Abstract
In clinical practice, point-of-care diagnostic testing has progressed rapidly in the last decade. For the field of wound care, there is a compelling need to develop rapid alternatives for bacterial identification in the clinical setting, where it generally takes over 24 hours to receive a positive identification. Even new molecular and biochemical identification methods require an initial incubation period of several hours to obtain a sufficient number of cells prior to performing the analysis. Here we report the use of an inexpensive, disposable electrochemical sensor to detect pyocyanin, a unique, redox-active quorum sensing molecule released by Pseudomonas aeruginosa, in wound fluid from patients with chronic wounds enrolled in the WE-HEAL Study. By measuring the metabolite excreted by the cells, this electrochemical detection strategy eliminates sample preparation, takes less than a minute to complete, and requires only 7.5 microliters of sample to complete the analysis. The electrochemical results were compared against 16S rRNA profiling using 454 pyrosequencing. Blind identification yielded 9 correct matches, 2 false negatives, and 3 false positives giving a sensitivity of 71% and specificity of 57% for detection of Pseudomonas. Ongoing enhancement and development of this approach with a view to develop a rapid point-of-care diagnostic tool is planned.
Keywords: Biosensors, Electrochemistry, Pseudomonas aeruginosa, Pyocyanin, Chronic wound, Wound fluid, Wound exudate
INTRODUCTION
For more than a century, the primary clinical identification method for bacterial infections has been plate cultures where bacteria are isolated and purified overnight using nutrient-based agar medium. Rapid, automated instrumentation has been widely regarded as the next step towards advancing bacterial identification. These instruments, however, still require a pure bacterial colony obtained from a plate culture and thus a lead time of at least 18–24 hours before any identification can be made (1, 2). Even novel molecular diagnostics such as the polymerase chain reaction require significant manpower and hours of processing, making them unfeasible as point-of-care tools in the clinic (3). There is an unmet need to develop rapid screening tools for identification of clinically-relevant bacterial species in the clinical setting. Pseudomonas aeruginosa is well recognized as a major organism contributing to healing delay in chronic wounds (4). For patients undergoing split thickness skin grafting of chronic venous leg ulcers, the presence of Pseudomonas aeruginosa is a predictor of skin graft outcome (5). In clinical environments, bacterial culture methods are often inadequate for fully analyzing the microbial content of biofilm (6). A rapid screen for Pseudomonas aeruginosa and other clinically-relevant bacteria would allow clinicians to promptly switch from broad-spectrum antibiotics to specific directed therapies, lowering hospital expenditures, minimizing drug resistance, and improving patient care outcomes (7).
Pseudomonas aeruginosa is a gram-negative, non-fermenting aerobic rod that is a common pathogen in nosocomial infections particularly in patients with ventilator-associated pneumonia, cystic fibrosis, chronic wounds, and burn wounds (8, 9). One of the major factors contributing to the pathogenicity of P. aeruginosa in the healthcare setting is its ability to form biofilm. Biofilm formation decreases clearance of the organism by resisting host immune responses and limits efficacy of antibiotics (10).
In 1981, Reyes et al. tested 835 strains of P. aeruginosa, all clinical isolates, and found that 98% of the strains produced pyocyanin (11), a redox-active quorum sensing molecule linked to biofilm formation (12, 13). The biosynthetic gene loci for pyocyanin and its precursor has been characterized in P. aeruginosa and related species (14). Other redox-active precursors exist on the biosynthesis pathway to pyocyanin and they include a variety of phenazine derivatives such as phenazine-1-carboxylic acid, 5-methylphenazine-1-carboxylic acid, phenazine-1-carboxamide, and 1-hydroxyphenazine. However, only pyocyanin exhibits a distinct, electrochemical signal when scanned using square-wave voltammetry (15). Additionally, pyocyanin is responsible for the characteristic blue-green color of Pseudomonas species and it acts both as a virulence factor and a quorum sensing molecule for P. aeruginosa, playing a role in bacterial respiration and as an antibacterial and anti-fungal agent.
Previous work has shown that the redox-active properties of pyocyanin can be harnessed as a unique electrochemical marker for the presence of P. aeruginosa (16–18). A recent study demonstrated utility of this detection strategy by identifying pyocyanin in liquid samples with the use of disposable electrochemical sensors (16, 19–21). The purpose of the current study was to evaluate the use of an inexpensive, disposable screen-printed electrode to screen wound fluid exudate samples obtained from patients with chronic wounds for the presence of P. aeruginosa. With the recent advances in bacterial biosensors (22, 23) and today’s new wave of antibiotic-resistant pathogens (24), there is an urgent and unmet need to develop platforms to rapidly identify Pseudomonas and other pathogens at the point of clinical care.
MATERIALS AND METHODS
Patients and biospecimens
This research was conducted through the Wound Etiology and Healing (WE-HEAL) Study, a biospecimen and data repository designed for studying chronic wounds approved by the George Washington University Institutional Review Board (041408). Subjects are eligible for this study if they have an open wound at the time of evaluation and are older than 18 years of age. All subjects gave written informed consent for collection of specimens and data.
For this experiment, 14 paired wound fluid and biofilm samples from 12 patients were selected for analysis (Table 1). This was a convenience sample selected based on availability of wound fluid and wound microbiome samples from the same collection date.
Table 1.
Demographic and clinical characteristics of patients (n=12) from whom wound fluid samples were tested. Wound size (mean ± SD) of all wounds with specimens collected (n=14).
| All patients n=12 |
Pseudomonas spp. positive on 16S rRNA n=6 |
Pseudomonas spp. negative on 16S rRNA n=6 |
p-value | ||
|---|---|---|---|---|---|
| Age (years, mean ± SD) | 50.76 (±17.14) | 49.85 (±11.57) | 51.67 (±22.55) | 0.8642 | |
| Male sex (n, %) | 8 (66%) | 4 (66%) | 4 (66%) | 1.00 | |
| Race | |||||
| African American (n, %) | 8 (66%) | 5 (83%) | 3 (50%) | ||
| Caucasian (n, %) | 3 (25%) | 1 (16.7%) | 2 (33.3%) | 0.3998 | |
| Asian (n, %) | 1 (8.3%) | 1 (16.7%) | |||
| Smoking | |||||
| Past | 5 | 2 | 3 | ||
| Never | 7 | 4 | 3 | 1.00 | |
| Current | 0 | 0 | 0 | ||
| Diabetes | 4 | 2 | 2 | 1.00 | |
| Renal disease | 2 | 1 | 1 | 1.00 | |
|
Wound surface area (cm2, mean ± SD) |
85.41 (±177.3) |
14.13 (±12.77) |
146.5 (±230.9) |
0.19 | |
Wound effluent collection
According to standard operating procedures for the WE-HEAL Study, wound effluent specimens were collected using the Levine technique (25). This technique has been well validated to ensure standardization throughout all specimens collected in the WE-HEAL Study. After collection, the swabs were immediately placed in 0.65 µm pore size centrifugal filters (Ultrafree-MC DV, Merck Millipore, MA, USA). Samples were centrifuged at 12000 rpm for 4 minutes to extract the wound exudate and remove cellular and fibrinous debris. Samples were stored at −80 °C until analysis.
Biofilm collection
According to standard operating procedures for the WE-HEAL Study, wound biofilm specimens were collected by swabbing the wound with a cotton swab also using the Levine technique (25, 26). Samples were then stored at −80 °C until analysis.
16S rRNA profiling by 454 pyrosequencing
Bacterial DNA for 16S sequencing was isolated from wound swabs. Wound swabs were resuspended in 1,200 µL of lysis buffer (20mM Tris-Cl, pH 8.0, 2mM EDTA, 1.2% Triton X-100) and vortexed thoroughly for 1 minute. Lysate (1,000 µL) was transferred into a lysing Matrix B tube (MP Biomedicals Cat # 6911-500, http://www.mpbio.com), vortexed, and centrifuged. Lysate was subsequently incubated at 75 °C for 10 minutes and treated with 200 mg/mL lysozyme and 20 mg/mL Proteinase K. DNA from the lysate was extracted twice using phenol-chloroform-isoamyl alcohol followed by ethanol precipitation.
DNA extracted from individual chronic wound swab samples was then amplified using PCR primers that target the V1-V3 regions of the 16S rRNA gene (27) and carry a unique 12bp barcode. 16S amplicons were generated using 100ng of extracted DNA, Platinum Taq polymerase (Life Technologies, CA) in the following cycling conditions: first cycle of 95 °C 5 min; 35 cycles of 95°C/30s, 55°C/30s, 72°C/30s; last cycle of 72°C 7 min. Amplicons were purified (QIAquick PCR purification kit, Qiagen, Valencia, CA), quantified fluorometrically (Tecan Group Mannedorf, Switzerland), normalized, and pooled. 454 sequencing was completed using Titanium chemistry (Roche, Branford, CT) following the manufacturer’s protocol.
Taxonomic classification was performed using the YAP package (28) which implements the mothur software (29) based on the 16S rRNA gene reference sequences from the Ribosomal Database Project (30). Biofilm specimens were considered to be positive for Pseudomonas spp. if any Pseudomonas reads were detected in the specimen regardless of relative abundance.
Pyocyanin electrochemical probe
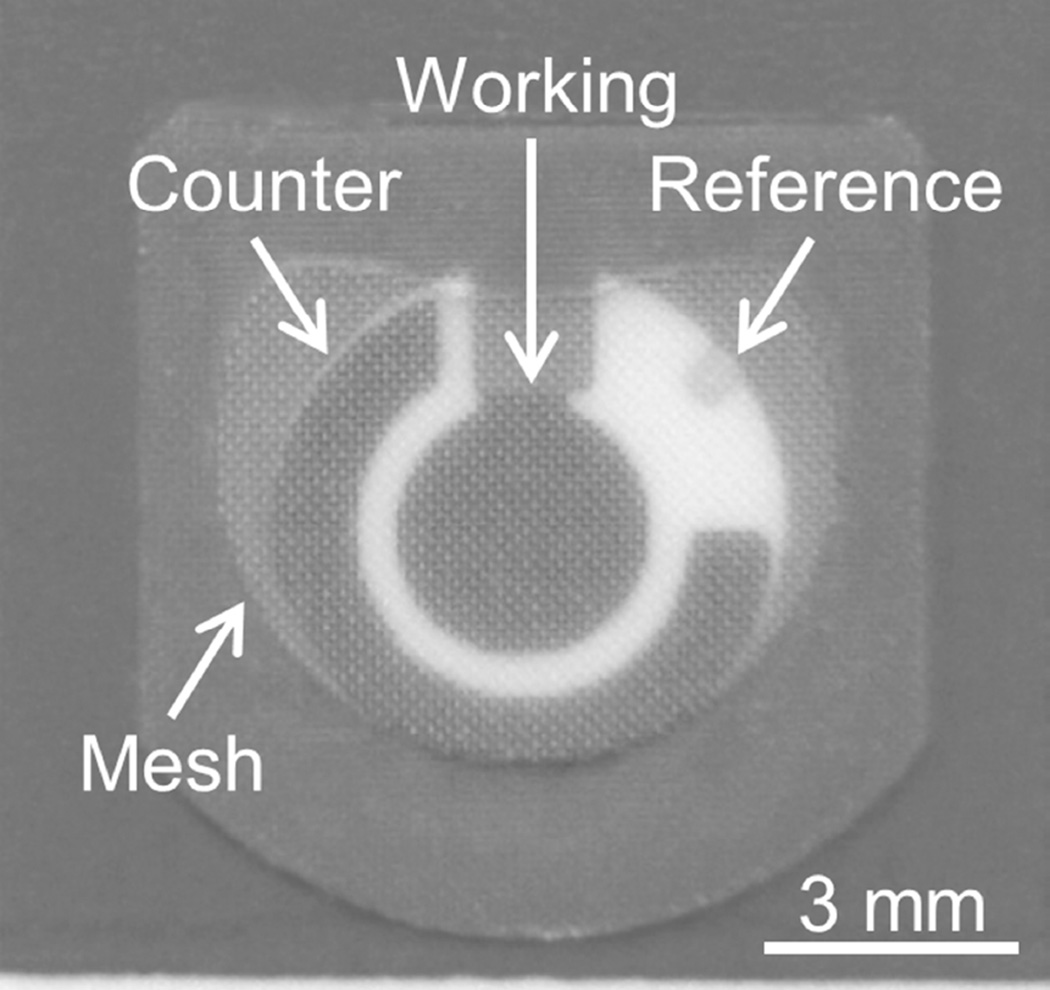
Disposable, screen-printed electrode sensors (TE100, Zensor, Taichung City, Taiwan) were used to detect the presence of pyocyanin in clinical samples (Figure 1). The sensors utilize a 3-electrode setup containing carbon-based working (3 mm diameter disk) and counter electrodes along with a Ag/AgCl reference electrode. All electrochemical measurements were recorded using a portable potentiostat (µStat 200, Dropsens, Parque Tecnológico de Asturias, Spain). The sensing surface was covered with polymeric membrane (DRP-MEMB, Dropsens, Parque Tecnológico de Asturias, Spain) to reduce the amount of sample volume required for analysis.
Figure 1.
Disposable, screen-printed electrode sensor with mesh modification for small-volume analysis.
For each test, 7.5 µL of wound exudate was pipetted into the detector well. 7.5 µL was determined to be the minimum fluid volume required for the sensor to successfully perform a measurement. The minimum sample volume was ascertained by placing increasing amounts of wound exudate collected from a patient that was diagnosed with a Pseudomonas infection on a sensor and running the instrument, starting from 5 µL. When less than 7.5 µL is used for analysis, there is insufficient fluid to electrically connect all three electrodes on the sensor. A lack of connection between the three electrodes during measurement results in voltammograms that have numerous, randomly distributed, sharp current spikes. For all of the experiments, square-wave voltammetric scans were performed at potentials ranging from −0.7 to 0.0 V at an amplitude voltage of 0.05 V, step voltage of 0.004 V, and a frequency of 15 Hz (Figures 2, S1–S5).
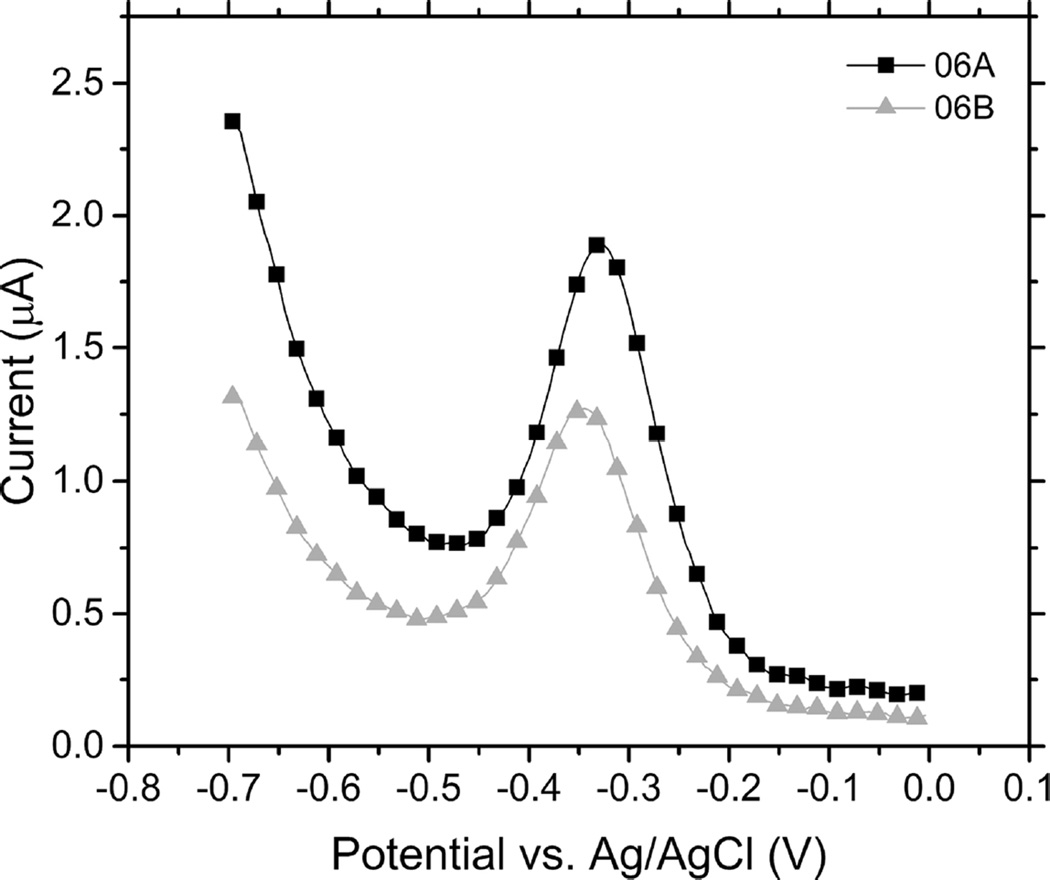
Figure 2.
Square-wave voltammograms of wound fluid exudate. Pyocyanin peak indicates the presence of Pseudomonas aeruginosa in the sample.
Each clinical sample was tested in duplicate with a new sensor used each time. The investigators (VS, AB, SM) were blinded to the microbiome 16S rRNA results at the time of the sensor detection experiment. The data was analyzed by two independent investigators (HS and EG) using OriginPro 9.1 (OriginLab Corporation, Northampton, MA). To quantify the amplitude of the peaks measured, a baseline was subtracted from the raw data to remove background signals observed in the measurements. Baselines were created for each data set using spline interpolation with 32 base points. The resulting baseline-subtracted data set was used to identify peaks in the current and to determine the maximum currents of those peaks (Figure S2). From these maximum current values, using a cutoff of 0.030 µA for the average of the two measurements, a binary determination was made for whether the probe was detecting pyocyanin (positive) or not (negative) (Table 2).
Table 2.
Experimental determinations (Positive/Negative) for whether clinical samples contained Pseudomonas aeruginosa based on peak currents obtained from electrochemical square-wave voltammograms.
| Sample | Peak current (µA) |
Determination using 0.030 µA average threshold cutoff |
16S rRNA sequencing results |
16S rRNA sequencing results (% relative abundance) |
|---|---|---|---|---|
| 01A | 0.0000 | Negative | Negative | 0 |
| 01B | 0.0000 | |||
| 02A | 0.1792 | Positive | Negative | 0 |
| 02B | 0.0000 | |||
| 03A | 0.0000 | Positive | Positive | 0.0479 |
| 03B | 0.0743 | |||
| 03C | 0.0094 | |||
| 04A | 0.1241 | Positive | Positive | 0.0558 |
| 04B | 0.0517 | |||
| 05A | 0.2263 | Positive | Positive | 0.0005 |
| 05B | 0.2309 | |||
| 06A | 1.3491 | Positive | Positive | 0.0027 |
| 06B | 0.9303 | |||
| 07A | 0.0195 | Negative | Positive | 0.0005 |
| 07B | 0.0163 | |||
| 08A | 0.0000 | Negative | Positive | 0.9779 |
| 08B | 0.0000 | |||
| 09A | 0.0090 | Negative | Negative | 0 |
| 09B | 0.0280 | |||
| 10A | 0.0000 | Negative | Negative | 0 |
| 10B | 0.0201 | |||
| 11A | 0.0757 | Positive | Negative | 0 |
| 12A | 0.0469 | Positive | Negative | 0 |
| 12B | 0.0539 | |||
| 13A | 0.0000 | Negative | Negative | 0 |
| 13B | 0.0000 | |||
| 14A | 0.6262 | Positive | Positive | 0.2478 |
STATISTICAL ANALYSIS
Data was analyzed using GraphPad Prism 5.03 (for Windows, GraphPad Software, San Diego California, USA). Fisher’s exact test and Chi-squared tests were used for categorical variables and Student’s t-test was used for continuous variables. Results are represented as mean ± SD. A p-value less than 0.05 indicate statistical significance; all significance tests were performed and interpreted in a two-sided manner.
Results obtained from the microbiome profile generated by 16S ribosomal RNA sequencing were reviewed and samples with any positive Pseudomonas reads were considered to test positive for Pseudomonas. These results were compared to the results from the pyocyanin detector. The sensitivity and specificity of the sensor were calculated.
RESULTS
Patient demographics
Paired wound effluent and biofilm samples were analyzed from 14 unique samples obtained from 12 patients (2 patients with serial samples collected at different time points were available). The mean age of patients was 50.18 years. Of the 14 samples subjected to microbiome profiling by 16S rRNA sequencing, 7 had detectable Pseudomonas spp. (sequencing positive). All 14 wounds were recalcitrant at the time of specimen collection.
There were no significant differences in age, sex, race, or comorbidities in the patients whose samples were positive for P. aeruginosa using 16S rRNA sequencing compared to those that were negative (Tables 1–2). Wounds that were positive for P. aeruginosa using 16S rRNA sequencing tended to be larger but this did not reach statistical significance.
Pyocyanin detector results
A positive test on the pyocyanin detector was considered to be an oxidation peak around −0.25 V vs. a Ag/AgCl reference electrode with a cutoff of 0.030 µA (15, 19). Data was analyzed as the mean of duplicates. Of the 14 samples, 8 tested positive using the pyocyanin detector cutoff of 0.030 µA.
Sensitivity and specificity of probe for detecting Pseudomonas spp
Sensitivity and specificity of the pyocyanin probe for detecting the samples that contained Pseudomonas spp. based on microbiome 16S sequencing are reported in Table 3. The probe tested positive in 5 out of 7 samples that were positive for Pseudomonas on 16S rRNA sequencing and was negative on 4 out of 7 samples with negative 16S rRNA results, giving a sensitivity of 71% (95% CI: 0.29–0.96) and specificity of 57% (95% CI: 0.18–0.90).
Table 3.
Sensitivity and specificity of pyocyanin probe compared to 16S rRNA sequencing for Pseudomonas spp. Data analyzed using Fisher’s exact test.
| 16S rRNA positive |
16S rRNA negative |
|
|---|---|---|
|
Pyocyanin sensor positive |
5 | 3 |
|
Pyocyanin sensor negative |
2 | 4 |
| Sensitivity (95% CI) |
Specificity (95% CI) |
Positive predictive value (95% CI) |
Negative predictive value (95% CI) |
|---|---|---|---|
| 0.71 (0.29–0.96) |
0.57 (0.18–0.90) |
0.63 (0.24–0.91) |
0.67 (0.22–0.96) |
DISCUSSION
The pyocyanin probe was simple to use and had high inter-observer agreement regarding interpretation of a positive result. When compared with a diagnostic gold-standard of 16S rRNA sequencing, the pyocyanin probe had a sensitivity of 71% and specificity of 57% indicating that it may be useful as a point-of-care test in screening for presence of Pseudomonas in human wound fluid.
One of the concerns raised about the utility of electrochemical probes for testing human samples is that there may be other molecules which may interfere with probe performance. Human wound samples often contain polymicrobial flora and this was indeed the case for the specimens reported here. The results reported showed no other redox peaks in the reference window for the pyocyanin probe. This indicates that despite the polymicrobial nature of human wound specimens, there do not appear to be other redox-active molecules that would impede the probe performance in a clinical setting. Previous work has shown that other bacteria such as Escherichia coli and Staphylococcus aureus also do not produce redox peaks in the reference window for pyocyanin (16). We did observe small potential shifts in the location of the pyocyanin peak in some samples. This may be attributed to differences in the salt and pH concentration of the sample media, and the limited stability of the Ag/AgCl quasi-reference electrode of the disposable sensor (16, 19).
We found that most of the samples containing P. aeruginosa tested pyocyanin positive using our electrochemical approach. By lowering the threshold for current peak identification, we could have improved the sensor’s sensitivity to 85.7% while decreasing specificity to 42.9%. Although still unclear if it is necessary, the detection limit of electrochemical sensors can be improved by switching to micro and nanofabricated electrodes, albeit with increased sensor cost (31). Nevertheless, future technological advances will lead to reduced costs and more sensitive electrochemical sensors, making this approach a practical option.
Our testing revealed some false negative results using the pyocyanin probe. While pyocyanin is a very specific molecule, produced only by Pseudomonas spp., the genes for producing the molecule are unique to P. aeruginosa (14). Furthermore, there exist environmental conditions in which pyocyanin production by P. aeruginosa is low. It is possible that the wound microenvironment may have impacted pyocyanin production in some of the clinical cases studied here, and thus merits further investigation using specimens from a larger cohort of patients with chronic wounds.
This study has several other limitations which merit discussion. The sample size is small because this was a pilot study designed to be hypothesis generating. Wound exudate is relatively challenging to collect at the bedside, and so as this probe is developed as a point-of-care testing device, further refinement will be necessary to enhance probe performance at low fluid volumes and improve clinical utility. Finally, while the 16S rRNA testing is a good gold standard test for determination of bacterial presence in the wound and relative abundance of specific bacteria relative to the entire microbiome profile of the specimen, 16S rRNA testing alone does not give information regarding the quantitative abundance of a particular bacterium in one specimen relative to other specimens. It is possible that some of the false positives and false negatives in this study are attributable to very low Pseudomonas abundance in the sample and further testing is needed to understand the clinical relevance of false positive and false negative tests as this device is further refined and developed with a view to clinical indications. In future studies, we intend to assess quantitative PCR with a universal 16S probe to extrapolate the count number to an estimation of bacterial abundance. Will plan to confirm the presence of pyocyanin, when larger sample volumes are available, using liquid chromatography with mass spectrometry (LC-MS).
CONCLUSIONS
Despite the availability of sophisticated, automated instrumentation, today’s standard clinical practices for bacterial identification still rely on plate culturing or more recently molecular diagnostics, but both require several hours before an identification can be made. Electrochemical sensors offer a simple and inexpensive alternative for immediate identification of bacteria presence based on the detection of bacterial quorum sensing molecules. Here, we demonstrate that an inexpensive, disposable electrochemical sensor can be used to rapidly screen for the presence of Pseudomonas aeruginosa in clinical wound effluent samples. The validation of this detector for use in the clinical setting suggests that this technology has potential for development as a rapid, point-of-care diagnostic for P. aeruginosa, allowing for better antimicrobial stewardship and improved patient care outcomes.
Supplementary Material
Acknowledgments
This work was supported by award R01NR013888 from the National Institute of Nursing Research and by award number UL1 TR000075 from the National Center for Advancing Translational Sciences (NCATS) and the National Institutes of Health through the Clinical and Translational Science Awards Program (CTSA). Funding was also provided by award 1542812 from the National Science Foundation. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Center for Advancing Translational Sciences, the National Institutes of Health, or the National Science Foundation. Dr. Goluch has a financial interest in QSM Diagnostics, Inc., which commercializes infection diagnostic technology.
REFERENCES
- 1.Cherkaoui A, Hibbs J, Emonet S, Tangomo M, Girard M, Francois P, et al. Comparison of two matrix-assisted laser desorption ionization-time of flight mass spectrometry methods with conventional phenotypic identification for routine identification of bacteria to the species level. J Clin Microbiol. 2010;48(4):1169–1175. doi: 10.1128/JCM.01881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holland RD, Wilkes JG, Rafii F, Sutherland JB, Persons CC, Voorhees KJ, et al. Rapid identification of intact whole bacteria based on spectral patterns using matrix-assisted laser desorption/ionization with time-of-flight mass spectrometry. Rapid Commun Mass Sp. 1996;10(10):1227–1232. doi: 10.1002/(SICI)1097-0231(19960731)10:10<1227::AID-RCM659>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 3.Meng J, Zhao S, Doyle MP, Mitchell SE, Kresovich S. Polymerase chain reaction for detecting Escherichia coli O157: H7. Int J Food Microbiol. 1996;32(1–2):103–113. doi: 10.1016/0168-1605(96)01110-5. [DOI] [PubMed] [Google Scholar]
- 4.Wolcott RD, Hanson JD, Rees EJ, Koenig LD, Phillips CD, Wolcott RA, et al. Analysis of the chronic wound microbiota of 2,963 patients by 16S rDNA pyrosequencing. Wound Repair Regen. 2015 doi: 10.1111/wrr.12370. [DOI] [PubMed] [Google Scholar]
- 5.Hogsberg T, Bjarnsholt T, Thomsen JS, Kirketerp-Moller K. Success rate of split-thickness skin grafting of chronic venous leg ulcers depends on the presence of Pseudomonas aeruginosa: a retrospective study. PLoS One. 2011;6(5):e20492. doi: 10.1371/journal.pone.0020492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rogers GB, Hoffman LR, Carroll MP, Bruce KD. Interpreting infective microbiota: the importance of an ecological perspective. Trends Microbiol. 2013;21(6):271–276. doi: 10.1016/j.tim.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trenholme GM, Kaplan RL, Karakusis PH, Stine T, Fuhrer J, Landau W, et al. Clinical impact of rapid identification and susceptibility testing of bacterial blood culture isolates. J Clin Microbiol. 1989;27(6):1342–1345. doi: 10.1128/jcm.27.6.1342-1345.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15(2):194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bodey GP, Bolivar R, Fainstein V, Jadeja L. Infections caused by Pseudomonas aeruginosa. Rev Infect Dis. 1983;5(2):279–313. doi: 10.1093/clinids/5.2.279. [DOI] [PubMed] [Google Scholar]
- 10.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 11.Reyes EA, Bale MJ, Cannon WH, Matsen JM. Identification of Pseudomonas aeruginosa by pyocyanin production on Tech agar. J Clin Microbiol. 1981;13(3):456–458. doi: 10.1128/jcm.13.3.456-458.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lau GW, Hassett DJ, Ran H, Kong F. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10(12):599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Whitehead NA, Barnard AML, Slater H, Simpson NJL, Salmond GPC. Quorum-sensing in Gram-negative bacteria. 2001 doi: 10.1111/j.1574-6976.2001.tb00583.x. [DOI] [PubMed] [Google Scholar]
- 14.Mavrodi DV, Bonsall RF, Delaney SM, Soule MJ, Phillips G, Thomashow LS. Functional analysis of genes for biosynthesis of pyocyanin and phenazine-1-carboxamide from Pseudomonas aeruginosa PAO1. J Bacteriol. 2001;183(21):6454–6465. doi: 10.1128/JB.183.21.6454-6465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bellin DL, Sakhtah H, Rosenstein JK, Levine PM, Thimot J, Emmett K, et al. Integrated circuit-based electrochemical sensor for spatially resolved detection of redox-active metabolites in biofilms. Nat Commun. 2014;5:3256. doi: 10.1038/ncomms4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webster TA, Sismaet HJ, Conte JL, Chan IP, Goluch ED. Electrochemical detection of Pseudomonas aeruginosa in human fluid samples via pyocyanin. Biosens Bioelectron. 2014;60:265–270. doi: 10.1016/j.bios.2014.04.028. [DOI] [PubMed] [Google Scholar]
- 17.Webster TA, Sismaet HJ, Chan IP, Goluch ED. Electrochemically monitoring the antibiotic susceptibility of Pseudomonas aeruginosa biofilms. Analyst. 2015;140(21):7195–7201. doi: 10.1039/c5an01358e. [DOI] [PubMed] [Google Scholar]
- 18.Sharp D, Gladstone P, Smith RB, Forsythe S, Davis J. Approaching intelligent infection diagnostics: Carbon fibre sensor for electrochemical pyocyanin detection. Bioelectrochemistry. 2010;77(2):114–119. doi: 10.1016/j.bioelechem.2009.07.008. [DOI] [PubMed] [Google Scholar]
- 19.Sismaet HJ, Webster TA, Goluch ED. Up-regulating pyocyanin production by amino acid addition for early electrochemical identification of Pseudomonas aeruginosa. Analyst. 2014;139(17):4241–4246. doi: 10.1039/c4an00756e. [DOI] [PubMed] [Google Scholar]
- 20.Farrow MJ, Hunter IS, Connolly P. Developing a real time sensing system to monitor bacteria in wound dressings. Biosensors (Basel) 2012;2(2):171–188. doi: 10.3390/bios2020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward AC, Connolly P, Tucker NP. Pseudomonas aeruginosa can be detected in a polymicrobial competition model using impedance spectroscopy with a novel biosensor. PLoS One. 2014;9(3):e91732. doi: 10.1371/journal.pone.0091732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ivnitski D, Abdel-Hamid I, Atanasov P, Wilkins E. Biosensors for detection of pathogenic bacteria. Biosens Bioelectron. 1999;14(7):599–624. doi: 10.1016/s0956-5663(99)00004-4. [DOI] [PubMed] [Google Scholar]
- 23.Lazcka O, Del Campo FJ, Munoz FX. Pathogen detection: a perspective of traditional methods and biosensors. Biosens Bioelectron. 2007;22(7):1205–1217. doi: 10.1016/j.bios.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 24.Boucher HW, Talbot GH, Bradley JS, Edwards JE, Gilbert D, Rice LB, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis. 2009;48(1):1–12. doi: 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- 25.Levine NS, Lindberg RB, Mason AD, Jr, Pruitt BA., Jr The quantitative swab culture and smear: A quick, simple method for determining the number of viable aerobic bacteria on open wounds. J Trauma. 1976;16(2):89–94. [PubMed] [Google Scholar]
- 26.Angel DE, Lloyd P, Carville K, Santamaria N. The clinical efficacy of two semi-quantitative wound-swabbing techniques in identifying the causative organism(s) in infected cutaneous wounds. Int Wound J. 2011;8(2):176–185. doi: 10.1111/j.1742-481X.2010.00765.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jeraldo P, Chia N, Goldenfeld N. On the suitability of short reads of 16S rRNA for phylogeny-based analyses in environmental surveys. Environ Microbiol. 2011;13(11):3000–3009. doi: 10.1111/j.1462-2920.2011.02577.x. [DOI] [PubMed] [Google Scholar]
- 28.Fouts DE, Szpakowski S, Purushe J, Torralba M, Waterman RC, MacNeil MD, et al. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS One. 2012;7(11):e48289. doi: 10.1371/journal.pone.0048289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75(23):7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, et al. The Ribosomal Database Project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37(Database issue):D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zevenbergen MA, Singh PS, Goluch ED, Wolfrum BL, Lemay SG. Stochastic sensing of single molecules in a nanofluidic electrochemical device. Nano Lett. 2011;11(7):2881–2886. doi: 10.1021/nl2013423. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.