Abstract
Neglected tropical diseases (NTDs) and other diseases of the developing world, such as malaria, attract research investments that are disproportionately low compared to their impact on human health worldwide. Therefore, pragmatic methods for launching new drug discovery programs have emerged that repurpose existing chemical matter as new drugs or new starting points for optimization. In this Digest we describe applications of different repurposing approaches for NTDs, and provide a means by which these approaches may be differentiated from each other. These include drug repurposing, target repurposing, target class repurposing, and lead repurposing.
Graphical abstract
I. Introduction
Neglected tropical diseases (NTDs) are defined by the World Health Organization (WHO) as “a diverse group of communicable diseases that prevail in tropical and subtropical conditions;” the official WHO list of NTDs is currently comprised of 17 infectious diseases.1 Alongside malaria, these diseases predominantly affect populations living in poverty, under poor living conditions and in close proximity with the vectors of disease-causing agents. Their effects are far-reaching and devastating: over 1 billion people in 149 countries suffer from one or more NTDs with millions of others at risk, and the economic repercussions of these diseases can be just as damaging as their health effects.1 These diseases are “neglected” primarily because there is no financial incentive to develop drugs for a patient population that cannot afford them. Consequently, noting that most drugs are developed by for-profit companies, there is little reason for these companies to invest in research and development for drugs that will not result in high financial returns.
Therefore, much of the drug discovery and hit-to-lead optimization for these diseases is performed in academic laboratories without the financial, personnel, and technical resources of a pharmaceutical company. With a view toward overcoming these limitations, a popular strategy for academic groups has been to “re-purpose” or reuse existing chemical matter, target knowledge, and other data from human or animal drug discovery campaigns in order to cut down on the time and cost of advancing a program from hit to lead to clinical candidate. Indeed, a number of the drugs currently in use for treating NTDs originated from low-throughput screens or repurposing of either human or veterinary drugs (Table 1). A recent review2 on approaches to drug discovery for malaria, HAT, and schistosomiasis highlights drug repurposing, drug repositioning, and drug rescue as strategies employed by NTD researchers; other terms employed in the field include “target repurposing”3 and “piggyback drug discovery.”4 Repurposing as a general strategy is therefore a well-established approach in the NTD drug discovery community. By systematizing the nomenclature for the many flavors of repurposing, we aim to enable the community to readily identify at the outset what type of information was available at the start of the campaign and the extent of optimization involved. A clearly defined, common vocabulary for these strategies will ease communication and collaboration in the field of NTD drug discovery.
Table 1.
Selected drugs currently employed in the treatment of NTDs discussed in this review, according to WHO.
| Disease | Drug | Discovery | Limitations |
|---|---|---|---|
| Malaria | Artemisinin | Natural product, found to have anti-malarial activity in the 1980s5 |
Emerging resistance6 |
| Human African Trypanosomiasis |
Suramin | Dye derivative, found to be anti-trypanosomal in the 1920s7 |
Treats Stage 1 only8 |
| Pentamidine | Part of a class of compounds discovered to treat equine trypanosomiasis in the 1940s7 |
Treats Stage 1 only8 | |
| Melarsoprol | Arsenicals discovered to cure infected laboratory animal models of trypanosomiasis; melarsoprol introduced in 19497 |
Fatal to 3–10% of patients; emerging resistance8 |
|
| Eflornithine | Originally an anti-cancer drug, in use for HAT since 19907 |
Complex treatment regimen8 |
|
| Chagas disease | Benznidazole | Empirical screening, introduced in 19719 |
Rare but severe side effects, emerging resistance9 |
| Nifurtimox | Empirical screening, introduced in 19659 |
GI and CNS side effects | |
| Lymphatic filariasis |
Albendazole | Originally developed to treat gut helminths in livestock, approved for use in humans in the 1980s10 |
Must be used in combination with either ivermectin or diethylcarbamazine citrate10 |
| Ivermectin | Found to be effective against canine hookworms and other nematodes, registered 198711 |
Long treatment regimen, unsuitable for use in areas co-endemic with Loa loa, emerging resistance12 |
|
| River blindness | Ivermectin | Discovered to be effective against Onchocerca in horses11 |
See “Ivermectin” above |
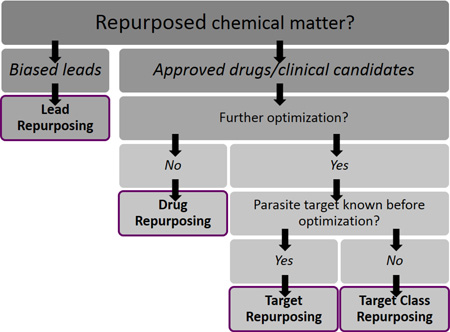
The many terms for repurposing strategies can be grouped into four major categories, which are a) drug repurposing, b) target repurposing, c) target class repurposing, and d) lead repurposing. Each has distinct advantages and disadvantages and may be appropriate in different project situations. These four approaches are characterized by the type of chemical matter that is being repurposed, the kind of information that is typically available at the starting point of a campaign, and the type of optimization that is required. For each strategy, the characteristics, advantages, and disadvantages are discussed, and case studies are provided to illustrate each one in practice.
II. Drug Repurposing
Drug repurposing is characterized chiefly by the lack of further optimization required for the repurposed chemical matter. In this approach, FDA-approved chemical entities for an initial indication are used for a second indication without any further structural modification of the compound at hand (although dosing and formulation modifications may be required). Drug repurposing is an established strategy used not only for neglected diseases, but other diseases as well, and is also referred to as drug repositioning, drug redirecting, and drug reprofiling.13 In for-profit settings, repurposing avoids the risk associated with the costs of drug discovery and development, up to (and often including) Phase I clinical trials. However, it offers great advantages for neglected diseases as well. Approved chemical matter has already been profiled in terms of safety and pharmacokinetics, giving an indication of tolerated human doses and any likely side effects. As a result, both the time and cost of drug development are drastically reduced using this approach. We describe below three examples of drug repurposing in various stages of progression along the drug discovery pipeline.
Case study 1: Eflornithine as a successfully repurposed drug for sleeping sickness
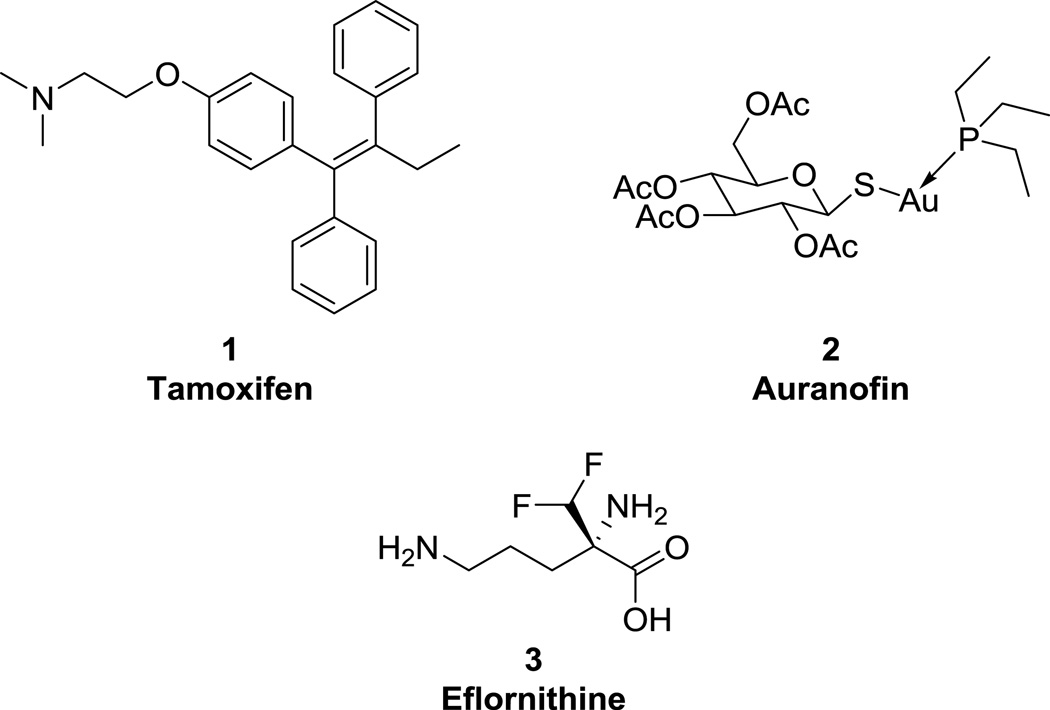
Human African trypanosomiasis (HAT), also known as sleeping sickness, is caused by two subspecies of the parasite Trypanosoma brucei (T.b. gambiense and T.b. rhodesiense). Prevalent in 36 countries in sub-Saharan Africa, HAT progresses from the lymph to the central nervous system, causing disruptions in sleeping patterns and death if left untreated.8 Originally developed as a cancer therapeutic (and now utilized as topical agent for hirsutism), eflornithine (3, Fig. 1), also known as difluoromethylornithine or DFMO, is an inhibitor of polyamine biosynthesis that was shown by Bacchi et al to inhibit the growth of trypanosomes by the same mechanism.14 In addition, eflornithine cured T.b. brucei infections in mice when given as a 1 or 2 percent solution in drinking water (defined as survival of greater than 30 days beyond that of untreated controls); the drug was also shown to be generally nontoxic.14
Figure 1.
Drugs that have been directly repurposed for NTDs.
Although in use as a single agent for many years, eflornithine requires large doses to be effective, has a complex mode of administration and high cost per patient, and is ineffective against T.b. rhodesiense.15 Recently, however, some of these drawbacks have been mitigated through the use of nifurtimox-eflornithine combination therapy (NECT). By combining eflornithine with nifurtimox (a Chagas disease therapeutic), the dose of eflornithine required, the complexity of administration, and the cost of the treatment are reduced.16 Importantly, eflornithine is effective against stage 2 HAT wherein the parasite crosses the blood-brain barrier, and NECT has become the most promising front-line treatment for second-stage T.b. gambiense infections.16
Case study 2: Tamoxifen as an anti-leishmanial treatment
310 million people are at risk of infection by Leishmania spp., which cause leishmaniasis in several forms, including the deadly visceral leishmaniasis (VL).17 The anti-leishmanial activity of tamoxifen (1, Fig. 1), an approved breast cancer drug used in the treatment of estrogen receptor-positive tumors, was first reported in 2007.18 Starting with the observation made by previous groups that tamoxifen was able to induce alkalinization of organelles in several cell lines,19 it was hypothesized that Leishmania parasites, which live in acidic vacuoles within the host cell and require low pH to survive, would be susceptible to tamoxifen via this mechanism. The drug was tested against the promastigote form of five species of Leishmania (including L. amazonensis), and against the intracellular amastigote of L. amazonensis, and was shown to have a cidal effect on all species with micromolar EC50 values. The mechanism of action was also investigated and was shown to be independent of host estrogen receptor modulation.18
In further studies, tamoxifen was evaluated in a mouse model of leishmaniasis.20 In a 15-day treatment of L. amazonensis-infected mice, tamoxifen reduced the parasite burden by 99% compared to untreated mice, outperforming meglumine antimonate, the standard treatment. Although a cure was not achieved (typical for leishmaniasis treatments), no toxicity was observed during or after treatment and symptoms were greatly alleviated. Tamoxifen was also shown to be effective in mouse and hamster models of L. braziliensis and L. chagasi, respectively, with 95–98% reduction in parasite load and 100% survival of treated animals 18 weeks post-infection.21
Case study 3: Repurposing auranofin for lymphatic filariasis and river blindness
Lymphatic filariasis (LF) and river blindness are NTDs caused by filariid nematodes that affect an estimated 145 million people and cause debilitating swelling and blindness, respectively.22, 23 The current standard of care for river blindness, caused by Onchocerca volvulus, includes ivermectin (itself an example of direct drug repurposing!), although treatment with this drug causes severe adverse reactions in patients that are co-infected with the parasite Loa loa; in addition, there is cause for concern about ivermectin resistance.12 Use of doxycycline to target symbiotic Wolbachia bacteria, which L. loa lacks, has been investigated as a complement or replacement to ivermectin, but is still limited for use in children and pregnant women.24 Simultaneously, in a 2015 library screen of over 2,000 FDA-approved compounds, the rheumatoid arthritis drug auranofin (2, Fig. 1) was found to directly kill adult Brugia spp. and Onchocerca ochengi, models for the causative agents of LF (Wuchereria bancrofti, Brugia malayi, and B. timori) and river blindness, respectively.25 Additionally, the drug was found to be ~43× selective over L. loa, which has promising implications for treatment in areas where co-infection is present. Auranofin was also tested in vivo in gerbil infection models of LF infection using B. pahangi and was found to reduce the worm burden by 58% and 91% in two different studies.25 Although auranofin is known to be metabolized fairly rapidly in vivo, gold plasma levels well above the in vitro worm IC50s were maintained in gerbils for two hours post-dose after two weeks of treatment. Finally, a series of experiments was performed to provide evidence that the parasitic target of auranofin is thioredoxin reductase.
In all of the above examples, FDA-approved compounds were directly repurposed as anti-parasitic agents without the need for further optimization, though the mechanism of action was not necessarily the same for the original indication and the new, anti-parasitic indication. Importantly in these cases, from a safety perspective, the original indications called for a long-term treatment, with tamoxifen being used for at least 5 years for cancer chemotherapy and auranofin being used for six months on average to treat rheumatoid arthritis.18, 25 This suggests that a short term NTD treatment would be within an acceptable therapeutic window for these agents. We note that the most common side effects of the repurposed drugs are minor in comparison to those of many existing NTD therapeutics. Although neither auranofin nor tamoxifen has yet to be approved for its respective NTD application, the demonstrated efficacy of these compounds in rodent models is a promising indication of their potential utility as anti-infective agents, and their FDA-approved status means that both the discovery and approval timelines for NTD indications would be significantly shortened.
III. Target Repurposing
Perhaps the most broadly used term of the four discussed in this paper, “target repurposing” has come to have slightly different meanings covering a range of repurposing strategies. However, true target repurposing projects begin with a defined parasitic target with a direct, established homolog in another species (human or otherwise). The chemical matter that targets the host protein is often an approved drug or clinical candidate, which is then used as a starting point to develop compounds that inhibit the parasitic target. In contrast to direct drug repurposing, target repurposing campaigns require medicinal chemistry optimization after the initial lead compound is identified, with the goals of improving selectivity for the parasite homolog as well as achieving disease-modifying efficacy for the given NTD.
Target repurposing offers several benefits over other strategies. The major advantage is that the parasitic target of the campaign is known, enabling structure-based drug design either through homology modeling or X-ray crystallography of the parasitic target, and simplifying potential mechanism-of-action studies. Although selectivity can be a challenge using this approach if the lead compounds have been optimized to act on the human target rather than the parasitic one, the wealth of information about the drug-target interactions that is typically available can help mitigate this concern. In addition, as with drug repurposing, the lead compounds typically have known toxicity, absorption-distribution-metabolism-excretion (ADME), and pharmacokinetic (PK) profiles. These advantages make target repurposing a powerful strategy for NTD drug discovery. On the other hand, target repurposing is limited by the required presence of parasite homologs; for example, kinetoplastid parasites such as T. brucei do not express G-protein coupled receptors, which represents a significant family of therapeutic targets in humans, precluding repurposing of inhibitors of this gene family. Another key limitation is the likelihood that an optimized compound repurposed from a human ortholog is unlikely to have high activity against the parasite homolog; this limitation can be difficult to overcome, and sometimes requires wholesale redevelopment of structure-activity relationships.
Case study 1: LeuRS and human African trypanosomiasis
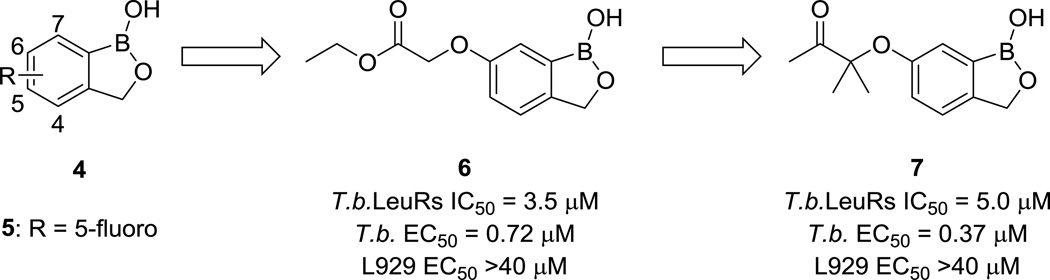
In a 2011 example of target repurposing, Ding and colleagues re-optimized benzoxaboroles (4, Fig. 2) into several sub-micromolar trypanosomal growth inhibitors that also had selectivity over L929 mouse lung fibroblast cells.26 Compound 5 (Fig. 2), in clinical trials as an anti-fungal agent, works by inhibiting the fungal LeuRS enzyme, which is responsible for translating the leucine RNA codons into the correct amino acid for protein synthesis.27 (It is worth noting that in this case, achieving selectivity over mammalian cells was not as challenging as it could have been, because the original target of 5 was a fungal enzyme.) The empty p-orbital of the boron atom accepts electrons from adenosine bases and forms an adduct with t-RNA, prohibiting further translation. Homology modeling guided the group to focus on C(6) as a site for modification, and they were able to improve the T. brucei LeuRS IC50 from 22.1 µM (H-substituted) to 1.6 µM with an ethyl ester at this position. Replacing the esters in the original analogs with ketones gave equipotent molecules with improved biological stability. These molecules were also potent in cellular assays, with T. brucei EC50 values as low as 0.37 µM, as shown in compound 7 (Fig. 2). The authors attribute the improved cellular potency to the fact that the benzoxaborole LeuRS inhibitors are covalent. Initial assays against mammalian cells indicate promising selectivity, pending further toxicity studies.
Figure 2.
Benzoxaborole compound progression. Efficacy against mouse lung fibroblast L929 cells was used as a measure of host cell toxicity.
Case study 2: Repurposing NMT inhibitors to develop anti-malarials
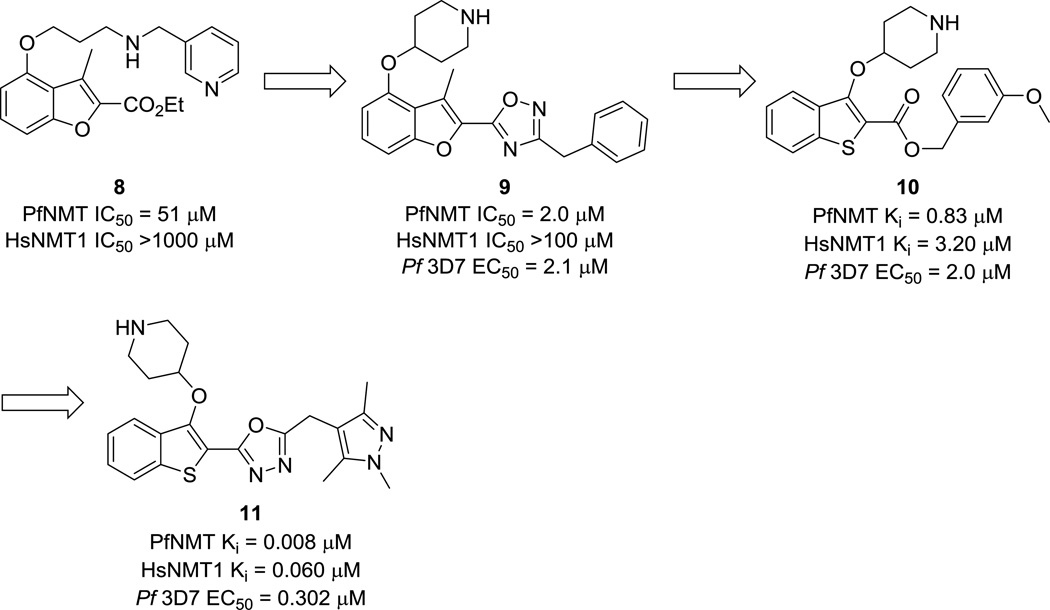
In another target repurposing program, the Leatherbarrow group repurposed a Roche anti-fungal N-myristoyltransferase (NMT) inhibitor as an anti-malarial scaffold. Caused by the parasite Plasmodium spp., malaria is the worldwide leading cause of parasitic morbidity and mortality.28 Although not one of the 17 official NTDs designated by the WHO, malaria is nonetheless primarily a disease of the developing world that is of comparatively little interest to for-profit drug discovery enterprises. There is evidence that NMT is an essential target in P. falciparum,29 and the enzyme has been shown to be essential in other NTD-causing pathogens as well.30 Compound 8 (Fig. 3) was shown to be moderately active against the parasitic target P. falciparium NMT (PfNMT) and had good selectivity against the human enzyme (HsNMT1). Medicinal chemistry optimization brought the campaign to lead compound 9 (Fig. 3), a single-digit-micromolar PfNMT inhibitor that displayed in vivo activity and was >100-fold selective over HsNMT1.29 SAR observations from this first campaign and the observed selectivity over HsNMT1 were later rationalized using a homology model of PfNMT built from P. vivax NMT (PvNMT).
Figure 3.
Structures of PfNMT inhibitors. Compounds were tested against the 3D7 strain of P. falciparum.
Further work by the group focused on producing more ligand-efficient lead compounds, which was accomplished using a scaffold-hopping strategy that led to 10 (Fig. 3).31 The m-methoxy substituent, while greatly beneficial for PvNMT potency, was sub-optimal against PfNMT. Finally, the metabolically-labile ester linkage was replaced with a more stable oxadiazole isostere, and the pendant methylpyrazole group was found to reduce lipophilicity and improve potency against PfNMT (compound 11, Fig. 3).32 This compound was also shown to be effective against drug-resistant P. falciparum strains and displays up to ~40-fold selectivity for the parasite over HepG2 cell lines.
Case study 3: Protein farnesyltransferase inhibitors to treat malaria
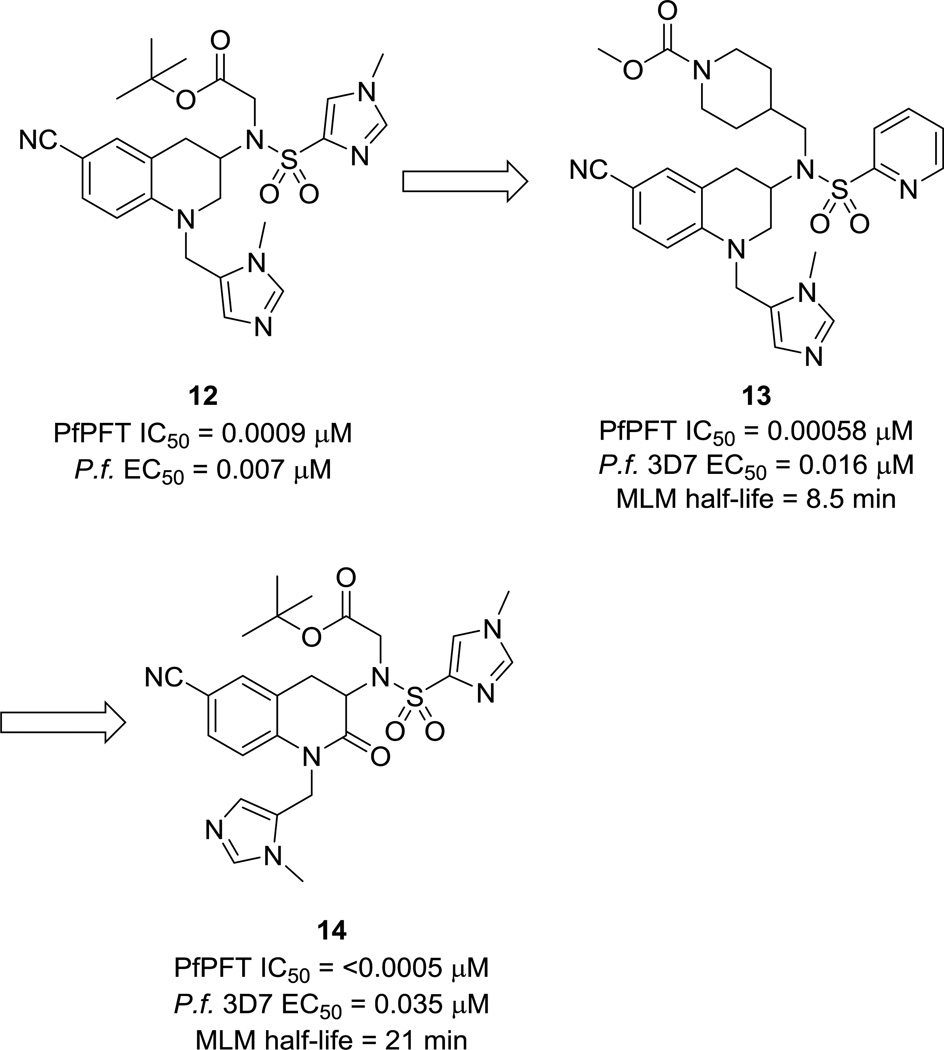
Protein farnesyl transferase (PFT) is an enzyme that transfers a farnesyl group as a post-translational modification onto specific proteins, including oncoproteins such as Ras GTPase.33 As such, PFT inhibitors have been well-developed for cancer therapeutics.34 P. falciparum lacks type I protein geranylgeranyltransferase, an enzyme found in mammalian cells that is similar in structure to PFT. P. falciparum does contain type II protein geranylgeranyltransferase, which acts on Rab GTPases. In mammalian cells treated with a PFT inhibitor (PFTI), proteins that are normally farnesylated can become geranylgeranylated by type I geranylgeranyltransferase. However, this rescue is not possible in P. falciparum and may account for the high toxicity of PFTIs to malarial cells.35 Based on this hypothesis, Nallan et al. tested the ability of a panel of PFTIs in preclinical and clinical development from several pharmaceutical companies to inhibit in vitro growth of P. falciparum parasites.35 Through this screen, a series of tetrahydroquinoline (THQ) PFTIs, typified by compound 12 (Fig. 4), was identified that displayed excellent potency in assays using the isolated PfPFT enzyme and whole-cell P. falciparum organisms. In addition, treatment with 12 eliminated parasitemia in 60% of mice with no observed toxicity in a mouse efficacy model of malaria.35
Figure 4.
P. falciparum PFT inhibitor progression.
Extensive medicinal chemistry optimization led to the identification of compound 13 (Fig. 4), which showed a good balance of potency and pharmacokinetic properties. In general, compounds with a 2-pyridyl substituent on the sulfonamide were less potent, but more orally bioavailable and had better Caco-2 permeability than compounds with the N-methyl-4-imidazole group.36 This optimization process was aided by the use of a homology model of the active site of PfPFT. Although most residues in the active site are conserved between Pf- and mammalian PFT, divergent SAR was observed with respect to compound efficacy (unpublished results).
Preclinical metabolism and pharmacokinetic studies were then conducted on a group of the most promising compounds from this campaign in order to further improve oral availability and clearance.37 Although rats treated with 13 showed significantly reduced parasitemia after just three days of treatment compared to controls in an efficacy study, injections were required every 8 hours in order for the treatment to be effective because of the rapid clearance of the compound. A metabolism study of 13 led to the identification of the N-dealkylated tetrahydroquinoline as the major metabolite. This led to the synthesis of 2-oxotetrahydroquinolines such as 14 (Fig. 4) in an effort to block metabolism via this pathway.38 Several of the compounds in this series showed significantly improved clearance over their matched THQ analogs. However, after a few years of work, PFTIs could not be obtained that had the proper collection of potency and adequate pharmacokinetic properties to warrant further development. Nevertheless, parasite PFTs remain a validated target for drug discovery.
The above case studies clearly demonstrate that it is possible to find parasitic enzyme inhibitors by repurposing chemical matter intended to hit human targets. In the absence of X-ray crystal structures of the parasitic targets, homology models can be a powerful tool for target repurposing campaigns and illustrate the advantages of both knowing the parasitic target and being able to compare it to a known homolog. Though a target repurposing project has yet to progress a compound to clinical candidacy, it is anticipated that the high quality lead compounds used in such campaigns should significantly shorten the time required to get to a quality candidate for the parasitic indication.
IV. Target Class Repurposing
Target class repurposing is distinct from target repurposing in that, while the specific parasitic target may not be known, the parasite is known either to express essential targets within a homologous target class, or to perform cellular functions that are homologous to those carried out by a certain target class. Therefore, most target class repurposing programs rely primarily on phenotypic assays. In the case of NTDs, the most obvious phenotype is parasite cell death or proliferation inhibition, though trademark aberrations in cell cycle can be used as a more refined tool. The ability to observe cellular activity is an advantage, in that this reflects both engagement with essential cellular processes, as well as cell penetration. However, a significant disadvantage of target class repurposing as compared to target repurposing is that the specific parasitic target is not directly proven, which can hinder more “rational design” approaches to optimization. Although the target(s) of action may be identified later, the bulk of target class repurposing campaigns are run without the benefit of structure-based drug design tools. On the other hand, the broad range of potential targets engaged in this approach may provide more opportunity to discover a novel mechanism of action, to find a target unique to the parasite, or to develop a compound that exhibits polypharmacology.
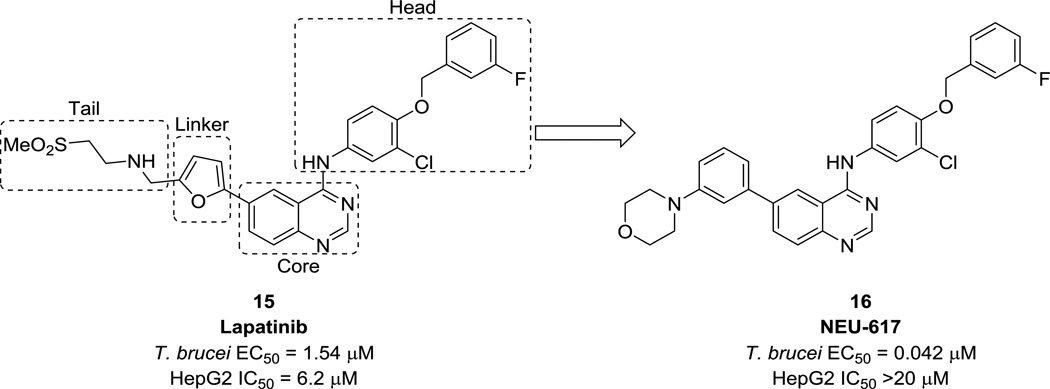
Case study 1: Tyrosine kinases in T. brucei
Patel et al. reported the optimization of lapatinib, a human EGFR tyrosine kinase inhibitor, as an anti-trypanosomal agent.39 Although T. brucei does not express receptor tyrosine kinases (RTKs), there is evidence of tyrosine phosphorylation in the parasite; inhibition of this process has a deleterious effect on parasite proliferation. Therefore, a set of nine human tyrosine kinase inhibitors were screened against T.b. brucei, resulting in the identification of lapatinib (15, Fig. 5) as a lead compound. Importantly, lapatinib also displayed oral efficacy in modifying the HAT disease state in mice.40 With some preliminary SAR around the tail region derived from the initial screening, optimization efforts were first focused in this area, and then on the linker and head regions of the molecule. These efforts resulted in NEU-617 (16, Fig. 5), a 42 nM inhibitor of T.b. brucei growth with >100-fold selectivity over HepG2 cells. This compound was also shown to be effective in reducing parasitemia in a mouse model of HAT, although issues related to toxicity and oral bioavailability were later discovered.
Figure 5.
Structures of lapatinib and NEU-617. Toxicity is indicated by inhibition of HepG2 human liver cells.
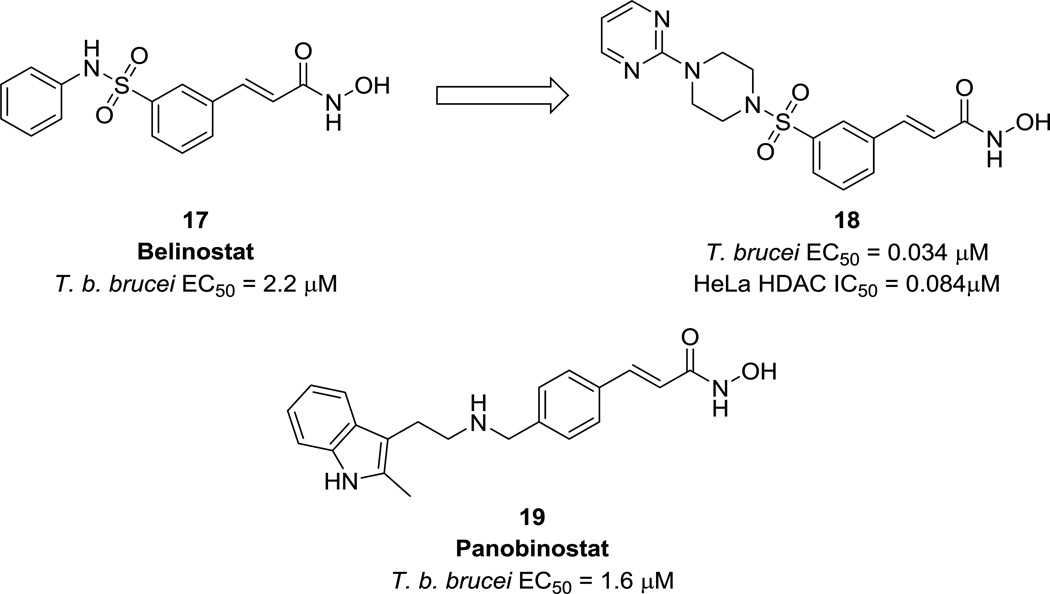
Case study 2. HDAC inhibitors for HAT
Histone deacetylases (HDACs) play a key role in gene regulation and expression in eukaryotic cells and are a major target class for cancer drug discovery.41 T. brucei is known to express four HDAC homologues, two of which are believed to be essential to parasite viability, that may be inhibited by repurposed human HDAC inhibitors.42, 43 In 2012, Kelly et al. reported an initial screening of representative HDAC inhibitors from a larger compound library, resulting in the discovery of belinostat (17, Fig. 6), a phase III clinical candidate for cancer, as a lead anti-trypanosomal compound.42 Other hydroxamic acid derivatives were synthesized, and analogs containing a sulfonepiperazine moiety, such as 18 (Fig. 6), were found to have sub-micromolar EC50s against trypanosomes. In addition, the authors noted little to no correlation between these compounds’ trypanocidal and human HDAC activities, indicating that mammalian HDAC inhibitor chemotypes can be optimized for anti-parasitic activity while avoiding acute host toxicity. This provides evidence that HDACs are amenable to a target class repurposing approach.
Figure 6.
Potency of HDAC inhibitors of against T. brucei.
In a second study, a set of HDAC inhibitors in clinical trials were assessed for trypanocidal activity against HAT (14 compounds total).43 Belinostat and panobinostat (19, Fig. 6) were identified as promising compounds with significant efficacy at physiologically relevant, tolerated doses. Even so, neither drug on its own was cidal to cultured parasites, nor did panobinostat exhibit any synergistic effects with current HAT drugs, indicating that they would not be suitable for direct drug repurposing. However, this work provides preliminary evidence for divergence of these compounds’ T. brucei and human HDAC activity driven by specific target interactions, rather than general metal-chelating properties. Provided that these compounds can be optimized to be cidal anti-T. brucei agents and the selectivity window can be widened, it should therefore be possible to develop a potent, selective anti-trypanosomal compound by repurposing HDAC inhibitors.
In sum, target class repurposing can be a highly productive approach to take in the absence of information about specific parasitic targets. Biological knowledge of parasitic enzymes and pathways involved in NTDs are limited in many ways due to the relatively low availability of resources devoted to basic molecular and cell biology research in these pathogens. However, these case studies demonstrate that it is possible to optimize both potency and selectivity even in the absence of biochemical assays or structural information about the compound target. As with target repurposing, compounds in advanced stages of development are used as starting points, and a target class repurposing program can still benefit from the advantages of repurposing these types of compounds to shorten the time required for the development of promising chemical entities for NTDs.
V. Lead Repurposing
In contrast to the three previously discussed approaches, a lead repurposing strategy does not seek to repurpose late-stage chemical matter (approved drugs or clinical candidates), but rather early-stage chemical matter. These campaigns typically begin with a high-throughput screen (HTS) of a class of targeted lead molecules, such as kinase- or protease-targeting inhibitors. However, lead repurposing has advantages over traditional (random) HTS, in that there is an abundance of information about the lead chemical matter that is not typically available in unbiased screens. Additionally, lead repurposing campaigns start with libraries of compounds specifically designed for drug-likeness and target family activity, providing better starting points as compared to unbiased library collections or natural products. Furthermore, by virtue of starting with a wider diversity of chemical matter, lead repurposing offers an advantage over target or target class repurposing in that it is more likely to yield a diverse variety of chemotypes to pursue for further development.
As with target class repurposing, phenotypic assays are typically used and the medicinal chemistry campaigns generated from the results of these screens are not usually enabled for structure-based drug design in the earliest stages of the program. Despite these challenges, lead repurposing campaigns have yielded some high-quality compounds and progressable chemotypes.
Case study 1: Human kinase inhibitors repurposed for T. brucei
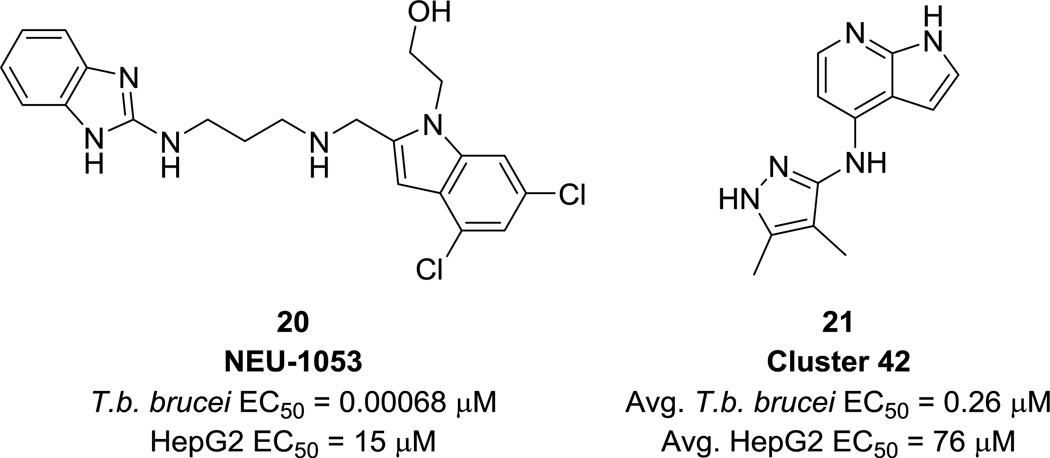
In 2014, we published the results of a kinase-focused HTS against T. brucei that yielded several promising chemotypes for development.44 Based on the knowledge that T. brucei expresses essential kinases and noting the success of several existing kinase inhibitor repurposing campaigns for HAT,39, 45–48 a set of 42,444 human kinase inhibitors, including the Published Kinase Inhibitor Set (PKIS),49 was tested against T. brucei. A subset of 797 compounds, grouped into 59 structural clusters, were found to have EC50 values <1 µM with >100-fold selectivity over HepG2 cells. The clusters were characterized and prioritized by potency, rate of action, cidal/static properties, and physicochemical properties, among others. In addition, three compounds were selected for mouse pharmacokinetic experiments and one, NEU-1053 (20, Fig. 7), cleared parasitemia in two out of four mice after one round of treatment and cured three out of four mice after a second round of treatment in an in vivo mouse model of HAT. High-priority clusters such as the one typified by 21 (Fig. 7) represent another promising starting point for ongoing optimization.
Figure 7.
Singleton NEU-1053 and a representative of a high-priority cluster discovered by HTS.
Case study 2: Protease lead repurposing for HAT
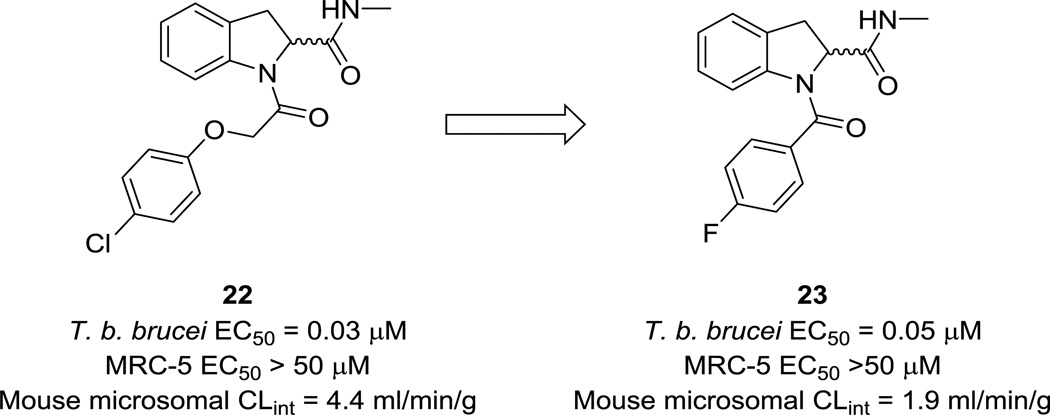
A lead repurposing program from Cleghorn et al. focused on protease inhibitors as anti-T. brucei compounds.50 A ~3400-member protease inhibitor library was constructed from a set of compounds with known protease activity that was subsequently filtered based on lead-like properties. Ninety-three of these compounds showed >50% cell proliferation inhibition at 30 µM; of these, a subset of compounds was hand-picked for a dose-response assay, resulting in the discovery of 22 (Fig. 8) as the most potent (EC50 = 27 nM) hit in a series of indoline-2-carboxamides. In addition, it was >1600-fold selective over mammalian cells and had excellent molecular weight, clogP, and polar surface area properties; it was therefore selected for further optimization. The major issue to resolve in this series was metabolic liability, and extensive SAR work led to the development of 23 (Fig. 8), which had improved microsomal stability and exposure, and demonstrated partial cure in a stage 2 mouse model of HAT. However, despite the selectivity over MRC-5 cells, toxicity was observed that prevented further progression of the series.
Figure 8.
Protease inhibitor optimization for HAT.
These examples clearly illustrate the benefits of lead repurposing over traditional, unbiased HTS. The kinase-targeted HTS resulted in many promising compound clusters, and further development of several high-priority clusters is ongoing, enabled by the wealth of SAR information available within a cluster. Additionally, ADME data was already available for several of the initial hits. Although the protease lead repurposing program ultimately resulted in a toxic compound, it only took one round of analog design to get to a molecule that was advanced enough to undergo animal efficacy studies that demonstrated proof-of-concept. As with other repurposing strategies, the time and resources required to go from hit to lead compound using lead repurposing are much reduced.
VI. Conclusion
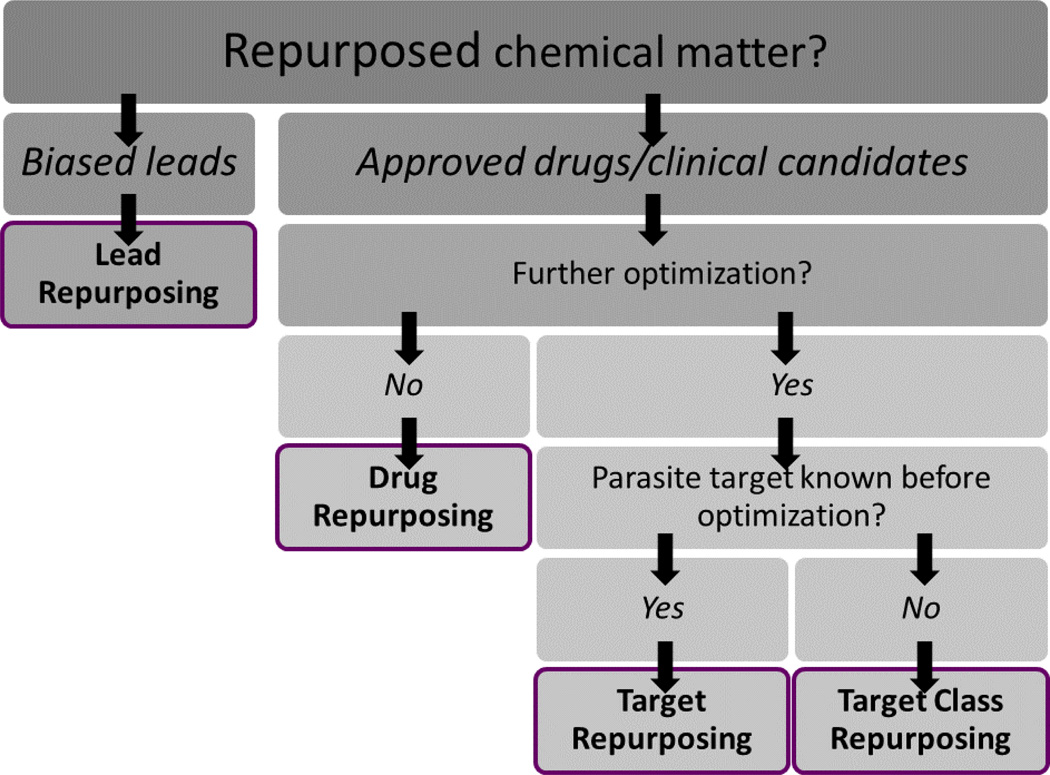
Repurposing strategies offer many benefits for drug discovery, especially for NTD and malaria drug discovery, where time, resources, and information are scarce. The possibilities for success are demonstrated by the popularity of repurposing approaches in the NTD scientific community. However, this popularity has also resulted in a plethora of disparate terms and phrases to describe the nuances of these strategies. It is our hope that the systematization of vocabulary for describing repurposing drug discovery strategies will foster better, clearer communication between researchers and enable better drug discovery in the future. We provide the diagram in Fig. 9 as a means to help the community differentiate between the approaches we have summarized in this Digest.
Figure 9.
Flow chart summarizing the four repurposing strategies highlighted in this Digest.
Acknowledgments
We acknowledge the National Institutes of Health (R56AI099476, R01AI114685) and the American Chemical Society Division of Medicinal Chemistry (Graduate Fellowship to DMK) for supporting this work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.World Health Organization Neglected Tropical Diseases. [November 24, 2015]; http://www.who.int/neglected_diseases/diseases/en/
- 2.Njoroge M, Njuguna NM, Mutai P, Ongarora DSB, Smith PW, Chibale K. Chem. Rev. 2014;114:11138. doi: 10.1021/cr500098f. [DOI] [PubMed] [Google Scholar]
- 3.Pollastri MP, Campbell RK. Future Med. Chem. 2011;3:1307. doi: 10.4155/fmc.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gelb MH, Van Voorhis WC, Buckner FS, Yokoyama K, Eastman R, Carpenter EP, Panethymitaki C, Brown KA, Smith DF. Mol. Biochem. Parasitol. 2003;126:155. doi: 10.1016/s0166-6851(02)00282-7. [DOI] [PubMed] [Google Scholar]
- 5.Hertweck C. Agnewandte Chem. Int. Edition. 2015;54:14622. doi: 10.1002/anie.201509828. [DOI] [PubMed] [Google Scholar]
- 6.World Health Organization Malaria Fact Sheet. [February 14, 2016]; http://www.who.int/mediacentre/factsheets/fs094/en/
- 7.Steverding D. Parasites and Vectors. 2010;3 doi: 10.1186/1756-3305-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.World Health Organization Human African Trypanosomiasis Fact Sheet. [November 24, 2015]; http://www.who.int/mediacentre/factsheets/fs259/en/
- 9.Bern C, Montgomery SP, Herwaldt BL, Rassi A, Jr, Marin-Neto JA, Dantas RO, Maguire JH, Acquatella H, Morillo C, Kirchoff LV, Gilman RH, Reyes PA, Salvatella R, Moore AC. J. Am. Med. Assoc. 2007;298:2171. doi: 10.1001/jama.298.18.2171. [DOI] [PubMed] [Google Scholar]
- 10.Horton J. Ann. Trop. Med. Parasitol. 2009;103:S33. doi: 10.1179/000349809X12502035776595. [DOI] [PubMed] [Google Scholar]
- 11.Molyneux D, Taylor HR. Trends in Parasitol. 2015;31:1. doi: 10.1016/j.pt.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Turner JD, Tendongfor N, Esum M, Johnston KL, Langley SR, Ford L, Faragher B, Specht S, Mand S, Hoerauf A, Enyong P, Wanji S, Taylor MJ. PLoS Negl. Trop. Dis. 2010;4:e660. doi: 10.1371/journal.pntd.0000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ashburn TT, Thor KB. Nat. Rev. Drug. Discov. 2004;3:673. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 14.Bacchi CJ, Nathan HC, Hunter SH, McCann PP, Sjoerdsma A. Science. 1980;210:332. doi: 10.1126/science.6775372. [DOI] [PubMed] [Google Scholar]
- 15.Kuzoe FAS. Acta Trop. 1993;54:153. doi: 10.1016/0001-706x(93)90089-t. [DOI] [PubMed] [Google Scholar]
- 16.Yun O, Priotto G, Tong J, Flevaud L, Chappuis F. PLoS Negl. Trop. Dis. 2010;4:e720. doi: 10.1371/journal.pntd.0000720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.World Health Organization Leishmaniasis Fact Sheet. [November 24, 2015]; http://www.who.int/mediacentre/factsheets/fs375/en/
- 18.Miguel DCY-Y, Jenicer KU, Andreoli Walter K, Mortara Renato A, Uliana Silvia RB. J. Antimicrob. Chemother. 2007;60:526. doi: 10.1093/jac/dkm219. [DOI] [PubMed] [Google Scholar]
- 19.Altan N, Chen Y, Schindler M, Simon SM. Proc. Natl. Acad. Sci. USA. 1999;96:4432. doi: 10.1073/pnas.96.8.4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miguel DCY-Y, Jenicer KU, Uliana Silvia RB. PLoS Negl. Trop. Dis. 2008:e249. doi: 10.1371/journal.pntd.0000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miguel DCZ-N, Rogeria C, Yokoyama-Yasunaka Jenicer KU, Katz Simone, Barbieri Clara L, Uliana Silvia RB. J. Antimicrob. Chemother. 2009;63:365. doi: 10.1093/jac/dkn509. [DOI] [PubMed] [Google Scholar]
- 22.World Health Organization Lymphatic filariasis Fact Sheet. [November 24, 2015]; http://www.who.int/mediacentre/factsheets/fs102/en/
- 23.World Health Organization Onchocerciasis Fact Sheet. [November 24, 2015]; http://www.who.int/mediacentre/factsheets/fs374/en/
- 24.Johnston KL, Ford L, Taylor MJ, Umareddy I, Townson S, Specht S, Pfarr K, Hoerauf A, Altmeyer R. Int. J. Parasitol. Drugs Drug Resist. 2014;4:278. doi: 10.1016/j.ijpddr.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bulman CA, Bidlow CM, Lustigman S, Cho-Ngwa F, Williams D, Rascon AA, Jr, Tricoche N, Samje M, Bell A, Suzuki B, Lim KC, Supakorndej N, Supakorndej P, Wolfe AR, Knudsen GM, Chen S, Wilson C, Ang K-H, Arkin M, Gut J, Franklin C, Marcellino C, McKerrow JH, Debnath A, Sakanari JA. PLoS Negl. Trop. Dis. 2015;9:e0003534. doi: 10.1371/journal.pntd.0003534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding DM Qingqing, Gao Guangwei, Zhao Yaxue, Wang Qing, Nare Bakela, Jacobs Robert, Rock Fernando, Alley Michael RK, Plattner Jacob J, Chen Guoqiang, Li Dawei, Zhou Huchen. J. Med. Chem. 2011;54:1276. doi: 10.1021/jm101225g. [DOI] [PubMed] [Google Scholar]
- 27.Rock FL, Mao W, Yaremchuk A, Tukalo M, Crepin T, Zhou H, Zhang Y-K, Hernandez V, Akama T, Baker SJ, Plattner JJ, Shapiro L, Martinis SA, Benkovic SJ, Cusack S, Alley MRK. Science. 2007;316:1759. doi: 10.1126/science.1142189. [DOI] [PubMed] [Google Scholar]
- 28.Drugs for Neglected Diseases Inititative - About Malaria. [November 25, 2015]; http://www.dndi.org/diseases-projects/diseases/malaria.html. [Google Scholar]
- 29.Yu Z, Brannigan JA, Moss DK, Brzozowski AM, Wilkinson AJ, Holder AA, Tate EW, Leatherbarrow RJ. J. Med. Chem. 2012;55:8879. doi: 10.1021/jm301160h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Price HP, Menon MR, Panethymitaki C, Goulding D, McKean PG, Smith DF. J. Biol. Chem. 2003;278:7206. doi: 10.1074/jbc.M211391200. [DOI] [PubMed] [Google Scholar]
- 31.Rackham MD, Brannigan JA, Moss DK, Yu Z, Wilkinson AJ, Holder AA, Tate EW, Leatherbarrow RJ. J. Med. Chem. 2013;56:371. doi: 10.1021/jm301474t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rackham MDB, James A, Rangachari K, Meister S, Wilkinson Anthony J, Holder Anthony A, Leatherbarrow Robin J, Tate Edward W. J. Med. Chem. 2014;57:2773. doi: 10.1021/jm500066b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yokoyama K, Goodwin GW, Ghomashchi F, Glomset J, Gelb MH. Biochem. Soc. Transcr. 1992;20:489. doi: 10.1042/bst0200489. [DOI] [PubMed] [Google Scholar]
- 34.Berndt N, Hamilton AD, Sebti SM. Nat. Rev. Cancer. 2011;11:775. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nallan L, Bauer KD, Bendale P, Rivas K, Yokoyama K, Horney CP, Pendyala PR, Floyd D, Lombardo LJ, Williams DK, Hamilton A, Sebti S, Windsor WT, Weber PC, Buckner FS, Chakrabarti D, Gelb MH, Van Voorhis WC. J. Med. Chem. 2005;48:3704. doi: 10.1021/jm0491039. [DOI] [PubMed] [Google Scholar]
- 36.Bendale P, Olepu S, Suryadevara PK, Bulbule V, Rivas K, Nallan L, Smart B, Yokoyama K, Ankala S, Pendyala PR, Floyd D, Lombardo LJ, Williams DK, Buckner FS, Chakrabarti D, Verlinde CLMJ, Van Voorhis WC, Gelb MH. J. Med. Chem. 2007;50:4585. doi: 10.1021/jm0703340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Voorhis WC, Rivas KL, Bendale P, Nallan L, Horney C, Barrett LK, Bauer KD, Smart BP, Ankala S, Hucke O, Verlinde CLMJ, Chakrabarti D, Strickland C, Yokoyama K, Buckner FS, Hamilton AD, Williams DK, Lombardo LJ, Floyd D, Gelb MH. Antimicrob. Agents Chemother. 2007;51:3659. doi: 10.1128/AAC.00246-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bulbule VJ, Rivas K, Verlinde CLMJ, Van Voorhis WC, Gelb MH. J. Med. Chem. 2008;51:384. doi: 10.1021/jm7013138. [DOI] [PubMed] [Google Scholar]
- 39.Patel G, Karver CE, Behera R, Guyett PJ, Sullenberger C, Edwards P, Roncal NE, Mensa-Wilmot K, Pollastri MP. J. Med. Chem. 2013;56:3820. doi: 10.1021/jm400349k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Behera R, Thomas SM, Mensa-Wilmot K. Antimicrob. Agents Chemother. 2014;58:2202. doi: 10.1128/AAC.01691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Minucci SP. Pier Giuseppe. Nat. Rev. Cancer. 2006;6:38. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 42.Kelly JMT, Martin C, Horn David, Loza Elinars, Kalvinsh Ivars, Bjorkling Fredrik. Bioorganic Med. Chem. Lett. 2012;22:1886. doi: 10.1016/j.bmcl.2012.01.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carrillo AKGW Armand, Guy R. Kiplin. Bioorganic Med. Chem. Lett. 2015;23:5151. doi: 10.1016/j.bmc.2014.12.066. [DOI] [PubMed] [Google Scholar]
- 44.Diaz RL-A, Sandra A, Seixas João D, Amata Emanuele, Devine William, Cordon-Obras Carlos, Rojas-Barros Domingo I, Jimenez Elena, Ortega Fatima, Crouch Sabrinia, Colmenarejo Gonzalo, Fiandor Jose Maria, Martin Jose Julio, Berlanga Manuela, Gonzalez Silvia, Manzano Pilar, Navarro Miguel, Pollastri Michael P. PLoS Negl. Trop. Dis. 2014;8:e3253. doi: 10.1371/journal.pntd.0003253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochiana SO, Pandarinath V, Wang Z, Kapoor R, Ondrechen MJ, Ruben L, Pollastri MP. Eur. J. Med. Chem. 2013;62:777. doi: 10.1016/j.ejmech.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seixas JD, Luengo-Arratta SA, Diaz R, Saldivia M, Rojas-Barros DI, Manzano P, Gonzalez S, Berlanga M, Smith TK, Navarro M, Pollastri MP. J. Med. Chem. 2014;57:4834. doi: 10.1021/jm500361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Urich R, Grimaldi R, Luksch T, Frearson JA, Brenk R, Wyatt PG. J. Med. Chem. 2014;57:7536. doi: 10.1021/jm500239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Merritt C, Silva LE, Tanner AL, Stuart K, Pollastri MP. Chem. Rev. 2014;114:11280. doi: 10.1021/cr500197d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elkins JM, Fedele V, Szklarz M, Azeez KRA, Salah E, Mikolajczyk J, Romanov S, Sepetov N, Huang X-P, Roth BL, Zen AAH, Fourches D, Muratov E, Tropsha A, Morris J, Teicher BA, Kunkel M, Polley E, Lackey KE, Atkinson FL, Overington JP, Bamborough P, Muller S, Price DJ, Willson TM, Dewry DH, Knapp S, Zuercher WJ. Nat. Biotechnol. 2015 doi: 10.1038/nbt.3374. In Press. [DOI] [PubMed] [Google Scholar]
- 50.Cleghorn LAT, Albrecht S, Stojanovski L, Simeons FRJ, Norval S, Kime R, Collie IT, De Rycker M, Campbell L, Hallyburton I, Frearson JA, Wyatt PG, Read KD, Gilbert IH. J. Med. Chem. 2015;58:7695. doi: 10.1021/acs.jmedchem.5b00596. [DOI] [PMC free article] [PubMed] [Google Scholar]