Abstract
Breast cancer, the leading cause of cancer among females, is supported by the presence of a rare subset of undifferentiated cells within the tumor, identified as breast cancer stem cells (BCSCs). BCSCs underlie the mechanisms of tumor initiation and sustenance and are implicated in the dissemination of the primary tumor to metastatic sites, as they have been found circulating in the blood of breast cancer patients. The discovery of BCSCs has generated a great amount of interest among the scientific community toward their isolation, molecular characterization, and therapeutic targeting. In this review, after summarizing the literature on molecular characterization of BCSCs and methodologies used for their isolation, we will focus on recent data supporting their molecular and functional heterogeneity. Additionally, following a synopsis of the latest approaches for BCSC targeting, we will specifically emphasize on the therapeutic use of naïve or engineered normal stem cells in the treatment of breast cancer and present contradictory findings challenging their safety.
Keywords: breast cancer stem cells, cancer stem cell heterogeneity, targeting cancer stem cells, circulating tumor cells, stem cell technology
Stemness and cancer development
Breast cancer represents the most common malignancy and leading cause of death among women. Recent findings in the field of breast cancer research have revealed that cancer stem cells (CSCs), a rare subpopulation of undifferentiated cells with embryonic characteristics within the tumor, are key contributors to the development and progression of the disease. The first report describing functional CSCs emerged more than 20 years ago when in 1994 Lapidot et al1 reported that a CD34+/CD38− cell can initiate acute myeloid leukemia after transplantation into severe combined immunodeficiency mice. In a series of following studies, the presence of CSCs was confirmed in breast,2 brain,3 liver,4 colorectal5 tumors, and melanomas.6,7 CSCs are characterized by their ability to self-renew and differentiate, display tumorigenic capability in immunosuppressed mice, and grow ex vivo as tumorspheres.8,9 It has been suggested that they originate from normal tissue stem cells undergoing somatic mutations and that cancer can be practically considered as a stem cell disease.10 Indeed, normal and CSCs share many similar phenotypic features. For instance, in breast cancer, the CD24−/lo/epithelial cell adhesion molecule (EpCAM)lo CSC phenotype resembles that of CD49f+/ESA−/lo normal human mammary stem cell11,12 and aldehyde dehydrogenase 1 (ALDH1), a marker of normal mammary stem cells, is a well-established breast cancer stem cell (BCSC) marker.13 It is proposed that despite their similarities, CSCs differ from their normal counterparts in their degree of dependence on their microenvironment. Although normal stem cells are entirely reliant on signals from their microenvironment in order to regulate their proliferative state, under normal or repair conditions, CSCs exhibit a more self-sufficient cell proliferation capacity. Furthermore, as will be described below, CSCs have been proposed to differ from their normal counterparts in the glycosylation patterns of their surface markers.14 Contrary to the perception that normal stem cells are the cells of cancer origin, several groups have reported that the luminal progenitor population, rather than a multipotent mammary stem cell, generates luminal and basal-like breast carcinomas.15–17 Interestingly, differentiated cells can also acquire self-renewal capacity through the action of genetic and/or epigenetic mechanisms.18 CSCs underlie the biological and functional heterogeneity within the tumor and support the preservation of the tumor cell population. The contribution of CSCs to tumor maintenance and heterogeneity is summarized in two principal theories: according to the hierarchical CSC model, only a small subset of tumor cells, capable of self-renewing and differentiating, display tumorigenic capability, while providing the tumor with all differentiated nontumorigenic progeny, thus maintaining the tumor hierarchy.19–22 In contrast, the stochastic or clonal evolution model suggests that all tumor cells contribute equally to tumor growth, as genetic mutations triggered by microenvironmental factors can give rise to undifferentiated cells that can adjust more effectively to the tumor microenvironment and evolve, promoting tumorigenesis.23–25 Recent data point toward a dynamic version of the CSC model, revealing a predominant significance of the tumor microenvironment on the phenotype of CSCs. More specifically, within the tumor hierarchy, nontumorigenic cells have the capacity to dedifferentiate and give rise to additional CSCs, resulting in fluctuations of the CSC subpopulation of a specific tumor over time. Interestingly, not all tumors are driven by rare CSC subpopulations but include rather large tumorigenic populations.26 Although normal somatic tissue cells have been shown to dedifferentiate following specific external stimuli, the dedifferentiation process can occur more readily in cancer cells due to their disrupted signaling pathways. It has been shown that exposure of differentiated colorectal cells to stromal factors can promote their CSC transformation. Analogous results have been obtained on mammary cancer11,27,28 (for a review see26,29). In addition to the dedifferentiation process, the tumor microenvironment is also crucial for preservation of the existing CSC subpopulation.18,26,30
The normal stem cell microenvironment or “niche” comprises a discrete dynamic domain within a tissue, with the ability to ensure tissue homeostasis under physiological and pathological conditions. It prevents the differentiation of stem cells and coordinates their behavior in homeostasis and repair. The deregulation of genetic homeostasis of the stroma can promote tumorigenesis. It is yet unclear whether in neoplastic tissues CSCs utilize and modulate their existing microenvironment or direct the formation of a new niche. Many soluble factors of the tumor microenvironment are involved in the maintenance of the CSC phenotype within a tumor. Transforming growth factor beta (TGF-beta), platelet-derived growth factor, fibroblast growth factor, insulin growth factor 1, epidermal growth factor, Wnt ligands, and hypoxia have been shown to participate in the preservation of the undifferentiated cell state of the CSC subpopulations.18 Similarly, CSCs can also secrete soluble factors, such as TGF-beta, which enhance the inflammatory and angiogenic properties of their surrounding microenvironment. Inhibiting the crosstalk between CSCs and their tumor microenvironment and discovering therapies targeting both hold great promise in the future of breast cancer treatment.31,32
Molecular and functional characteristics of CSCs
CSCs have been detected in primary and metastatic tumors, where they sustain the tumor mass, preserve its heterogeneity, and drive metastasis.9,18–21,23–26,29,30,33 They have also been found in the peripheral blood of breast cancer patients,34–39 where they promote systemic dissemination and the bone marrow of early stage breast cancer patients.40 According to Liu et al,41 CSCs constitute a highly heterogeneous population with specific hierarchical patterns that are composed of various subcategories, such as precancerous stem cells, and primary, migrating, and chemoradioresistant CSCs, with distinct role in tumor formation and cancer progression. The term “horizontal hierarchy of CSCs” has been proposed to describe the complex network of heterogeneous CSCs and their interactions.41 Specific molecular markers have been employed for the designation of the CSC subpopulation, the majority of which are expressed in both solid tumors and circulating tumor cells (CTCs) in the bloodstream of breast cancer patients.42 Nevertheless, molecular analysis of CTCs has revealed that they can exhibit distinct phenotypic profiles compared to their tumor of origin.43,44
The resistance of CSCs to conventional chemotherapy underlies the recurrence of the initial tumor and associates with poor clinical outcome.45 The biological properties of CSCs render them resistant to conventional chemotherapy. CSCs are characterized by elevated levels of ATP-binding cassette (ABC) transporters, membrane transporters that pump cytotoxic drugs and dye out of the cells (reviewed in46,47). It has also been shown that high levels of the anti-apoptotic protein Bcl-2 were observed in CD44+/CD24−/low BCSCs.48 Moreover, a number of signaling pathways as Wnt/β-catenin, Notch, and NF-κB that contribute to the survival and self-renewal of CSCs have been associated with the resistance of CSCs to conventional chemotherapy. Finally, CSCs display an enhanced DNA damage response under hypoxic conditions, which is mediated by the activation of two major signaling pathways that regulate the cell cycle, ataxia telangiectasia mutated and ataxia telangiectasia mutated-and Rad3-related.46,47
One major impediment in breast cancer treatment is cancer recurrence after a prolonged disease-free interval, referred to as tumor dormancy. It has been reported that one in five clinically disease-free patients after surgical removal of their primary tumor developed cancer after 5–25 years.49 Interestingly, many aspects of the biological behavior of CSCs, as quiescence and immune escape, reflect the biological mechanisms involved in tumor dormancy, indicating that CSC and dormant cell populations exhibit a significant overlap.49 Surviving cytotoxic treatment and hostile microenvironments, evasion of antitumor immune responses, and regulation from microenvironmental signals as angiogenic factors are some of the traits shared by both CSCs and dormant cells pointing at an at least partial overlap of these two subpopulations.49 Inactivation of MYC oncogene, a key regulator of tumor cell dormancy, resulted in production of stem-like cells with differentiating as well as oncogenic potential.50 Additionally, dormancy-promoting microenvironments (eg, TGF-beta signaling) support the preservation of the CSC subpopulation.51,52
CSCs also display epithelial-to-mesenchymal transition (EMT) traits, which promote their migratory potential. EMT is critical for organogenesis during embryonic development and involves the transformation of nonmotile epithelial cells to mesenchymal migratory cells.53 A number of extracellular factors (epidermal growth factor, fibroblast growth factor, TGF-beta, Notch, Hedgehog, and Wnt ligands),54 microRNAs,55 and epigenetic regulators56 have been implicated in the induction of EMT. The most common EMT-inducing transcription factors are Snail, Twist, Slug, Zeb1, Zeb2, and FoxC2.57 Activating the EMT pathway triggers the elevation of mesenchymal markers (eg, fibronectin, N-cadherin, vimentin, alpha-smooth muscle actin) and the loss or downregulation of epithelial markers (epithelial keratins, E-cadherin, occludins, claudins, and desmoplakin), which promote motility and invasiveness of EMT undergoing cells. Although EMT represents a physiological step in normal development, in cancer it results in metastasis, increased invasiveness,58–60 and chemoresistance.61 The induction of EMT has been associated with cancer stemness, as it can trigger the production of undifferentiated cells with CSC properties, capable of escaping the primary tumor and entering the bloodstream.27,59,62,63 Specifically, immortalized human mammary epithelial cells undergoing EMT express stem cell markers and exhibit an increased ability to form mammospheres. The expression of EMT markers has been associated with stem-like cell population isolated from mouse or human mammary glands or mammary carcinomas27 and the basal-like phenotype in breast cancer.64 Numerous EMT-inducing transcription factors (eg, FoxC2, Snail, Twist, Zeb, and p53) have been correlated with the expression of stem cell markers and metastatic phenotype in human mammary carcinoma cells and breast cancer cell lines.27,65,66 Finally, blockade of autocrine TGF-beta signaling has been shown to inhibit stem cell phenotype of murine breast cancer cells.67
The size of the CSC fraction within a tumor is directly associated with neoplastic grading and disease clinicopathology.68 In order to eliminate the carcinogenic effect of CSCs, research has focused on their effective isolation, detailed molecular characterization, and targeting via the development of novel therapeutic strategies. These schemes involve the inhibition of specific survival and differentiation pathways and are currently being assessed on a preclinical and clinical level.42,46,69 New advances and improved methodologies are continuously being introduced in the constantly evolving field of CSC therapy to facilitate a more selective and efficient targeting of the CSC subpopulation.
Isolation of BCSCs
CD44, CD24, and ALDH1 are the most commonly used biomarkers to identify the BCSC fraction. The expression of the surface glycoprotein CD44 combined with low expression or absence of the surface glycoprotein CD24 (CD44+/CD24−/low) typifies BCSCs, as first demonstrated by Al-Hajj et al.2 BCSCs are also typically characterized by elevated activity of ALDH1. Increased expression levels of ALDH1 in the tumors of breast cancer patients correlate with poor prognosis and overall survival.13 In addition to CD44, CD24, and ALDH1, other proteins have also been implicated in the characterization of BCSCs (Table 1). These include the epithelial cell adhesion molecule EpCAM, a6 and b1 integrin subunits CD49f and CD29, respectively, integrin-associated protein CD47, MET receptor tyrosine kinase, human epidermal growth factor receptor 2 (HER2), transmembrane glycoprotein CD133 (prominin-1), transcription factors Sox2 and Bmi1, and p53 tumor suppressor protein (for a detailed review see42). The expression of Ca2+-dependent cell–cell adhesion glycoprotein P-cadherin has been correlated to the expression of BCSC markers, confers resistance to X-ray induced cell death and designates cells with an intermediate EMT status associated with a transitional phenotype.71 Many of the CTCs detected in metastatic breast cancer patients display an intermediate EMT status,72 while numerous studies have revealed the coexpression of EMT (Twist1, Akt2, Snail, Zeb1, Tg2, and N-cadherin) and stem cell markers in breast cancer CTCs (BCTCs).36,73–76 The expression of the ABCG2 transporter (CD338) also identifies the tumor-initiating luminal progenitor subpopulation of BRCA1-mutated cells and is proposed as a novel antigen for cell sorting.77 The zinc finger transcription factor and Hedgehog effector GLI1 is important for the regulation of stemness in breast cancer as it is involved in the regulation of cellular differentiation pathways.78
Table 1.
Markers for BCSC characterization
| Markers | Localization | Isolation | References |
|---|---|---|---|
| ALDH1 | Cytoplasm | + | 13 |
| CD24 | Cell surface | + | 2 |
| CD44 | Cell surface | + | 2 |
| EpCAM | Cell surface | + | 42 |
| MUC1 | Cell surface | + | 76 |
| P-cadherin | Cell surface | 70, 71 | |
| N-cadherin | Cell surface | 74 | |
| CD49f | Cell surface | 42 | |
| CD29 | Cell surface | 42 | |
| CD47 | Cell surface | 42 | |
| CD133 | Cell surface | 42, 74 | |
| CD338 | Cell surface | 77 | |
| MET | Cell surface | 42, 35 | |
| HER2 | Cell surface | 42 | |
| Keratins | Cytoplasm | 79 | |
| Vimentin | Cytoplasm/cell surface | + | 58, 72, 93 |
| Sox2 | Nucleus/cytoplasm | 42 | |
| Bmi1 | Nucleus/cytoplasm | 42 | |
| p53 | Nucleus/cytoplasm | 42 | |
| GLI1 | Nucleus/cytoplasm | 78 | |
| Twist1 | Nucleus/cytoplasm | 73, 75 | |
| Akt2 | Nucleus/cytoplasm | 75, 76 | |
| Snail | Nucleus/cytoplasm | 73 | |
| Zeb1 | Nucleus/cytoplasm | 76 | |
| Tg2 | Nucleus/cytoplasm | 76 |
Note: The localization of proteins expressed in BCSCs and used for their isolation is shown.
Abbreviations: ALDH1, aldehyde dehydrogenase 1; BCSC, breast cancer stem cell; EpCAM, epithelial cell adhesion molecule; HER2, human epidermal growth factor receptor 2.
One of the least interventional ways to monitor the effects of therapy on the progress of the disease is the use of the so-called liquid biopsy, that is, the collection, enumeration, and characterization of BCTCs, cell-free DNA, or exosomes from the peripheral blood of breast cancer patients. Liquid biopsies are more advantageous in comparison to conventional biopsies (reviewed in79), as the latter may often result in inefficient tumor sampling and increased patient risk. Moreover, conventional biopsies reflect the genetic profile of the disease only at the specific time point when the biopsy is performed, thus failing to reflect the dynamic genetic pattern of tumors during the course of the disease or in response to specific drugs. In contrast, due to their noninvasive nature, liquid biopsies enable the longitudinal monitoring of pharmacodynamics in cancer therapy.80 Even though liquid biopsies are constantly gaining popularity in the field of breast cancer treatment, technical challenges and result inconsistencies among various isolation and detection techniques have arisen regarding the detection rate of CTCs, frequency of positive patients, and association between CTCs and survival rate.81–83 Additionally, due to the scarcity of CTCs, blood samples from breast cancer patients do not always reflect the entire population of CTCs in patient blood.84
The earliest detection method of BCTCs is Ficoll density centrifugation, which is widely applied for the isolation of peripheral blood mononuclear cells from whole blood (Figure 1). Subsequently, BCTCs included in the isolated fragment of peripheral blood mononuclear cells can be immunofluorescently labeled with specific antibodies.34 Since the expression of epithelial markers is frequently detected in BCTCs, many techniques for their isolation involve the use of an antibody specific for an epithelial marker, such as EpCAM and MUC1. The most widely used EpCAM-based isolation techniques involve the use of the US Food and Drug Administration-approved Cellsearch™ system (Veridex LLC, Raritan, NJ, USA),85 the herringbone chip,86 the AdnaTest breast cancer detection kit,87 fluorescence activated cell sorting analysis,36,88,89 and the use of microfluidic technology.90 Recently, a novel methodology for the in vivo isolation of BCTCs has been developed by Saucedo-Zeni et al91 who have used a medical Seldinger guidewire conjugated to a monoclonal antibody against EpCAM to capture EpCAM-positive BCTCs from the blood circulation, after a 30-minute insertion in the vein of the patient. Positive selection based on epithelial markers such as EpCAM results in the exclusion of mesenchymal CTCs at the final stages of EMT, which lack epithelial traits, but yet exhibit an elevated metastatic potential. For this scope, negative selection techniques have been developed, which are based on the removal of non-CTCs. The elimination of peripheral blood mononuclear cells, which are CD45-positive cells, via the use of microbeads conjugated to anti-CD45 antibodies, has been used for the enrichment of CTCs from whole blood.92 Apart from negative selection, positive selection using cell-surface vimentin, a mesenchymal marker not expressed in blood cells, has been recently used for the isolation of mesenchymal CTCs from metastatic breast cancer patients. The summation of cell-surface vimentin and CellSearch methods has been proposed as an efficient approach for the detection of CTCs.93 The RosetteSep™ Human CD45 Depletion Cocktail is one of the commercially available kits designed to enrich epithelial CTCs via the crosslinking of unwanted cells to red blood cells, so that they pellet along red blood cells during Ficoll density centrifugation. Notably, the RosetteSep™ system (StemCell Technology, Vancouver, Canada) is frequently combined with the epithelial immuno SPOT (EPISPOT) assay, which has been used to detect proteins released in the peripheral blood from single epithelial cancer cells thus detecting and characterizing viable CTCs and disseminated tumor cells in the bone marrow in cancer patients.94 The CTC-iChip microfluidic system can also enrich CTCs after removal of white and red blood cells with the use of magnetic beads.95,96 Finally, exploiting the physicochemical properties of BCTCs, the isolation by size of epithelial tumor cells (ISET) methodology, which is based on the filtration, is used for the isolation of CTCs due to their large size compared to leukocytes.97,98
Figure 1.
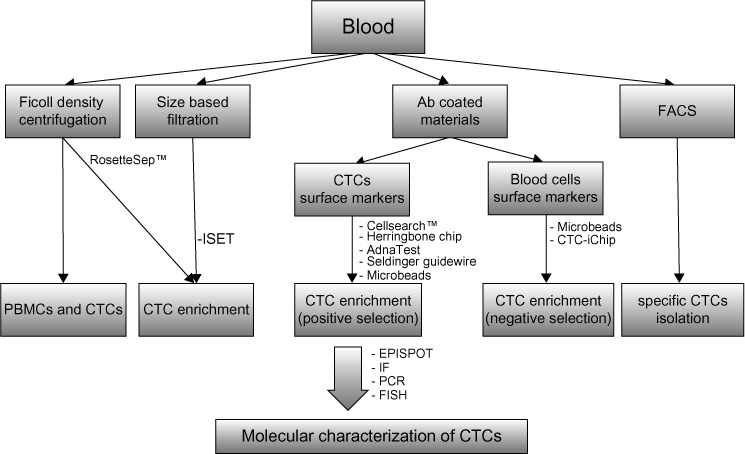
Methods for the isolation and molecular characterization of BCTCs.
Abbreviations: Ab, antibody; BCTC, breast cancer circulating tumor cell; CTC, circulating tumor cell; EPISPOT, epithelial immuno SPOT; FACS, fluorescence activated cell sorting; IF, immunofluorescence; ISET, isolation by size of epithelial tumor cells, FISH, fluorescence in situ hybridization; PBMC, peripheral blood mononuclear cell; PCR, polymerase chain reaction.
Apart from the peripheral blood, BCSCs have also been isolated directly from primary or metastatic tumors of breast cancer patients. Using a semiautomated dissociation methodology, followed by magnetic cell sorting and cDNA amplification, transcriptome analysis has been performed on isolated CD44+/CD24−/CD45− BCSCs from primary estrogen receptor α-positive tumors.99 Also, BCSCs from primary tumor derived tissue cultures have been isolated via sorting CD44+CD24−/dim cell population by catcher tube-based cell sorter in combination with the flow cytometer and then propagated ex vivo as nonadherent mammospheres.100
Molecular and functional heterogeneity of BCSCs
Unraveling the molecular signature of BCSCs will promote a better insight into the biology of breast cancer, will enable the discovery of new therapeutic targets, lead to the improvement of current therapies, and will provide new tools toward understanding and preventing disease relapse. The use of specific markers for the characterization of the BCSC subpopulation has revealed that BCSCs display a significant degree of heterogeneity and can be classified into subcategories. These CSC subsets have been found to exhibit distinct tumorigenic capacity and discrete functions within the neoplastic cell population (Table 2).
Table 2.
Molecular and functional heterogeneity of BCSCs
| Molecular signature | Significance/properties | References |
|---|---|---|
| ALDH1+ | High tumorigenic potential | 101 |
| Epithelial-like, proliferative | ||
| Expression of MET genes | ||
| Located in tumor interior | ||
| EpCAM+/ALDH1+/HER2+/EGFR+/HPSE+/Notch1+ | Brain metastasis | 104 |
| High expression levels of vimentin and keratins 8 and 18 | ||
| No detectable levels of keratins 19 and 20 | ||
| Expression of the stem cell phenotype CD44+/CD24low | ||
| ALDH1+/CD44+/CD24− | Very high tumorigenic potential, rare | 13, 101 |
| CD44+/CD24− | High tumorigenic potential | 101, 102 |
| Mesenchymal-like, quiescent | ||
| Expression of EMT genes | ||
| Located in tumor periphery | ||
| CD44+/CD24low | High tumorigenic potential | 102 |
| Epithelial-like | ||
| Expression of keratins 5/6 | ||
| Low expression of vimentin, keratin 17, and E-cadherin | ||
| CD44−/CD24+ | Bone tropism | 105 |
| Expression of low molecular weight keratins, EMA, and vimentin | ||
| Bone tropism signature (88 upregulated and 22 downregulated genes) | ||
| ER−/CD44+/CD49fhi/CD133 | High tumorigenic potential | 103 |
| High levels of keratins 18 and 14 | ||
| EpCAM+CD44+CD47+MET+ | Escaping immune response | 35 |
| Critical in bone, lung, and liver metastases | ||
| Associated with disease progression |
Abbreviations: ALDH1, aldehyde dehydrogenase 1; BCSC, breast cancer stem cell; EMA, epithelial membrane antigen; EGFR, epidermal growth factor receptor; EMT, epithelial-to-mesenchymal transition; EpCAM, epithelial cell adhesion molecule; ER, estrogen receptor; HER2, human epidermal growth factor receptor 2; HPSE, heparanase MET, mesenchymal-to-epithelial transition.
A recent study101 has revealed that human BCSCs can be generally divided into two subcategories, one expressing the ALDH1+ and the other displaying the CD44+/CD24− phenotype. These have been shown to interconvert from one type to another, presumably depending on the tumor phase and requirements. While CD44+/CD24−cells express EMT genes, display a quiescent phenotype and are localized in the tumor periphery, possibly promoting tumor spreading, ALDH1+ cells are located in the tumor interior, display an epithelial profile expressing MET genes, and exhibit a proliferative phenotype, most likely supporting tumor growth. Microarray analysis revealed that a large number of genes are reciprocally expressed among these subpopulations. These cells are suggested to resemble their normal epithelial (EpCAM+/CD49f+) and mesenchymal (EpCAM−/CD49f+) type mammary stem cells. BCSCs expressing an overlapping phenotype (ALDH1+/CD44+/CD24−) were characterized by a very low detection frequency within the tumor mass and exhibited a more aggressive profile and greater metastatic potential.101 Interestingly, as few as 20 ALDH1+/CD44+/CD24− cells were able to generate tumors in immunosuppressed mice.13
Functional and phenotypic heterogeneity can also exist within the CD44+ compartment alone. Using a triple negative cell line, Leth-Larsen et al102 demonstrated that CD44high cells could be subdivided into two categories, CD44+/CD24− (mesenchymal type cells) and CD44+/CD24low (epithelioid cells). CD44+/CD24− displayed a mesenchymal morphology and expressed higher levels of vimentin, keratin 17, and E-cadherin, and lower levels of keratins 5/6, in comparison to epithelial-type CD44+/CD24low cells. Interestingly, epithelial-like CD44+/CD24low rather than mesenchymal-like CD44+/CD24− cells were able to form mammospheres ex vivo and retained the ability to form tumors when inoculated into the mammary fat pad of immunodeficient mice and showed greater resistance to conventional chemotherapy (doxorubicin, methotrexate, and paclitaxel).
In estrogen receptor α-negative breast tumors CD44pos CD49fhiCD133/2hi, xenograft initiating cells exhibit elevated tumorigenicity, and self-renewal capacity (expressing high levels of Sox2, Bmi-1, and/or Nanog and displaying CpG island hypermethylation) as well as high levels of the luminal marker keratin 18 and the basal marker keratin 14. These cells generate molecular and functional heterogeneity, giving rise to nontumorigenic progeny, an ability maintained after many in vivo passages, while ex vivo, they displayed elevated tumorsphere forming capability. CD44posCD49fhiCD133/2hi cells were primarily observed immunohistochemically in disordered clusters.103
A specific subset of cells competent for brain metastasis has been identified in the blood of breast cancer patients and cultivated ex vivo.104 Apart from the stem cell markers ALDH1 and HER2, these cells were also positive for the expression of epidermal growth factor receptor (EGFR), heparanase (HPSE), and Notch1 and negative for the expression of EpCAM (EpCAM−/ALDH1+/HER2+/EGFR+/HPSE+/Notch1+). Reverse transcription polymerase chain reaction analysis revealed high expression levels of vimentin, keratins 8 and 18, and expression of the stem cell phenotype CD44+/CD24low, while no detectable levels of keratins 19 and 20 were observed. When injected intracardially or into the tail vein of immunodeficient mice, these cells metastasized to the brain in a time period of 6 weeks, and generated tumors closely resembling the histology of the neoplastic tissue of their patient of origin.104
A bone tropism signature for BCTCs was also proposed.105 Specifically, the authors monitored the CD44+/CD24− subpopulation by in vivo luciferase imaging, which colonized subcutaneous human bone implants after injection in non-obese diabetic/severe combined immunodeficiency mice. Interestingly, the bone-isolated CSCs-like cells displayed a “phenotypic switch” expressing the CD44−CD24+ phenotype probably resulting from microenvironmental interactions. These cells expressed low molecular weight keratins, epithelial membrane antigen, and vimentin and initiated tumor formation after injection in secondary mice. Transcriptional analysis of CD44−CD24+ revealed a distinct bone tropism signature of 88 upregulated and 22 downregulated genes,105 which can delineate bone metastatic breast cancers and distinguish them from other types of metastatic breast cancer.
Mouse xenograft analysis of CTCs has also revealed that CTCs from primary human luminal breast cancer contain specific clones that generated bone, lung, and liver metastases in mice, which expressed EpCAM, CD44, CD47, and MET. CD47, known as the “don’t eat me” molecule, inhibits phagocytosis and contributes to immune evasion106–108 and the hepatocyte growth factor receptor MET tyrosine kinase has been shown to exhibit a migratory and invasive role in many types of cancer.109 When EpCAM+CD44+CD47+MET+ cells were transplanted into the femoral medullar cavity of mice, bone metastasis was observed after 8 months. Consequently, a molecular signature (EpCAM+CD44+CD47+MET+) designating a BCTC subset capable of metastasis initiation and indicative of poor prognosis has been proposed.35 Indeed, EpCAM+CD44+CD47+MET+ cells are detected more frequently in patients with disease progression, in contrast to that of bulk EpCAM+ CTCs. The fact that the expression of CD47 is observed in patient bone metastases and corresponding bone metastases in mice but not primary tumors indicates that CD47 expression is acquired after metastatic dissemination.
Stem cell technology and breast cancer therapy
Targeting BCSCs
One of the principal objectives of breast cancer treatment is the eradication of BCSCs, which display resistance to conventional chemotherapy and underlie tumor recurrence. Failure of conventional chemotherapy to eradicate the BCSC subpopulation results in BCSC-enriched residual tumors, which display a more mesenchymal and aggressive phenotype.110 Thus, simultaneous targeting of CSCs and non-CSCs holds a great promise toward the development of more efficient therapeutic methodologies. Indeed, combined chemo- and BCSC-targeting therapy is currently being evaluated on a clinical basis.42 The fact that CSCs exhibit phenotypic similarities with normal stem cells, however, raises the question of selective targeting of cancer versus normal stem cells. CSCs could be distinguished from their normal counterparts because they carry cancer-specific glycans. These probably originate from altered glycosylation of normal stem cell glycoproteins during their malignant transformation and are introduced as CSC-specific glycans.14
The targeting of BCSCs involves the disruption of BCSC survival signaling pathways (ie, Notch, HER2, Hedgehog, Wnt, PI3K/Akt/mToR, interleukin 8, TGF-beta); induction of differentiation with the use of small inhibitors as salinomycin, histone deacetylase inhibitors, all trans retinoic acid, and small hairpin RNA lentivirus particles; targeting of CSC metabolic pathways; and the use of microRNAs, cancer immunotherapy, drugs involved in the treatment of noncancer diseases, and nanotechnology (Table 3) (for a review see42). Nanodrugs can easily accumulate within tumor sites due to their enhanced vascular permeability. Biodegradable polymeric micelles loaded with paclitaxel and functionalized with anti-CD44 antibodies have been used in breast cancer cell lines. In order to examine the effectiveness of nanomedicines on the CSC subpopulation, an in vitro fluorescent CSC model was developed that enables the visualization and posttreatment assessment of biological performance of CSCs.111 Although targeting BCSCs holds a great promise in the treatment of breast cancer and is widely tested on a basic research level, a disproportionally limited number of clinical trials evaluating the effect of treatment on the expression of BCSC biomarkers are in progress. In a recent clinical trial, interrupting the Notch pathway with the administration of the γ-secretase inhibitor MK-0752 resulted in decreased levels of CD44+/CD24−, ALDH+ cell populations in the tumor biopsies.112 Ongoing clinical trials evaluating the effect of Hedgehog, CXCR1/2, EGFR/HER2, AKT, and angiogenesis inhibitors on the CSC subpopulation of breast cancer patients in addition to the clinical testing of novel CSC vaccines will provide further insight on their clinical applicability and efficacy (see42 and references therein).
Table 3.
Applications of stem cell technology in breast cancer therapy
The use of antibiotics for the targeting of CSCs is a novel approach in the field of breast cancer. In a recent study, Lamb et al113 demonstrated that MCF-7 mammospheres exhibit a marked overexpression of >60 mitochondrial-related proteins, nine of which were markedly upregulated. The authors tested whether mitochondrial biogenesis is required for the survival and proliferation of CSCs with the use of Food and Drug Administration-approved antibiotics, which target mitochondria. Mitochondrial targeting of normal cells by specific classes of antibiotics results from the “endosymbiotic theory of mitochondrial evolution” and in normal cells it exerts mild side effects, which are well tolerated in most patients. It was shown that the survival of CSCs is largely dependent on the biogenesis of mitochondria and that specific Food and Drug Administration-approved antibiotics, such as erythromycins, tetracyclines, glycylcyclines, and chloramphenicol, can be used to specifically eliminate cancer cells grown as mammospheres in 12 different cancer cell lines across eight different tumor types, including breast cancer. Intriguingly, the authors propose the treatment of cancer as an infectious disease and highlight the role of antibiotics in the prevention of the disease relapse, emphasizing on the fact that many of these drugs are nontoxic to normal cells, thus reducing the side effects of anticancer therapy.113
One of these promising antibiotics is doxycycline, a member of the tetracycline class, with excellent pharmacokinetics. It has been shown that doxycycline treatment significantly reduced the expression of many key protein targets functionally associated with mitochondrial metabolism, glycolysis, EMT, protein synthesis, and the DNA damage response, as well as inflammation and protein degradation, in human breast cancer cells.114 In particular, DNA-PK, an enzyme thought to confer resistance in cancer cells, was dramatically downregulated by doxycycline.114 Doxycycline is relatively attractive as a new anticancer agent with low toxic side effects. It has a long half-life systemically and has been used successfully for the long-term treatment of patients with urinary tract infections, prostatitis, or acne, for extended periods of time. More importantly, it has been shown that doxycycline enhances the culturing efficiency, survival, and self-renewal of human pluripotent stem cells.115,116 Precisely, through the direct activation of the PI3K-AKT intracellular pathway, it dramatically enhances the expandability of human embryonic stem and induced pluripotent stem cells.
Additionally, salinomycin has been shown to reduce the fraction of CSCs by >100-fold compared to paclitaxel and promote mammary tumor growth arrest in mice.117 Combination therapy targeting both BCSCs and breast cancer cells via the co-delivery of salinomycin and doxorubicin displayed a twofold in vivo breast tumor suppression compared to single drug therapy.118
Recently, the establishment of ex vivo cultures of CTCs from the blood of breast cancer patients has enabled the examination of drug sensitivity of cultured cells, revealing new potential therapeutic targets, activation of specific signaling pathways (including stem cell related signatures), and constituted the basis for the future design of novel individualized therapeutic strategies.119
Normal stem cell-mediated delivery of anticancer drugs
One of the most recent innovative approaches in breast cancer therapy is the recruitment of normal stem cells for the eradication of tumor cells. A number of studies have shown that mesenchymal stem cells (MSCs) have the inherent ability to migrate toward sites of inflammation, injury, ischemia and, most importantly, tumor microenvironments (a property termed “tumor tropism”). The migratory mechanisms of MSCs toward cancer sites are not yet unraveled, although cytokine signaling (stromal cell-derived factor 1, hepatocyte growth factor, vascular endothelial growth factor (VEGF), platelet-derived growth factor, and monocyte chemoattractant protein 1) is a key regulator of this behavior.120 The use of MSCs for the delivery of anticancer drugs is highly selective and can overcome the obstacle of limited drug half-life as MSCs can be engineered to continually secrete the drug of choice.121 Interestingly, not only engineered but also unmodified or “naïve” MSCs exhibit antitumor activity in various mouse models of cancer, as they secrete tumoricidal factors.122 Umbilical cord, adipose and amniotic fluid, and neural tissue stem cells have been recruited for targeting breast cancer cells (Table 3).
Umbilical cord MSCs
Genetically engineered human umbilical cord mesenchymal stem cells (HUMSCs) that overexpress the tumor suppressor gene follistatin displayed enhanced tumoricidal activity.123 Coculture of HUMSCs and MDA-MB231 has been shown to result in apoptosis of the latter, either through direct cellular contact or by a novel phenomenon that includes the internalization of HUMSC into MDA-MB231. Moreover, injection of HUMSCs in primary and metastatic breast cancer sites in animal models displayed satisfactory results as indicated by fading or absent in vivo bioluminescence signals of tumor cells.124 It has been suggested that in vitro and in vivo inhibition of BCSC growth by HUMSCs is associated with cell cycle arrest, induction of tumor cell apoptosis, and suppression of PI3K and AKT protein kinases.125
Adipose stem cells
The use of adipose stem cells (ADSCs) has also been recruited in the fight against breast cancer. In animal models of human breast cancer, it has been shown that the injection of ADSCs peripheral to the tumor leads to tumor withdrawal and total recovery within 6 months.126 In spite of the therapeutic potential of ADSCs, recent reports have revealed that they can possibly support breast cancer progression. Specifically, white adipose tissue-derived progenitor cells, which are used widely for soft tissue reconstruction, have been suggested to promote local breast cancer growth and metastasis.127 In murine models, human adipose tissue-derived stromal/stem cells from abdominal lipoaspirates were reported to underlie metastasis of MDA-MB-231 breast tumor xenografts to various organs. Moreover, MDA-MB-231 tumors that were coinjected in mice with adipose tissue-derived stromal/stem cells displayed partial EMT, expression of matrix metalloproteinase-9 (MMP-9), and increased angiogenesis.128 Adipose tissue progenitor cells induced EMT in luminal breast cancer cells and were shown to promote metastasis of breast cancer.129
Amniotic fluid stem cells
Genetically engineered human amniotic fluid-derived stem cells expressing cytosine deaminase and thymidine kinase, which convert nontoxic prodrugs into cytotoxic metabolites, have been shown to inhibit the growth of breast cancer cells in cellular and xenograft mouse models. Specifically, in the presence of prodrugs, they hindered the growth of MDA-MB-231 human breast cancer cells and reduced the tumor size in BALB/c nude mouse MDA-MB-231 xenografts, whereas their use did not disturb the normal architecture of breast tissues. Thus, they have been suggested as effective vehicles for the selective targeting of breast tumors.130 Human amniotic membrane-derived epithelial stem cells also display anticancer activity in BALB/c nude mice bearing metastatic breast cancer xenografts. When mice have been treated with human amniotic membrane-derived epithelial stem cells, tumor progression was inhibited, whereas no side effects were observed. Human amniotic membrane-derived epithelial stem cells are proposed as a safe and effective breast cancer targeting therapy.131
Neural stem cells
Neural stem cells genetically modified for the expression of a tumor selective and secretable variant of TNF-related apoptosis-inducing ligand (TRAIL) migrated toward brain metastasis after implantation or injection in the circulation, suppressed metastatic tumor growth, and prolonged the survival of mice bearing metastatic breast tumors in a breast-to-brain metastasis mouse model.132 Intracranial injection of neural stem cells secreting anti-HER2 antibody in a mouse model of breast cancer brain metastases has been shown to improve survival,133 while genetically engineered human neural stem cells with rabbit carboxyl esterase can target brain metastasis from breast cancer.134 Finally, neural stem cells transduced to express Escherichia coli cytosine deaminase, which exhibits a prodrug converting capacity, and human interferon-beta, which inhibits tumor growth via apoptosis, have also been found to target ductal breast cancer cells in cellular and xenograft models.135
Adverse effects and clinical limitations
One of the major concerns on the use of pathotropic stem cells for the treatment of cancer is their ability to secrete signaling molecules that could modify the tumor microenvironment and contribute to tumor invasiveness, growth, and angiogenesis. As mentioned above, ADSCs have been shown to enhance the metastatic potential of breast cancer cells promoting tumor growth. The secretion of interleukin 6 and CCL5 by MSCs increased the growth and metastasis of breast cancer cells.136,137 Thus, the pro-neoplastic properties of normal stem cells within a deregulated tumor microenvironment should be taken into consideration prior to the development of any therapeutic strategy. Furthermore, the route of administration and cell concentration must be determined for an optimal therapeutic result. Due to the above limitations, clinical trials examining the effect of normal stem cell-mediated therapy for the specific treatment of breast cancer are rather lacking.
Conclusion
Undoubtedly, the discovery of CSCs has initiated a new era in the field of tumor biology and provided the basis for the development of novel therapeutic strategies against cancer. It is now beginning to emerge that BCSCs comprise a highly dynamic and heterogeneous subpopulation, with compartmentalized properties and specific functions within the tumor and the ability to act coordinately in response to stimuli from the tumor environment. The phenotypic and functional characterization of BCSCs combined with the use of advanced isolation technologies and targeting schemes will enable the efficient eradication of the entire BCSC population.
Recent advances in the field of breast cancer research, as the ex vivo culture of BCTCs, use of antibiotics, nanomedicine, and the employment of normal stem cells for the elimination of BCSCs, have promoted the establishment of new individualized treatment schemes and are paving the way toward a more patient-friendly therapeutic approach. However, the safety of newly developed methodologies, such as the introduction of normal stem cells into an already genetically destabilized environment, should be critically evaluated and standardized, in view of the fact that they can trigger disease relapse. Despite current impediments, the development of stem cell-based therapy, in combination with conventional chemotherapy, still remains a significant tool for breast cancer scientists and holds a promising future in the treatment of breast cancer.
Remaining questions and future directions
Although our knowledge on CSCs has largely expanded in the last decades, many aspects of their biology and behavior still remain elusive. Apart from the origination of CSCs, which still remains one of the most fundamental and intriguing questions in breast cancer research, a better understanding of the interplay between the tumor microenvironment and CSCs will promote the development of safer and more specific therapeutic approaches targeting the tumor microenvironment, while predicting their effects on interacting tumor cells. Also, defining the molecular markers and metabolic pathways that distinguish normal and CSCs will ensure the specific targeting of the latter and minimize therapy side effects. Finally, uncovering the molecular markers implicated in CSC quiescence will shed light on the mechanisms of the biological resistance of CSCs and pave the way for novel clinical applications.
From a practical point of view, an important challenge to be resolved is the isolation of the total population of BCTCs from the blood of patients. Thus, new isolation technologies based on the physicochemical properties and specific markers of BCSCs need to be developed. This will allow their elimination from the circulation and consequently will diminish the risk of metastasis for adjuvant patients. Characterizing BCSCs will promote the generation of new molecular signatures applied in liquid biopsy, which will significantly contribute to the treatment and follow-up care of breast cancer patients.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645–648. doi: 10.1038/367645a0. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.Yang ZF, Ngai P, Ho DW, et al. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47(3):919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 5.Dalerba P, Dylla SJ, Park IK, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007;104(24):10158–10163. doi: 10.1073/pnas.0703478104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schatton T, Murphy GF, Frank NY, et al. Identification of cells initiating human melanomas. Nature. 2008;451(7176):345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456(7222):593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McDermott SP, Wicha MS. Targeting breast cancer stem cells. Mol Oncol. 2010;4(5):404–419. doi: 10.1016/j.molonc.2010.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Laterra J. Cancer stem cells: distinct entities or dynamically regulated phenotypes? Cancer Res. 2012;72(3):576–580. doi: 10.1158/0008-5472.CAN-11-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaffer CL, Brueckmann I, Scheel C, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A. 2011;108(19):7950–7955. doi: 10.1073/pnas.1102454108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eirew P, Stingl J, Raouf A, et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat Med. 2008;14(12):1384–1389. doi: 10.1038/nm.1791. [DOI] [PubMed] [Google Scholar]
- 13.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karsten U, Goletz S. What makes cancer stem cell markers different? Springerplus. 2013;2(1):301. doi: 10.1186/2193-1801-2-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proia TA, Keller PJ, Gupta PB, et al. Genetic predisposition directs breast cancer phenotype by dictating progenitor cell fate. Cell Stem Cell. 2011;8(2):149–163. doi: 10.1016/j.stem.2010.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim E, Vaillant F, Wu D, et al. Aberrant luminal progenitors as the candidate target population for basal tumor development in BRCA1 mutation carriers. Nat Med. 2009;15(8):907–913. doi: 10.1038/nm.2000. [DOI] [PubMed] [Google Scholar]
- 17.Molyneux G, Geyer FC, Magnay FA, et al. BRCA1 basal-like breast cancers originate from luminal epithelial progenitors and not from basal stem cells. Cell Stem Cell. 2010;7(3):403–417. doi: 10.1016/j.stem.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 18.Castano Z, Fillmore CM, Kim CF, McAllister SS. The bed and the bugs: interactions between the tumor microenvironment and cancer stem cells. Semin Cancer Biol. 2012;22(5–6):462–470. doi: 10.1016/j.semcancer.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 20.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 2009;138(5):822–829. doi: 10.1016/j.cell.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 21.Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22(3):457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dick JE. Looking ahead in cancer stem cell research. Nat Biotechnol. 2009;27(1):44–46. doi: 10.1038/nbt0109-44. [DOI] [PubMed] [Google Scholar]
- 23.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 24.Campbell LL, Polyak K. Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle. 2007;6(19):2332–2338. doi: 10.4161/cc.6.19.4914. [DOI] [PubMed] [Google Scholar]
- 25.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP. The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol. 2012;13(2):e83–e89. doi: 10.1016/S1470-2045(11)70257-1. [DOI] [PubMed] [Google Scholar]
- 27.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheel C, Eaton EN, Li SH, et al. Paracrine and autocrine signals induce and maintain mesenchymal and stem cell states in the breast. Cell. 2011;145(6):926–940. doi: 10.1016/j.cell.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owens TW, Naylor MJ. Breast cancer stem cells. Front Physiol. 2013;4:225. doi: 10.3389/fphys.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borovski T, De Sousa EMF, Vermeulen L, Medema JP. Cancer stem cell niche: the place to be. Cancer Res. 2011;71(3):634–639. doi: 10.1158/0008-5472.CAN-10-3220. [DOI] [PubMed] [Google Scholar]
- 31.Meads MB, Gatenby RA, Dalton WS. Environment-mediated drug resistance: a major contributor to minimal residual disease. Nat Rev Cancer. 2009;9(9):665–674. doi: 10.1038/nrc2714. [DOI] [PubMed] [Google Scholar]
- 32.DeNardo DG, Brennan DJ, Rexhepaj E, et al. Leukocyte complexity predicts breast cancer survival and functionally regulates response to chemotherapy. Cancer Discov. 2011;1(1):54–67. doi: 10.1158/2159-8274.CD-10-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sampieri K, Fodde R. Cancer stem cells and metastasis. Semin Cancer Biol. 2012;22(3):187–193. doi: 10.1016/j.semcancer.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 34.Theodoropoulos PA, Polioudaki H, Agelaki S, et al. Circulating tumor cells with a putative stem cell phenotype in peripheral blood of patients with breast cancer. Cancer Lett. 2010;288(1):99–106. doi: 10.1016/j.canlet.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 35.Baccelli I, Schneeweiss A, Riethdorf S, et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat Biotechnol. 2013;31(6):539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 36.Giordano A, Gao H, Anfossi S, et al. Epithelial-mesenchymal transition and stem cell markers in patients with HER2-positive metastatic breast cancer. Mol Cancer Ther. 2012;11(11):2526–2534. doi: 10.1158/1535-7163.MCT-12-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aktas B, Tewes M, Fehm T, Hauch S, Kimmig R, Kasimir-Bauer S. Stem cell and epithelial-mesenchymal transition markers are frequently overexpressed in circulating tumor cells of metastatic breast cancer patients. Breast Cancer Res. 2009;11(4):R46. doi: 10.1186/bcr2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raimondi C, Gradilone A, Naso G, et al. Epithelial-mesenchymal transition and stemness features in circulating tumor cells from breast cancer patients. Breast Cancer Res Treat. 2011;130(2):449–455. doi: 10.1007/s10549-011-1373-x. [DOI] [PubMed] [Google Scholar]
- 39.Mego M, Gao H, Lee BN, et al. Prognostic value of EMT-circulating tumor cells in metastatic breast cancer patients undergoing high-dose chemotherapy with autologous hematopoietic stem cell transplantation. J Cancer. 2012;3:369–380. doi: 10.7150/jca.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Balic M, Lin H, Young L, et al. Most early disseminated cancer cells detected in bone marrow of breast cancer patients have a putative breast cancer stem cell phenotype. Clin Cancer Res. 2006;12(19):5615–5621. doi: 10.1158/1078-0432.CCR-06-0169. [DOI] [PubMed] [Google Scholar]
- 41.Liu HG, Chen C, Yang H, Pan YF, Zhang XH. Cancer stem cell subsets and their relationships. J Transl Med. 2011;9:50. doi: 10.1186/1479-5876-9-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiotaki R, Polioudaki H, Theodoropoulos PA. Cancer stem cells in solid and liquid tissues of breast cancer patients: characterization and therapeutic perspectives. Curr Cancer Drug Targets. 2015;15(3):256–269. doi: 10.2174/1568009615666150211102503. [DOI] [PubMed] [Google Scholar]
- 43.Fernandez SV, Bingham C, Fittipaldi P, et al. TP53 mutations detected in circulating tumor cells present in the blood of metastatic triple negative breast cancer patients. Breast Cancer Res. 2014;16(5):445. doi: 10.1186/s13058-014-0445-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fehm T, Becker S, Duerr-Stoerzer S, et al. Determination of HER2 status using both serum HER2 levels and circulating tumor cells in patients with recurrent breast cancer whose primary tumor was HER2 negative or of unknown HER2 status. Breast Cancer Res. 2007;9(5):R74. doi: 10.1186/bcr1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X, Lewis MT, Huang J, et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J Natl Cancer Inst. 2008;100(9):672–679. doi: 10.1093/jnci/djn123. [DOI] [PubMed] [Google Scholar]
- 46.Chen K, Huang YH, Chen JL. Understanding and targeting cancer stem cells: therapeutic implications and challenges. Acta Pharmacol Sin. 2013;34(6):732–740. doi: 10.1038/aps.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abdullah LN, Chow EK. Mechanisms of chemoresistance in cancer stem cells. Clin Transl Med. 2013;2(1):3. doi: 10.1186/2001-1326-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Madjd Z, Mehrjerdi AZ, Sharifi AM, Molanaei S, Shahzadi SZ, Asadi-Lari M. CD44+ cancer cells express higher levels of the anti-apoptotic protein Bcl-2 in breast tumours. Cancer Immun. 2009;9:4. [PMC free article] [PubMed] [Google Scholar]
- 49.Kleffel S, Schatton T. Tumor dormancy and cancer stem cells: Two sides of the same coin? Adv Exp Med Biol. 2013;734:145–179. doi: 10.1007/978-1-4614-1445-2_8. [DOI] [PubMed] [Google Scholar]
- 50.Shachaf CM, Kopelman AM, Arvanitis C, et al. MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature. 2004;431(7012):1112–1117. doi: 10.1038/nature03043. [DOI] [PubMed] [Google Scholar]
- 51.Schober M, Fuchs E. Tumor-initiating stem cells of squamous cell carcinomas and their control by TGF-beta and integrin/focal adhesion kinase (FAK) signaling. Proc Natl Acad Sci U S A. 2011;108(26):10544–10549. doi: 10.1073/pnas.1107807108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tobar N, Villar V, Santibanez JF. ROS-NFkappaB mediates TGF-beta1-induced expression of urokinase-type plasminogen activator, matrix metalloproteinase-9 and cell invasion. Mol Cell Biochem. 2010;340(1–2):195–202. doi: 10.1007/s11010-010-0418-5. [DOI] [PubMed] [Google Scholar]
- 53.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7(2):131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 54.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342(6159):1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 55.Singh R, Mo YY. Role of microRNAs in breast cancer. Cancer Biol Ther. 2013;14(3):201–212. doi: 10.4161/cbt.23296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jovanovic J, Ronneberg JA, Tost J, Kristensen V. The epigenetics of breast cancer. Mol Oncol. 2010;4(3):242–254. doi: 10.1016/j.molonc.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14(6):818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 59.Ansieau S. EMT in breast cancer stem cell generation. Cancer Lett. 2013;338(1):63–68. doi: 10.1016/j.canlet.2012.05.014. [DOI] [PubMed] [Google Scholar]
- 60.Drasin DJ, Robin TP, Ford HL. Breast cancer epithelial-to-mesenchymal transition: examining the functional consequences of plasticity. Breast Cancer Res. 2011;13(6):226. doi: 10.1186/bcr3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dave B, Mittal V, Tan NM, Chang JC. Epithelial-mesenchymal transition, cancer stem cells and treatment resistance. Breast Cancer Res. 2012;14(1):202. doi: 10.1186/bcr2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PloS One. 2008;3(8):e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voon DC, Wang H, Koo JK, et al. Runx3 protects gastric epithelial cells against epithelial-mesenchymal transition-induced cellular plasticity and tumorigenicity. Stem Cells. 2012;30(10):2088–2099. doi: 10.1002/stem.1183. [DOI] [PubMed] [Google Scholar]
- 64.Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008;68(4):989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- 65.Mani SA, Yang J, Brooks M, et al. Mesenchyme Forkhead 1 (FOXC2) plays a key role in metastasis and is associated with aggressive basal-like breast cancers. Proc Natl Acad Sci U S A. 2007;104(24):10069–10074. doi: 10.1073/pnas.0703900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mizuno H, Spike BT, Wahl GM, Levine AJ. Inactivation of p53 in breast cancers correlates with stem cell transcriptional signatures. Proc Natl Acad Sci U S A. 2010;107(52):22745–22750. doi: 10.1073/pnas.1017001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu J, Liao S, Diop-Frimpong B, et al. TGF-beta blockade improves the distribution and efficacy of therapeutics in breast carcinoma by normalizing the tumor stroma. Proc Natl Acad Sci U S A. 2012;109(41):16618–16623. doi: 10.1073/pnas.1117610109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pece S, Tosoni D, Confalonieri S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140(1):62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 69.McCubrey JA, Davis NM, Abrams SL, et al. Targeting breast cancer initiating cells: advances in breast cancer research and therapy. Adv Biol Regul. 2014;56:81–107. doi: 10.1016/j.jbior.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 70.Vieira AF, Ricardo S, Ablett MP, et al. P-cadherin is coexpressed with CD44 and CD49f and mediates stem cell properties in basal-like breast cancer. Stem Cells. 2012;30(5):854–864. doi: 10.1002/stem.1075. [DOI] [PubMed] [Google Scholar]
- 71.Ribeiro AS, Paredes J. P-Cadherin linking breast cancer stem cells and invasion: A promising marker to identify an “intermediate/metastable” EMT state. Front Oncol. 2014;4:371. doi: 10.3389/fonc.2014.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Polioudaki H, Agelaki S, Chiotaki R, et al. Variable expression levels of keratin and vimentin reveal differential EMT status of circulating tumor cells and correlation with clinical characteristics and outcome of patients with metastatic breast cancer. BMC Cancer. 2015;15:399. doi: 10.1186/s12885-015-1386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Papadaki MA, Kallergi G, Zafeiriou Z, et al. Co-expression of putative stemness and epithelial-to-mesenchymal transition markers on single circulating tumour cells from patients with early and metastatic breast cancer. BMC Cancer. 2014;14:651. doi: 10.1186/1471-2407-14-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bock C, Rack B, Huober J, Andergassen U, Jeschke U, Doisneau-Sixou S. Distinct expression of cytokeratin, N-cadherin and CD133 in circulating tumor cells of metastatic breast cancer patients. Future Oncol. 2014;10(10):1751–1765. doi: 10.2217/fon.14.58. [DOI] [PubMed] [Google Scholar]
- 75.Barriere G, Riouallon A, Renaudie J, Tartary M, Rigaud M. Mesenchymal and stemness circulating tumor cells in early breast cancer diagnosis. BMC Cancer. 2012;12:114. doi: 10.1186/1471-2407-12-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Krawczyk N, Meier-Stiegen F, Banys M, Neubauer H, Ruckhaeberle E, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in circulating tumor cells of breast cancer patients. Bio Med Res Int. 2014;2014:415721. doi: 10.1155/2014/415721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Leccia F, Del Vecchio L, Mariotti E, et al. ABCG2, a novel antigen to sort luminal progenitors of BRCA1-breast cancer cells. Mol Cancer. 2014;13:213. doi: 10.1186/1476-4598-13-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fernandez-Zapico ME. GLI1 finds a new role in cancer stem cell biology. EMBO Mol Med. 2013;5(4):483–485. doi: 10.1002/emmm.201302505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lianidou ES, Markou A. Circulating tumor cells in breast cancer: detection systems, molecular characterization, and future challenges. Clin Chem. 2011;57(9):1242–1255. doi: 10.1373/clinchem.2011.165068. [DOI] [PubMed] [Google Scholar]
- 80.Brock G, Castellanos-Rizaldos E, Hu L, Cottichia C, Skog J. Liquid biopsy for cancer screening, patient stratification and monitoring. Transl Cancer Res. 2015;4(3):280–290. [Google Scholar]
- 81.Paterlini-Brechot P, Benali NL. Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett. 2007;253(2):180–204. doi: 10.1016/j.canlet.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 82.Nagaiah G, Abraham J. Circulating tumor cells in the management of breast cancer. Clin Breast Cancer. 2010;10(3):209–216. doi: 10.3816/CBC.2010.n.028. [DOI] [PubMed] [Google Scholar]
- 83.Alunni-Fabbroni M, Sandri MT. Circulating tumour cells in clinical practice: Methods of detection and possible characterization. Methods. 2010;50(4):289–297. doi: 10.1016/j.ymeth.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 84.Hong B, Zu Y. Detecting circulating tumor cells: current challenges and new trends. Theranostics. 2013;3(6):377–394. doi: 10.7150/thno.5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Riethdorf S, Fritsche H, Muller V, et al. Detection of circulating tumor cells in peripheral blood of patients with metastatic breast cancer: a validation study of the CellSearch system. Clin Cancer Res. 2007;13(3):920–928. doi: 10.1158/1078-0432.CCR-06-1695. [DOI] [PubMed] [Google Scholar]
- 86.Stott SL, Hsu CH, Tsukrov DI, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107(43):18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fehm T, Hoffmann O, Aktas B, et al. Detection and characterization of circulating tumor cells in blood of primary breast cancer patients by RT-PCR and comparison to status of bone marrow disseminated cells. Breast Cancer Res. 2009;11(4):R59. doi: 10.1186/bcr2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Danila DC, Pantel K, Fleisher M, Scher HI. Circulating tumors cells as biomarkers: progress toward biomarker qualification. Cancer J. 2011;17(6):438–450. doi: 10.1097/PPO.0b013e31823e69ac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Reuben JM, Lee BN, Gao H, et al. Primary breast cancer patients with high risk clinicopathologic features have high percentages of bone marrow epithelial cells with ALDH activity and CD44(+)CD24lo cancer stem cell phenotype. Eur J Cancer. 2011;47(10):1527–1536. doi: 10.1016/j.ejca.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nora Dickson M, Tsinberg P, Tang Z, Bischoff FZ, Wilson T, Leonard EF. Efficient capture of circulating tumor cells with a novel immunocytochemical microfluidic device. Biomicrofluidics. 2011;5(3):34119–3411915. doi: 10.1063/1.3623748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Saucedo-Zeni N, Mewes S, Niestroj R, et al. A novel method for the in vivo isolation of circulating tumor cells from peripheral blood of cancer patients using a functionalized and structured medical wire. Int J Oncol. 2012;41(4):1241–1250. doi: 10.3892/ijo.2012.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Balic M, Dandachi N, Lin H, Datar RH. Cancer metastasis: advances in the detection and characterization of disseminated tumour cells facilitate clinical translation. Natl Med J India. 2005;18(5):250–255. [PubMed] [Google Scholar]
- 93.Satelli A, Brownlee Z, Mitra A, Meng QH, Li S. Circulating tumor cell enumeration with a combination of epithelial cell adhesion moleculeand cell-surface vimentin-based methods for monitoring breast cancer therapeutic response. Clin Chem. 2015;61(1):259–266. doi: 10.1373/clinchem.2014.228122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Alix-Panabieres C. EPISPOT assay: detection of viable DTCs/CTCs in solid tumor patients. Recent Results Cancer Res. 2012;195:69–76. doi: 10.1007/978-3-642-28160-0_6. [DOI] [PubMed] [Google Scholar]
- 95.Ozkumur E, Shah AM, Ciciliano JC, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5(179):179ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Karabacak NM, Spuhler PS, Fachin F, et al. Microfluidic, marker-free isolation of circulating tumor cells from blood samples. Nat Protoc. 2014;9(3):694–710. doi: 10.1038/nprot.2014.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vona G, Sabile A, Louha M, et al. Isolation by size of epithelial tumor cells: a new method for the immunomorphological and molecular characterization of circulating tumor cells. Am J Pathol. 2000;156(1):57–63. doi: 10.1016/S0002-9440(10)64706-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Farace F, Massard C, Vimond N, et al. A direct comparison of CellSearch and ISET for circulating tumour-cell detection in patients with metastatic carcinomas. Br J Cancer. 2011;105(6):847–853. doi: 10.1038/bjc.2011.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hardt O, Wild S, Oerlecke I, et al. Highly sensitive profiling of CD44+/CD24− breast cancer stem cells by combining global mRNA amplification and next generation sequencing: evidence for a hyperactive PI3K pathway. Cancer Lett. 2012;325(2):165–174. doi: 10.1016/j.canlet.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 100.Phuc PV, Khuong TTT, Dong LV, Truong KD, Tran G, Phan NK. Isolation and characterization of breast cancer stem cells from malignant tumours in Vietnamese women. JCAB. 2010;4(12):163–169. [Google Scholar]
- 101.Liu S, Cong Y, Wang D, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2014;2(1):78–91. doi: 10.1016/j.stemcr.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Leth-Larsen R, Terp MG, Christensen AG, et al. Functional heterogeneity within the CD44 high human breast cancer stem cell-like compartment reveals a gene signature predictive of distant metastasis. Mol Med. 2012;18:1109–1121. doi: 10.2119/molmed.2012.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meyer MJ, Fleming JM, Lin AF, Hussnain SA, Ginsburg E, Vonderhaar BK. CD44posCD49fhiCD133/2hi defines xenograft-initiating cells in estrogen receptor-negative breast cancer. Cancer Res. 2010;70(11):4624–4633. doi: 10.1158/0008-5472.CAN-09-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang L, Ridgway LD, Wetzel MD, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5(180):180ra148. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.D’Amico L, Patane S, Grange C, et al. Primary breast cancer stem-like cells metastasise to bone, switch phenotype and acquire a bone tropism signature. Br J Cancer. 2013;108(12):2525–2536. doi: 10.1038/bjc.2013.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Willingham SB, Volkmer JP, Gentles AJ, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109(17):6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Majeti R, Chao MP, Alizadeh AA, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–299. doi: 10.1016/j.cell.2009.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chao MP, Alizadeh AA, Tang C, et al. Therapeutic antibody targeting of CD47 eliminates human acute lymphoblastic leukemia. Cancer Res. 2011;71(4):1374–1384. doi: 10.1158/0008-5472.CAN-10-2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11(12):834–848. doi: 10.1038/nrm3012. [DOI] [PubMed] [Google Scholar]
- 110.Creighton CJ, Li X, Landis M, et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A. 2009;106(33):13820–13825. doi: 10.1073/pnas.0905718106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gener P, Gouveia LP, Sabat GR, et al. Fluorescent CSC models evidence that targeted nanomedicines improve treatment sensitivity of breast and colon cancer stem cells. Nanomedicine. 2015;11(8):1883–1892. doi: 10.1016/j.nano.2015.07.009. [DOI] [PubMed] [Google Scholar]
- 112.Schott AF, Landis MD, Dontu G, et al. Preclinical and clinical studies of gamma secretase inhibitors with docetaxel on human breast tumors. Clin Cancer Res. 2013;19(6):1512–1524. doi: 10.1158/1078-0432.CCR-11-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lamb R, Ozsvari B, Lisanti CL, et al. Antibiotics that target mitochondria effectively eradicate cancer stem cells, across multiple tumor types: treating cancer like an infectious disease. Oncotarget. 2015;6(7):4569–4584. doi: 10.18632/oncotarget.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lamb R, Fiorillo M, Chadwick A, et al. Doxycycline down-regulates DNA-PK and radiosensitizes tumor initiating cells: Implications for more effective radiation therapy. Oncotarget. 2015;6(16):14005–14025. doi: 10.18632/oncotarget.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chang MY, Oh B, Rhee YH, Lee SH. Doxycycline supplementation allows for the culture of human ESCs/iPSCs with media changes at 3-day intervals. Stem Cell Res. 2015;15(3):608–613. doi: 10.1016/j.scr.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 116.Chang MY, Rhee YH, Yi SH, et al. Doxycycline enhances survival and self-renewal of human pluripotent stem cells. Stem Cell Reports. 2014;3(2):353–364. doi: 10.1016/j.stemcr.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gupta PB, Onder TT, Jiang G, et al. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138(4):645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim YJ, Liu Y, Li S, et al. Co-eradication of breast cancer cells and cancer stem cells by cross-linked multilamellar liposomes enhances tumor treatment. Mol Pharm. 2015;12(8):2811–2822. doi: 10.1021/mp500754r. [DOI] [PubMed] [Google Scholar]
- 119.Yu M, Bardia A, Aceto N, et al. Cancer therapy. Ex vivo culture of circulating breast tumor cells for individualized testing of drug susceptibility. Science. 2014;345(6193):216–220. doi: 10.1126/science.1253533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Momin EN, Vela G, Zaidi HA, Quinones-Hinojosa A. The oncogenic potential of mesenchymal stem cells in the treatment of cancer: directions for future research. Curr Immunol Rev. 2010;6(2):137–148. doi: 10.2174/157339510791111718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Shah K. Mesenchymal stem cells engineered for cancer therapy. Adv Drug Deliv Rev. 2012;64(8):739–748. doi: 10.1016/j.addr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Qiao L, Xu Z, Zhao T, et al. Suppression of tumorigenesis by human mesenchymal stem cells in a hepatoma model. Cell Res. 2008;18(4):500–507. doi: 10.1038/cr.2008.40. [DOI] [PubMed] [Google Scholar]
- 123.Ohta N, Ishiguro S, Kawabata A, et al. Human umbilical cord matrix mesenchymal stem cells suppress the growth of breast cancer by expression of tumor suppressor genes. PloS One. 2015;10(5):e0123756. doi: 10.1371/journal.pone.0123756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chao KC, Yang HT, Chen MW. Human umbilical cord mesenchymal stem cells suppress breast cancer tumourigenesis through direct cell-cell contact and internalization. J Cell Mol Med. 2012;16(8):1803–1815. doi: 10.1111/j.1582-4934.2011.01459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ma Y, Hao X, Zhang S, Zhang J. The in vitro and in vivo effects of human umbilical cord mesenchymal stem cells on the growth of breast cancer cells. Breast Cancer Res Treat. 2012;133(2):473–485. doi: 10.1007/s10549-011-1774-x. [DOI] [PubMed] [Google Scholar]
- 126.Illouz YG. Breast cancer treatment by adipose-derived stem cells: an experimental study. J Stem Cells. 2014;9(4):211–217. [PubMed] [Google Scholar]
- 127.Bertolini F, Petit JY, Kolonin MG. Stem cells from adipose tissue and breast cancer: hype, risks and hope. Br J Cancer. 2015;112(3):419–423. doi: 10.1038/bjc.2014.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rowan BG, Gimble JM, Sheng M, et al. Human adipose tissue-derived stromal/stem cells promote migration and early metastasis of triple negative breast cancer xenografts. PloS One. 2014;9(2):e89595. doi: 10.1371/journal.pone.0089595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Orecchioni S, Gregato G, Martin-Padura I, et al. Complementary populations of human adipose CD34+ progenitor cells promote growth, angiogenesis, and metastasis of breast cancer. Cancer Res. 2013;73(19):5880–5891. doi: 10.1158/0008-5472.CAN-13-0821. [DOI] [PubMed] [Google Scholar]
- 130.Kang NH, Hwang KA, Yi BR, et al. Human amniotic fluid-derived stem cells expressing cytosine deaminase and thymidine kinase inhibits the growth of breast cancer cells in cellular and xenograft mouse models. Cancer Gene Ther. 2012;19(6):412–419. doi: 10.1038/cgt.2012.15. [DOI] [PubMed] [Google Scholar]
- 131.Kang NH, Yi BR, Lim SY, et al. Human amniotic membrane-derived epithelial stem cells display anticancer activity in BALB/c female nude mice bearing disseminated breast cancer xenografts. Int J Oncol. 2012;40(6):2022–2028. doi: 10.3892/ijo.2012.1372. [DOI] [PubMed] [Google Scholar]
- 132.Bagci-Onder T, Du W, Figueiredo JL, Martinez-Quintanilla J, Shah K. Targeting breast to brain metastatic tumours with death receptor ligand expressing therapeutic stem cells. Brain. 2015;138(Pt 6):1710–1721. doi: 10.1093/brain/awv094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kanojia D, Balyasnikova IV, Morshed RA, et al. Neural stem cells secreting anti-HER2 antibody improve survival in a preclinical model of HER2 overexpressing breast cancer brain metastases. Stem Cells. 2015;33(10):2985–2994. doi: 10.1002/stem.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Seol HJ, Jin J, Seong DH, et al. Genetically engineered human neural stem cells with rabbit carboxyl esterase can target brain metastasis from breast cancer. Cancer Lett. 2011;311(2):152–159. doi: 10.1016/j.canlet.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 135.Yi BR, Hwang KA, Aboody KS, Jeung EB, Kim SU, Choi KC. Selective antitumor effect of neural stem cells expressing cytosine deaminase and interferon-beta against ductal breast cancer cells in cellular and xenograft models. Stem Cell Res. 2014;12(1):36–48. doi: 10.1016/j.scr.2013.09.010. [DOI] [PubMed] [Google Scholar]
- 136.Hall B, Dembinski J, Sasser AK, Studeny M, Andreeff M, Marini F. Mesenchymal stem cells in cancer: tumor-associated fibroblasts and cell-based delivery vehicles. Int J Hematol. 2007;86(1):8–16. doi: 10.1532/IJH97.06230. [DOI] [PubMed] [Google Scholar]
- 137.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449(7162):557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]


