAG. The full-length EmGFP coding sequence was amplified from the pcDNA6.2-EmGFP vector (V355–20, Invitrogen) and cloned into the pQCXIP vector using the restriction enzymes NotI/BamHI to generate pQCXIP-EmGFP. The primers used for amplifying the above cDNAs are summarized in Table 2.
Table 2.
Primers used for plasmid construction, real-time quantitative PCR and genomic DNA sequencing.
| Gene | Forward Primer (5′ - 3′) | Reverse primer (5′ - 3′) |
|---|---|---|
| Plasmid construction | ||
| Syncytin-2 cDNA | 5′-CGCACTCGAGCACGCCACCATGGGCCTGCTCCTGCTGGTTCTCATTCTC-3′ | 5′-ATTCTGCAGTCACTAGAAGGGTGACTCTTGAATATTGCGAGG-3′ |
| MFSD2A-FLAG cDNA | 5′-CTAGCTAGCCTCGAGGCCACCATGGCCAAAGGAGAAG-3′ | 5′-GCTCTAGACTACTTGTCATCGTCATCCTTGTAGTCGATGTCATGATCTTTATAATCACCGTCATGGTCTTTGTAGTCATTAAGTTTGTGCCCCAG-3′ |
| EmGFP cDNA | 5′-GAGCGGCCGCAGCCACCATGGTGAGCAAGGGCGAGGA-3′ | 5′-GCGGGGATCCTCTATTACTTGTACAGCTCGTCCA-3′ |
| Real-time quantitative PCR | ||
| Syncytin-2 | 5′-CCAAAGGAACCTGAAAATCC-3′ | 5′-TTAGGGATTCTTAGTCAGCC-3′ |
| GAPDH | 5′-AAGGTGAAGGTCGGAGTCAAC-3′ | 5′-GGGGTCATTGATGGCAACAATA-3′ |
| Genomic DNA sequencing | ||
| HERV-FRD | LF: 5′-AACTTAGGCACATTAGCATT-3′ | LR: 5′-AACTTAGGCACATTAGCATT-3′ |
| syncytin-2 ORF | SF: 5′-TGTACACCACCAGGAGTTCC-3′ | SR: 5′-TGGGTCTTGGCCTCTTGCTA-3′ |
| syncytin-2-S1 | — | 5′-GCTGTAGAGCTGAGGTTGGA-3′ |
| syncytin-2-S2 | 5′-CAGACTTACCAAACATACACCC-3′ | — |
| syncytin-2-S3 | 5′-ACCCCAGACATCTTCATAGC-3′ | — |
The 21 human syncytin-2 mutants, including 10 mutants (N133Q, N146Q, N177Q, N220Q, N241Q, N247Q, N312Q, N332Q, N443Q and N523Q) removing potential N-glycosylation sites, 1 naturally occurring SNP (N118S) introducing a new potential N-glycosylation site, and another 10 naturally occurring SNPs (C46R, A359T, T367M, R417H, L432V, V483I, L503F, I518V, T522M and S536L) located in predicted important functional domains were generated on the WT phCMV-syncytin-2 expression vector template using the QuikChange II XL site-directed mutagenesis kit (200514, Stratagene) following the manufacturer's instructions and verified by sequencing. The forward and reverse primers used for mutagenesis are summarized in Table 2. All constructs were sequenced to confirm that only the targeted mutations had occurred. The putative N-glycosylation sites in human syncytin-2 proteins were obtained from the UniProt database.
Cell lines
293T and HeLa cells were purchased from the American Type Culture Collection. Cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Life Technologies), 100 µg/ml streptomycin and 100 U/ml penicillin, and maintained in 95% air and 5% carbon dioxide at 37°C.
Establishment of stable GFP-expressing 293T and HeLa cell lines
293T and HeLa cells were transfected with pQCXIP-EmGFP using Lipofectamine 2000 (15338–100, Invitrogen) according to the manufacturer's instructions. Transfected cells were subjected to puromycin (P8833; Sigma Chemical Co.) selection (1.5 μg/ml for 293 T cells and 0.125 μg/ml for HeLa cells), and the antibiotics-resistant clones were pooled for studies. The expression of EmGFP was confirmed under a fluorescence microscope.
Fusion assay of syncytin-2
Cell-cell fusion mediated by syncytin-2 and its mutants was monitored 36 h after expression plasmid transfection by tracing EmGFP-expressing 293T or HeLa cells. Fusion events were traced by fluorescence microscopy as the formation of green multinucleated syncytia in transfected cells.38 For the negative control, cells were transfected with phCMV empty vector.
Real-time quantitative-PCR
Total RNA was extracted and purified from cultured cells using TRIzol reagent (15596–018, Invitrogen) according to the manufacturer's instructions. RNA concentration was determined with a NanoDrop 2000 UV-Vis spectrophotometer (Thermo Scientific). Two micrograms of total RNA were reverse transcribed into single-stranded cDNA using Superscript II reverse transcriptase (18064022, Invitrogen). Real-time quantitative PCR was performed using SYBR Premix Ex Taq kit (DRR081A, Takara) with the Real-time PCR System (ABI PRISM 7500 Real-time PCR System, Applied Biosystems). Specific primers are indicated in Table 2. Data were analyzed using ΔCt method and normalized to GAPDH expression. All data were based on experiments performed at least in triplicates.
Western blotting and co-immunoprecipitation
The cells were homogenized in whole cell lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol, 0.5% Triton X-100). A total of 30 μg/well of proteins were resolved by SDS-PAGE and transferred electrophoretically onto nitrocellulose membranes (Pall Corporation). After incubation with blocking buffer (5% fat-free milk in phosphate-buffered saline containing 0.5% Tween 20), membranes were incubated overnight at 4°C with specific primary antibodies against syncytin-2 (P60508, Abgent) and GAPDH (ab37187, Abcam), followed by incubation with appropriate secondary antibody. The blot was developed using the enhanced chemiluminescence (Pierce Chemical Co.). PNGase F N-glycanase (P0704S, Sigma Chemical Co.) was used to remove N-glycans in the protein samples following the manufacturer's instructions.
For the co-immunoprecipitation assay, HeLa cells were co-transfected with 4 μg WT or mutant human syncytin-2 constructs and 4 μg pcDNA3.1-MFSD2A-FLAG in 60 mm culture dishes. Control experiments were performed by transfecting cells with appropriate empty vectors, phCMV empty vector or pcDNA3.1-GFP-FLAG. After transfection for 48 h, the cells were lysed by suspension in ice-cold lysis buffer (50 mM Tris pH 7.5, 150 mM NaCl, 10% glycerol and 0.5% Triton X-100) supplemented with protease inhibitor cocktail (1mM phenylmethylsulfonylfluoride, 10 μg/ml aprotinin and 1mM sodium orthovanadate) (04693132001, Roche Diagnostics) for 1 h. After centrifugation at 12,000 g for 20 min, the cell lysates containing FLAG-tagged MFSD2A proteins were immunoprecipitated with anti-FLAG M2 agarose beads (F2426, Sigma Chemical Co.) at 4°C overnight. After centrifuging and washing for 3 times, samples were subjected to Western blotting analysis with an anti-syncytin-2 antibody.
Mass spectrometry analysis
Immunoprecipitation of the concentrated proteins was carried out with protein A sepharose beads (10–1141, Invitrogen) essentially as described by the manufacturer. 293T cells were transfected with phCMV empty vector, phCMV-syncyntin-2 WT, phCMV-syncytin-2 N312Q and phCMV-syncytin-2 T522M plasmids, respectively. After transfection for 48 h, the cells were lysed by suspension in ice-cold lysis buffer for 1 h. Lysates were collected after centrifugation at 12,000 g for 20 min and immunoprecipitated with 3 μg anti-syncytin-2 antibody. Following an overnight incubation, samples were added to 30 μl of protein A bead slurry and rotated for 4 hours at 4°C. After centrifuging and washing 3 times, samples were separated on an SDS-PAGE resolving gel and subjected to direct mass spectrometry (MS) analysis. Using the LTQ-orbitrap XL mass spectrometer (Thermo Fisher Scientific), 4 samples were run and analyzed by Scaffold (Proteome Software). The peptide identities were accepted when the Peptide Prophet algorithm specified probabilities were at >95.0%. Argot2 (Annotation Retrieval of Gene Ontology Terms; http://www.medcomp.medicina.unipd.it/Argot2) was used to identify the functional classifications of the total and specific expressed proteins. DAVID (DAVID Bioinformatics Resources 6.7; http://david.abcc.ncifcrf.gov/tools.jsp) was used to perform functional classification of the differentially expressed proteins (the fold change was greater than or equal to 1.5 and the spectrum count number was not less than 4). Cytoscape was used to draw programs.
Genomic DNA sequencing of the human syncytin-2 coding region
All PCR amplification reactions (50 μl) were performed using the TaKaRa LA Taq standard conditions. The 9.5 kb ERV-FRD locus including flanking non-retroviral sequences in chromosome 6, LTRs, gag, pol and env ORF was amplified by long-distance PCR according to the reported method11 using sense primer LF and antisense primer LR. Each reaction tube contained a maximum of 50 ng of DNA, 250 nM of each primer, 1.2 mM MgCl2, 500 μM dNTPs, and 5 units of TaKaRa LA Taq DNA polymerase in 50 μl of PCR buffer. Cycling conditions were 3 min at 94°C, followed by 30 cycles of PCR. Each cycle consisted of 30 sec at 94°C, 40 sec at 58°C, and a 10 min elongation step at 68°C. The 30th cycle included an additional 10 min elongation step at 68°C.
For template, 0.5 μl of the 9.5 kb long-distance amplification product was used to amplify the full-length syncytin-2 env gene using sense primer SF and antisense primer SR. The amplification fragment was sequenced using antisense primer S1, sense primer S2 and sense primer S3, which are located at 655–674 bp, 466–487 bp and 967–986 bp in the coding region of syncytin-2, respectively. At least 2 independent readings on each strand were performed. The primers used for amplifying and sequencing the full-length syncytin-2 ORF are indicated in Table 2.
A chi-square test was used to analyze the difference of the frequency of T522M between normal pregnant women and sPE patients. The analysis was conducted using Statistical Package for Social Science software (SPSS for Windows version 10.0; SPSS Inc.), and P < 0.05 was considered significant.
Abbreviations
- ASCT
alanine/serine/cysteine/threonine transporter
- CTBs
cytotrophoblast cells
- CTM
cytoplasmic domain
- DMEM
Dulbecco's modified Eagle's medium
- FBS
fetal bovine serum
- env
envelope
- FP
fusion peptide
- gag
group-specific antigen gene
- HERV
human endogenous retrovirus
- HR
heptad repeat region
- LTR
long terminal repeat
- MAF
minor allele frequency
- MFSD2A
major facilitator superfamily domain containing 2
- ISD
immunosuppressive domain
- ORF
open reading frame
- PE
pre-eclampsia
- pol
polymerase
- SNP
single-nucleotide polymorphism
- SP
signal peptide
- STB
syncytiotrophoblast
- ST-I
syncytiotrophoblast layer I
- ST-II
syncytiotrophoblast layer II
- SU
surface protein
- TM
transmembrane protein
- TMD
transmembrane domain
- WT
wide-type
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We thank Dr. Hungwen Chen for providing the pTM1-syncytin-2 plasmid, Dr. Francois Mallet for providing the phCMV-syncytin-1 plasmid, and Drs. Tommaso Dragani and Francesca Colombo for providing the pEF6-MFSD2A 830 plasmid.
Funding
This work was supported by the National Key Basic Research Program of China (2011CB944400), the Major Program of National Natural Science Foundation of China (NSFC) (81490741) and a grant from NSFC (31271603). H. Y. Lin is a recipient of National Excellent Young Scientist supported by NSFC (81322008).
Supplemental Material
Supplemental Material may be downloaded here: publisher's website
References
- 1.Sibai B, Dekker G, Kupferminc M. Pre-eclampsia. Lancet 2005; 365:785-99; PMID:15733721; http://dx.doi.org/ 10.1016/S0140-6736(05)71003-5 [DOI] [PubMed] [Google Scholar]
- 2.Blond JL, Beseme F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J Virol 1999; 73:1175-85; PMID:9882319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al.. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000; 403:785-9; PMID:10693809; http://dx.doi.org/ 10.1038/35001608 [DOI] [PubMed] [Google Scholar]
- 4.Blaise S, de Parseval N, Benit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc Natl Acad Sci U S A 2003; 100:13013-8; PMID:14557543; http://dx.doi.org/ 10.1073/pnas.2132646100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frendo JL, Olivier D, Cheynet V, Blond JL, Bouton O, Vidaud M, Rabreau M, Evain-Brion D, Mallet F. Direct Involvement of HERV-W Env Glycoprotein in Human Trophoblast Cell Fusion and Differentiation. Mol Cell Biol 2003; 23:3566-74; PMID:12724415; http://dx.doi.org/ 10.1128/MCB.23.10.3566-3574.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J Virol 2000; 74:3321-9; PMID:10708449; http://dx.doi.org/ 10.1128/JVI.74.7.3321-3329.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jern P, Coffin JM. Effects of retroviruses on host genome function. Annu Rev Genet 2008; 42:709-32; PMID:18694346; http://dx.doi.org/ 10.1146/annurev.genet.42.110807.091501 [DOI] [PubMed] [Google Scholar]
- 8.Voisset C, Blancher A, Perron H, Mandrand B, Mallet F, Paranhos-Baccala G. Phylogeny of a novel family of human endogenous retrovirus sequences, HERV-W, in humans and other primates. AIDS Res Hum Retroviruses 1999; 15:1529-33; PMID:10580403; http://dx.doi.org/ 10.1089/088922299309810 [DOI] [PubMed] [Google Scholar]
- 9.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta 2012; 33:663-71; PMID:22695103; http://dx.doi.org/ 10.1016/j.placenta.2012.05.005 [DOI] [PubMed] [Google Scholar]
- 10.Bonnaud B, Bouton O, Oriol G, Cheynet V, Duret L, Mallet F. Evidence of selection on the domesticated ERVWE1 env retroviral element involved in placentation. Mol Biol Evol 2004; 21:1895-901; PMID:15254254; http://dx.doi.org/ 10.1093/molbev/msh206 [DOI] [PubMed] [Google Scholar]
- 11.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci U S A 2004; 101:1731-6; PMID:14757826; http://dx.doi.org/ 10.1073/pnas.0305763101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dupressoir A, Marceau G, Vernochet C, Benit L, Kanellopoulos C, Sapin V, Heidmann T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc Natl Acad Sci U S A 2005; 102:725-30; PMID:15644441; http://dx.doi.org/ 10.1073/pnas.0406509102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci U S A 2009; 106:12127-32; PMID:19564597; http://dx.doi.org/ 10.1073/pnas.0902925106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupressoir A, Vernochet C, Harper F, Guegan J, Dessen P, Pierron G, Heidmann T. A pair of co-opted retroviral envelope syncytin genes is required for formation of the two-layered murine placental syncytiotrophoblast. Proc Natl Acad Sci U S A 2011; 108:E1164-73; PMID:22032925; http://dx.doi.org/ 10.1073/pnas.1112304108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heidmann O, Vernochet C, Dupressoir A, Heidmann T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: a new “syncytin” in a third order of mammals. Retrovirology 2009; 6:107; PMID:19943933; http://dx.doi.org/ 10.1186/1742-4690-6-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cornells G, Heidmann O, Bernard-Stoecklin S, Véron G, Reynaud K, Mulot B, Dupressoir A, Heidmann T. Identification of syncytin-car-1, an endogenous retroviral envelope gene involved in placentation and conserved in Carnivora: a syncytin in a new superorder of placental mammals. Retrovirology 2011; 8:P13; http://dx.doi.org/ 10.1186/1742-4690-8-S2-P13 [DOI] [Google Scholar]
- 17.Cornelis G, Heidmann O, Degrelle SA, Vernochet C, Lavialle C, Letzelter C, Bernard-Stoecklin S, Hassanin A, Mulot B, Guillomot M, et al.. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc Natl Acad Sci U S A 2013; 110:E828-37; PMID:23401540; http://dx.doi.org/ 10.1073/pnas.1215787110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cornelis G, Vernochet C, Malicorne S, Souquere S, Tzika AC, Goodman SM, Catzeflis F, Robinson TJ, Milinkovitch MC, Pierron G, et al.. Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc Natl Acad Sci U S A 2014; 111:E4332-41; PMID:25267646; http://dx.doi.org/ 10.1073/pnas.1412268111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Redelsperger F, Cornelis G, Vernochet C, Tennant BC, Catzeflis F, Mulot B, Heidmann O, Heidmann T, Dupressoir A, et al.. Capture of syncytin-Mar1, a fusogenic endogenous retroviral envelope gene involved in placentation in the Rodentia squirrel-related clade. J Virol 2014; 88:7915-28; PMID:24789792; http://dx.doi.org/ 10.1128/JVI.00141-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornelis G, Vernochet C, Carradec Q, Souquere S, Mulot B, Catzeflis F, Nilsson MA, Menzies BR, Renfree MB, Pierron G, et al.. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc Natl Acad Sci U S A 2015; 112:E487-96; PMID:25605903; http://dx.doi.org/ 10.1073/pnas.1417000112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malassine A, Handschuh K, Tsatsaris V, Gerbaud P, Cheynet V, Oriol G, Mallet F, Evain-Brion D. Expression of HERV-W Env glycoprotein (syncytin) in the extravillous trophoblast of first trimester human placenta. Placenta 2005; 26:556-62; PMID:15993705; http://dx.doi.org/ 10.1016/j.placenta.2004.09.002 [DOI] [PubMed] [Google Scholar]
- 22.Muir A, Lever AM, Moffett A. Human endogenous retrovirus-W envelope (syncytin) is expressed in both villous and extravillous trophoblast populations. J Gen Virol 2006; 87:2067-71; PMID:16760410; http://dx.doi.org/ 10.1099/vir.0.81412-0 [DOI] [PubMed] [Google Scholar]
- 23.Lee X, Keith JC Jr., Stumm N, Moutsatsos I, McCoy JM, Crum CP, Genest D, Chin D, Ehrenfels C, Pijnenborg R, et al.. Downregulation of placental syncytin expression and abnormal protein localization in pre-eclampsia. Placenta 2001; 22:808-12; PMID:11718567; http://dx.doi.org/ 10.1053/plac.2001.0722 [DOI] [PubMed] [Google Scholar]
- 24.Holder BS, Tower CL, Abrahams VM, Aplin JD. Syncytin 1 in the human placenta. Placenta 2012; 33:460-6; PMID:22381536; http://dx.doi.org/ 10.1016/j.placenta.2012.02.012 [DOI] [PubMed] [Google Scholar]
- 25.Lavillette D, Marin M, Ruggieri A, Mallet F, Cosset FL, Kabat D. The Envelope Glycoprotein of Human Endogenous Retrovirus Type W Uses a Divergent Family of Amino Acid Transporters/Cell Surface Receptors. J Virol 2002; 76:6442-52; PMID:12050356; http://dx.doi.org/ 10.1128/JVI.76.13.6442-6452.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kekuda R, Prasad PD, Fei YJ, Torres-Zamorano V, Sinha S, Yang-Feng TL, Leibach FH, Ganapathy V. Cloning of the sodium-dependent, broad-scope, neutral amino acid transporter Bo from a human placental choriocarcinoma cell line. J Biol Chem 1996; 271:18657-61; PMID:8702519; http://dx.doi.org/ 10.1074/jbc.271.31.18657 [DOI] [PubMed] [Google Scholar]
- 27.Marin M, Lavillette D, Kelly SM, Kabat D. N-Linked Glycosylation and Sequence Changes in a Critical Negative Control Region of the ASCT1 and ASCT2 Neutral Amino Acid Transporters Determine Their Retroviral Receptor Functions. J Virol 2003; 77:2936-45; PMID:12584318; http://dx.doi.org/ 10.1128/JVI.77.5.2936-2945.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cariappa R, Heath-Monnig E, Smith CH. Isoforms of amino acid transporters in placental syncytiotrophoblast: plasma membrane localization and potential role in maternal/fetal transport. Placenta 2003; 24:713-26; PMID:12852862; http://dx.doi.org/ 10.1016/S0143-4004(03)00085-7 [DOI] [PubMed] [Google Scholar]
- 29.Potgens AJ, Drewlo S, Kokozidou M, Kaufmann P. Syncytin: the major regulator of trophoblast fusion? Recent developments and hypotheses on its action. Hum Reprod Update 2004; 10:487-96; PMID:15333590; http://dx.doi.org/ 10.1093/humupd/dmh039 [DOI] [PubMed] [Google Scholar]
- 30.Kudo Y, Boyd CAR. Changes in Expression and Function of Syncytin and its Receptor, Amino Acid Transport System B0 (ASCT2), in Human Placental Choriocarcinoma BeWo Cells During Syncytialization. Placenta 2002; 23:536-41; PMID:12175968; http://dx.doi.org/ 10.1053/plac.2002.0839 [DOI] [PubMed] [Google Scholar]
- 31.Green BJ, Lee CS, Rasko JE. Biodistribution of the RD114/mammalian type D retrovirus receptor, RDR. J Gene Med 2004; 6:249-59; PMID:15026986; http://dx.doi.org/ 10.1002/jgm.517 [DOI] [PubMed] [Google Scholar]
- 32.Hayward MD, Potgens AJ, Drewlo S, Kaufmann P, Rasko JE. Distribution of human endogenous retrovirus type W receptor in normal human villous placenta. Pathology 2007; 39:406-12; PMID:17676482; http://dx.doi.org/ 10.1080/00313020701444572 [DOI] [PubMed] [Google Scholar]
- 33.Malassine A, Blaise S, Handschuh K, Lalucque H, Dupressoir A, Evain-Brion D, Heidmann T. Expression of the fusogenic HERV-FRD Env glycoprotein (syncytin 2) in human placenta is restricted to villous cytotrophoblastic cells. Placenta 2007; 28:185-91; PMID:16714059; http://dx.doi.org/ 10.1016/j.placenta.2006.03.001 [DOI] [PubMed] [Google Scholar]
- 34.Esnault C, Priet S, Ribet D, Vernochet C, Bruls T, Lavialle C, Weissenbach J, Heidmann T. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc Natl Acad Sci U S A 2008; 105:17532-7; PMID:18988732; http://dx.doi.org/ 10.1073/pnas.0807413105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vargas A, Moreau J, Landry S, LeBellego F, Toufaily C, Rassart E, Lafond J, Barbeau B. Syncytin-2 plays an important role in the fusion of human trophoblast cells. J Mol Biol 2009; 392:301-18; PMID:19616006; http://dx.doi.org/ 10.1016/j.jmb.2009.07.025 [DOI] [PubMed] [Google Scholar]
- 36.Chen CP, Chen LF, Yang SR, Chen CY, Ko CC, Chang GD, Chen H. Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biol Reprod 2008; 79:815-23; PMID:18650494; http://dx.doi.org/ 10.1095/biolreprod.108.069765 [DOI] [PubMed] [Google Scholar]
- 37.Renard M, Varela PF, Letzelter C, Duquerroy S, Rey FA, Heidmann T. Crystal structure of a pivotal domain of human syncytin-2, a 40 million years old endogenous retrovirus fusogenic envelope gene captured by primates. J Mol Biol 2005; 352:1029-34; PMID:16140326; http://dx.doi.org/ 10.1016/j.jmb.2005.07.058 [DOI] [PubMed] [Google Scholar]
- 38.Chang C, Chen PT, Chang GD, Huang CJ, Chen H. Functional characterization of the placental fusogenic membrane protein syncytin. Biol Reprod 2004; 71:1956-62; PMID:15269105; http://dx.doi.org/ 10.1095/biolreprod.104.033340 [DOI] [PubMed] [Google Scholar]
- 39.Drewlo S, Leyting S, Kokozidou M, Mallet F, Potgens AJ. C-Terminal truncations of syncytin-1 (ERVWE1 envelope) that increase its fusogenicity. Biol Chem 2006; 387:1113-20; PMID:16895482; http://dx.doi.org/ 10.1515/BC.2006.137 [DOI] [PubMed] [Google Scholar]
- 40.Cheynet V, Ruggieri A, Oriol G, Blond JL, Boson B, Vachot L, Verrier B, Cosset FL, Mallet F. Synthesis, assembly, and processing of the Env ERVWE1/syncytin human endogenous retroviral envelope. J Virol 2005; 79:5585-93; PMID:15827173; http://dx.doi.org/ 10.1128/JVI.79.9.5585-5593.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aguilar PS, Baylies MK, Fleissner A, Helming L, Inoue N, Podbilewicz B, Wang H, Wong M. Genetic basis of cell-cell fusion mechanisms. Trends Genet 2013; 29:427-37; PMID:23453622; http://dx.doi.org/ 10.1016/j.tig.2013.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moyes D, Griffiths DJ, Venables PJ. Insertional polymorphisms: a new lease of life for endogenous retroviruses in human disease. Trends Genet 2007; 23:326-33; PMID:17524519; http://dx.doi.org/ 10.1016/j.tig.2007.05.004 [DOI] [PubMed] [Google Scholar]
- 43.Yu H, Liu T, Zhao Z, Chen Y, Zeng J, Liu S, Zhu F. Mutations in 3′-long terminal repeat of HERV-W family in chromosome 7 upregulate syncytin-1 expression in urothelial cell carcinoma of the bladder through interacting with c-Myb. Oncogene 2014; 33:3947-58; PMID:24013223; http://dx.doi.org/ 10.1038/onc.2013.366 [DOI] [PubMed] [Google Scholar]
- 44.Montefiori DC, Robinson WE Jr., Mitchell WM. Role of protein N-glycosylation in pathogenesis of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 1988; 85:9248-52; PMID:3264072; http://dx.doi.org/ 10.1073/pnas.85.23.9248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Samleerat T, Braibant M, Jourdain G, Moreau A, Ngo-Giang-Huong N, Leechanachai P, Hemvuttiphan J, Hinjiranandana T, Changchit T, Warachit B, et al.. Characteristics of HIV type 1 (HIV-1) glycoprotein 120 env sequences in mother-infant pairs infected with HIV-1 subtype CRF01_AE. J Infect Dis 2008; 198:868-76; PMID:18700833; http://dx.doi.org/ 10.1086/591251 [DOI] [PubMed] [Google Scholar]
- 46.Go EP, Chang Q, Liao HX, Sutherland LL, Alam SM, Haynes BF, Desaire H. Glycosylation site-specific analysis of clade C HIV-1 envelope proteins. J Proteome Res 2009; 8:4231-42; PMID:19610667; http://dx.doi.org/ 10.1021/pr9002728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohgimoto S, Shioda T, Mori K, Nakayama EE, Hu H, Nagai Y. Location-specific, unequal contribution of the N glycans in simian immunodeficiency virus gp120 to viral infectivity and removal of multiple glycans without disturbing infectivity. J Virol 1998; 72:8365-70; PMID:9733886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheynet V, Oriol G, Mallet F. Identification of the hASCT2-binding domain of the Env ERVWE1/syncytin-1 fusogenic glycoprotein. Retrovirology 2006; 3:41; PMID:16820059; http://dx.doi.org/ 10.1186/1742-4690-3-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chivers PT, Laboissiere MC, Raines RT. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J 1996; 15:2659-67; ;PMID:8654363 [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi A, Day NK, Luangwedchakarn V, Good RA, Haraguchi S. A retroviral-derived immunosuppressive peptide activates mitogen-activated protein kinases. J Immunol 2001; 166:6771-5; PMID:11359835; http://dx.doi.org/ 10.4049/jimmunol.166.11.6771 [DOI] [PubMed] [Google Scholar]
- 51.Mangeney M, Renard M, Schlecht-Louf G, Bouallaga I, Heidmann O, Letzelter C, Richaud A, Ducos B, Heidmann T. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc Natl Acad Sci U S A 2007; 104:20534-9; PMID:18077339; http://dx.doi.org/ 10.1073/pnas.0707873105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gauster M, Moser G, Orendi K, Huppertz B. Factors involved in regulating trophoblast fusion: potential role in the development of preeclampsia. Placenta 2009; 30 Suppl A:S49-54; ;PMID:19027159; http://dx.doi.org/ 10.1016/j.placenta.2008.10.011 [DOI] [PubMed] [Google Scholar]
- 53.Vargas A, Toufaily C, LeBellego F, Rassart E, Lafond J, Barbeau B. Reduced expression of both syncytin 1 and syncytin 2 correlates with severity of preeclampsia. Reprod Sci 2011; 18:1085-91; PMID:21493955; http://dx.doi.org/ 10.1177/1933719111404608 [DOI] [PubMed] [Google Scholar]
- 54.Knerr I, Beinder E, Rascher W. Syncytin, a novel human endogenous retroviral gene in human placenta: Evidence for its dysregulation in preeclampsia and HELLP syndrome. Am J Obstet Gynecol 2002; 186:210-3; PMID:11854637; http://dx.doi.org/ 10.1067/mob.2002.119636 [DOI] [PubMed] [Google Scholar]
- 55.Knerr I. Transcriptional effects of hypoxia on fusiogenic syncytin and its receptor ASCT2 in human cytotrophoblast BeWo cells and in ex vivo perfused placental cotyledons. Am J Obstet Gynecol 2003; 189:583-8; PMID:14520239; http://dx.doi.org/ 10.1067/S0002-9378(03)00538-6 [DOI] [PubMed] [Google Scholar]
- 56.Kudo Y, Boyd CAR, Sargent IL, Redman CWG. Hypoxia alters expression and function of syncytin and its receptor during trophoblast cell fusion of human placental BeWo cells: implications for impaired trophoblast syncytialisation in pre-eclampsia. Biochimica et Biophysica Acta (BBA) - Molecular Basis of Disease 2003; 1638:63-71; http://dx.doi.org/ 10.1016/S0925-4439(03)00043-7 [DOI] [PubMed] [Google Scholar]
- 57.Knerr I, Huppertz B, Weigel C, Dotsch J, Wich C, Schild RL, Beckmann MW, Rascher W. Endogenous retroviral syncytin: compilation of experimental research on syncytin and its possible role in normal and disturbed human placentogenesis. Mol Hum Reprodu 2004; 10:581-8; PMID:15181178; http://dx.doi.org/ 10.1093/molehr/gah070 [DOI] [PubMed] [Google Scholar]
- 58.Huang Q, Chen H, Wang F, Brost BC, Li J, Gao Y, Li Z, Gao Y, Jiang SW. Reduced syncytin-1 expression in choriocarcinoma BeWo cells activates the calpain1-AIF-mediated apoptosis, implication for preeclampsia. Cell Mol Life Sci 2014; 71:3151-64; PMID:24413738; http://dx.doi.org/ 10.1007/s00018-013-1533-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhuang XW, Li J, Brost BC, Xia XY, Chen HB, Wang CX, Jiang SW. Decreased expression and altered methylation of syncytin-1 gene in human placentas associated with preeclampsia. Curr Pharm Des 2014; 20:1796-802; PMID: 23888950; http://dx.doi.org/ 10.2174/13816128113199990541 [DOI] [PubMed] [Google Scholar]
- 60.Chen CP, Wang KG, Chen CY, Yu C, Chuang HC, Chen H. Altered placental syncytin and its receptor ASCT2 expression in placental development and pre-eclampsia. BJOG 2006; 113:152-8; PMID:16411991; http://dx.doi.org/ 10.1111/j.1471-0528.2005.00843.x [DOI] [PubMed] [Google Scholar]
- 61.Langbein M, Strick R, Strissel PL, Vogt N, Parsch H, Beckmann MW, Schild RL. Impaired cytotrophoblast cell-cell fusion is associated with reduced Syncytin and increased apoptosis in patients with placental dysfunction. Mol Reprod Dev 2008; 75:175-83; PMID:17546632; http://dx.doi.org/ 10.1002/mrd.20729 [DOI] [PubMed] [Google Scholar]
- 62.ACOG practice bulletin . Diagnosis and management of preeclampsia and eclampsia. Number 33, January 2002. Obstet Gynecol 2002; 99:159-67; PMID:16175681; http://dx.doi.org/ 10.1016/S0029-7844(01)01747-1 [DOI] [PubMed] [Google Scholar]
- 63.Spinola M, Falvella FS, Colombo F, Sullivan JP, Shames DS, Girard L, Spessotto P, Minna JD, Dragani TA. MFSD2A is a novel lung tumor suppressor gene modulating cell cycle and matrix attachment. Mol Cancer 2010; 9:62; PMID:20236515; http://dx.doi.org/ 10.1186/1476-4598-9-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


