Abstract
Photosynthetic water oxidation is catalyzed by the Mn4CaO5 cluster of photosystem II. The assembly of the Mn4O5Ca requires light and involves a sequential process called photoactivation. This process harnesses the charge-separation of the photochemical reaction center and the coordination environment provided by the amino acid side chains of the protein to oxidize and organize the incoming manganese ions to form the oxo-bridged metal cluster capable of H2O-oxidation. Although most aspects of this assembly process remain poorly understood, recent advances in the elucidation of the crystal structure of the fully assembled cyanobacterial PSII complex help in the interpretation of the rich history of experiments designed to understand this process. Moreover, recent insights on the structure and stability of the constituent ions of the Mn4CaO5 cluster may guide future experiments. Here we consider the literature and suggest possible models of assembly including one involving single Mn2+ oxidation site for all Mn but requiring ion relocation.
Keywords: photosystem II, water oxidation, manganese, oxygen evolution, photoactivation, EPR, oxo bridge
Introduction
A decline in the photosynthetic activity of oxygenic photosynthetic organisms due to light stress has been described as photoinhibition (Björkman, 1981; Osmond, 1981; Powles and Björkman, 1981; Ohad et al., 1984). The primary damage occurs within the reaction center of Photosystem II (PSII). It is distinct from the concurrent oxidative damage to the machinery of protein synthesis, which compounds the problem since de novo protein synthesis is necessary for the replacement of damaged PSII proteins (Adir et al., 2003; Lupínková and Komenda, 2004; Nishiyama et al., 2004; Edelman and Mattoo, 2008). The precise mechanism of PSII photoinhibition in vivo remains under debate (Adir et al., 2003; Edelman and Mattoo, 2008; Vass and Cser, 2009). Despite this uncertainty, it is evident that the D1 reaction center protein is the primary target for photodamage and this leads to an increased turnover rate of D1, in comparison to other PSII proteins, upon exposure to high light intensities (Ohad et al., 1984). To cope with light stress, all oxygenic photosynthetic organisms have developed protective mechanisms both to minimize the effects of exposure to excess light and to efficiently repair the damage when it occurs. Overall, the efficiency of photosynthetic electron transfer decreases markedly only when the rate of damage exceeds the rate of repair. A crucial phase of the de novo biogenesis of PSII, as well as the damage repair process, is the assembly of the Mn4CaO5 complex. This involves the oxidative assembly of Mn2+ and Ca2+ ions into the coordination environment of the PSII water-oxidation complex (WOC) in a light-driven process called photoactivation (for previous reviews, see Ono, 2001; Burnap, 2004; Dismukes et al., 2005).
PSII damage and D1 replacement
Replacement of damaged D1
The entire process of PSII damage-repair cycle can be described as follows: (i) damage occurring to PSII, (ii) signaling of this damage, (iii) monomerization of PSII dimer and partial disassembly of PSII monomer, (iv) degradation of D1 and insertion of a newly synthesized D1 into PSII sub-complex, and (v) reassembly of holoenzyme and photoactivation of the Mn4CaO5 cluster (Aro et al., 1993; Koivuniemi et al., 1995; Nixon et al., 2005; Figure 1). We briefly outline some features of the overall PSII assembly and repair process to place the assembly of the Mn4CaO5 cluster in context. For more comprehensive information the reader is advised to examine several recent review articles (Nixon et al., 2010; Becker et al., 2011; Nickelsen and Rengstl, 2013; Heinz et al., 2016).
Figure 1.
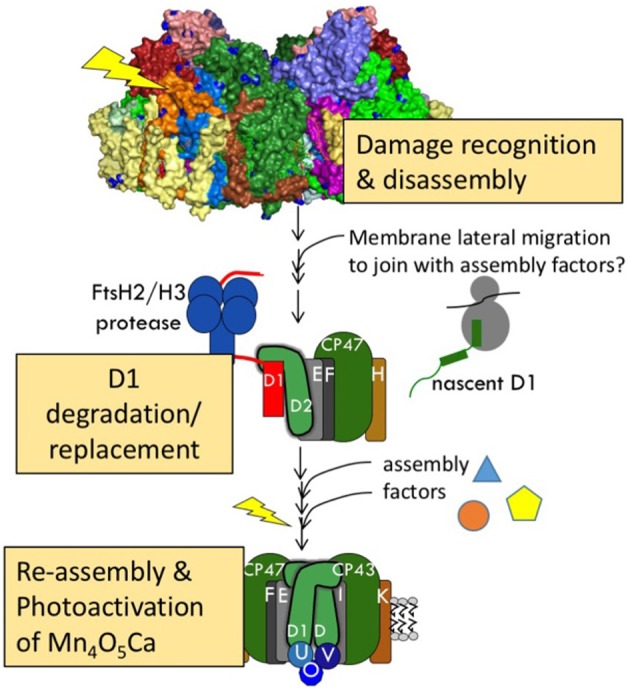
Schematic repair pathway for photodamaged PSII. The process can be divided into the three main phases: (1) damage recognition and partial disassembly of photodamage PSII complexes, (2) D1 degradation and replacement, and (3) reassembly of the subunits and light-driven assembly (photoactivation) of the Mn4O5Ca metal cluster.
Monomerization of dimeric PSII has been suggested to result from the detachment or rearrangement of PsbO, one of three luminal extrinsic subunits of PSII (Nixon et al., 2010). The basis of this assessment is the failure to accumulate dimeric PSII in a mutant of Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) lacking PsbO (Komenda et al., 2010). In plants and green algae, it has also been proposed that PSII core phosphorylation might trigger disassembly of PSII dimer to form monomer by acting alone or in conjunction with PsbO (Puthiyaveetil and Kirchhoff, 2013). Detachment of CP43 from PSII monomer leads to the formation of so-called RC47 complex which is a pivotal sub-complex for further replacement of damaged D1 during PSII repair (Komenda et al., 2005). Given the fact that PsbO functions as PSII manganese-stabilizing protein and CP43 participates with D1 in ligating the Mn4CaO5 cluster, it is conceivable that photodamage to Mn4CaO5 cluster might cause the detachment of these two subunits. It is also interesting to note that the assembly and disassembly of the Mn4O5Ca regulates the coupling of the phycobilisome to the cyanobacterial PSII reaction center such that centers without an intact metal cluster are not efficiently coupled with respect to energy transfer from the phycobilisome (Hwang et al., 2008).
Radioactive pulse-chase experiments (Komenda and Barber, 1995) showed that translation inhibitors slow D1 degradation, suggesting that D1 degradation and new D1 synthesis are synchronized. Increased turnover of D1 could be a generalized response to damage-promoting light conditions, with all D1 copies prone to increased probability of replacement or there could be a specific targeting mechanism that replaces only damaged D1 copies. Intuitively, a targeting mechanism seems more likely. However, despite good circumstantial evidence, direct evidence for the specific targeting of PSII centers with damaged D1 has not been obtained, mainly because it is technically difficult to separately track damaged and undamaged forms of D1 through the replacement process. Recently, targeting has been inferred from experiments where cells are allowed to express two alternative forms of the D1 protein in the same cell, with one wild-type form and the other a light-sensitive form. The analysis indicates that only the light-sensitive version of D1 and not the wild-type version is turned over very rapidly (Nagarajan and Burnap, 2014).
FtsH proteases play an important role in degradation of damaged D1 during PSII repair (Mann et al., 2000; Bailey et al., 2002; Silva et al., 2003). Mutants lacking FtsH proteases display impaired D1 degradation and thus accumulate damaged D1 (Bailey et al., 2002; Silva et al., 2003; Komenda et al., 2006; Kato et al., 2009). Additionally, the AAA-type protease, FtsH is crucial for the degradation of D1 protein (Silva et al., 2003; Nixon et al., 2005). Without it efficient repair ceases. How newly synthesized D1 subunit is integrated into the RC47 sub-complex is still a matter of debate. Studies in chloroplasts have led to the conclusion that D1 replacement occurs co-translationally (Zhang et al., 1999, 2000). Following initiation of psbA mRNA translation, nascent D1 protein is targeted to the thylakoid membrane by the chloroplast signal recognition particle (cpSRP54) and then released after interacting with a putative SRP receptor (Zhang and Aro, 2002). It was demonstrated that cpSRP54 can be efficiently crosslinked to nascent D1 chains that are still attached to ribosomes (Nilsson et al., 1999; Nilsson and van Wijk, 2002). Polypeptide chain elongation of the docked complex results in precursor D1 (pD1, see below) insertion into the thylakoid translocation channel (cpSecY) (Zhang et al., 2001). During translocation, the transmembrane domains of nascent pD1 appear to interact with existing PSII sub-complexes containing D2, PsbI, and cytochrome b559, CP47, but lacking CP43 (van Wijk et al., 1996, 1997; Zhang and Aro, 2002) and subsequently incorporate into PSII complex. Pulse-labeling studies indicated that this association already exists before the synthesis of the pD1 protein is complete (Zhang et al., 1999, 2000).
There is still uncertainty about where the repair of damaged PSII takes place and it is worth noting that PSII assembly for repair and PSII de novo assembly appear to involve distinct PSII assembly pathways (reviewed in Heinz et al., 2016). Regions of connection between the plasma membrane and thylakoid membrane appear to be sites of PSII assembly (Klinkert et al., 2004; Schottkowski et al., 2009; Nickelsen et al., 2011; Stengel et al., 2012; Heinz et al., 2016). These studies have led to the suggestion that regions of the thylakoid membrane are differentiated by being specifically enriched in assembly proteins, which are designated PratA-defined membranes (PDMs) being especially relevant to the photoassembly of the Mn4CaO5 as discussed below. The idea of localized region of the thylakoid membrane enriched in assembly factors fits with the report that FtsH proteins are localized in thylakoids (Komenda et al., 2006; Krynická et al., 2014) in distinct patches that are less enriched in chlorophyll (Sacharz et al., 2015). Thus, from an ultrastructural perspective, a reasonable working hypothesis is that the repair processes, including photoactivation, are located in discreet regions of the thylakoid enriched in the factors facilitating reassembly.
Processing of the D1 carboxy terminus
D1 protein is synthesized in a precursor form (pD1) with a carboxyl-terminal extension (C-terminal) whose length and sequence vary among different organisms (Diner et al., 1988b; Seibert et al., 1989; Nixon et al., 1992; Anbudurai et al., 1994; Shestakov et al., 1994; Ivleva et al., 2000; Zhang and Aro, 2002). The pD1 protein is subsequently cleaved on the carboxyl side of residue Ala344, resulting in the removal of the extension (Nixon and Diner, 1992; Nixon et al., 1992), which is carried out by carboxy terminal protease (CtpA), which is dedicated to this post-translational processing (Diner et al., 1988a; Seibert et al., 1989; Nixon et al., 1992; Taguchi et al., 1995; Trost et al., 1997; Ivleva et al., 2000). In plants, an extension consisting of 9 residues is cleaved in a single proteolytic step, whereas in Synechocystis 6803 a 16 amino acid extension is removed in two steps (Komenda et al., 2007; Satoh and Yamamoto, 2007). Although the extension is not essential for assembly of functional PSII complex (Nixon et al., 1992; Satoh and Yamamoto, 2007), it is required for optimal photosynthetic performance implying that it might plays an important role in PSII repair (Diner, 2001). For example, Synechocystis mutants lacking the C-terminal extension exhibit decreased fitness and are more susceptible to photodamage (Ivleva et al., 2000; Kuviková et al., 2005). D1 maturation is a prerequisite for assembly of the Mn4CaO5 cluster (Diner et al., 1991; Nixon et al., 1992) and binding of the PSII extrinsic proteins (Roose and Pakrasi, 2004), thus is essential for oxygen evolution activity (Taylor et al., 1988). The extension must be cleaved before the Mn4CaO5 cluster can be functionally assembled (Nixon et al., 1992; Anbudurai et al., 1994; Komenda et al., 2007), suggesting the C-terminus of the mature D1 polypeptide is involved in assembly of the Mn4CaO5 cluster. In recent X-ray structures (Umena et al., 2011), Ala344 is shown to coordinate the Mn(2) and the Ca atom of Mn4CaO5 through its backbone α-carboxyl moiety. These assignments are consistent with mutational analysis that had originally led to this suggestion (Diner et al., 1991; Nixon et al., 1992)
Accessory proteins for PSII assembly and repair
Numerous accessory proteins are being discovered to have roles in the assembly, maturation and repair of the PSII complex (Shestakov et al., 1994; Inagaki et al., 2001; Yamamoto, 2001; Kashino et al., 2002; Silva et al., 2003; Roose and Pakrasi, 2004; Keren et al., 2005; Chen et al., 2006; Komenda et al., 2006; Nowaczyk et al., 2006; Park et al., 2007). All full accounting of these is beyond the scope of this review and for the most recent summary of the numerous assembly factors the reader should consult (Heinz et al., 2016). Biochemical approaches (e.g., Nowaczyk et al., 2006; Mamedov et al., 2007) and genetic analyses (e.g., Klinkert et al., 2004; Liu et al., 2011b), have led to the identification of proteins facilitating the assembly of PSII that could be of specific relevance to the process of photoactivation, most notably, PratA and Psb27. Deletion of pratA results in a dramatic decrease in the accumulation of PSII in Synechocystis and a defect in the processing of the D1 C-terminus by CtpA. Moreover, PratA interacts with the D1 C-terminus and may bind Mn2+ possibly facilitating the assembly of the Mn4O5Ca (Klinkert et al., 2004; Schottkowski et al., 2009). Psb27 is found to bind to forms of the PSII complex thought to represent assembly and/or disassembly intermediates (Roose and Pakrasi, 2004; Nowaczyk et al., 2006; Mamedov et al., 2007; Liu et al., 2011a,b) and deletion of the protein affects photoactivation of the complex (Roose and Pakrasi, 2007). Thus, Psb27 and PratA are especially good candidates for facilitating photoactivation of the Mn4CaO5. Indeed, there is good reason to believe that the published in vitro assembly experiments are missing assembly cofactors, which may explain why the yield of active PSII centers produced by in vitro photoactivation of Mn4CaO5 clusters by biochemical methods is invariably lower than intact cells as discussed below.
Mechanism of photoactivation
Coordinating residues of Mn4CaO5 cluster
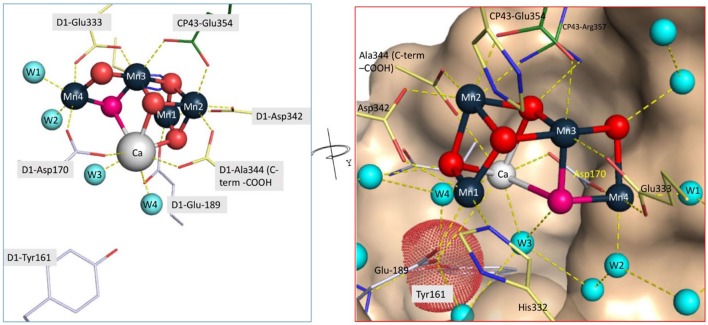
According to 1.9 Å PSII crystal structure (Umena et al., 2011), Mn4O5Ca cluster coordinated by one nitrogen ligand from D1-His332 and six carboxylate ligands from D1-Asp170, D1-Glu189, D1-Glu333, D1-Asp342, D-Ala344, CP43-Glu354 (Figure 2). Three of them, D1-Glu333, D1-Asp342, and CP43-Glu354, form μ–carboxylate bridges between Mn (Mn(1)–Mn(2) (Asp342), Mn(2)–Mn(3) (CP43-Glu354), and Mn(3)–Mn(4) (Glu333)). D1-Asp170 and the C-terminal carboxylate group of D1-Ala344 bridge Ca with Mn(4) and Mn(2), respectively. The Mn(4) has been referred to as the “dangler manganese” (Peloquin et al., 2000) because it is located outside the semi-cubic cluster formed by the other four metals of the cluster, Ca, Mn(1), Mn(2), and Mn(3). Both D1-Glu189 and D1-His332 serve as monodentate ligands to Mn(1). The D1-Asp170 plays an especially crucial role during the assembly process since it helps form the so-called “high affinity site” involved in the initial photooxidation of Mn2+ (Nixon and Diner, 1992).
Figure 2.
Coordination environment of the assembled Mn4CaO5 H2O-oxidation complex of PSII. The high affinity site of Mn2+ binding and photooxidation during the initial phase of the assembly process minimally involves D1-Asparate170 (Nixon and Diner, 1992; Campbell et al., 2000) located in the vicinity of Mn4 in the final complex. The initial state of the complex for photoassembly appears to involve the binding of one Mn2+ at the high affinity site (Ono and Mino, 1999) together with one Ca2+ ion that modulates the ligand environment of the Mn2+ possibly via the formation of a bridging water or hydroxide, although the presence of the Ca2+ does not appreciably change the binding affinity of the Mn2+ at the high affinity site (Tyryshkin et al., 2006). The C-terminal polypeptide backbone carboxylate of D1-Alanine344, which is available only following proteolytic cleavage of the precursor form of the D1 protein (pD1), is also critical for the assembly process, although it too does not markedly alter the binding of Mn2+ at the high affinity site (Nixon et al., 1992; Cohen et al., 2007). Figures developed upon 3D coordinates (PDB 4UB6) of the published X-ray diffraction model (Umena et al., 2011).
Two-quantum model of photoactivation
The assembly of the metals of the Mn4O5Ca requires light to induce charge separation to oxidize and strongly bind the Mn ions. It is important to note that the assembly is an oxidative process that involves removal of electrons from the Mn ions and the formation of oxo-bridges between the metals of the cluster with the bridging oxygen atoms (shown in red, Figure 2) derived from water molecular coordinated to the metal ions. The oxidative assembly utilizes the same light-driven charge separation events within the photochemical reaction center that subsequently drive photosynthetic electron transfer in the fully functional enzyme. Apart from the definition of the Mn-binding site characteristics and some very well-defined kinetic features that govern the development of H2O-oxidation activity, photoactivation remains poorly understood. The quantum efficiency of photoactivation is very low, typically in the range of ~1%, which is much lower than for photosynthetic water oxidation in the assembled PSII (>90%) even in intact systems (Cheniae and Martin, 1971a,b; Cheniae and Martin, 1972; Radmer and Cheniae, 1971; Ono and Inoue, 1982, 1983). The kinetic model of photoactivation, termed as “two-quantum series model” (Radmer and Cheniae, 1971), was originally observed during photoactivation as a function of either light intensity or flash interval using fixed numbers of Xe light flashes (Cheniae and Martin, 1971a,b; Cheniae and Martin, 1972; Radmer and Cheniae, 1971). These pioneering studies showed that the quantum efficiency for photoactivation is low at low light intensities, reached a maximum at intermediate intensities, and were again low at high light intensities. Equivalently, the quantum efficiency is low when saturating, single turnover flashes are given at long intervals, maximum at intermediate flash frequencies (~1 per second), and were again low when the flashes are given with short intervals between flashes. From these features, Cheniae derived a minimal model, the so-called two-quantum model that postulated the light-induced Mn assembly with at least one unstable chemical intermediate as depicted in Figure 3. The first photoevent involves the high quantum yield photooxidation of a single Mn2+ to Mn3+ ion (Ono and Mino, 1999) at the unique high affinity Mn-binding site (see below). The resultant Mn3+ species (B) can spontaneously convert to C in the dark with a 100–150 mshalf-time, with a kinetic constant designated kR in the scheme in Figure 3. A second quantum of light must be absorbed to convert the nascent complex into the first stable intermediate D as shown in Figure 3 as C⇒D. The formation of a labile intermediate, t1∕2 ~1–2 s, accounted for the optimum in light intensity or, alternatively, flash frequency, utilized for the assembly process. Photoactivation using saturating single turnover flashes is optimal with flash spacing of ~1 s, which is enough time to allow the dark rearrangement to occur (kR), but short enough to minimize the decay of the intermediate(s). If, however, the flash interval is too long, the second flash is not in time to trap forward progress and the reactants decay (kD1, KD2, Figure 3). The molecular nature of the process occurring during this dark rearrangement period (B→C) is not clear, and its understanding is key to understanding the overall molecular mechanism. After the initial two Mn are photoligated, subsequent Mn appear to be added with high quantum yield.
Figure 3.
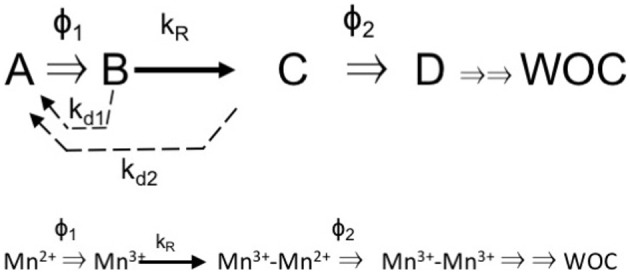
Kinetic scheme of basic two-quantum mechanism. Double arrows indicate light-activated processes with the quantum efficiencies Φ1 and Φ2 representing the first and second photooxidative events in the assembly sequence, kR representing the still ill-defined “dark” rearrangement, and kD1, kD2 representing the decay of intermediates. After the initial two Mn are photoligated, subsequent Mn appear to be added with high quantum yield.
Of the many examples providing experimental support for the two quantum mechanism, perhaps the most striking are the experiments of Miyao, which showed a minimal two quantum requirement in an experiment where as few as five flashes restored nearly 20% of the maximal activity (Miyaotokutomi and Inoue, 1992). This amounts to several percent assembly per flash, which is remarkable given that the typical per flash yield is often on the order of 1% or even lower. That experiment and others also showed that the instability of the intermediates could be minimized by preventing the back-reaction of the electrons from the acceptor side of the PSII reaction center (Miyao and Inoue, 1991; Miyaotokutomi and Inoue, 1992). This also fits with another early result showing that the intermediates of assembly are highly sensitive to reductant (Ono and Inoue, 1987) and fits with the concept that the formation of state C (eligible for utilizing the second quantum) occurs with low frequency and/or once formed, the quantum yield of photooxidation of the second Mn2+ occurs with low quantum yield (also see Miller and Brudvig, 1989, for relevant model).
It has been speculated the rearrangement (kR) involves a protein conformational change required for the binding and subsequent photooxidation of the second Mn2+ (Chen et al., 1995a; Ananyev and Dismukes, 1996b; Qian et al., 1999; Burnap, 2004). However, if (B → C) is indeed a protein structural change, then it is unlikely a large scale conformational rearrangement since carboxy terminal ligands are already close to high affinity site ligand D1-Asp170 during the first photooxidation (A⇒B) (Cohen et al., 2007). Also, whether the dark unstable intermediate is B or C (or both) remains unresolved. Given this uncertainty, Figure 3 shows both decays are possible (kD1 and kD2) (Miller and Brudvig, 1989). The development of a highly sensitive and fast Clark-type oxygen electrode (Ananyev and Dismukes, 1996b) led to the assignment of additional photoactivation intermediates and has provided alternative parameter estimates for the kinetic components (Ananyev and Dismukes, 1996a,b, 1997; Zaltsman et al., 1997; Baranov et al., 2000, 2004). At the same time, the use of this apparatus makes comparisons difficult because to the different illumination regimes. Most of the original experiments utilized single turnover Xe flashes for actinic illumination. In contrast, the photoactivation studies using the fast Clark-type oxygen electrode employed 30 ms red LED pulses promote optimum yields of assembled center (Ananyev and Dismukes, 1996b). This relatively long duration of the LED light pulses allows greater mixing of different assembly states because of the possibility of having multiple “hits” per center per pulse. That said, the 30 ms duration of the pulse is relatively short with respect to the t1∕2 ~150 ms of the B → C rearrangement and therefore the majority of those centers in the initial state that were excited (i.e., those undergoing A⇒B), will not be ready to utilize the second quantum and would thus the LED pulse would be effectively similar to a single turnover flash distributed in time over the population of centers. Variations and refinements of the original two-quantum model have been advanced based upon alternative techniques for illumination and O2 detection during photoactivation (Miller and Brudvig, 1989; Meunier et al., 1995; Ananyev and Dismukes, 1996b; Zaltsman et al., 1997; Hwang and Burnap, 2005). The multiflash experiments of Hwang and Burnap (2005) using staggered Xe single turnover flashes revealed a new kinetic intermediate, more rapid rearrangement, although where it is in the sequence could not be established owing to high miss factor (low quantum efficiency) and the associated de-phasing of the assembly during the flash induced assembly process.
Are the complicated “two-quantum” kinetics of photoactivation an artifact of in vitro experimental procedures?
Many of the insights into the mechanism, including the nature of the cofactor requirements, were from experiments performed in vitro using biochemical techniques, including detergent solublization, that yield simplified PSII preparations. Such preparations allowed a range of information from the better definition of the affinity constants for the cofactors (Tamura and Cheniae, 1986, 1987; Miller and Brudvig, 1989; Tamura et al., 1989; Ananyev and Dismukes, 1996a,b, 1997; Zaltsman et al., 1997; Baranov et al., 2000, 2004) to the comparative efficiency of artificial electron acceptors (Miyao and Inoue, 1991; Miyaotokutomi and Inoue, 1992). However, the most efficient in vitro protocols explicitly involve the remove of extrinsic proteins or involve procedures that would also cause the loss of extrinsic proteins, although the authors may not have evaluated the degree to which this loss may have occurred. Importantly, the simplified preparations also likely lack the multiple assembly factors that are now identified. Since photoactivation is a low quantum yield process, even in vivo, it has been important for technical reasons to maximize the in vitro efficiency to estimate its kinetic parameters. For example, a Mn2+ concentration dependence assays require modest increments in yield, which can only be experimentally distinguished if the procedures provide materials with sufficiently high rates of O2 evolution to allow discrimination beyond to envelope of experimental errors. Thus, in the development of the procedures, removal of the extrinsic proteins provided researchers with a system that satisfied these requirements. The mutational loss of the extrinsic proteins increases the quantum efficiency of photoactivation (Burnap et al., 1996; Shen et al., 1998) as do mutations that weaken the binding of the extrinsic proteins (Qian et al., 1997, 1999). This appears to be due greater access of the Mn2+ ions to their site of photooxidation on the donor side of PSII (Chu et al., 1994a). However, this begs the question of how much the “natural” physiological kinetics are distorted by the loss of the extrinsic proteins and what other factors that might be removed or inactivated in the process. Indeed, it is almost certain that important assembly factors may have been absent in many of the defining photoactivation experiments. From this standpoint, it is clear that the in vitro photoactivation experiments have been decidedly non-physiological. Then what are the implications for the kinetics that have defined the mechanism to date? Here it is worth noting that the basic two quantum mechanism was first discovered using intact cells and chloroplasts. This includes experiments in samples that were not extracted with chemical reductant to remove the Mn4O5Ca. For example, these include using intact chloroplasts from leaves grown under intermittent light to promote de-etiolation, but remaining un-photoactivated (Ono and Inoue, 1982, 1983) and cyanobacterial cells grown under conditions of Mn-deficiency, and dark grown (with glucose) Chlorella cells (Cheniae and Martin, 1973). These samples are likely have the full complement of extrinsic proteins and assembly factors. Similarly, the extraction of whole cyanobacterial cells and chloroplasts with the hydroxylamine probably preserves many if not most assembly factors as operational. This probably explains the fact that nearly 100% of PSII centers become reactivated by photoactivation in these more intact preparations, but in vitro preparations typically have considerably lower total yields. The more “physiological” preparations still exhibit (1) low quantum efficiency and (2) the requirement for optimal flash spacing. However, inspection of data involving dark grown Chlorella indicates that photoactivation occurs more quickly, although optimal flash spacing is still required. Clearly, the role of assembly factors needs to be further pursued. One might imagine in this case that a closely associated assembly factor, like PratA, provides Mn2+ ions at critical times during the formation of intermediates and thereby mitigates potential losses due to intermediate decay. Except for one instance where the Psb27 mutant was analyzed (Roose and Pakrasi, 2007), a careful side-by-side comparisons of photoactivation in mutants and wild-type remain to be performed.
High affinity binding site
Biochemical preparations of PSII that have been depleted of their Mn4O5Ca have been used to test the binding affinity for Mn2+. The principal finding is that a single binding site, termed the high affinity site, dominates the kinetics. The dissociation constant for Mn2+ at this site is estimated to be in the range of 0.1–2 μM (Hsu et al., 1987; Diner, 2001) and is strongly pH dependent (pKa 6–7) (Ono and Mino, 1999). As described below, these are accurate, but essentially, non-equilibrium assays. In one of the few examples of an estimate of the true equilibrium binding constant, a significantly higher value of 40–50 μM was estimated. In this case, binding was allowed to occur in the dark and the samples were frozen to −20°C, where diffusion was eliminated and an EPR binding photoxoidation signal could be detected, thereby giving a “snapshot” of the amount of photooxidizable Mn2+ bound at equilibrium (Tyryshkin et al., 2006). Notably, the binding affinity was found to be independent of the binding of Ca2+. Almost all other assays have been performed utilizing the ability of Mn2+ to donate electrons to photochemically generated . Therefore, the high affinity site has been largely defined biochemically based upon the combination of affinity and the ability to be photooxidized by during charge separation, rather that equilibrium binding assays alone (see older review Debus, 1992, for still up-to-date discussion). -This high affinity/efficient oxidation site remains intact in the mutant without processing of D1 carboxy terminus (Nixon et al., 1992), although subtle differences in the affinity characteristics of these mutants are observed when the biphasic kinetics of the binding/oxidation are fully taken into account (Cohen et al., 2007). On the other hand, the access of Mn2+ to this site is significantly increased in the carboxyterminal processing mutants as well as mutants lacking extrinsic proteins (Chu et al., 1994a; Semin et al., 2015). Mutagenesis of D1-Asp170 has shown that the residue clearly has the strongest effect upon on the affinity of Mn2+ and the ability to assemble a fully functional Mn4CaO5 cluster (Boerner et al., 1992; Diner and Nixon, 1992; Nixon and Diner, 1992; Chu et al., 1994b; Whitelegge et al., 1995; Campbell et al., 2000; Cohen et al., 2007). This is consistent with the crystal structures of PSII (Umena et al., 2011; Suga et al., 2015), which have shown that Asp170 is a ligand to the Mn(4) of assembled (intact) Mn4CaO5 cluster (Figure 2). Notably, the other carboxyl O of the D1-Asp170 side chain provides a monodentate ligand to the adjacent Ca2+ ion of the assembled cluster, which probably relates to the Ca2+ requirement photoactivation, as discussed below. Other residues, notably the other main amino acid ligand to Mn(4), D1-Glu333 affect the affinity characteristics of the high affinity binding site, but none as decisively as mutations of D1-Asp170 (Cohen et al., 2007).
D1-Glu333 is a ligand to Mn(4) presumptive the first photo-oxidized Mn2+ at the high affinity binding site (Umena et al., 2011). In all mutants of Glu333, substantial fractions of PSII complexes lack photooxidizable Mn ions in vivo (Chu et al., 1995), showing that Glu333 influences the assembly or stability of the Mn4CaO5 cluster. Nevertheless, mutations of Glu333 do not display the large changes in Mn2+ affinity compared to D1-Asp170 mutations, at least as measured using single turn-over methods to assay affinity (Nixon and Diner, 1994; Cohen et al., 2007). One possibility to accommodate these observations is that Glu333 provides some coordination of Mn2+ ions at the high affinity site, but play an even greater role in the subsequent assembly process. Pulsed electron-electron double resonance (PELDOR) experiments have recently provided the evidence to support this hypothesis that the high-affinity Mn2+ site is located at the position denoted by Mn(4) in the crystal structure and the first photooxidized Mn2+ bound to the apo-WOC is coordinated with axial ligands Asp170 and Glu333 in the D1 protein (Asada and Mino, 2015). These results thus substantiate and extend the initial assessments of the high affinity site based upon site-directed mutagenesis, yet deepen the puzzle about the seeming modest influence this axial ligand has to the affinity/photooxidation characteristics of the site. For the C-terminal residues D1-Ala344, neither mutations in the C-terminal region of D1 nor the processing of the C-terminal extension (Ala344stop and Ser345Pro mutants) has a large influence on the ability to bind and oxidize the first Mn2+ in the assembly of the cluster (Nixon et al., 1992). These observations indicate that these C-terminal residues do not participate in the coordination of the first bound Mn, though they certainly must contribute to the coordination of those bound later on in the assembly process (Diner, 2001). In the recent crystal structure of PS II, His337 residue is sufficiently close to Mn4CaO5 cluster and engage in H-bonding interactions with the μ3-oxo bridge connecting Mn(1), Mn(2), and Mn(3) (Umena et al., 2011).
Trapping intermediates of photoassembly
Britt and coworkers (Campbell et al., 2000) provided the first direct EPR spectral evidence for the initial photooxidized intermediate formed at the high affinity site in Synechocystis 6803 PSII core complexes. Conventional perpendicular-mode EPR in X-band is used to detect spin transitions in half integer spin systems which satisfy the selection rules ΔMs = ± 1. Accordingly, the first light induced Mn3+ species, due to the integer spin S = 2 of Mn3+, is an EPR-silent species for perpendicular polarization EPR spectroscopy at X-band frequencies. X-band parallel polarization EPR spectroscopy, however, can be used to investigate integral spin systems with S ≥ 1 where the spin transitions satisfy the selection rules ΔMs = ± 2 and higher. This latter technique is therefore well-suited to examine the coordination environment of this Mn3+ intermediate (high spin S = 2). A six-line signal with a hyperfine splitting of ~45 G that was only visible in parallel mode. This signal clearly arises from Mn3+ as it closely resembles that observed for Mn3+ in superoxide dismutase (Campbell et al., 1999). The parallel mode EPR spectrum of this photooxidation species consists of six well-resolved transitions split by a relatively small 55Mn hyperfine coupling (44 G). The Mn3+ parallel mode EPR signal gives an axial zero-field splitting value of D ≈−2.5 cm−1 and a rhombic zero-field splitting value of |E| ≈ 0.269 cm−1. The negative D value for this d4 ion is indicative of either an octahedral Mn3+ geometry or a five-coordinate square-pyramidal Mn3+ geometry. In contrast to wild-type, a different parallel polarization EPR signal of Mn3+ ion without a resolved hyperfine structure was observed in Asp170His mutant, suggesting a modified coordination environment of Mn3+. In the case of Asp170Glu mutant, instead of a parallel mode Mn3+ signal, a perpendicular mode signal generated by Mn4+ ion was detectable (Campbell et al., 2000), which indicates an impact of glutamate on the redox property of the photo-oxidized Mn2+ ion. As noted previously (Hoganson et al., 1989), coordination by oxo anions as would have effect of lowering the redox potential of the Mn2+ ion into the range that the oxidizing potential of .
The weak EPR signal found by Dismukes et al. in the dark apo-PSII samples upon binding of Mn2+ in the absence of Ca2+ is characterized by six-line 55Mn hyperfine structure and geffe = 8.3, which indicates a high-spin electronic ground state (S = 5/2) of Mn2+ bound in a low-symmetry environment (Ananyev and Dismukes, 1997). This signal is likely arise from a Mn2+ bound in the high-affinity site. Dismukes et al. (Ananyev et al., 1999) later suggested using competitive inhibition studies that the first species that initiated photoactivation (Ananyev et al., 1999) is hydroxide of Mn2+, [MnOH]+, bound to the apo-WOC at high affinity site. Subsequent work by the same group would provide evidence that the hydroxide formation was modulated by Ca2+ (Tyryshkin et al., 2006).
Role of Ca2+ and the Ca2+ bound intermediate
Ca2+ is an indispensable cofactor of the water-splitting Mn4CaO5 cluster. As noted in the previous section, biophysical studies attempting to trap early intermediates showed that Ca2+ exerts pronounced, and possibly physiologically significant effects upon the structure of Mn ions undergoing photooxidation at the high affinity site. However, from the biochemical perspective, the roles of Ca2+ ion in the process of photoactivation initially appeared contradictory: A requirement for Ca2+ in photoactivation was also noted using cyanobacterial preparations (Pistorius and Schmid, 1984). Ono and Inoue (1983) proposed that photoactivation occurs in one stage with Ca2+ essential for the assembly process itself using isolated intact chloroplasts depleted of Mn. According to the one-stage model the first-order rate constant for the assembly of O2-evolving centers is dependent on the extent of occupancy of both Mn2+ and Ca2+ bound to their specific binding sites during photoactivation. Later experiments seemed to indicate that the Ca2+-binding site is “created” during the photoassembly (Shinohara et al., 1992). For example, Tamura and Cheniae (1987) found that only light and Mn2+ were essential for Mn re-ligation to the apo-WOC-PSII, but Ca2+ addition was required for maximal expression of water oxidation activity by the photoligated Mn. In other words, it appeared that Ca2+ was not required for proper assembly, but was needed as a cofactor that readily diffused into its site of action after the assembly of the Mn cluster was completed and, once in place, activated its catalytic function. However, this conclusion was later modified to account for the complicating effects of the artificial electron acceptor used in the assay (Chen et al., 1995a). Ultimately, it was thus concluded that Ca2+ is indeed absolutely required during the assembly of functional clusters, not simply being added in after assembly (Chen et al., 1995a). The same work provides what may be another important clue about the role of Ca2+. It was found that photoactivation of PSII membranes in the absence of Ca2+ led to the formation of inactive PSII with more than four Mn ion per PSII center (5–10 non-functional Mn per PSII). Thus, when Ca2+ is left out of the photoactivation medium, binding and photooxidation of many more Mn2+ to the apo-WOC-PSII protein occurs, but no O2 evolution activity is observable (Chen et al., 1995a). Manganese bound in this way could be released with reductant indicating that it was bound oxidatively, but it clearly cannot bind to specific protein binding sites. Instead, probably resembles amorphous oxides which are multinucleate metal-oxo deposits produced by inorganic processes (Sauer and Yachandra, 2002). This suggests that one role for Ca2+ is to guide assembly or simply block Mn2+ oxidation at the Ca2+ site, which prevents “inappropriately assembled” Mn (Chen et al., 1995a). Interestingly, mutants that are defective in processing the D1 carboxy terminus also seem to assemble centers with excess Mn (Seibert et al., 1989). Recent X-ray crystallographic studies (Umena et al., 2011) provides a structural explanation for the Seibert result. The PSII structure reveals that mature D1 C-terminal residue Ala344 ligate the Ca2+ and Mn(2) of Mn4CaO5 cluster, thus without the availability of the mature C-terminus Ala344, Ca2+ cannot bind and the destructive photoligation of Mn2+ to inappropriate sites can proceed. Additionally, competition between Ca2+ and Mn2+ for each other's binding sites has been indicated by many studies (Cheniae and Martin, 1971b; Radmer and Cheniae, 1971; Ono and Inoue, 1983; Tamura and Cheniae, 1987; Miller and Brudvig, 1989; Chen et al., 1995a,b; Ananyev and Dismukes, 1996a; Zaltsman et al., 1997). Since Sr2+ can substitute of Ca2+ in PSII in vivo, albeit with impaired H2O-oxidation activity (Boussac et al., 2004), it would be interesting to see how photoactivation occurs with this substitution. However, only limited information is currently available (Ananyev et al., 2001).
The effect of Ca2+ on the formation of the first photoactivation intermediate, corresponding to a photooxidized mononuclear Mn3+ species bound to apo-WOC-PSII, was investigated by EPR spectroscopy (Tyryshkin et al., 2006). In the absence of Ca2+, the Mn3+ species was found to be generated as two forms in a pH-dependent equilibrium: an EPR-invisible low-pH form and an EPR-visible high-pH form. Note, these spectra of Mn3+ species were acquired in parallel mode and EPR invisible vs. visible is attributable to changes in the influences in ligand environment rather than the spin state selection rules noted above. The conversion between the visible and invisible forms occurs by deprotonation of an ionizable ligand bound to Mn3+, postulated to be a H2O molecule: [Mn3+(OH2)]↔[Mn3+(OH−)]. The EPR-visible high-pH form exhibits a strong pH effect (pH 6.5–9) on Mn3+ spectral parameters, including the rhombicity (δ) derived from center field position (geff), the 55Mn hyperfine coupling (AZ), and the signal intensity. A pH-induced protein conformational change was proposed to account for the observed significant changes in the symmetry of the ligand field at the Mn3+ site. On the other hand, the EPR-detectable Mn3+ induced in the presence of Ca2+, exhibits a greatly weakened pH dependence of its ligand-field symmetry with reduced variation of rhombicity δ and 55Mn hyperfine coupling AZin the pH range of 6.5–9.0. Moreover, the addition of Ca2+ moves both geff and AZ to a range of values observed at alkaline pH ≥ 9 without added Ca2+, indicating that Ca2+ binding exerts an influence on the coordination shell of Mn3+ species equivalent to the alkaline pH effect in the absence of Ca2+. Therefore, it was proposed that Ca2+ binding induces a second ionization of the bridging hydroxo ligand bound to Mn3+ resulting in the formation of a bridging oxide ion ([Mn3+(OH−)-Ca2+] ↔ [Mn3+(O2−)-Ca2+]). The proton ionization of the water ligand is postulated to be controlled by a nearby base B−, which serves as an immediate proton acceptor with a pKa that depends upon the occupancy of the Ca2+ effector site. Looking at the current crystal structure and assuming a similar, although almost certainly not identical, spatial configuration of the Ca and Mn, the inferred oxo bridge would join these ions with D1-Asp170, Glu333 and the carboxyl terminus in proximity of one another.
Other inorganic cofactors
There is a long debated role of inorganic carbon in photosynthetic water oxidation. Recently, it has been demonstrate that bicarbonate () can act as a mobile acceptor and transporter of protons produced by photosynthetic water oxidation PSII (Koroidov et al., 2014). Bicarbonate also seem to have an impact on photoassembly of Mn4CaO5 cluster (reviewed in Dasgupta et al., 2008). The proposed roles of bicarbonate in facilitating assembly of Mn4CaO5 cluster during PSII repair include acceleration of the binding and photooxidation of the first Mn2+ at the high affinity Mn site, putatively by increase the location concentration of Mn2+ and even direct ligation to Mn2+ (Baranov et al., 2000, 2004; Dasgupta et al., 2007). Bicarbonate has been found not an essential constituent of the WOC of PSII based on the most recent PSII crystal structure (Umena et al., 2011), making the direct ligation seem unlikely. However, this does not necessarily mean that the possibility of a weakly bound at the donor side affecting the PSII repair has been excluded. It is also important to note that high concentrations of Cl− also enhance photoactivation in vitro, but this appears to be an effect distinct from the known effects on the activation of the photosynthetic water oxidation catalytic activity. Instead, it more likely relates to the stabilization of the Mn4CaO5 in the absence of the extrinsic proteins in the studied photoactivation reactions (Miyao and Murata, 1985; Miyaotokutomi and Inoue, 1992). The catalytic activation properties of Cl− are likely to be exerted indirectly upon water and/or proton movement during H2O-oxidation given it binding locations in the second ligation sphere of the assembled Mn4O5Ca.
Possible models of assembly
Although the two-quantum model remains solidly at the foundation of our understanding of photoactivation, the molecular mechanisms that give rise to these kinetic features remain almost completely unresolved. A crucial question is what molecular processes gives rise to the so-called dark rearrangement. As discussed, consistent models would require occupancy of the high affinity site by Mn2+ and the presence Ca2+ at a nearby site, and the Ca2+ probably needs to be present during the initial photooxidation of the Mn2+. This first photooxidation (A⇒B) likely occurs with high quantum efficiency, yet the photooxidation of the second Mn2+ (C⇒D) occurring after the rearrangement occurs with low quantum efficiency. There are essentially two general alternative hypotheses accounting for the low quantum yield of C⇒D that depend, in part, on the nature of the rearrangement. First, the rearrangement after A⇒B forms a binding site for the second Mn2+ that is not optimal for electron transfer to , perhaps because it is further away from the high affinity site and because of this, charge recombination effectively competes with the low probability of photooxidation of the second Mn2+ at the new site. Please note that the charge recombination may actually be the ‘rearrangement’ and could be important for removing mis-assembled clusters (Hwang et al., 2007). Second, the rearrangement is again a slow process, but this time leads to formation of efficient site for Mn2+ photooxidation, but its initial product, the first Mn3+ fails to convert efficiently into a stable chemical product. This could happen if, for example, a requisite oxo bridge forms inefficiently and the new Mn3+ diffuses away or is re-reduced. Based upon the finding that the C-terminus of D1 is already in a conformation close to the final configuration in the fully assembled enzyme (Cohen et al., 2007), then the dark rearrangement is unlikely to be a major rearrangement of the D1 polypeptide backbone. With this constraint, we imagine these two alternative assembly mechanisms as follows. One alternative is that the slow rearrangement corresponds to slow oxo bridge formation chemistry that is kinetically very sluggish, occurring with the rate constant kR after the initial photooxidation. Further, once the first Mn3+ is produced at the high affinity site, the new binding site for the second Mn2+ is at a more distal site where the quantum yield of its photooxidation lower. In this case, the first Mn3+ at the high affinity site is already engaged oxo linkage with the Ca2+ (Tyryshkin et al., 2006) and one of its other coordination positions occupied by a water must deprotonate to form a stable linkage with the second Mn2+. According to this hypothesis, the slow formation of the second oxo corresponds to kR and the low quantum yield of C⇒D is due to the more distal location from . However, one problem with this model is that each of the subsequent two Mn additions would seem to have to also occur with low quantum yields, since the high affinity site remains occupied. However, these later photoligations are thought to occur with high quantum efficiency, although the evidence even for that remains sparse. A second alternative to a primarily protein rearrangement is a rearrangement of the ions. As mentioned above, the high affinity site has been largely defined biochemically based upon the combination of affinity and the ability to be photooxidized by during charge separation. Perhaps the high affinity site remains the site of oxidation for each of the photooxidations for the assembly reactions and the resultant Mn3+ ions migrate to their final locations. In this model, the relocation of the Mn ion, vacating the high affinity site, and its coordination into its new site accounts for the “dark rearrangement.” However, the dissociation of the Mn3+ dissociates from its initial binding and oxidation site presents a problem because of the likely increase in ligand field stabilization energy. This would not be a problem for Mn2+, which has no associated LFSE. On the other hand, Mn3+ should bind with a substantially higher affinity owing to the acquisition of LFSE in the higher oxidation state. There may be meahisms to alleviate this problem such as a redox disproportionation of bound Mn3+-Mn2+ to (Mn2+-Mn3+ so the original Mn3+ is now weakly bound (no LFSE) and can exchange to another site. The EPR observations consistent with oxo bridge formation Ca2+ ion accompanying the photooxidation of Mn2+ ions at the high affinity site suggest a templating function for Ca2+ during assembly. Based upon the crystal structure, the Ca2+ would be captured between D1-Asp170 and the carboxyterminal carboxylate of the D1 protein. As noted, there is good evidence that Ca2+ binding induces a second ionization of the bridging hydroxo ligand bound to Mn3+ resulting in the formation of a bridging oxide ion ([Mn3+(OH−)-Ca2+] ↔ [Mn3+(O2−)-Ca2+]) (Tyryshkin et al., 2006). If the Mn3+ leaves the high affinity site, then Ca2+ may tether the new Mn with an oxo (or hydroxo) bridge and facilitate the movement to the next site. Moreover, the water molecules coordinated Ca2+ may be provided as the substrate for additional μ-oxo bridge formation. In this way, Ca2+ functions in a manner similar to proposals for its role in catalytic water oxidation and substrate exchange though the positioning of coordinated waters (Vrettos et al., 2001; Hillier and Wydrzynski, 2008; Rappaport et al., 2011; Cox and Messinger, 2013). According to electrostatic calculations, there are substantial differences in the redox potentials for each of the four spatially distributed Mn ions of the assembled Mn4CaO5 cluster due to their specific coordination and large electrostatic environments (Amin et al., 2015). Based upon these electrostatic calculations of Amin et al. (2015), it is likely that Mn3+ ions are thermodynamically more stable at coordination positions elsewhere within the partially assembled cluster in comparison to their primary site of oxidation at the high affinity site. It also fits with a variety of biochemical studies showing that one pair of Mn ions, presumably a binuclear di-μ-oxo bridged unit, in the assembled cluster is more stable and has different accessibility to external reductants than the other pair (Frankel and Bricker, 1989; Mei and Yocum, 1991, 1992; Riggs et al., 1992). In this model, the rearrangement time may correspond to the relocation of the first Mn3+ ion to exit the high affinity site and relocate to another site, perhaps guided via a nascent oxo with the Ca2+.
Author contributions
HB researched the topic and wrote the initial draft of the manuscript. RB added materialand made figures, performed additional research and edited the draft.
Funding
The work was generously funded by a grant form the National Science Foundation (MCB-1244586).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Aparna Nagarajan for providing the initial sketch of Figure 1. We also thank reviewer 2 for the pointing out the issue with our proposed rearrangement model regarding ligand field stabilization energy and suggesting redox disproportionation as a possible mechanism. The work was funded by the National Science Foundation MCB-1244586.
References
- Adir N., Zer H., Shochat S., Ohad I. (2003). Photoinhibition - a historical perspective. Photosyn. Res. 76, 343–370. 10.1023/A:1024969518145 [DOI] [PubMed] [Google Scholar]
- Amin M., Vogt L., Szejgis W., Vassiliev S., Brudvig G. W., Bruce D., et al. (2015). Proton-coupled electron transfer during the S-state transitions of the oxygen-evolving complex of photosystem II. J. Phys. Chem. B. 119, 7366–7377. 10.1021/jp510948e [DOI] [PubMed] [Google Scholar]
- Ananyev G. M., Dismukes G. C. (1996a). Assembly of the tetra Mn site of photosynthetic water oxidation by photoactivation: Mn stoichiometry and detection of a new intermediate. Biochemistry 35, 4102–4109. 10.1021/bi952667h [DOI] [PubMed] [Google Scholar]
- Ananyev G. M., Dismukes G. C. (1996b). High-resolution kinetic studies of the reassembly of the tetra-manganese cluster of photosynthetic water oxidation: proton equilibrium, cations, and electrostatics. Biochemistry 35, 14608–14617. 10.1021/bi960894t [DOI] [PubMed] [Google Scholar]
- Ananyev G. M., Dismukes G. C. (1997). Calcium induces binding and formation of a spin-coupled dimanganese(II,II) center in the apo-water oxidation complex of photosystem II as precursor to the functional tetra-Mn/Ca cluster. Biochemistry 36, 11342–11350. 10.1021/bi970626a [DOI] [PubMed] [Google Scholar]
- Ananyev G. M., Murphy A., Abe Y., Dismukes G. C. (1999). Remarkable affinity and selectivity for Cs+ and uranyl () binding to the manganese site of the apo-water oxidation complex of photosystem II. Biochemistry 38, 7200–7209. 10.1021/bi990023u [DOI] [PubMed] [Google Scholar]
- Ananyev G. M., Zaltsman L., Vasko C., Dismukes G. C. (2001). The inorganic biochemistry of photosynthetic oxygen evolution/water oxidation. Biochim. Biophys. Acta 1503, 52–68. 10.1016/S0005-2728(00)00215-2 [DOI] [PubMed] [Google Scholar]
- Anbudurai P. R., Mor T. S., Ohad I., Shestakov S. V., Pakrasi H. B. (1994). The ctpA gene encodes the C-terminal processing protease for the D1 protein of the photosystem II reaction center complex. Proc. Natl. Acad. Sci. U.S.A. 91, 8082–8086. 10.1073/pnas.91.17.8082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aro E. M., Virgin I., Andersson B. (1993). Photoinhibition of photosystem II. Inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143, 113–134. 10.1016/0005-2728(93)90134-2 [DOI] [PubMed] [Google Scholar]
- Asada M., Mino H. (2015). Location of the High-Affinity Mn(2+) Site in Photosystem II Detected by PELDOR. J. Phys. Chem. B 119, 10139–10144. 10.1021/acs.jpcb.5b03994 [DOI] [PubMed] [Google Scholar]
- Bailey S., Thompson E., Nixon P. J., Horton P., Mullineaux C. W., Robinson C., et al. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277, 2006–2011. 10.1074/jbc.M105878200 [DOI] [PubMed] [Google Scholar]
- Baranov S. V., Ananyev G. M., Klimov V. V., Dismukes G. C. (2000). Bicarbonate accelerates assembly of the inorganic core of the water- oxidizing complex in manganese-depleted photosystem II: a proposed biogeochemical role for atmospheric carbon dioxide in oxygenic photosynthesis. Biochemistry 39, 6060–6065. 10.1021/bi992682c [DOI] [PubMed] [Google Scholar]
- Baranov S. V., Tyryshkin A. M., Katz D., Dismukes G. C., Ananyev G. M., Klimov V. V. (2004). Bicarbonate is a native cofactor for assembly of the manganese cluster of the photosynthetic water oxidizing complex. Kinetics of reconstitution of O2 evolution by photoactivation. Biochemistry 43, 2070–2079. 10.1021/bi034858n [DOI] [PubMed] [Google Scholar]
- Becker K., Cormann K. U., Nowaczyk M. M. (2011). Assembly of the water-oxidizing complex in photosystem II. J. Photochem. Photobiol. B. Biol. 104, 204–211. 10.1016/j.jphotobiol.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Björkman O. (1981). Responses to different quantum flux densities, in Physiological Plant Ecology I: Responses to the Physical Environment, eds Lange O. L., Nobel P. S., Osmond C. B., Ziegler H. (Berlin; Heidelberg: Springer; ), 57–107. 10.1007/978-3-642-68090-8_4 [DOI] [Google Scholar]
- Boerner R. J., Nguyen A. P., Barry B. A., Debus R. J. (1992). Evidence from directed mutagenesis that aspartate 170 of the D1 polypeptide influences the assembly and-or stability of the manganese cluster in the photosynthetic water-splitting complex. Biochemistry 31, 6660–6672. 10.1021/bi00144a005 [DOI] [PubMed] [Google Scholar]
- Boussac A., Rappaport F., Carrier P., Verbavatz J. M., Gobin R., Kirilovsky D., et al. (2004). Biosynthetic Ca2+/Sr2+ exchange in the photosystem II oxygen evolving enzyme of Thermosynechococcus elongatus. J. Biol. Chem. 279, 22809–22819. 10.1074/jbc.M401677200 [DOI] [PubMed] [Google Scholar]
- Burnap R. L. (2004). D1 protein processing and Mn cluster assembly in light of the emerging photosystem II structure. Phys. Chem. Chem. Phys. 6, 4803–4809. 10.1039/b407094a [DOI] [Google Scholar]
- Burnap R. L., Qian M., Pierce C. (1996). The manganese-stabilizing protein (MSP) of photosystem II modifies the in vivo deactivation and photoactivation kinetics of the H2O-oxidation complex in Synechocystis sp. PCC6803. Biochemistry 35, 874–882. 10.1021/bi951964j [DOI] [PubMed] [Google Scholar]
- Campbell K. A., Force D. A., Nixon P. J., Dole F., Diner B. A., Britt R. D. (2000). Dual-mode EPR detects the initial intermediate in photoassembly of the photosystem II Mn cluster: the influence of amino acid residue 170 of the D1 polypeptide on Mn coordination. Biochemistry 122, 3754–3761. 10.1021/ja000142t [DOI] [Google Scholar]
- Campbell K. A., Yikilmaz E., Grant C. V., Gregor W., Miller A.-F., Britt R. D. (1999). Parallel polarization EPR characterization of the Mn(III) center of oxidized manganese superoxide dismutase. J. Am. Chem. Soc. 121, 4714–4715. 10.1021/ja9902219 [DOI] [Google Scholar]
- Chen C., Kazimir J., Cheniae G. M. (1995a). Calcium modulates the photoassembly of photosystem II (Mn)4 clusters by preventing ligation of nonfunctional high valency states of manganese. Biochemistry 34, 13511–13526. [DOI] [PubMed] [Google Scholar]
- Chen G. X., Blubaugh D. J., Homann P. H., Golbeck J. H., Cheniae G. M. (1995b). Superoxide contributes to the rapid inactivation of specific secondary donors of the photosystem II reaction center during photodamage of manganese depleted photosystem II membranes. Biochemistry 3470, 2317–2332. 10.1021/bi00007a028 [DOI] [PubMed] [Google Scholar]
- Chen H., Zhang D., Guo J., Wu H., Jin M., Lu Q., et al. (2006). A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol. Biol. 61, 567–575. 10.1007/s11103-006-0031-x [DOI] [PubMed] [Google Scholar]
- Cheniae G. M., Martin I. F. (1971a). Effects of hydroxylamine on photosystem II. I. Factors affecting the decay of O2 evolution. Plant Physiol. 47, 568–575. 10.1104/pp.47.4.568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheniae G. M., Martin I. F. (1971b). Photoactivation of the manganese catalyst of O2 evolution. I. Biochemical and kinetic aspects. Biochim. Biophys. Acta 253, 167–181. 10.1016/0005-2728(71)90242-8 [DOI] [PubMed] [Google Scholar]
- Cheniae G. M., Martin I. F. (1972). Effects of hydroxylamine on photosystem II. II Photoreversal of the NH2OH destruction of O2 evolution. Plant Physiol. 50, 87–94. 10.1104/pp.50.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheniae G. M., Martin I. F. (1973). Absence of oxygen-evolving capacity in dark-grown Chlorella: the photoactivation of oxygen evolving centers. Photochem. Photobiol. 17, 441–459. 10.1111/j.1751-1097.1973.tb06378.x [DOI] [Google Scholar]
- Chu H.-A., Nguyen A. P., Debus R. J. (1994a). Site-directed mutagenesis of photosynthetic oxygen evolution: increased binding or photooxidation of manganese in the absence of the extrinsic 33-kDa polypeptide in vivo. Biochemistry 33, 6150–6157. 10.1021/bi00186a014 [DOI] [PubMed] [Google Scholar]
- Chu H.-A., Nguyen A. P., Debus R. J. (1994b). Site-directed mutagenesis of photosynthetic oxygen evolution: instability or inefficient assembly of the manganese cluster in vivo. Biochemistry 33, 6137–6149. 10.1021/bi00186a013 [DOI] [PubMed] [Google Scholar]
- Chu H. A., Nguyen A. P., Debus R. J. (1995). Amino acid residues that influence the binding of manganese or calcium to photosystem II. 2. The carboxy terminal domain of the D1 polypeptide. Biochemistry 3496, 5859–5882. 10.1021/bi00017a017 [DOI] [PubMed] [Google Scholar]
- Cohen R. O., Nixon P. J., Diner B. A. (2007). Participation of the C-terminal region of the D1-polypeptide in the first steps in the assembly of the Mn4Ca cluster of photosystem II. J. Biol. Chem. 282, 7209–7218. 10.1074/jbc.M606255200 [DOI] [PubMed] [Google Scholar]
- Cox N., Messinger J. (2013). Reflections on substrate water and dioxygen formation. Biochim. Biophys. Acta 1827, 1020–1030. 10.1016/j.bbabio.2013.01.013 [DOI] [PubMed] [Google Scholar]
- Dasgupta J., Ananyev G. M., Dismukes G. C. (2008). Photoassembly of the water-oxidizing complex in photosystem II. Coord. Chem. Rev. 252, 347–360. 10.1016/j.ccr.2007.08.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta J., Tyryshkin A. M., Dismukes G. C. (2007). ESEEM spectroscopy reveals carbonate and an N-donor protein-ligand binding to Mn2+ in the photoassembly reaction of the Mn4Ca cluster in photosystem II. Angew. Chem. Int. Ed Engl. 46, 8028–8031. 10.1002/anie.200702347 [DOI] [PubMed] [Google Scholar]
- Debus R. J. (1992). The manganese and calcium ions of photosynthetic oxygen evolution. Biochim. Biophys. Acta 1102, 269–352. 10.1016/0005-2728(92)90133-M [DOI] [PubMed] [Google Scholar]
- Diner B. A. (2001). Amino acid residues involved in the coordination and assembly of the manganese cluster of photosystem II. Proton-coupled electron transport of the redox-active tyrosines and its relationship to water oxidation. Biochim. Biophys. Acta 1503, 147–163. 10.1016/S0005-2728(00)00220-6 [DOI] [PubMed] [Google Scholar]
- Diner B. A., Nixon P. J. (1992). The rate of reduction of oxidized redox-active tyrosine, Z+, by exogenous Mn2+ Is slowed in a site-directed mutant, at aspartate 170 of polypeptide D1 of photosystem II, inactive for photosynthetic oxygen evolution. Biochim. Biophys. Acta 1101, 134–138. 10.1016/0005-2728(92)90196-9 [DOI] [Google Scholar]
- Diner B. A., Nixon P. J., Farchaus J. W. (1991). Site-directed mutagenesis of photosynthetic reaction centers. Curr. Opin. Struct. Biol. 1, 546–554. 10.1016/S0959-440X(05)80076-4 [DOI] [Google Scholar]
- Diner B. A., Ries D. F., Cohen B. N., Metz J. G. (1988a). Carboxyl-terminal processing of polypeptide D of the photosystem II reaction center of Scenedesmus obliquus is necessary for the assembly of the oxygen-evolving complex. J. Biol. Chem. 263, 8972–8980. [PubMed] [Google Scholar]
- Diner B. A., Ries D. F., Cohen B. N., Metz J. G. (1988b). COOH-terminal processing of polypeptide D1 of the photosystem II reaction center of Scenedesmus obliquus is necessary for the assembly of the oxygen-evolving complex. J. Biol. Chem. 263, 8972–8980. [PubMed] [Google Scholar]
- Dismukes C. G., Ananyev G., Watt R. (2005). Photo-assembly of the catalytic manganese cluster, in Photosystem II, eds Wydrzynski T., Satoh K., Freeman J. (Dordrecht: Springer; ), 609–626. [Google Scholar]
- Edelman M., Mattoo A. K. (2008). D1-protein dynamics in photosystem II: the lingering enigma. Photosyn. Res. 98, 609–620. 10.1007/s11120-008-9342-x [DOI] [PubMed] [Google Scholar]
- Frankel L. K., Bricker T. M. (1989). Epitope mapping of the monoclonal antibody FAC2 on the apoprotein of Cpa-1 in photosystem II. FEBS Lett. 257, 279–282. 10.1016/0014-5793(89)81552-2 [DOI] [PubMed] [Google Scholar]
- Heinz S., Liauw P., Nickelsen J., Nowaczyk M. (2016). Analysis of photosystem II biogenesis in cyanobacteria. Biochim. Biophy. Acta 1857, 274–287. 10.1016/j.bbabio.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Hillier W., Wydrzynski T. (2008). O18-Water exchange in photosystem II: substrate binding and intermediates of the water splitting cycle. Coord. Chem. Rev. 252, 306–317. 10.1016/j.ccr.2007.09.004 [DOI] [Google Scholar]
- Hoganson C. W., Ghanotakis D. F., Babcock G. T., Yocum C. F. (1989). Manganese ion reduces redox activated tyrosine in manganese-depleted photosystem II preparations. Photosyn. Res. 22, 285–294. 10.1007/BF00048306 [DOI] [PubMed] [Google Scholar]
- Hsu B. D., Lee J. Y., Pan R. L. (1987). The high-affinity binding-site for manganese on the oxidizing side of photosystem-Ii. Biochim. Biophys. Acta 890, 89–96. 10.1016/0005-2728(87)90072-7 [DOI] [PubMed] [Google Scholar]
- Hwang H. J., Burnap R. L. (2005). Multiflash experiments reveal a new kinetic phase of photosystem II manganese cluster assembly in Synechocystis sp PCC6803 in vivo. Biochemistry 44, 9766–9774. 10.1021/bi050069p [DOI] [PubMed] [Google Scholar]
- Hwang H. J., McLain A., Debus R. J., Burnap R. L. (2007). Photoassembly of the manganese cluster in mutants perturbed in the high affinity Mn-binding site of the H2O-oxidation complex of photosystem II. Biochemistry 46, 13648–13657. 10.1021/bi700761v [DOI] [PubMed] [Google Scholar]
- Hwang H. J., Nagarajan A., McLain A., Burnap R. L. (2008). Assembly and disassembly of the photosystem II manganese cluster reversibly alters the coupling of the reaction center with the light-harvesting phycobilisome. Biochemistry 47, 9747–9755. 10.1021/bi800568p [DOI] [PubMed] [Google Scholar]
- Inagaki N., Maitra R., Satoh K., Pakrasi H. B. (2001). Amino acid residues that are critical for in vivo catalytic activity of CtpA, the carboxyl-terminal processing protease for the D1 protein of photosystem II. J. Biol. Chem. 14, 14 10.1074/jbc.M102600200 [DOI] [PubMed] [Google Scholar]
- Ivleva N. B., Shestakov S. V., Pakrasi H. B. (2000). The carboxyl-terminal extension of the precursor D1 protein of photosystem II is required for optimal photosynthetic performance of the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 124, 1403–1412. 10.1104/pp.124.3.1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashino Y., Lauber W. M., Carroll J. A., Wang Q., Whitmarsh J., Satoh K., et al. (2002). Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012. 10.1021/bi026012+ [DOI] [PubMed] [Google Scholar]
- Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. (2009). The variegated mutants lacking chloroplastic FtsHs are defective in D1 degradation and accumulate reactive oxygen species. Plant Physiol. 151, 1790–1801. 10.1104/pp.109.146589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren N., Ohkawa H., Welsh E. A., Liberton M., Pakrasi H. B. (2005). Psb29, a conserved 22-kD protein, functions in the biogenesis of Photosystem II complexes in Synechocystis and Arabidopsis. Plant Cell 17, 2768–2781. 10.1105/tpc.105.035048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert B., Ossenbühl F., Sikorski M., Berry S., Eichacker L., Nickelsen J. (2004). PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 279, 44639–44644. 10.1074/jbc.M405393200 [DOI] [PubMed] [Google Scholar]
- Koivuniemi A., Aro E. M., Andersson B. (1995). Degradation of the D1- and D2-proteins of photosystem II in higher plants is regulated by reversible phosphorylation. Biochemistry 34, 16022–16029. 10.1021/bi00049a016 [DOI] [PubMed] [Google Scholar]
- Komenda J., Barber J. (1995). Comparison of psbO and psbH deletion mutants of Synechocystis PCC 6803 indicates that degradation of D1 protein is regulated by the QB site and dependent on protein synthesis. Biochemistry 34, 9625–9631. 10.1021/bi00029a040 [DOI] [PubMed] [Google Scholar]
- Komenda J., Barker M., Kuviková S., de Vries R., Mullineaux C. W., Tichy M., et al. (2006). The FtsH protease slr0228 is important for quality Control of photosystem II in the thylakoid membrane of Synechocystis sp. PCC 6803. J. Biol. Chem. 281, 1145–1151. 10.1074/jbc.M503852200 [DOI] [PubMed] [Google Scholar]
- Komenda J., Knoppová J., Krynická V., Nixon P. J., Tichý M. (2010). Role of FtsH2 in the repair of Photosystem II in mutants of the cyanobacterium Synechocystis PCC 6803 with impaired assembly or stability of the CaMn4 cluster. Biochim. Biophys. Acta 1797, 566–575. 10.1016/j.bbabio.2010.02.006 [DOI] [PubMed] [Google Scholar]
- Komenda J., Kuviková S., Granvogl B., Eichacker L. A., Diner B. A., Nixon P. J. (2007). Cleavage after residue Ala352 in the C-terminal extension is an early step in the maturation of the D1 subunit of Photosystem II in Synechocystis PCC 6803. Biochim. Biophys. Acta 1767, 829–837. 10.1016/j.bbabio.2007.01.005 [DOI] [PubMed] [Google Scholar]
- Komenda J., Tichý M., Eichacker L. A. (2005). The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 46, 1477–1483. 10.1093/pcp/pci159 [DOI] [PubMed] [Google Scholar]
- Koroidov S., Shevela D., Shutova T., Samuelsson G., Messinger J. (2014). Mobile hydrogen carbonate acts as proton acceptor in photosynthetic water oxidation. Proc. Natl. Acad. Sci. U.S.A. 111, 6299–6304. 10.1073/pnas.1323277111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krynická V., Tichý M., Krafl J., Yu J., Kana R., Boehm M., et al. (2014). Two essential FtsH proteases control the level of the Fur repressor during iron deficiency in the cyanobacterium Synechocystis sp. PCC 6803. Mol. Microbiol. 94, 609–624. 10.1111/mmi.12782 [DOI] [PubMed] [Google Scholar]
- Kuviková S., Tichý M., Komenda J. (2005). A role of the C-terminal extension of the photosystem II D1 protein in sensitivity of the cyanobacterium Synechocystis PCC 6803 to photoinhibition. Photochem. Photobiol. Sci. 4, 1044–1048. 10.1039/b506059a [DOI] [PubMed] [Google Scholar]
- Liu H., Huang R. Y., Chen J., Gross M. L., Pakrasi H. B. (2011a). Psb27, a transiently associated protein, binds to the chlorophyll binding protein CP43 in photosystem II assembly intermediates. Proc. Natl. Acad. Sci. U.S.A. 108, 18536–18541. 10.1073/pnas.1111597108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Roose J. L., Cameron J. C., Pakrasi H. B. (2011b). A genetically tagged Psb27 protein allows purification of two consecutive photosystem II (PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium. J. Biol. Chem. 286, 24865–24871. 10.1074/jbc.M111.246231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupinkova L., Komenda J. (2004). Oxidative modifications of the Photosystem II D1 protein by reactive oxygen species: from isolated protein to cyanobacterial cells. Photochem. Photobiol. 79, 152–162. 10.1111/j.1751-1097.2004.tb00005.x [DOI] [PubMed] [Google Scholar]
- Mamedov F., Nowaczyk M. M., Thapper A., Rogner M., Styring S. (2007). Functional characterization of monomeric photosystem II core preparations from Thermosynechococcus elongatus with or without the Psb27 protein. Biochemistry 46, 5542–5551. 10.1021/bi7000399 [DOI] [PubMed] [Google Scholar]
- Mann N. H., Novac N., Mullineaux C. W., Newman J., Bailey S., Robinson C. (2000). Involvement of an FtsH homologue in the assembly of functional photosystem I in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 479, 72–77. 10.1016/S0014-5793(00)01871-8 [DOI] [PubMed] [Google Scholar]
- Mei R., Yocum C. F. (1991). Calcium retards NH2OH inhibition of O2 evolution activity by stabilization of Mn2+ binding to Photosystem II. Biochemistry 30, 7863–7842. 10.1021/bi00245a025 [DOI] [PubMed] [Google Scholar]
- Mei R., Yocum C. F. (1992). Comparative properties of hydroquinone and hydroxylamine reduction of the Ca2+stablilized O2-evolving complex of photosystem II: reductant-dependent Mn2+ formation and activity inhibition. Biochemistry 31, 8449–8454. 10.1021/bi00151a009 [DOI] [PubMed] [Google Scholar]
- Meunier P. C., Burnap R. L., Sherman L. A. (1995). Modelling of the S-state mechanism and Photosystem II manganese photoactivation in cyanobacteria. Photosyn. Res. 47, 61–76. 10.1007/BF00017754 [DOI] [PubMed] [Google Scholar]
- Miller A. F., Brudvig G. W. (1989). Manganese and calcium requirements for reconstitution of oxygen evolution activity in manganese-depleted photosystem II membranes. Biochemistry 28, 8181–8190. 10.1021/bi00446a033 [DOI] [PubMed] [Google Scholar]
- Miyao M., Inoue Y. (1991). An improved procedure for photoactivation of photosynthetic oxygen evolution - effect of artificial electron-acceptors on the photoactivation yield of NH2OH -treated wheat photosystem II membranes. Biochim. Biophys. Acta 1056, 47–56. 10.1016/S0005-2728(05)80071-4 [DOI] [Google Scholar]
- Miyao M., Murata N. (1985). The Cl- effect on photosynthetic oxygen evolution: interaction of Cl- with 18-kDa, 24-kDa and 33-kDa proteins. FEBS Lett. 180, 303–308. 10.1016/0014-5793(85)81091-7 [DOI] [Google Scholar]
- Miyaotokutomi M., Inoue Y. (1992). Improvement by benzoquinones of the quantum yield of photoactivation of photosynthetic oxygen evolution - direct evidence for the 2-quantum mechanism. Biochemistry 31, 526–532. 10.1021/bi00117a032 [DOI] [PubMed] [Google Scholar]
- Nagarajan A., Burnap R. L. (2014). Parallel expression of alternate forms of psbA2 gene provides evidence for the existence of a targeted D1 repair mechanism in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1837, 1417–1426. 10.1016/j.bbabio.2014.02.022 [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Rengstl B. (2013). Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 609–635. 10.1146/annurev-arplant-050312-120124 [DOI] [PubMed] [Google Scholar]
- Nickelsen J., Rengstl B., Stengel A., Schottkowski M., Soll J., Ankele E. (2011). Biogenesis of the cyanobacterial thylakoid membrane system–an update. FEMS Microbiol. Lett. 315, 1–5. 10.1111/j.1574-6968.2010.02096.x [DOI] [PubMed] [Google Scholar]
- Nilsson R., Brunner J., Hoffman N. E., van Wijk K. J. (1999). Interactions of ribosome nascent chain complexes of the chloroplast- encoded D1 thylakoid membrane protein with cpSRP54. EMBO J. 18, 733–742. 10.1093/emboj/18.3.733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R., van Wijk K. J. (2002). Transient interaction of cpSRP54 with elongating nascent chains of the chloroplast-encoded D1 protein; ‘cpSRP54 caught in the act’. FEBS Lett. 524, 127–133. 10.1016/S0014-5793(02)03016-8 [DOI] [PubMed] [Google Scholar]
- Nishiyama Y., Allakhverdiev S. I., Yamamoto H., Hayashi H., Murata N. (2004). Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry 43, 11321–11330. 10.1021/bi036178q [DOI] [PubMed] [Google Scholar]
- Nixon P. J., Barker M., Boehm M., de Vries R., Komenda J. (2005). FtsH-mediated repair of the photosystem II complex in response to light stress. J. Exp. Bot. 56, 357–363. 10.1093/jxb/eri021 [DOI] [PubMed] [Google Scholar]
- Nixon P. J., Diner B. A. (1992). Aspartate 170 of the photosystem II reaction center polypeptide D1 is involved in the assembly of the oxygen evolving manganese cluster. Biochemistry 31, 942–948. 10.1021/bi00118a041 [DOI] [PubMed] [Google Scholar]
- Nixon P. J., Diner B. A. (1994). Analysis of water oxidation mutants constructed in the cyanobacterium Synechocystis sp. PCC 6803. Biochem. Soc. Trans. 22, 338–343. 10.1042/bst0220338 [DOI] [PubMed] [Google Scholar]
- Nixon P. J., Michoux F., Yu J., Boehm M., Komenda J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16. 10.1093/aob/mcq059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon P. J., Trost J. T., Diner B. A. (1992). Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water oxidizing manganese cluster in photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: assembly requires a free carboxyl group at C terminal position 344. Biochemistry 31, 10859–10871. 10.1021/bi00159a029 [DOI] [PubMed] [Google Scholar]
- Nowaczyk M. M., Hebeler R., Schlodder E., Meyer H. E., Warscheid B., Rögner M. (2006). Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18, 3121–3131. 10.1105/tpc.106.042671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohad I., Kyle D. J., Arntzen C. J. (1984). Membrane protein damage and repair: removal and replacement of inactivated 32-kilodalton polypeptides in chloroplast membranes. J. Cell Biol. 99, 481–485. 10.1083/jcb.99.2.481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T. (2001). Metallo-radical hypothesis for photoassembly of (Mn)4-cluster of photosynthetic oxygen evolving complex. Biochim. Biophys. Acta 1503, 40–51. 10.1016/S0005-2728(00)00226-7 [DOI] [PubMed] [Google Scholar]
- Ono T. A., Inoue Y. (1982). Photoactivation of the water oxidation system in isolated intact chloroplasts prepared from wheat triticum-aestivum leaves grown under intermittent flash illumination. Plant Physiol. 69, 1418–1422. 10.1104/pp.69.6.1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T. A., Inoue Y. (1983). Requirement of divalent cations for photoactivation of the latent water oxidation system in intact chloroplasts from flashed leaves. Biochim. Biophys. Acta 723, 191–201. 10.1016/0005-2728(83)90119-6 [DOI] [Google Scholar]
- Ono T. A., Inoue Y. (1987). Reductant-sensitive intermediates involved in multi-quantum process of photoactivation of latent oxygen-evolving system. Plant Cell Physiol. 28, 1293–1300. [Google Scholar]
- Ono T. A., Mino H. (1999). Unique binding site for Mn2+ ion responsible for reducing an oxidized YZ tyrosine in manganese-depleted photosystem II membranes. Biochemistry 38, 8778–8785. 10.1021/bi982949s [DOI] [PubMed] [Google Scholar]
- Osmond C. B. (1981). Photorespiration and photoinhibition. Biochim. Biophys. Acta 639, 77–98. 10.1016/0304-4173(81)90006-9 [DOI] [Google Scholar]
- Park S., Khamai P., Garcia-Cerdan J. G., Melis A. (2007). REP27, a tetratricopeptide repeat nuclear-encoded and chloroplast- localized protein, functions in D1/32-kD reaction center protein turnover and photosystem II repair from photodamage. Plant Physiol. 143, 1547–1560. 10.1104/pp.107.096396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin J. M., Campbell K. A., Randall D. W., Evanchik M. A., Pecoraro V. L., Armstrong W. H., et al. (2000). 55Mn ENDOR of the S2-state multiline EPR signal of photosystem II: implications on the structure of the tetranuclear Mn cluster. J. Am. Chem. Soc. 122, 10926–10942. 10.1021/ja002104f [DOI] [Google Scholar]
- Pistorius E. K., Schmid G. H. (1984). Effect of Mn2+ and Ca2+ on O2 evolution and on the variable fluorescence yield associated with Photosystem II in preparations of Anacystis nidulans. FEBS Lett. 171, 173–178. 10.1016/0014-5793(84)80482-2 [DOI] [Google Scholar]
- Powles S. B., Björkman O. (1981). Photoinhibition of photosynthesis: effect on chlorophyll fluorescence at 77K in intact leaves and in chloroplast membranes of Nerium oleander. Planta 156, 97–107. 10.1007/BF00395424 [DOI] [PubMed] [Google Scholar]
- Puthiyaveetil S., Kirchhoff H. (2013). A phosphorylation map of the photosystem II supercomplex C2S2M2. Front. Plant Sci. 4:459. 10.3389/fpls.2013.00459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian M., Al-Khaldi S. F., Putnam-Evans C., Bricker T. M., Burnap R. L. (1997). Photoassembly of the photosystem II (Mn)4 cluster in site directed mutants impaired in the binding of the manganese stabilizing protein. Biochemistry 36, 15244–15252. [DOI] [PubMed] [Google Scholar]
- Qian M., Dao L., Debus R. J., Burnap R. L. (1999). Impact of mutations within the putative Ca2+-binding lumenal interhelical a-b loop of the photosystem II D1 protein on the kinetics of photoactivation and H2O-oxidation in Synechocystis sp. PCC6803. Biochemistry 38, 6070–6081. 10.1021/bi982331i [DOI] [PubMed] [Google Scholar]
- Radmer R., Cheniae G. M. (1971). Photoactivation of the manganese catalyst of O2 evolution. II. A two quantum mechanism. Biochim. Biophys. Acta 253, 182–186. 10.1016/0005-2728(71)90243-X [DOI] [PubMed] [Google Scholar]
- Rappaport F., Ishida N., Sugiura M., Boussac A. (2011). Ca2+ determines the entropy changes associated with the formation of transition states during water oxidation by Photosystem II. Energy Environ. Sci. 4, 2520–2524. 10.1039/c1ee01408k [DOI] [Google Scholar]
- Riggs P. J., Mei R., Yocum C. F., Penner H. J. E. (1992). Reduced derivatives of the manganese cluster in the photosynthetic oxygen-evolving complex. J. Am. Chem. Soc. 114, 10650–10651. 10.1021/ja00052a079 [DOI] [Google Scholar]
- Roose J. L., Pakrasi H. B. (2004). Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into photosystem II. J. Biol. Chem. 279, 45417–45422. 10.1074/jbc.M408458200 [DOI] [PubMed] [Google Scholar]
- Roose J. L., Pakrasi H. B. (2007). The PSB27 protein facilitates manganese cluster assembly in photosystem II. J. Biol. Chem. 44, 45417–45422. 10.1074/jbc.M408458200 [DOI] [PubMed] [Google Scholar]
- Sacharz J., Bryan S. J., Yu J., Burroughs N. J., Spence E. M., Nixon P. J., et al. (2015). Sub-cellular location of FtsH proteases in the cyanobacterium Synechocystis sp. PCC 6803 suggests localised PSII repair zones in the thylakoid membranes. Mol. Microbiol. 96, 448–462. 10.1111/mmi.12940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K., Yamamoto Y. (2007). The carboxyl-terminal processing of precursor D1 protein of the photosystem II reaction center. Photosyn. Res. 94, 203–215. 10.1007/s11120-007-9191-z [DOI] [PubMed] [Google Scholar]
- Sauer K., Yachandra V. K. (2002). A possible evolutionary origin for the Mn-4 cluster of the photosynthetic water oxidation complex from natural MnO2 precipitates in the early ocean. Proc. Natl. Acad. Sci. U.S.A. 99, 8631–8636. 10.1073/pnas.132266199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schottkowski M., Gkalympoudis S., Tzekova N., Stelljes C., Schünemann D., Ankele E., et al. (2009). Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 284, 1813–1819. 10.1074/jbc.M806116200 [DOI] [PubMed] [Google Scholar]
- Seibert M., Tamura N., Inoue Y. (1989). Lack of photoactivation capacity in scenedesmus-obliquus Lf-1 results from loss of half the high-affinity manganese-binding site - relationship to the unprocessed D1 protein. Biochim. Biophys. Acta 974, 185–191. 10.1016/S0005-2728(89)80371-8 [DOI] [Google Scholar]
- Semin B. K., Podkovirina T. E., Davletshina L. N., Timofeev K. N., Ivanov I.I, Rubin, A. B. (2015). The extrinsic PsbO protein modulates the oxidation/reduction rate of the exogenous Mn cation at the high-affinity Mn-binding site of Mn-depleted PSII membranes. J. Bioenerg. Biomembr. 47, 361–367. 10.1007/s10863-015-9618-8 [DOI] [PubMed] [Google Scholar]
- Shen J. R., Qian M., Inoue Y., Burnap R. L. (1998). Functional characterization of Synechocystis sp. PCC 6803 delta psbU and delta psbV mutants reveals important roles of cytochrome c-550 in cyanobacterial oxygen evolution. Biochemistry 37, 1551–1558. 10.1021/bi971676i [DOI] [PubMed] [Google Scholar]
- Shestakov S. V., Anbudurai P. R., Stanbekova G. E., Gadzhiev A., Lind L. K., Pakrasi H. B. (1994). Molecular cloning and characterization of the ctpA gene encoding a carboxyl terminal processing protease. Analysis of a spontaneous photosystem II deficient mutant strain of the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 269, 19354–19359. [PubMed] [Google Scholar]
- Shinohara K., Ono T. A., Inoue Y. (1992). Photoactivation of oxygen-evolving enzyme in dark-grown pine cotyledons relationship between assembly of photosystem II proteins and integration of manganese and calcium. Plant Cell Physiol. 33, 281–289. [Google Scholar]
- Silva P., Thompson E., Bailey S., Kruse O., Mullineaux C. W., Robinson C., et al. (2003). FtsH is involved in the early stages of repair of photosystem II in Synechocystis sp PCC 6803. Plant Cell 15, 2152–2164. 10.1105/tpc.012609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengel A., Gügel I. L., Hilger D., Rengstl B., Jung H., Nickelsen J. (2012). Initial steps of photosystem II de novo assembly and preloading with manganese take place in biogenesis centers in Synechocystis. Plant Cell 24, 660–675. 10.1105/tpc.111.093914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suga M., Akita F., Hirata K., Ueno G., Murakami H., Nakajima Y., et al. (2015). Native structure of photosystem II at 1.95 A resolution viewed by femtosecond X-ray pulses. Nature 517, 99–103. 10.1038/nature13991 [DOI] [PubMed] [Google Scholar]
- Taguchi F., Yamamoto Y., Satoh K. (1995). Recognition of the structure around the site of cleavage by the carboxyl terminal processing protease for D1 precursor protein of the photosystem II reaction center. J. Biol. Chem. 270, 10711–10716. [DOI] [PubMed] [Google Scholar]
- Tamura N., Cheniae G. (1987). Photoactivation of the water-oxidizing complex in photosystem II membranes depleted of Mn and extrinsic proteins. I. Biochemical and kinetic characterization. Biochim. Biophys. Acta 890, 179–194. 10.1016/0005-2728(87)90019-3 [DOI] [Google Scholar]
- Tamura N., Cheniae G. M. (1986). Requirements for the photoligation of Mn2+ in PSII membranes and the expression of water-oxidizing activity of the polynuclear Mn-catalyst. Feder. Eur. Biochem. Soc. Lett. 200, 231–236. 10.1016/0014-5793(86)80544-0 [DOI] [Google Scholar]
- Tamura N., Inoue Y., Cheniae G. (1989). Photoactivation of the water-oxidizing complex in photosystem II membranes depleted of Mn, Ca, and extrinsic proteins. II. Studies on the function of Ca2+. Biochim. Biophys. Acta 976, 173–181. 10.1016/S0005-2728(89)80227-0 [DOI] [Google Scholar]
- Taylor M. A., Nixon P. J., Todd C. M., Barber J., Bowyer J. R. (1988). Characterisation of the D1 protein in a photosystem II mutant (LF-1) of Scenedesmus obliquus blocked on the oxidising side Evidence supporting non-processing of D1 as the cause of the lesion. FEBS Lett. 235, 109–116. 10.1016/0014-5793(88)81243-2 [DOI] [Google Scholar]
- Trost J. T., Chisholm D. A., Jordan D. B., Diner B. A. (1997). The D1 C-terminal processing protease of photosystem II from Scenedesmus obliquus. Protein purification and gene characterization in wild type and processing mutants. J. Biol. Chem. 272, 20348–20356. 10.1074/jbc.272.33.20348 [DOI] [PubMed] [Google Scholar]
- Tyryshkin A. M., Watt R. K., Baranov S. V., Dasgupta J., Hendrich M. P., Dismukes G. C. (2006). Spectroscopic evidence for Ca2+ involvement in the assembly of the Mn4Ca cluster in the photosynthetic water-oxidizing complex. Biochemistry 45, 12876–12889. 10.1021/bi061495t [DOI] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J. R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Ångstrom. Nature 473, U55–U65. 10.1038/nature09913 [DOI] [PubMed] [Google Scholar]
- van Wijk K. J., Andersson B., Aro E. M. (1996). Kinetic resolution of the incorporation of the D1 protein into photosystem II and localization of assembly intermediates in thylakoid membranes of spinach chloroplasts. J. Biol. Chem. 271, 9627–9636. 10.1074/jbc.271.16.9627 [DOI] [PubMed] [Google Scholar]
- van Wijk K. J., Roobol-Boza M., Kettunen R., Andersson B., Aro E. M. (1997). Synthesis and assembly of the D1 protein into photosystem II: processing of the C terminus and identification of the initial assembly partners and complexes during photosystem II repair. Biochemistry 36, 6178–6186. 10.1021/bi962921l [DOI] [PubMed] [Google Scholar]
- Vass I., Cser K. (2009). Janus-faced charge recombinations in photosystem II photoinhibition. Trends Plant Sci. 14, 200–205. 10.1016/j.tplants.2009.01.009 [DOI] [PubMed] [Google Scholar]
- Vrettos J. S., Limburg J., Brudvig G. W. (2001). Mechanism of photosynthetic water oxidation: combining biophysical studies of photosystem II with inorganic model chemistry. Biochim. Biophys. Acta 1503, 229–245. 10.1016/S0005-2728(00)00214-0 [DOI] [PubMed] [Google Scholar]
- Whitelegge J. P., Koo D., Diner B. A., Domian I., Erickson J. M. (1995). Assembly of the Photosystem II oxygen evolving complex is inhibited in psbA site directed mutants of Chlamydomonas reinhardtii. Aspartate 170 of the D1 polypeptide. J. Biol. Chem. 270, 225–235. 10.1074/jbc.270.1.225 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y. (2001). Quality control of photosystem ii. Plant Cell Physiol. 42, 121–128. 10.1093/pcp/pce022 [DOI] [PubMed] [Google Scholar]
- Zaltsman L., Ananyev G. M., Bruntrager E., Dismukes G. C. (1997). Quantitative kinetic model for photoassembly of the photosynthetic water oxidase from its inorganic constituents: requirements for manganese and calcium in the kinetically resolved steps. Biochemistry 36, 8914–8922. 10.1021/bi970187f [DOI] [PubMed] [Google Scholar]
- Zhang L., Aro E. M. (2002). Synthesis, membrane insertion and assembly of the chloroplast-encoded D1 protein into photosystem II. FEBS Lett. 512, 13–18. 10.1016/S0014-5793(02)02218-4 [DOI] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen V., Suorsa M., Aro E. M. (2001). A SecY homologue is involved in chloroplast-encoded D1 protein biogenesis. J. Biol. Chem. 276, 37809–37814. 10.1074/jbc.M105522200 [DOI] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen V., van Wijk K. J., Aro E. M. (1999). Co-translational assembly of the D1 protein into photosystem II. J. Biol. Chem. 274, 16062–16067. 10.1074/jbc.274.23.16062 [DOI] [PubMed] [Google Scholar]
- Zhang L., Paakkarinen V., van Wijk K. J., Aro E. M. (2000). Biogenesis of the chloroplast-encoded D1 protein: regulation of translation elongation, insertion, and assembly into photosystem II. Plant Cell 12, 1769–1782. 10.1105/tpc.12.9.1769 [DOI] [PMC free article] [PubMed] [Google Scholar]