Abstract
To investigate the function and regulation mechanism of ATP-binding cassette, subfamily G, member 2 (ABCG2) in retinoblastoma cancer stem cells (RCSCs), a long-term culture of RCSCs from WERI-Rb1 cell line was successfully established based on the high expression level of ABCG2 on the surface of RCSCs. To further explore the molecular mechanism of ABCG2 on RCSCs, a microRNA that specifically targets ABCG2 was predicted. Subsequently, miR-3163 was selected and confirmed as the ABCG2-regulating microRNA. Overexpression of miR-3163 led to a significant decrease in ABCG2 expression. Additionally, ABCG2 loss-of-function induced anti-proliferation and apoptosis-promoting functions in RCSCs, and multidrug resistance to cisplatin, carboplatin, vincristine, doxorubicin, and etoposide was greatly improved in these cells. Our data suggest that miR-3163 has a significant impact on ABCG2 expression and can influence proliferation, apoptosis, and drug resistance in RCSCs. This work may provide new therapeutic targets for retinoblastoma.
Keywords: Retinoblastoma, RCSCs, ABCG2, miR-3163
INTRODUCTION
Retinoblastoma (RB), the most common eye tumor found in children, is caused by the inactivation of both alleles of the retinoblastoma 1 (Rb1) gene (1). Chemoreduction using systemic chemotherapy combined with local therapy (photoablation, cryotherapy or thermotherapy) has become a therapeutic mainstay for retinoblastoma and has improved patients’ prognosis and chance of eye preservation. Nonetheless, chemotherapeutic drug resistance is common in retinoblastoma, resulting in the increased incidence of unsuccessful treatment.
Tumors have long been viewed as a population in which all cells have an equal propensity to form new tumors. This is known as the conventional stochastic model. However, the hierarchical model claims that there is a small subset of cancer stem cells (CSCs) responsible for tumor initiation and growth. Cancer stem cells have been identified by a specific cell-surface marker in several solid tumors including breast cancer (2), brain tumors (3), prostate cancer (4), and lung cancer (5). Most markers used for CSC isolation are selected from normal stem cells or other malignancies.
Previous reports have demonstrated that stem cells expressing embryonic and neuronal stem cell markers are present in human RB (6,7,8). Our research group has successfully designed culture methods for selecting and expanding stem-like cancer cells in RB (RCSCs) (9). Interestingly, the cultured cells also express retinal development–related genes. In past years, one emerging hypothesis postulates that the development of drug-resistant tumors is sustained by a self-renewing subpopulation termed “putative CSCs.” However, a method to cure RB by regulating RCSCs has yet to be developed.
RB usually occurs in children younger than five years, and the majority of patients relapse and become drug-resistant. Various types of ATP-binding cassette (ABC) Transporters, especially ABCG2, contribute to drug resistance in cancers such as retinoblastoma by pumping chemotherapy drugs out of cancer cells (10). ATP-binding cassette, subfamily G, member 2 (ABCG2), is a member of the ABC transporter family and was originally cloned from doxorubicin-resistant human MCF-7 breast cancer cells and named breast cancer resistance protein (BCRP) (11). To date, ABCG2 overexpression has been observed in many tumor types, including bladder cancer, lung cancer, leukemia, and several squamous cell carcinomas (12,13,14), where it functions to pump a wide variety of endogenous and exogenous compounds out of cells (15,16). Elevated expression of ABCG2 in vitro causes resistance to anticancer drugs, including topotecan, irinotecan, mitoxantrone, and doxorubicin (17). Furthermore, ABCG2 expression was shown to be a molecular determinant of the side population cell phenotype, a characteristic reminiscent of stem cells (18).
MicroRNAs are a large group of short, noncoding RNAs that act on their mRNA targets by complementary Watson-Crick base pairing. They also control posttranscriptional regulation of target genes through translation inhibition or mRNA cleavage (19,20). Thousands of miRNAs have been identified within human genome, and they may regulate thousands of protein-coding genes involved in almost all critical biological processes. There is also accumulating evidence supporting the hypothesis that miRNAs are involved in the regulation of drug metabolism and disposition (21,22). Some miRNAs directly target the 3'-untranslated region (3'UTR) of genes encoding drug-metabolizing enzymes and/or drug transporters (23,24). Other miRNAs act on the 3'UTR of transcriptional regulators (e.g., xenobiotic receptors) of drug-metabolizing enzymes and drug transporters (25). As a result, these miRNAs may determine the final protein expression levels of enzymes or transporters, modulate the capacity of drug metabolism and disposition, and affect the response of cells to xenobiotic drugs. However, the identity and molecular mechanisms of microRNAs acting on RCSCs are still poorly understood.
In this study, we successfully isolated RCSCs cultured from the WERI-Rb1 cell line expressing stem cell-like genes such as OCT4 and NANOG. Furthermore, miR-3163, a novel microRNA whose function is poorly understood in tumorigenesis and development, was identified as the potential microRNA that post-transcriptionally regulates ABCG2 expression upon binding to its 3'UTR. In addition, through gain-of-function studies, the function of miR-3163 in RCSCs was investigated. Our results show that overexpression of miR-3163 inhibits cell proliferation, promotes apoptosis, and improves multidrug resistance. These findings provide new insight into RCSCs and offer a new therapeutic strategy for RB.
MATERIALS AND METHODS
Cell culture
The human retinoblastoma cell line WERI-Rb1 was purchased from the ATCC Company (Manassas, VA, USA) and maintained in RPMI-1640 medium (Hyclone, Logan, UT, USA) with 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA). The total number of WERIRb1 cells was amplified to 108. Subsequently, the cells were resuspended to a concentration of 1×107/mL and incubated with anti-ABCG2 antibody at 4°C for 20 minutes in sterile conditions. IgG2a antibody was used as the isotype control. Cells were washed with PBS, 5 μL of goat anti-mouse FITC secondary antibody was added, and the samples were maintained at 4°C for 20 minutes. Cells were then washed 3 to 5 times before FACS sorting. After the sorting, ABCG2 (+) cells were cultured with chemically defined serum-free culture medium, while ABCG2 (-) cells were cultured with RPMI 1640 culture medium containing 10% FBS. The medium was changed every 3 d. Expression of ABCG2 was detected after sorting by flow cytometry. All of the cells used in this study were placed in a 37°C humidified incubator with 5% CO2 and observed using an inverted microscope every other day.
Plasmids and reagents
The luciferase Reporter-ABCG2 3'UTR (pmiR-reporter-luciferase plasmid containing the ABCG2 3'UTR) plasmid was commercially constructed (Ribo Company, Guangzhou, China). Mutation of miR-3163 binding sites on the Reporter-ABCG2 3'UTR was carried out commercially (Transgene Company, Beijing, China). The drugs cisplatin, mitoxantrone and topotecan were all purchased from AbMole BioScience (Shanghai, China).
Transfection with microRNA nucleotide fragments
MirVana™ Mimics are chemically modified, synthetic nucleic acids designed to mimic mature miRNAs. These products provide a means to functionally study the role of specific miRNAs within cellular systems or to validate the role of miRNAs in regulating target genes. RCSCs were transfected with miR-3163 Scramble or Mimic according to the Lipofectamine protocol (Invitrogen, Carlsbad, CA, USA). Briefly, 10 μL of nucleotide fragment stock solution was diluted in 500 μL of serum-free DMEM for each well, and 4 μL of Lipofectamine RNAiMAX reagent was added to the diluted DNA and incubated for 30 minutes at room temperature. Then, approximately 500 μL of the DNA-Lipofectamine RNAiMAX complex was added to each well of RCSCs, which were 70% confluent. The cells were incubated at 37°C in a CO2 incubator for 24 hours, and cells were then harvested for real-time PCR or Western blot analysis.
RT-PCR analysis
Total RNA was extracted from RCSCs using RNAzol (Molecular Research Center, Cincinnati, OH, USA) following the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using the appropriate primers with reverse transcriptase (Promega Corp., Fitchurg, WI, USA). The primers used were as follows: ABCG2 (93 bp), forward, 5'-ACGAACGGATTAACAGGGTCA-3' and reverse, 5'-CTCCAGACACACCACGGAT-3'; Nanog (116 bp), forward, 5'-TTTGTGGGCCTGAAGAAAACT-3' and reverse, 5'-AGGGCTGTCCTGAATAAGCAG-3'. Oct4 (156 bp), forward, 5'-GTGTTCAGCCAAAAGACCATCT-3' and reverse, 5'-GGCCTGCATGAGGGTTTCT'. GAPDH (101 bp), forward, 5'-ACAACTTTGGTATCGTGGAAGG-3' and reverse, 5'-GCCATCACGCCACAGTTTC'. All PCR reactions were conducted in duplicate, and cells cultured in triplicate were tested. All experiments were repeated with separate cultures.
Western blot analysis
Protein was extracted from cells, and protein concentrations were determined using the BCA Protein Assay (Thermo Scientific, Somerset, NJ, USA). Protein samples were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and electrotransferred onto polyvinylidene difluoride membrane (all from Invitrogen). The membrane was incubated with antibodies against ABCG2 (sc-58224, Santa Cruz Biotechnology, Santa Cruz, CA, USA), OCT4 (sc-5279, Santa Cruz Biotechnology), NANOG (sc-33759, Santa Cruz Biotechnology) and GAPDH (sc-25778, Santa Cruz Biotechnology). Proteins of interest were detected with horseradish peroxidase-conjugated secondary antibodies (Zhongshan Jinqiao Company, Beijing, China) and were developed using the enhanced chemiluminescence Plus kit (Amersham, Freiburg, Germany).
Cell viability assay
Cells were seeded in 96-well plates at a density of 2,000 cells per well. The absorption of the cells was measured at different time points using a CCK-8 kit (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. The data were obtained from three separate experiments of three replications each.
Cell apoptosis assay
Apoptosis assays were performed using an Annexin V-FITC apoptosis detection kit (Biosea Company, Beijing, China) according to the manufacturer’s instructions. Briefly, cells (105 cells/well) were collected and resuspended in binding buffer. Annexin V-FITC and PI were added, and the reaction was incubated in the dark for 15 minutes. Cells were analyzed using a FACScan flow cytometer.
IC50 (half effective inhibitory concentration) detection via CCK-8 assay
Cells were seeded in 96-well plates with 100 μL of cell suspension at a density of 1 × 104/mL. The cells were cultured until they reached approximately 70% confluence, transfected, and then placed in incubator. The cells were treated with cisplatin, mitoxantrone and topotecan prior to the determination of IC50. Then, at different time points, 10 μL of CCK-8 solution was added to the appropriate wells, and the plates were incubated for 2 hours. Absorbance at 450 nm was measured by a microplate reader, and IC50 was calculated according to the formula provided in the instructions.
Luciferase assay
HeLa cells were co-transfected with wild-type or mutant ABCG2 3'UTR-luciferase reporter plasmid, along with a miRNA expression plasmid, using Lipofectamine 2,000 reagent (Invitrogen). Luciferase activity was determined using the Dual Luciferase Reporter Assay System (Promega, Madison, WI, USA) 48 hours after transfection, according to the manufacturer’s protocol. Transfection was conducted in triplicate and repeated once with a separate culture. Renilla luciferase activities were normalized to corresponding firefly luciferase activities and then different treatments or groups were compared.
ABCG2 ATPase assay
Crude membrane protein (100 μg protein/mL) from different groups of RCSCs was incubated at 37°C in the presence or absence of beryllium fluoride (0.2 mM/L beryllium sulfate and 2.5 mM/L sodium fluoride) in ATPase assay buffer (50 mM/L KCl, 5 mM/L NaN3, 2 mM/L EGTA, 10 mM/L MgCl2, 1 mM/L DTT, pH 6.8) for 10 minutes. The specific ATPase activity was recorded as beryllium fluoride–sensitive ATPase activity.
Statistical analysis
All values are expressed as the mean ± SD. Differences between mean results of all assays were compared by analysis of variance (ANOVA) or Student’s t-test using the SPSS (Statistical Package for the Social Sciences) statistical software package (SPSS, Inc., Chicago, IL, USA). The threshold for statistical significance was set at P < 0.05.
RESULTS
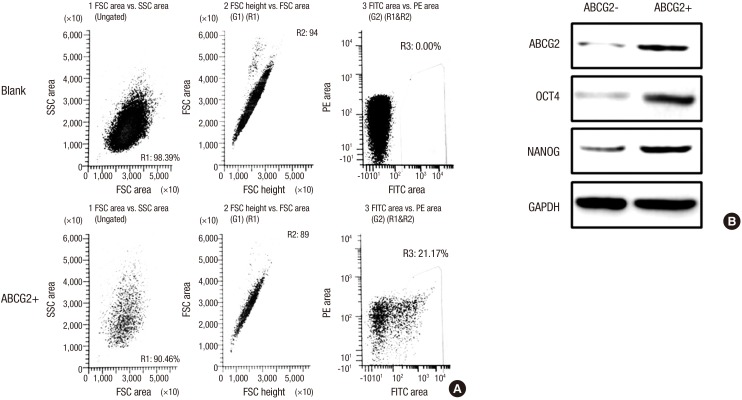
Identification of RCSCs in the WERI-Rb1 retinoblastoma cell line
Several tumors and tumor cell lines contain populations of cells with stem-like characteristics. To determine whether RCSC cells exist in the human WERI-Rb1 cell line, we stained WERI-Rb1 cells with the RCSC surface marker ABCG2 and analyzed them by flow cytometry (26). The ABCG2 positive cells (ABCG2+), hereafter referred to as RCSCs, and the ABCG negative cells (ABCG2-) were sorted by flow cytometry (Fig. 1A). The ABCG2+ and ABCG2- cells were cultured, and the expression levels of stem cell markers OCT4 and NANOG were detected. We found an increased expression of OCT4 and NANOG in RCSCs compared with ABCG2- cells, which suggests that RCSCs were successfully cultured from the WERI-Rb1 cell line (Fig. 1B).
Fig. 1.
Identification of RCSCs in the WERI-Rb1 retinoblastoma cell line. (A) ABCG2 positive (ABCG2+) and negative (ABCG2-) cells were sorted by flow cytometry based on expression of the RCSC surface marker ABCG2. For FACS sorting, the blank group was incubated with IgG2a antibody, whereas the ABCG2+ group was incubated with ABCG2 antibody. The area of R3 was the ABCG2+ cells sorted for further research, the rest cells was ABCG2- cells. (B) Expression levels of ABCG2, OCT4 and NANOG were detected in ABCG2+ and ABCG2- cells by Western blot.
miRNA prediction and expression of miR-3163 in RCSCs
To investigate which miRNA might target ABCG2, we first searched the miRNA database TargetScan, which predicted miR-3163 to be the potential miRNA targeting ABCG2. miR-3163 was considered to be a candidate for regulating ABCG2 because its binding site is conserved in humans, mice, rats and others (Fig. 2A). Next, we detected the miR-3163 expression level in ABCG2+ and ABCG2- cells. qRT-PCR revealed an almost complete absence of miR-3136 expression in ABCG2+ cells, whereas the expression of miR-3163 in ABCG2- cells was much higher (Fig. 2B).
Fig. 2.
miRNA prediction and expression of miR-3163 in RCSCs. (A) Putative miR-3163-binding sites on the ABCG2 3'UTR with potential complementary residues shown in black. (B) Expression levels of miR-3163 were detected in ABCG2+ and ABCG2- cells by qRT-PCR.
* P < 0.05 compared with the control. The data represent the results of three independent experiments.
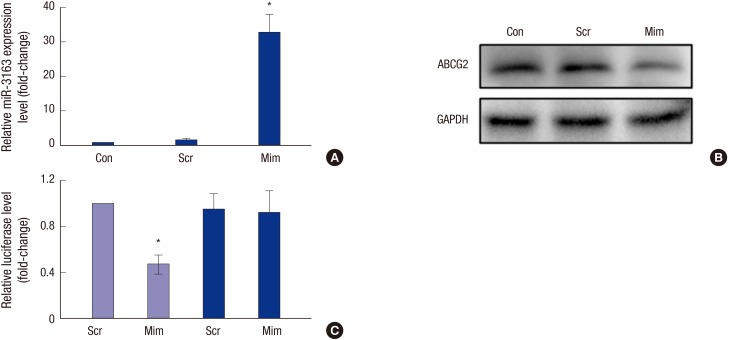
Verification of miRNA target sites
To study the function of miR-3163 in RCSCs, we overexpressed miR-3163 in RCSCs by transfecting them with Scramble or miR-3163 Mimics. Results of this assay showed that miR-3163 was significantly upregulated by miR-3163 Mimics (Fig. 3A). Western blot assays indicated that there was abundant ABCG2 protein in RCSCs, but expression decreased in RCSCs overexpressing miR-3163 (Fig. 3B). The miR-Report luciferase reporter was constructed to determine whether miR-3163 could directly target the 3'UTR of ABCG2. The reporter was co-transfected with miR-3163 Mimics for overexpression and Scramble (Scr) for control. Luciferase assays indicated that miR-3163 overexpression significantly reduced luciferase activity in the wild-type ABCG2 3'UTR reporter but not in the mutant (Fig. 3C).
Fig. 3.
Verification of miRNA target sites. (A) miR-3163 could be overexpressed by Mimics (Mim) as measured by qRT-PCR. Scramble (Scr) was used as the negative control. (B) Western blot analysis showing the protein levels of ABCG2 in RCSCs treated with Scramble or miR-3163 Mimics. GAPDH was used as a loading control. (C) Luciferase activity was measured in HeLa cells co-transfected with miR-3163 and either wide-type Reporter ABCG2 3'UTR (grey) or mutated Reporter ABCG2 3’UTR (black). Renilla activity was used as an internal control.
* P < 0.05 compared with the control. The data represent the results of three independent experiments.
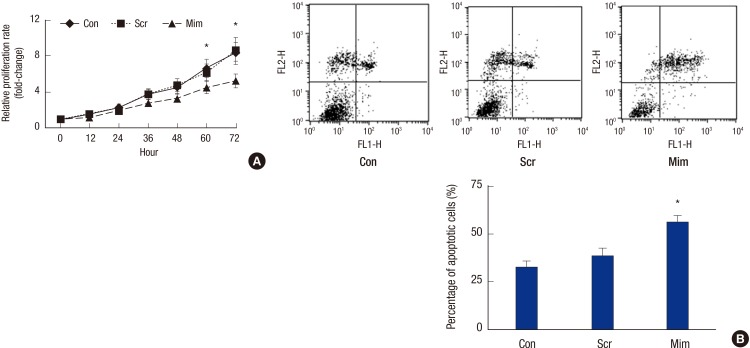
Effects of miR-3163 on RCSC proliferation and apoptosis
To study whether miR-3163 indeed participates in the proliferation of RCSCs, we performed a CCK-8 assay to verify the proliferation rate. A lower proliferation rate was observed in RCSCs transfected with miR-3163 Mimics using the CCK-8 assay (Fig. 4A). Flow cytometry analysis was employed to study whether miR-3163 participates in RCSC apoptosis triggered by cisplatin. RCSCs treated with miR-3163 Mimics were more susceptible to cisplatin-induced apoptosis compared to those treated with Scramble (Fig. 4B).
Fig. 4.
Effects of miR-3163 on RCSC proliferation and apoptosis.
(A) The effect of miR-3163 on RCSC growth was detected by CCK-8 assay at the indicated time points. (B) Representative images of flow cytometry analysis showing Annexin V/PI-stained RCSC cells with Scramble or miR-205 Mimics in the presence of cisplatin. Results from three independent experiments quantifying the percentage of Annexin V-positive cells were counted.
* P < 0.05 compared with the control. The data represent the results of three independent experiments.
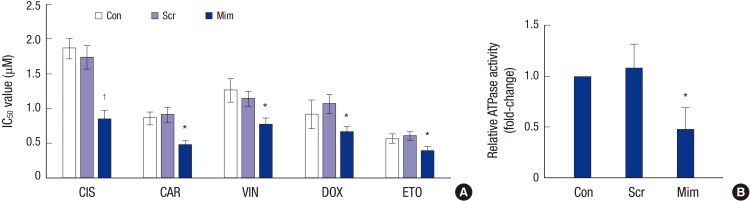
Effects of miR-3163 on RCSC multidrug sensitivity
Drug resistance, especially multidrug resistance, often leads to treatment failure in RB and is a major obstacle in the treatment of lymphoma. A CCK-8 assay was used to detect the resistance of RCSCs to multidrug treatment. The results showed that after transfection of Scramble and miR-3163 Mimics, multidrug resistance declined significantly for cisplatin, carboplatin, vincristine, doxorubicin and etoposide treatment in the miR-3163 overexpression group. This suggests that miR-3163 plays important roles in multidrug resistance (Table 1, Fig. 5A). In addition, the ABCG2 ATPase assay was used to determine the level of ATPase activity in different groups. Results showed that when miR-3163 was overexpressed in RCSCs, the ATPase activity of ABCG2 was significantly downregulated compared to RCSCs with Scramble (Fig. 5B).
Table 1. Comparison of IC50 for different drugs (μM).
| Drugs | Control | Scramble | Mimics |
|---|---|---|---|
| Cisplatin | 1.87 ± 0.15 | 1.74 ± 0.17 | 0.85 ± 0.13 |
| Carboplatin | 0.86 ± 0.09 | 0.91 ± 0.11 | 0.48 ± 0.06 |
| Vincristine | 1.27 ± 0.17 | 1.14 ± 0.11 | 0.77 ± 0.10 |
| Doxorubicin | 0.92 ± 0.21 | 1.07 ± 0.14 | 0.66 ± 0.09 |
| Etoposide | 0.57 ± 0.07 | 0.61 ± 0.06 | 0.39 ± 0.07 |
Fig. 5.
Effects of miR-3163 on RCSC multidrug resistance.
(A) The effect of miR-3163 on RCSC multidrug resistance was detected by CCK-8 assay of cells treated with different drugs. CIS, cisplatin; CAR, carboplatin; VIN, vincristine; DOX, doxorubicin; ETO, etoposide. (B) The effect of miR-3163 on RCSC ATPase activity was detected by ABCG2 ATPase assay.
* P < 0.05 compared with the control. † P < 0.05 compared with the control. The data represent the results of three independent experiments.
DISCUSSION
Current therapies target rapidly dividing cells that comprise the bulk of the tumor but fail to eradicate the CSCs, which subsequently re-initiate the malignancy. It is likely that these residual CSCs are able to survive in a dormant state for many years after remission due to their marked resistance. Although not all types of cancers subscribe to CSC theory, it provides a cellular mechanism that could account for the metastasis and chemoresistance of RB. CSCs have been identified in many solid tumors, including brain, breast, pancreas, prostate, melanoma, colon and ovarian cancers. However, compared to other solid tumors, RB stem cell research is impeded by the shortage of fresh samples and difficulty in isolating cells from RB lesions, which contain a mixture of tumor cells and necrotic and calcified tissues.
Therefore, a proper in vitro model is required to study stem cell-like retinoblastoma cells and to design future therapeutic approaches. Here, we report that a culture of RCSCs from WERIRb1 was established. These cells maintain their cancer stem cell-like properties, including the capacity for self-renewal, proliferation, differentiation, tumorigenicity and chemoresistance. Moreover, the expression of cell surface markers, such as ABCG2, was studied on these cultured cells. Further research on surface markers that can be used to identify retinoblastoma cancer stem-like cells (RCSC) may be carried out based on these results.
BCRP/ABCG2, P-gp and multidrug resistance-associated proteins (MRPs) all have ABC domains, which use ATP hydrolysis to pump drugs stored inside cancer cells outside of the cell membrane, thereby facilitating drug resistance (27). Determining how to reverse multidrug resistance mediated by drug-resistance proteins is a problem that must be solved. RNAi using DNA constructs encoding siRNAs complementary to the ABC domain of BCRP/ABCG2 has shown that gene silencing of multidrug-resistant proteins is possible (28,29).
Several studies have found that miRs are globally downregulated in different cancers, with a correlation between the degree of differentiation and the global expression levels of miRs. It has been suggested that global downregulation promotes cell transformation and tumorigenesis (30,31). Hamfjord et al. (32) found that global downregulation did not occur in the colorectal adenocarcinomas of their cohort, even though a substantial number of individual miRNAs were downregulated in the adenocarcinomas relative to the normal samples. The same group identified a significant number of uniformly downregulated miRs including the little-known miR-3163. miR-3163 was first found by Stark et al. (33) in 2010, and its function in tumorigenesis and development is still unclear.
In this paper, a microRNA that specifically targets ABCG2 was predicted. miR-3163 was identified and confirmed to be the ABCG2-regulating microRNA. ABCG2 expression decreased significantly upon overexpression of miR-3163. miR-3163 gain-of-function led to anti-proliferation and promotion of apoptosis in RCSCs. Additionally, multidrug resistance to cisplatin, carboplatin, vincristine, doxorubicin and etoposide was greatly improved in these cells. Our data suggest that miR-3163 has a significant impact on ABCG2 expression, which subsequently influences the proliferation, apoptosis and drug resistance of RCSCs. Together, these results may provide new therapeutic targets for RB.
Footnotes
DISCLOSURE: The authors have no potential conflicts of interest to disclose.
AUTHOR CONTRIBUTION: Experiment design: Jia M, Wei Z. Conducting experiment: Jia M, Wei Z, Liu P. Data analysis: Jia M, Zhao X. Approval of final manuscript: all authors.
References
- 1.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusimano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 4.Collins AT, Berry PA, Hyde C, Stower MJ, Maitland NJ. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res. 2005;65:10946–10951. doi: 10.1158/0008-5472.CAN-05-2018. [DOI] [PubMed] [Google Scholar]
- 5.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, De Maria R. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 6.Seigel GM, Hackam AS, Ganguly A, Mandell LM, Gonzalez-Fernandez F. Human embryonic and neuronal stem cell markers in retinoblastoma. Mol Vis. 2007;13:823–832. [PMC free article] [PubMed] [Google Scholar]
- 7.Seigel GM, Campbell LM, Narayan M, Gonzalez-Fernandez F. Cancer stem cell characteristics in retinoblastoma. Mol Vis. 2005;11:729–737. [PubMed] [Google Scholar]
- 8.Balla MM, Vemuganti GK, Kannabiran C, Honavar SG, Murthy R. Phenotypic characterization of retinoblastoma for the presence of putative cancer stem-like cell markers by flow cytometry. Invest Ophthalmol Vis Sci. 2009;50:1506–1514. doi: 10.1167/iovs.08-2356. [DOI] [PubMed] [Google Scholar]
- 9.Zhong X, Li Y, Peng F, Huang B, Lin J, Zhang W, Zheng J, Jiang R, Song G, Ge J. Identification of tumorigenic retinal stem-like cells in human solid retinoblastomas. Int J Cancer. 2007;121:2125–2131. doi: 10.1002/ijc.22880. [DOI] [PubMed] [Google Scholar]
- 10.Doyle L, Ross DD. Multidrug resistance mediated by the breast cancer resistance protein BCRP (ABCG2) Oncogene. 2003;22:7340–7358. doi: 10.1038/sj.onc.1206938. [DOI] [PubMed] [Google Scholar]
- 11.Doyle LA, Yang W, Abruzzo LV, Krogmann T, Gao Y, Rishi AK, Ross DD. A multidrug resistance transporter from human MCF-7 breast cancer cells. Proc Natl Acad Sci USA. 1998;95:15665–15670. doi: 10.1073/pnas.95.26.15665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diestra JE, Condom E, Del Muro XG, Scheffer GL, Pérez J, Zurita AJ, Muñoz-Seguí J, Vigués F, Scheper RJ, Capellá G, et al. Expression of multidrug resistance proteins P-glycoprotein, multidrug resistance protein 1, breast cancer resistance protein and lung resistance related protein in locally advanced bladder cancer treated with neoadjuvant chemotherapy: biological and clinical implications. J Urol. 2003;170:1383–1387. doi: 10.1097/01.ju.0000074710.96154.c9. [DOI] [PubMed] [Google Scholar]
- 13.Xie J, Jin B, Li DW, Shen B, Cong N, Zhang TZ. Dong P. ABCG2 regulated by MAPK pathways is associated with cancer progression in laryngeal squamous cell carcinoma. Am J Cancer Res. 2014;4:698–709. [PMC free article] [PubMed] [Google Scholar]
- 14.Tsunoda S, Okumura T, Ito T, Kondo K, Ortiz C, Tanaka E, Watanabe G, Itami A, Sakai Y, Shimada Y. ABCG2 expression is an independent unfavorable prognostic factor in esophageal squamous cell carcinoma. Oncology. 2006;71:251–258. doi: 10.1159/000106787. [DOI] [PubMed] [Google Scholar]
- 15.Polgar O, Robey RW, Bates SE. ABCG2: structure, function and role in drug response. Expert Opin Drug Metab Toxicol. 2008;4:1–15. doi: 10.1517/17425255.4.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Miwa M, Tsukahara S, Ishikawa E, Asada S, Imai Y, Sugimoto Y. Single amino acid substitutions in the transmembrane domains of breast cancer resistance protein (BCRP) alter cross resistance patterns in transfectants. Int J Cancer. 2003;107:757–763. doi: 10.1002/ijc.11484. [DOI] [PubMed] [Google Scholar]
- 17.Brangi M, Litman T, Ciotti M, Nishiyama K, Kohlhagen G, Takimoto C, Robey R, Pommier Y, Fojo T, Bates SE. Camptothecin resistance: role of the ATP-binding cassette (ABC), mitoxantrone-resistance half-transporter (MXR), and potential for glucuronidation in MXR-expressing cells. Cancer Res. 1999;59:5938–5946. [PubMed] [Google Scholar]
- 18.Song J, Chang I, Chen Z, Kang M, Wang CY. Characterization of side populations in HNSCC: highly invasive, chemoresistant and abnormal Wnt signaling. PLoS One. 2010;5:e11456. doi: 10.1371/journal.pone.0011456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 21.Choudhuri S, Cui Y, Klaassen CD. Molecular targets of epigenetic regulation and effectors of environmental influences. Toxicol Appl Pharmacol. 2010;245:378–393. doi: 10.1016/j.taap.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gomez A, Ingelman-Sundberg M. Epigenetic and microRNA-dependent control of cytochrome P450 expression: a gap between DNA and protein. Pharmacogenomics. 2009;10:1067–1076. doi: 10.2217/pgs.09.56. [DOI] [PubMed] [Google Scholar]
- 23.Wang F, Xue X, Wei J, An Y, Yao J, Cai H, Wu J, Dai C, Qian Z, Xu Z, et al. hsa-miR-520h downregulates ABCG2 in pancreatic cancer cells to inhibit migration, invasion, and side populations. Br J Cancer. 2010;103:567–574. doi: 10.1038/sj.bjc.6605724. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24.Guo L, Liu Y, Bai Y, Sun Y, Xiao F, Guo Y. Gene expression profiling of drug-resistant small cell lung cancer cells by combining microRNA and cDNA expression analysis. Eur J Cancer. 2010;46:1692–1702. doi: 10.1016/j.ejca.2010.02.043. [DOI] [PubMed] [Google Scholar]
- 25.Takagi S, Nakajima M, Kida K, Yamaura Y, Fukami T, Yokoi T. MicroRNAs regulate human hepatocyte nuclear factor 4alpha, modulating the expression of metabolic enzymes and cell cycle. J Biol Chem. 2010;285:4415–4422. doi: 10.1074/jbc.M109.085431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohan A, Kandalam M, Ramkumar HL, Gopal L, Krishnakumar S. Stem cell markers: ABCG2 and MCM2 expression in retinoblastoma. Br J Ophthalmol. 2006;90:889–893. doi: 10.1136/bjo.2005.089219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Abbott BL, Colapietro AM, Barnes Y, Marini F, Andreeff M, Sorrentino BP. Low levels of ABCG2 expression in adult AML blast samples. Blood. 2002;100:4594–4601. doi: 10.1182/blood-2002-01-0271. [DOI] [PubMed] [Google Scholar]
- 28.Ee PL, He X, Ross DD, Beck WT. Modulation of breast cancer resistance protein (BCRP/ABCG2) gene expression using RNA interference. Mol Cancer Ther. 2004;3:1577–1583. [PubMed] [Google Scholar]
- 29.Xie N, Mou L, Yuan J, Liu W, Deng T, Li Z, Jing Y, Hu Z. Modulating drug resistance by targeting BCRP/ABCG2 using retrovirus-mediated RNA interference. PLoS One. 2014;9:e103463. doi: 10.1371/journal.pone.0103463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585:2087–2099. doi: 10.1016/j.febslet.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 31.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 32.Hamfjord J, Stangeland AM, Hughes T, Skrede ML, Tveit KM, Ikdahl T, Kure EH. Differential expression of miRNAs in colorectal cancer: comparison of paired tumor tissue and adjacent normal mucosa using high-throughput sequencing. PLoS One. 2012;7:e34150. doi: 10.1371/journal.pone.0034150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stark MS, Tyagi S, Nancarrow DJ, Boyle GM, Cook AL, Whiteman DC, Parsons PG, Schmidt C, Sturm RA, Hayward NK. Characterization of the Melanoma miRNAome by Deep Sequencing. PLoS One. 2010;5:e9685. doi: 10.1371/journal.pone.0009685. [DOI] [PMC free article] [PubMed] [Google Scholar]