Abstract
Despite the availability of an effective vaccine, hepatitis B virus (HBV) infection remains a major health problem, with more than 350 million chronically infected people worldwide and over 1 million annual deaths due to cirrhosis and liver cancer. HBV mutations are primarily generated due both to a lack of proofreading capacity by HBV polymerase and to host immune pressure, which is a very important factor for predicting disease progression and therapeutic outcomes. Several types of HBV precore/core (preC/C) mutations have been described to date. The host immune response against T cells drives mutation in the preC/C region. Specifically, preC/C mutations in the MHC class II restricted region are more common than in other regions and are significantly related to hepatocellular carcinoma. Certain mutations, including preC G1896A, are also significantly related to HBeAg-negative chronic infection. This review article mainly focuses on the HBV preC/C mutations that are related to disease severity and on the HBeAg serostatus of chronically infected patients.
Keywords: Hepatitis B virus infection, Precore/core mutations, Hepatocellular carcinoma, HBeAg serostatus, Disease severity
Core tip: The presence of several distinct types of mutations in HBV infections has been shown to contribute to the progression of liver disease in chronically infected patients. Although the relationships between single mutation types in the preC/C region and clinical severity have rarely been studied to date, it was recently reported that some preC/C mutations, particularly in the MHC class II restricted region, are significantly correlated with hepatocellular carcinoma. Several preC/C mutations, including preC G1896A, are also related to HBeAg sero-negative status, which can affect the disease progression of chronic patients. Mutations such as I97F/L or P135Q, which inhibit core nucleocapsid formation, also contribute to disease progression by evading host innate immunity. In addition, the P5H/L/T mutation may lead to hepatocarcinogenesis by inducing the ER stress-ROS axis. In this review, we mainly focus on the clinical implications of the reported preC/C mutations.
INTRODUCTION
Despite the availability of an effective vaccine, hepatitis B virus (HBV) infection remains a major public health concern in most countries, but particularly in endemic areas such as China and South Korea. Globally, there are more than 350 million people who are chronically infected with HBV, which annually causes over 1 million deaths due to serious liver diseases, such as cirrhosis and hepatocellular carcinoma (HCC)[1].
HBV is an enveloped Hepadnavirus belonging to the Hepadnaviridae family. HBV has an incomplete double-stranded DNA genome that is approximately 3.2 kb in length and contains 4 overlapping open reading frames (ORFs) encoding the polymerase (P), core (C), surface antigen (S), and X protein[2,3]. Based on an intergroup divergence of > 8% in its complete genome sequence, HBV strains are classified into 8 genotypes, designated A-H, which correlate strongly with the ethnicity of infected patients[4-8]. There is increasing evidence that specific HBV genotypes may play significant roles in causing different disease profiles during chronic hepatitis B (CHB) infection[9-12]. Notably, an extraordinary prevalence of the C2 genotype has been reported in South Korea[13-15]. This genotype is more prone to mutations and is associated with more severe liver disease and poorer antiviral responses compared to genotype B[16,17]. In actuality, several types of HBV mutations that are rarely, if ever, encountered in other areas have been found in South Korea. These mutations were demonstrated via molecular epidemiologic and functional studies to be related to the disease progression of chronic patients[14,15,18-34].
Over the past decade, increasing attention has been focused on variant HBV strains that contribute to the clinical severity of liver diseases, especially HCC. To date, certain mutation patterns of HBV, such as the precore (preC) mutation at nucleotide 1896 (G1896A) or the double mutation in the basal core promoter (BCP) region at nucleotides 1762 (A→T) and 1764 (G→A), have been widely studied in the context of clinical severity[35-42]. Recently, several types of naturally occurring mutations in the pre-surface antigen (preS), S and X regions (i.e., the preS1 start codon deletion[28,43], the preS2 deletion[28], W4P/R in preS1[29,32], sW182* in S[28,31,44], and V5M in X[23]), which are related to clinical severity, have also been described.
HBcAg AND HBeAg STRUCTURE AND THEIR VARIANTS
The HBV C protein antigen (HBcAg), the major structural protein of the nucleocapsid, is 183 residues long, of which the N-terminal 149 residues are the assembly domain[45-47]. The preC/C ORF is transcribed and translated into a precore/core fusion protein. During entry into the endoplasmic reticulum (ER), 19 residues of the 29-residue preC region are cleaved off by a signal peptidase, generating a 22-kDa protein. When transported into the Golgi compartment, additional amino acids are removed from the C-terminus by intra-Golgi proteases to form the HBe antigen, leading to a final, heterogeneous secreted protein of 15-18 kDa[48-50]. The secreted HBeAg is regarded as a marker of productive infection in clinical practice. Although the biological function of the HBeAg remains unsolved, it has been suggested that the HBeAg may contribute to HBV replication and modulate the host immune system as a type of tolerogen[51-53].
Most preC/C mutations are generated during HBeAg seroconversion in chronic HBV infections[54-56]. Such mutations can affect HBeAg serostatus and antigenicity, HBV nucleocapsid structure and stability, and the packaging of pregenomic RNA into the nucleocapsid[57]. HBcAg is the principal target of the host immune response and especially of CD4 and CD8 T cell attack[58,59]. Thus, mutations in the preC/C region are mainly distributed in the MHC-restricted region and can induce persistent HBV infections[27,60,61].
Several mutations in the preC/C region have been described, and the most frequently reported mutations in the literature are listed in Table 1[27,61-80]. Except in cases of nucleotide deletions, most mutations in the preC/C region are point mutations[65,80]. Most of the reported mutations in the preC region are associated with reduced HBeAg levels or reduced HBV replication in patient sera. In the C region, mutations are mainly located in immuno-active regions (MHC class I + II) rather than in immuno-inactive regions.
Table 1.
Mutations in the hepatitis B virus preC/C region reported by previous literatures
| Regions | Type of mutation |
Mutations |
HBV genotype | MHC class | Ref. | |||
| Amino acid | Changes | Nucelotide | Changes | |||||
| preCore | AS | 1 | M1L/T/I | 1814-1816 | A1814T/C, T1815C, G1816T/A | A | [69-72] | |
| AS | 2 | Q2Stop | 1817 | C1817T | A | [71, 73] | ||
| Ins | Insertion TT | 1821 | 1821-1825 | [74] | ||||
| Ins | Insertion TT | 1825 | 1825-1826 | [71] | ||||
| Del | T deletion | 1839 | 1839del | [72] | ||||
| Ins | Insertion T | 1839 | 1839-1840 | [71] | ||||
| AS | 15 | P15S | 1856 | C1856T | [75] | |||
| NC | 1858 | T1858C | A/B/E | [76] | ||||
| AS | 17 | V17F | 1862 | G1862T | A1 | [95] | ||
| AS | 26 | W26R | 1889 | T1889A | B/C | [77] | ||
| Ins | 36 | Insertion 36nt | 1895 | 1895-1896 | [78] | |||
| AS | 28 | W28Stop | 1896 | G1896A | B/C/C2/D | [29, 61, 77, 79, 80] | ||
| AS | 29 | G29D | 1899 | G1899A | C2 | [29] | ||
| Core | AS | 5 | P5T/L/H | 1913-1915 | C1913A, C1914A/T, G1915A/C | B/C2 | II | [29, 61, 81] |
| P5R | 1914 | C1914G | A/D | [79] | ||||
| NC | 1915 | G1915A/C, A1915T | B/C | [77, 82] | ||||
| AS | 12 | T12S | 1934 | A1934T | A/C/D | II | [83] | |
| AS | 21 | S21H | 1961-1962 | T1961C, C1962A | A/C/D | I | [68, 83] | |
| AS | 27 | V/I27I/V | 1979 | G1979A, A1979G | A/B/C/D | I | [68, 77, 83] | |
| NC | 1981 | C1981A | A/D | [68] | ||||
| AS | 32 | D32N/H | 1994 | G1994A/C | C2 | Immuno-inactive | [29] | |
| AS | 34 | NC | 2002 | C2002A | A/D | Immuno-inactive | [68] | |
| AS | 35 | S35L | 2004 | C2004T | A/D | Immuno-inactive | [68] | |
| AS | 43 | E43K | 2027 | G2027A | C2 | Immuno-inactive | [29] | |
| NC | 2029 | G2029A | A/D | [68] | ||||
| AS | 45 | NC | 2035 | T2035A/G | A/D | Immuno-inactive | [68] | |
| AS | 48 | NC | 2044 | T2044C | B/C | Immuno-inactive | [77] | |
| AS | 49 | NC | 2047 | A2047C | A/D | Immuno-inactive | [68] | |
| AS | 50 | P50Y/H/A | 2048-2049 | C2048T/G, C2049A | B/C/C2 | II | [29, 84] | |
| AS | 55 | L55I | 2063 | C2063A | A/D | II | [68] | |
| AS | 59 | I59F | 2075 | A2075T | A/C/D | II | [83] | |
| NC | 2077 | T2077A/C | B/C | [77] | ||||
| AS | 60 | L60V | 2078 | C2078G | II | [85] | ||
| NC | 2080 | C2080A | A/D | [68] | ||||
| Core | AS | 64 | E/K64D | 2090-2092 | A2090G, A2092T/C | A/C/D | II | [68, 83] |
| AS | 65 | L65V | 2093 | C2093G | A/D | II | [68] | |
| AS | 67 | T67N | 2100 | C2100A | A/C/D | II | [68, 83] | |
| AS | 77 | E77Q | 2129 | G2129C | B/C | Immuno-inactive | [84] | |
| NC | 2131 | A2131G | A/D | [68] | ||||
| AS | 78 | NC | 2134 | C2134T | B/C | Immuno-inactive | [77] | |
| Del | 105nt deletion | 2134-2238 | 2134-2238 | [86] | ||||
| AS | 79 | NC | 2137 | A2137G/T/C | B/C | Immuno-inactive | [77] | |
| AS | 83 | E83D | 2149 | A2149T/C | C2 | II | [29] | |
| Del | 105nt deletion | 2150-2254 | 2150-2254 | [86] | ||||
| AS | 87 | S87R | 2161 | C2161G | B/C | II | [77] | |
| AS | 89 | NC | 2167 | T2167C | A/D | I and II | [68] | |
| AS | 92 | NC | 2176 | T2176C | B/C | I and II | [77] | |
| AS | 95 | L95I | 2183 | C2183A | A/D | I and II | [68] | |
| AS | 97 | I97F/L | 2189-2191 | A2189T/C, C2191T | A/B/C/C2/D | II | [29, 77, 83, 85] | |
| NC | 2191 | C2191A/T | A/D | [68] | ||||
| AS | 100 | L100I | 2198 | C2198A | A/C/C2/D | II | [29, 83] | |
| AS | 101 | NC | 2201 | T2201C | B/C | II | [77] | |
| Del | 130nt deletion | 2204-2333 | 2204-2333 | [86] | ||||
| AS | 107 | NC | 2221 | C2221T | B/C | Immuno-inactive | [77] | |
| AS | 113 | E113Q | 2237 | G2237C | A/D | Immuno-inactive | [68] | |
| AS | 115 | NC | 2245 | C2245T | B/C | Immuno-inactive | [77] | |
| AS | 117 | NC | 2251 | G2251A | B/C | II | [77] | |
| AS | 119 | NC | 2257 | G2257A | A/D | II | [68] | |
| AS | 120 | NC | 2260 | G2260A | B/C | II | [77] | |
| AS | 126 | NC | 2278 | T2278A | A/D | II | [68] | |
| AS | 130 | P130S/T | 2288 | C2288T/A | B/C | II | [74, 77] | |
| NC | 2290 | C2290T | B/C | [77] | ||||
| AS | 131 | A131P/N/G | 2291-2293 | G2291C/A, C2292A/G, T2293C | A/C/C2/D | I and II | [29, 83] | |
| NC | 2293 | C2293T | A/D | [68] | ||||
| AS | 134 | NC | 2302 | A2302G | A/D | I | [68] | |
| AS | 135 | P135Q/S/A | 2303-2304 | C2303T/G, C2304A | A/B/C/D | I | [61, 68, 83] | |
| AS | 142 | NC | 2326 | A2326T | A/D | I and II | [68] | |
| AS | 145 | NC | 2335 | A2335G | A/D | I and II | [68] | |
| AS | 149 | V149I | 2345 | G2345A | B/C | I and II | [84] | |
| AS | 181 | S181P/H | 2441-2442 | T2441C, C2442A | C2 | Immuno-inactive | [29] | |
| AS | 182 | Q182K/Stop | 2444-2445 | C2444A/T, A2445G | C2 | Immuno-inactive | [29] | |
AS: Amino acid substitution; Del: Deletion; Ins: Insertion; MHC: Major histocompatibility complex; NC: No change.
preC/C MUTATIONS RELATED TO CLINICAL SEVERITY OR HBeAg SEROSTATUS
Certain preC/C mutations have been associated with significant virological or clinical events, such as the failure to form a nucleocapsid, liver disease progression, or HBeAg seroconversion.
preC/C mutations related to HBeAg serostatus
As patients with HBeAg-negative CHB respond poorly to conventional interferon-alpha therapy, they should be treated differently from those with HBeAg-positive CHB[81-83]. Thus, preC/C mutation analysis can provide valuable information for the management of patients with HBeAg-negative CHB. Basal core promoter mutations (BCP, nt1742-1849) that suppress the production of preC mRNA at the transcriptional level may contribute to the defective synthesis of HBeAg in vivo[35]. However, the most frequently occurring mutations that are responsible for an HBeAg-negative hepatitis B profile are mutations that occur within the preC region and that inhibit translation of the protein due to frameshift mutations or premature stop codons (Table 1). Among the mutations reported thus far (Table 1), the mutation that is most often responsible for defective HBeAg secretion is a point mutation, namely a G to A transition at nucleotide 1896 (G1896A) that changes the 28th codon of preC from tryptophan (UGG) into a translational stop codon (UAG). The reduced HBeAg level can also have an important effect on HBV replication and can thereby influence liver disease progression, particularly in fulminant hepatitis and acute exacerbation of CHB[66]. Several studies have reported a positive association between the severity of liver disease and the occurrence of G1896A mutations[27,41,70]. However, it has also been reported that there is no correlation between this mutation and liver disease[72-74]. The discrepancy between different findings may be due to various factors, including HBV genotype, the geographical location or race/ethnicity of patients, host immune competence, and co-infection with other viruses, such as HIV or HCV[5,30,84-86].
Most published studies have reported that HBeAg-negative CHB due to the preC G1896A mutation is only common in non-A genotypes. In patients with genotype A, preC start codon mutations (A1814C/T, T1815C/A) that lead to a failure of HBeAg production have been frequently found[87], but such mutations are very rare in those infected with non-A genotypes. Recently, Mayaphi et al[61] reported that 24% of patients with sub-genotype A1 had preC start codon mutations, suggesting that this mutation, rather than the G1896A mutation, may contribute to HBeAg-negative CHB infection in patients with sub-genotype A1.
The G1862T mutation, which leads to a valine-to-phenylalanine amino acid substitution at residue 17 of the preC region, can affect the expression of HBeAg by interfering with signal peptidase cleavage[88]. This mutation prevails in HBV genotype A and particularly in sub-genotype A1; together with preC start codon mutations, G1862T is responsible for the HBeAg-negative serostatus and much lower viremia titers in patients infected with HBV genotype A. Moreover, Saha et al[77] recently reported that all tested HBV/A1 isolates from Eastern India harbored the G1862T mutation irrespective of HBeAg status, supporting the idea that this mutation might represent a natural variation in HBV/A1 rather than an adaptive mutation.
Mutations in the C-terminus of the preC/C region alter the biosynthesis, transportation, and secretion of HBeAg[73,79,89]. Such mutations lead to the cytoplasmic accumulation of the HBeAg proprotein (p22) in hepatocytes, resulting in decreased HBV replication in patient sera due to down-regulated HBV DNA replication and HBcAg capsid polymerization[73]. Recently, Wu et al[73] reported that, together with the G1896A mutation, the C2304A mutation, which causes a glutamine-to-proline substitution at residue 135 of HBcAg (P135Q), is a predictor of spontaneous HBeAg seroconversion following long-term immune-tolerance development within chronic HBV genotype B-infected subjects. Based on a functional study, this mutation was significantly associated with increased cytoplasmic accumulation of the 22-kDa HBeAg proprotein (p22), decreased mature 17-kDa HBeAg (p17) secretion, and a decreased number of HBV capsid particles in Huh7 hepatoma cells, suggesting that this mutation may be associated with spontaneous HBeAg seroconversion in chronic HBV genotype B-infected patients via the decreased secretion of the 17-kDa mature HBeAg (p17)[57,90-92].
Recently, we reported that a total of 5 preC/C mutations (G1896A in preC plus 4 in the C region, namely E43K, P50A/H/Y, A131G/N/P and S181H/P) were found to be significantly related to HBeAg serostatus in chronic patients infected with sub-genotype C2[27]. Of those, interestingly, 2 mutations (D32N/H, and E43K) were related to the HBeAg-positive serostatus that was first introduced in the report. These 2 mutations were not located in the regions encoding T or B cell epitopes, suggesting that they may be induced by mechanisms other than immune evasion. Notably, none of the above-described 5 preC/C mutations that were related to HBeAg-negative serostatus was significantly related to HCC, although the G1896A mutation tended towards association with HCC (P = 0.093). These findings raise questions regarding the actual pathogenetic implications of HBeAg-defective mutants. Prospective studies on mutations in the preC/C region and their molecular mechanisms as they relate to the progression of liver disease would provide a better understanding of the clinical relevance of preC/C mutations in relation to HBeAg-negative serostatus. The preC/C mutations that have been reported to cause a change in the HBeAg serostatus of CHB patients are summarized in Table 2.
Table 2.
Mutations in the hepatitis B virus preC/C region leading to change of HBeAg serostatus in chronic patients
| Regions |
Mutations |
HBV genotype | MHC class | HBeAg serostatus (P value) | Ref. | |||
| Amino acid | Changes | Nucleotide | Changes | |||||
| preCore | 28 | W28Stop | 1896 | G1896A | A/B/C/C2/D | N (n = 14) vs P (n = 3) (P = 0.004) | [27] | |
| eAg Seroconverters (n = 14 of 29, 48.3%) | [73] | |||||||
| N (43.4%) vs P (11.9%) (P = 0.001) | [72] | |||||||
| N (61.5%) vs P (10.9%) with HIV/HBV (P ≤ 0.0001) | [77] | |||||||
| Core | 5 | P5R | 1914 | C1914G | A/D | II | N (23.4%) vs P (4.7%) (P = 0.001) | [72] |
| 32 | D32N/H | 1994 | G1994A/C | C2 | Immuno-inactive | N (n = 2) vs P (n = 7) (P = 0.074) | [27] | |
| 43 | E43K | 2027 | G2027A | C2 | Immuno-inactive | N (n = 1) vs P (n = 7) (P = 0.024) | [27] | |
| 50 | P50Y/H/A | 2048-2049 | C2048T/G, C2049A | C2 | II | N (n = 5) vs P (n = 0) (P = 0.020) | [27] | |
| 131 | A131P/N/G | 2291-2293 | G2291C/A, C2292A/G, T2293C | C2 | II | N (n = 4) vs P (n = 0) (P = 0.039) | [27] | |
| 135 | P135Q | 2304 | C2304A | B | I | eAg Seroconverters (n = 18 of 29, 62.1%) | [73] | |
| 181 | S181P/H | 2441-2442 | T2441C, C2442A | C2 | Immuno-inactive | N (n = 4) vs P (n = 0) (P = 0.039) | [27] | |
MHC: Major histocompatibility complex; N: Negative; P: Positive.
HBcAg mutations related to clinical severance
As HBcAg is the principal target of the host immune response and particularly of CD4 and CD8 T cell attack, the mutations in this region are mainly distributed in the MHC restricted region and may induce persistent HBV infection[58,59,78]. Indeed, a positive relationship between the frequency of HBcAg and the progression of liver disease has been reported[93-96]. We recently reported that the mutation rate in the MHC class II restricted region (M2RR) (2.7% vs 1.9%, P = 0.024), but not in the MHC class I restricted region (M1RR) (2.4% vs 1.8%, P = 0.3), was significantly higher in HCC patients infected with sub-genotype C2 than in non-HCC patients with the same HBV genotype[27]. Furthermore, the difference between HCC and non-HCC patients with respect to the hotspot region in the M2RR (residues 81-105) was also pronounced (5.6% vs 2.6%, P = 0.002). In genotype A1-infected patients, HBcAg mutations were also most commonly found in the M2RR, suggesting that mutations in the M2RR of HBcAg that aid in immune escape can contribute to persistent HBV infection and influence disease progression[97].
We recently introduced 5 mutations in the HBcAg region (P5H/L/T, E83D, I97F/L, L100I and Q182K/Stop) that have been significantly associated with HCC patients compared to patients at other stages of the disease, such as liver cirrhosis (LC) and chronic hepatitis (CH)[27]. Notably, 4 of the 5 HCC-related HBcAg mutations, namely P5H/L/T, E83D, I97F/L and L100I, were located in the M2RR. I97F/L, which was previously known to lead to defects in HBcAg assembly[98,99], was most frequently found in HCC patients (13/35 HCC patients, 37.1%). The frequency of this mutation (16%) was also higher among sub-genotype A1 variants in South Africa[61]. Intrahepatic expression of HBcAg induced a robust IFN response in mice that facilitated control of the viral infection[100]. However, a recent study reported that the majority of mice that received a capsid assembly-deficient HBV mutant with the Y132A mutation in HBcAg failed to elicit the appropriate HBV-specific immune responses for eliminating hepatitis B surface antigenemia, suggesting that nucleocapsid formation is important for triggering a proper antiviral immune response, perhaps via the induction of innate immunity against HBV[101]. Thus, the I97F/L mutation, which is responsible for defective nucleocapsid assembly, may partially contribute to the progress of severe liver disease by failing to elicit a proper host immune response against HBV infection. We have previously reported that the lower level of HBV DNA in patients infected with mutated strains in preC/C region than in those with wild strains were found[27], suggesting preC/C mutations could lead to inhibition of HBV replication, generally. But, the identification of mutation types affecting HBV replication should also be done via functional study in the future.
Mutations in residue 5 of HBcAg (P5H/L/T or P5R) are reported to be significantly more frequent in HCC patients relative to a reference group, not only in Korean patients infected with sub-genotype C2[27] but also in Indian patients infected with genotypes A or D[72]. Recently, we demonstrated that P5H/L/T mutations in HBV genotype C2 can elicit the ER stress-ROS axis in hepatocytes[75]. This response then leads to inflammatory cytokine production, TGF-β secretion, apoptosis and HBsAg secretion, all of which are related to liver disease progression. The resulting prolonged inflammation, liver damage and increased HBsAg secretion induced by these mutations may contribute to the progression of liver disease in chronic patients.
Xie et al[70] recently reported that 5 mutation sites in the preC/C region, namely 1915, 2134, 2221, 2245, and 2288, were identified as statistically significant independent predictors of HCC survival by multivariate survival analysis. Of these, only the C2288A mutation in HBcAg (P130T) results in an amino acid change, while the other 4 mutations are silent. However, further validation in other populations and functional studies will be required to elucidate the mechanism by which these mutations, particularly the 4 silent mutations, affect HCC progression. The preC/C mutations that have been previously reported as associated with liver disease progression in chronic patients and particularly in HCC patients are summarized in Table 3.
Table 3.
Mutations in the hepatitis B virus preC/C region related to clinical severance
| Regions |
Mutations |
HBV genotype | MHC class | Clinical significance (P value) | Ref. | |||
| Amino acid | Changes | Nucleotide | Changes | |||||
| preCore | 28 | W28Stop | 1896 | G1896A | C2 | HCC (n = 12) vs Mild (n = 5) (P = 0.093) | [27] | |
| A/D | HCC (P < 0.05), FHF (45.0%) vs AVH (17.4%) (P = 0.038) | [72] | ||||||
| Core | 5 | P5T/L/H | 1913-1915 | C1913A, C1914A/T, G1915A/C | C2 | II | HCC (n = 5) vs Mild (n = 0) (P = 0.02) | [27] |
| NC | 1915 | G1915A/C | C3 | HCC (P = 0.020) | [70] | |||
| P5R | 1914 | C1914G | C4 | HCC (32.2%) vs ASC (5.0%)/CHB (7.8%)/LC (10.3%) (P < 0.05) | [72] | |||
| 78 | NC | 2134 | C2134T | C5 | Immuno-inactive | HCC (P = 0.001) | [70] | |
| 83 | E83D | 2149 | A2149T/C | C6 | II | HCC (n = 4) vs Mild (n = 0) (P = 0.039) | [27] | |
| 92 | NC | 2176 | T2176C | C7 | I and II | HCC (P = 0.139) | [70] | |
| 97 | I97F/L | 2189-2191 | A2189T/C, C2191T | C8 | II | HCC (n = 13) vs Mild (n = 4) (P = 0.024) | [27, 73, 93] | |
| 100 | L100I | 2198 | C2198A | C9 | II | HCC (n = 6) vs Mild (n = 1) (P = 0.046) | [27] | |
| 107 | NC | 2221 | C2221T | C10 | Immuno-inactive | HCC (P = 0.001) | [70] | |
| 115 | NC | 2245 | C2245T | C11 | Immuno-inactive | HCC (P = 0.007) | [70] | |
| 130 | P130T | 2288 | C2288A | C12 | I and II | HCC (P = 0.022) | [70] | |
| 182 | Q182K/Stop | 2444-2445 | C2444A/T, A2445G | C13 | Immuno-inactive | HCC (n = 4) vs Mild (n = 0) (P = 0.039) | [27] | |
ASC: Asymptomatic S antigen carrier; AVH: Acute viral hepatitis; CHB: Chronic hepatitis B; FHF: Fulminant hepatic failure; HCC: Hepatocellular carcinoma; LC: Liver cirrhosis; MHC: Major histocompatibility Complex; NC: No change.
CONCLUSION
Mutations in the preC/C region can affect HBeAg serostatus, HBV replication, nucleocapsid formation, and even pgRNA encapsidation[27,45,48,49,74]. Despite disparities in the infecting genotypes and in patient factors such as co-infection with HIV and patient ethnicity and geographical location, several preC/C mutation types that affect disease progression in chronic patients have been identified[61,77,78,87]. In general, mutations such as G1896A, which decrease or cause a loss of HBeAg, may contribute to disease progression via persistent HBV infection[21,102,103]. Immune-escape mutations in the M2RR of HBcAg, particularly in the hotspot region comprising amino acid residues 81-105, may contribute to persistent HBV infection that is related to disease progression[53,73,79,104]. Mutations such as I97F/L and P135Q, which inhibit core nucleocapsid formation, may also contribute to disease progression by evading host innate immunity[27,73,98,99]. The P5H/L/T mutation was shown to lead to hepatocarcinogenesis by inducing the ER stress-ROS axis (Figure 1). However, to better understand the clinical relevance of preC/C mutations, large, worldwide prospective studies on the clinical relevance of the different types of preC/C mutations are required, as are functional studies that relate the mutations to liver disease progression.
Figure 1.
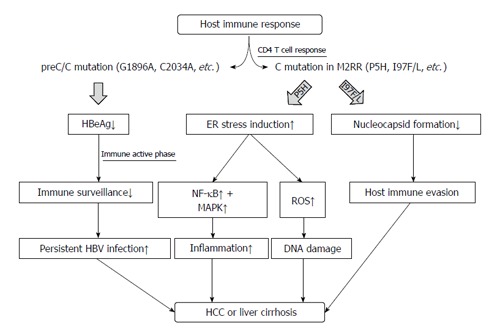
Schematic representation indicating role of mutations in the hepatitis B virus preC/C region in the disease progression of chronic patients. HBV: Hepatitis B virus; HCC: Hepatocellular carcinoma; ER: Endoplasmic reticulum; NF-κB: Nuclear factor kappa B.
Footnotes
Supported by A National Research Foundation grant of Ministry of Science, ICT and Future Planning, South Korea, No. NRF-2015R1C1A1A02037267; and a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute, the Ministry of Health and Welfare, South Korea, No. HI14C0955.
Conflict-of-interest statement: No conflict of interest.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: February 17, 2016
First decision: March 7, 2016
Article in press: April 7, 2016
P- Reviewer: Otsuka M, Park YM, Shimizu Y S- Editor: Gong ZM L- Editor: A E- Editor: Wang CH
References
- 1.Lee WM. Hepatitis B virus infection. N Engl J Med. 1997;337:1733–1745. doi: 10.1056/NEJM199712113372406. [DOI] [PubMed] [Google Scholar]
- 2.Delius H, Gough NM, Cameron CH, Murray K. Structure of the hepatitis B virus genome. J Virol. 1983;47:337–343. doi: 10.1128/jvi.47.2.337-343.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liang TJ. Hepatitis B: the virus and disease. Hepatology. 2009;49:S13–S21. doi: 10.1002/hep.22881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kidd-Ljunggren K, Miyakawa Y, Kidd AH. Genetic variability in hepatitis B viruses. J Gen Virol. 2002;83:1267–1280. doi: 10.1099/0022-1317-83-6-1267. [DOI] [PubMed] [Google Scholar]
- 5.Sunbul M. Hepatitis B virus genotypes: global distribution and clinical importance. World J Gastroenterol. 2014;20:5427–5434. doi: 10.3748/wjg.v20.i18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guirgis BS, Abbas RO, Azzazy HM. Hepatitis B virus genotyping: current methods and clinical implications. Int J Infect Dis. 2010;14:e941–e953. doi: 10.1016/j.ijid.2010.03.020. [DOI] [PubMed] [Google Scholar]
- 7.Akuta N, Kumada H. Influence of hepatitis B virus genotypes on the response to antiviral therapies. J Antimicrob Chemother. 2005;55:139–142. doi: 10.1093/jac/dkh533. [DOI] [PubMed] [Google Scholar]
- 8.Miyakawa Y, Mizokami M. Classifying hepatitis B virus genotypes. Intervirology. 2003;46:329–338. doi: 10.1159/000074988. [DOI] [PubMed] [Google Scholar]
- 9.Chan HL, Wong ML, Hui AY, Hung LC, Chan FK, Sung JJ. Hepatitis B virus genotype C takes a more aggressive disease course than hepatitis B virus genotype B in hepatitis B e antigen-positive patients. J Clin Microbiol. 2003;41:1277–1279. doi: 10.1128/JCM.41.3.1277-1279.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vivekanandan P, Abraham P, Sridharan G, Chandy G, Daniel D, Raghuraman S, Daniel HD, Subramaniam T. Distribution of hepatitis B virus genotypes in blood donors and chronically infected patients in a tertiary care hospital in southern India. Clin Infect Dis. 2004;38:e81–e86. doi: 10.1086/383144. [DOI] [PubMed] [Google Scholar]
- 11.Osiowy C, Giles E, Trubnikov M, Choudhri Y, Andonov A. Characterization of Acute and Chronic Hepatitis B Virus Genotypes in Canada. PLoS One. 2015;10:e0136074. doi: 10.1371/journal.pone.0136074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toan NL, Song le H, Kremsner PG, Duy DN, Binh VQ, Koeberlein B, Kaiser S, Kandolf R, Torresi J, Bock CT. Impact of the hepatitis B virus genotype and genotype mixtures on the course of liver disease in Vietnam. Hepatology. 2006;43:1375–1384. doi: 10.1002/hep.21188. [DOI] [PubMed] [Google Scholar]
- 13.Cho JH, Yoon KH, Lee KE, Park DS, Lee YJ, Moon HB, Lee KR, Choi CS, Cho EY, Kim HC. Distribution of hepatitis B virus genotypes in Korea. Korean J Hepatol. 2009;15:140–147. doi: 10.3350/kjhep.2009.15.2.140. [DOI] [PubMed] [Google Scholar]
- 14.Kim H, Kim BJ. Association of preS/S Mutations with Occult Hepatitis B Virus (HBV) Infection in South Korea: Transmission Potential of Distinct Occult HBV Variants. Int J Mol Sci. 2015;16:13595–13609. doi: 10.3390/ijms160613595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim H, Jee YM, Song BC, Shin JW, Yang SH, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ, et al. Molecular epidemiology of hepatitis B virus (HBV) genotypes and serotypes in patients with chronic HBV infection in Korea. Intervirology. 2007;50:52–57. doi: 10.1159/000096313. [DOI] [PubMed] [Google Scholar]
- 16.Li X, Wang L, Zhong Y, Wong VW, Xu Z, Liu Y, Li Q, Xin S, Zhao J, Xu D. Hepatitis B virus (HBV) subgenotypes C2 and B2 differ in lamivudine- and adefovir-resistance-associated mutational patterns in HBV-infected Chinese patients. J Clin Microbiol. 2010;48:4363–4369. doi: 10.1128/JCM.01518-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen YM, Wu SH, Qiu CN, Yu DJ, Wang XJ. Hepatitis B virus subgenotype C2- and B2-associated mutation patterns may be responsible for liver cirrhosis and hepatocellular carcinoma, respectively. Braz J Med Biol Res. 2013;46:614–622. doi: 10.1590/1414-431X20133032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song BC, Kim H, Kim SH, Cha CY, Kook YH, Kim BJ. Comparison of full length sequences of hepatitis B virus isolates in hepatocellular carcinoma patients and asymptomatic carriers of Korea. J Med Virol. 2005;75:13–19. doi: 10.1002/jmv.20230. [DOI] [PubMed] [Google Scholar]
- 19.Song BC, Kim SH, Kim H, Ying YH, Kim HJ, Kim YJ, Yoon JH, Lee HS, Cha CY, Kook YH, et al. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J Med Virol. 2005;76:194–202. doi: 10.1002/jmv.20354. [DOI] [PubMed] [Google Scholar]
- 20.Kim H, Jee Y, Mun HS, Park JH, Yoon JH, Kim YJ, Lee HS, Hyun JW, Hwang ES, Cha CY, et al. Characterization of two hepatitis B virus populations in a single Korean hepatocellular carcinoma patient with an HBeAg-negative serostatus: a novel X-Gene-deleted strain with inverted duplication sequences of upstream enhancer site II. Intervirology. 2007;50:273–280. doi: 10.1159/000103915. [DOI] [PubMed] [Google Scholar]
- 21.Kim H, Jee Y, Mun HS, Song BC, Park JH, Hyun JW, Hwang ES, Cha CY, Kook YH, Kim BJ. Comparison of full genome sequences between two hepatitis B virus strains with or without preC mutation (A1896) from a single Korean hepatocellular carcinoma patient. J Microbiol Biotechnol. 2007;17:701–704. [PubMed] [Google Scholar]
- 22.Kim H, Jee YM, Song BC, Hyun JW, Mun HS, Kim HJ, Oh EJ, Yoon JH, Kim YJ, Lee HS, et al. Analysis of hepatitis B virus quasispecies distribution in a Korean chronic patient based on the full genome sequences. J Med Virol. 2007;79:212–219. doi: 10.1002/jmv.20789. [DOI] [PubMed] [Google Scholar]
- 23.Kim HJ, Park JH, Jee Y, Lee SA, Kim H, Song BC, Yang S, Lee M, Yoon JH, Kim YJ, et al. Hepatitis B virus X mutations occurring naturally associated with clinical severity of liver disease among Korean patients with chronic genotype C infection. J Med Virol. 2008;80:1337–1343. doi: 10.1002/jmv.21219. [DOI] [PubMed] [Google Scholar]
- 24.Mun HS, Lee SA, Jee Y, Kim H, Park JH, Song BC, Yoon JH, Kim YJ, Lee HS, Hyun JW, et al. The prevalence of hepatitis B virus preS deletions occurring naturally in Korean patients infected chronically with genotype C. J Med Virol. 2008;80:1189–1194. doi: 10.1002/jmv.21208. [DOI] [PubMed] [Google Scholar]
- 25.Lee SA, Mun HS, Kim H, Lee HK, Kim BJ, Hwang ES, Kook YH, Kim BJ. Naturally occurring hepatitis B virus X deletions and insertions among Korean chronic patients. J Med Virol. 2011;83:65–70. doi: 10.1002/jmv.21938. [DOI] [PubMed] [Google Scholar]
- 26.Mun HS, Lee SA, Kim H, Hwang ES, Kook YH, Kim BJ. Novel F141L pre-S2 mutation in hepatitis B virus increases the risk of hepatocellular carcinoma in patients with chronic genotype C infections. J Virol. 2011;85:123–132. doi: 10.1128/JVI.01524-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim DW, Lee SA, Hwang ES, Kook YH, Kim BJ. Naturally occurring precore/core region mutations of hepatitis B virus genotype C related to hepatocellular carcinoma. PLoS One. 2012;7:e47372. doi: 10.1371/journal.pone.0047372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H, Lee SA, Kim DW, Lee SH, Kim BJ. Naturally occurring mutations in large surface genes related to occult infection of hepatitis B virus genotype C. PLoS One. 2013;8:e54486. doi: 10.1371/journal.pone.0054486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee SA, Kim KJ, Kim DW, Kim BJ. Male-specific W4P/R mutation in the pre-S1 region of hepatitis B virus, increasing the risk of progression of liver diseases in chronic patients. J Clin Microbiol. 2013;51:3928–3936. doi: 10.1128/JCM.01505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim BJ. Hepatitis B virus mutations related to liver disease progression of Korean patients. World J Gastroenterol. 2014;20:460–467. doi: 10.3748/wjg.v20.i2.460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim H, Lee SA, Won YS, Lee H, Kim BJ. Occult infection related hepatitis B surface antigen variants showing lowered secretion capacity. World J Gastroenterol. 2015;21:1794–1803. doi: 10.3748/wjg.v21.i6.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee SA, Kim H, Won YS, Seok SH, Na Y, Shin HB, Inn KS, Kim BJ. Male-specific hepatitis B virus large surface protein variant W4P potentiates tumorigenicity and induces gender disparity. Mol Cancer. 2015;14:23. doi: 10.1186/s12943-015-0303-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H, Hong SH, Lee SA, Gong JR, Kim BJ. Development of Fok-I based nested polymerase chain reaction-restriction fragment length polymorphism analysis for detection of hepatitis B virus X region V5M mutation. World J Gastroenterol. 2015;21:13360–13367. doi: 10.3748/wjg.v21.i47.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H, Gong JR, Lee SA, Kim BJ. Discovery of a Novel Mutation (X8Del) Resulting in an 8-bp Deletion in the Hepatitis B Virus X Gene Associated with Occult Infection in Korean Vaccinated Individuals. PLoS One. 2015;10:e0139551. doi: 10.1371/journal.pone.0139551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tseng TC, Liu CJ, Yang HC, Chen CL, Yang WT, Tsai CS, Kuo SF, Verbree FC, Su TH, Wang CC, et al. Higher proportion of viral basal core promoter mutant increases the risk of liver cirrhosis in hepatitis B carriers. Gut. 2015;64:292–302. doi: 10.1136/gutjnl-2014-306977. [DOI] [PubMed] [Google Scholar]
- 36.Asim M, Malik A, Sarma MP, Polipalli SK, Begum N, Ahmad I, Khan LA, Husain SA, Akhtar N, Husain S, et al. Hepatitis B virus BCP, Precore/core, X gene mutations/genotypes and the risk of hepatocellular carcinoma in India. J Med Virol. 2010;82:1115–1125. doi: 10.1002/jmv.21774. [DOI] [PubMed] [Google Scholar]
- 37.Liao Y, Hu X, Chen J, Cai B, Tang J, Ying B, Wang H, Wang L. Precore mutation of hepatitis B virus may contribute to hepatocellular carcinoma risk: evidence from an updated meta-analysis. PLoS One. 2012;7:e38394. doi: 10.1371/journal.pone.0038394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu CJ, Chen BF, Chen PJ, Lai MY, Huang WL, Kao JH, Chen DS. Role of hepatitis B viral load and basal core promoter mutation in hepatocellular carcinoma in hepatitis B carriers. J Infect Dis. 2006;193:1258–1265. doi: 10.1086/502978. [DOI] [PubMed] [Google Scholar]
- 39.Chu CM, Lin CC, Chen YC, Jeng WJ, Lin SM, Liaw YF. Basal core promoter mutation is associated with progression to cirrhosis rather than hepatocellular carcinoma in chronic hepatitis B virus infection. Br J Cancer. 2012;107:2010–2015. doi: 10.1038/bjc.2012.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Ding HG. Key role of hepatitis B virus mutation in chronic hepatitis B development to hepatocellular carcinoma. World J Hepatol. 2015;7:1282–1286. doi: 10.4254/wjh.v7.i9.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Park YM, Jang JW, Yoo SH, Kim SH, Oh IM, Park SJ, Jang YS, Lee SJ. Combinations of eight key mutations in the X/preC region and genomic activity of hepatitis B virus are associated with hepatocellular carcinoma. J Viral Hepat. 2014;21:171–177. doi: 10.1111/jvh.12134. [DOI] [PubMed] [Google Scholar]
- 42.Clementi M, Manzin A, Paolucci S, Menzo S, Bangarelli P, Carloni G, Bearzi I. Hepatitis B virus preC mutants in human hepatocellular carcinoma tissues. Res Virol. 1993;144:297–301. doi: 10.1016/s0923-2516(06)80044-0. [DOI] [PubMed] [Google Scholar]
- 43.Lee SA, Kim KJ, Kim H, Choi WH, Won YS, Kim BJ. Hepatitis B virus preS1 deletion is related to viral replication increase and disease progression. World J Gastroenterol. 2015;21:5039–5048. doi: 10.3748/wjg.v21.i16.5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee SA, Kim K, Kim H, Kim BJ. Nucleotide change of codon 182 in the surface gene of hepatitis B virus genotype C leading to truncated surface protein is associated with progression of liver diseases. J Hepatol. 2012;56:63–69. doi: 10.1016/j.jhep.2011.06.028. [DOI] [PubMed] [Google Scholar]
- 45.Schödel F, Peterson D, Zheng J, Jones JE, Hughes JL, Milich DR. Structure of hepatitis B virus core and e-antigen. A single precore amino acid prevents nucleocapsid assembly. J Biol Chem. 1993;268:1332–1337. [PubMed] [Google Scholar]
- 46.Pumpens P, Grens E. Hepatitis B core particles as a universal display model: a structure-function basis for development. FEBS Lett. 1999;442:1–6. doi: 10.1016/s0014-5793(98)01599-3. [DOI] [PubMed] [Google Scholar]
- 47.Crowther RA, Kiselev NA, Böttcher B, Berriman JA, Borisova GP, Ose V, Pumpens P. Three-dimensional structure of hepatitis B virus core particles determined by electron cryomicroscopy. Cell. 1994;77:943–950. doi: 10.1016/0092-8674(94)90142-2. [DOI] [PubMed] [Google Scholar]
- 48.Ou JH, Laub O, Rutter WJ. Hepatitis B virus gene function: the precore region targets the core antigen to cellular membranes and causes the secretion of the e antigen. Proc Natl Acad Sci USA. 1986;83:1578–1582. doi: 10.1073/pnas.83.6.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Milich DR, Jones JE, Hughes JL, Price J, Raney AK, McLachlan A. Is a function of the secreted hepatitis B e antigen to induce immunologic tolerance in utero? Proc Natl Acad Sci USA. 1990;87:6599–6603. doi: 10.1073/pnas.87.17.6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kramvis A, Kew MC. Relationship of genotypes of hepatitis B virus to mutations, disease progression and response to antiviral therapy. J Viral Hepat. 2005;12:456–464. doi: 10.1111/j.1365-2893.2005.00624.x. [DOI] [PubMed] [Google Scholar]
- 51.Milich DR, Chen MK, Hughes JL, Jones JE. The secreted hepatitis B precore antigen can modulate the immune response to the nucleocapsid: a mechanism for persistence. J Immunol. 1998;160:2013–2021. [PubMed] [Google Scholar]
- 52.Jean-Jean O, Levrero M, Will H, Perricaudet M, Rossignol JM. Expression mechanism of the hepatitis B virus (HBV) C gene and biosynthesis of HBe antigen. Virology. 1989;170:99–106. doi: 10.1016/0042-6822(89)90356-5. [DOI] [PubMed] [Google Scholar]
- 53.Chen M, Sällberg M, Hughes J, Jones J, Guidotti LG, Chisari FV, Billaud JN, Milich DR. Immune tolerance split between hepatitis B virus precore and core proteins. J Virol. 2005;79:3016–3027. doi: 10.1128/JVI.79.5.3016-3027.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen MH, Keeffe EB. Are hepatitis B e antigen (HBeAg)-positive chronic hepatitis B and HBeAg-negative chronic hepatitis B distinct diseases? Clin Infect Dis. 2008;47:1312–1314. doi: 10.1086/592571. [DOI] [PubMed] [Google Scholar]
- 55.Liaw YF. HBeAg seroconversion as an important end point in the treatment of chronic hepatitis B. Hepatol Int. 2009;3:425–433. doi: 10.1007/s12072-009-9140-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bozkaya H, Ayola B, Lok AS. High rate of mutations in the hepatitis B core gene during the immune clearance phase of chronic hepatitis B virus infection. Hepatology. 1996;24:32–37. doi: 10.1002/hep.510240107. [DOI] [PubMed] [Google Scholar]
- 57.Schlicht HJ, Wasenauer G. The quaternary structure, antigenicity, and aggregational behavior of the secretory core protein of human hepatitis B virus are determined by its signal sequence. J Virol. 1991;65:6817–6825. doi: 10.1128/jvi.65.12.6817-6825.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matsumura S, Yamamoto K, Shimada N, Okano N, Okamoto R, Suzuki T, Hakoda T, Mizuno M, Higashi T, Tsuji T. High frequency of circulating HBcAg-specific CD8 T cells in hepatitis B infection: a flow cytometric analysis. Clin Exp Immunol. 2001;124:435–444. doi: 10.1046/j.1365-2249.2001.01561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jung MC, Diepolder HM, Spengler U, Wierenga EA, Zachoval R, Hoffmann RM, Eichenlaub D, Frösner G, Will H, Pape GR. Activation of a heterogeneous hepatitis B (HB) core and e antigen-specific CD4+ T-cell population during seroconversion to anti-HBe and anti-HBs in hepatitis B virus infection. J Virol. 1995;69:3358–3368. doi: 10.1128/jvi.69.6.3358-3368.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yim SY, Um SH, Young Jung J, Kim TH, Kim JD, Keum B, Seo YS, Yim HJ, Jeen YT, Lee HS, et al. Clinical significance of hepatitis B virus precore and core promoter variants in Korean patients with chronic hepatitis B. J Clin Gastroenterol. 2015;49:61–68. doi: 10.1097/MCG.0000000000000052. [DOI] [PubMed] [Google Scholar]
- 61.Mayaphi SH, Martin DJ, Mphahlele MJ, Blackard JT, Bowyer SM. Variability of the preC/C region of hepatitis B virus genotype A from a South African cohort predominantly infected with HIV. J Med Virol. 2013;85:1883–1892. doi: 10.1002/jmv.23695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Santantonio T, Jung MC, Miska S, Pastore G, Pape GR, Will H. Prevalence and type of pre-C HBV mutants in anti-HBe positive carriers with chronic liver disease in a highly endemic area. Virology. 1991;183:840–844. doi: 10.1016/0042-6822(91)91022-9. [DOI] [PubMed] [Google Scholar]
- 63.Raimondo G, Schneider R, Stemler M, Smedile V, Rodino G, Will H. A new hepatitis B virus variant in a chronic carrier with multiple episodes of viral reactivation and acute hepatitis. Virology. 1990;179:64–68. doi: 10.1016/0042-6822(90)90274-u. [DOI] [PubMed] [Google Scholar]
- 64.Okamoto H, Yotsumoto S, Akahane Y, Yamanaka T, Miyazaki Y, Sugai Y, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Hepatitis B viruses with precore region defects prevail in persistently infected hosts along with seroconversion to the antibody against e antigen. J Virol. 1990;64:1298–1303. doi: 10.1128/jvi.64.3.1298-1303.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fiordalisi G, Cariani E, Mantero G, Zanetti A, Tanzi E, Chiaramonte M, Primi D. High genomic variability in the pre-C region of hepatitis B virus in anti-HBe, HBV DNA-positive chronic hepatitis. J Med Virol. 1990;31:297–300. doi: 10.1002/jmv.1890310410. [DOI] [PubMed] [Google Scholar]
- 66.Laskus T, Persing DH, Nowicki MJ, Mosley JW, Rakela J. Nucleotide sequence analysis of the precore region in patients with fulminant hepatitis B in the United States. Gastroenterology. 1993;105:1173–1178. doi: 10.1016/0016-5085(93)90964-e. [DOI] [PubMed] [Google Scholar]
- 67.Wakita T, Kakumu S, Shibata M, Yoshioka K, Ito Y, Shinagawa T, Ishikawa T, Takayanagi M, Morishima T. Detection of pre-C and core region mutants of hepatitis B virus in chronic hepatitis B virus carriers. J Clin Invest. 1991;88:1793–1801. doi: 10.1172/JCI115500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Carman WF, Thomas HC. Genetic variation in hepatitis B virus. Gastroenterology. 1992;102:711–719. doi: 10.1016/0016-5085(92)90125-i. [DOI] [PubMed] [Google Scholar]
- 69.Li J, Tong S, Vitvitski L, Zoulim F, Trépo C. Rapid detection and further characterization of infection with hepatitis B virus variants containing a stop codon in the distal pre-C region. J Gen Virol. 1990;71(Pt 9):1993–1998. doi: 10.1099/0022-1317-71-9-1993. [DOI] [PubMed] [Google Scholar]
- 70.Xie Y, Liu S, Zhao Y, Zhang L, Zhao Y, Liu B, Guo Z. Precore/Core Region Mutations in Hepatitis B Virus DNA Predict Postoperative Survival in Hepatocellular Carcinoma. PLoS One. 2015;10:e0133393. doi: 10.1371/journal.pone.0133393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhat RA, Ulrich PP, Vyas GN. Molecular characterization of a new variant of hepatitis B virus in a persistently infected homosexual man. Hepatology. 1990;11:271–276. doi: 10.1002/hep.1840110218. [DOI] [PubMed] [Google Scholar]
- 72.Malik A, Singhal DK, Albanyan A, Husain SA, Kar P. Hepatitis B virus gene mutations in liver diseases: a report from New Delhi. PLoS One. 2012;7:e39028. doi: 10.1371/journal.pone.0039028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wu JF, Ni YH, Chen HL, Hsu HY, Chang MH. The impact of hepatitis B virus precore/core gene carboxyl terminal mutations on viral biosynthesis and the host immune response. J Infect Dis. 2014;209:1374–1381. doi: 10.1093/infdis/jit638. [DOI] [PubMed] [Google Scholar]
- 74.Carman WF, Jacyna MR, Hadziyannis S, Karayiannis P, McGarvey MJ, Makris A, Thomas HC. Mutation preventing formation of hepatitis B e antigen in patients with chronic hepatitis B infection. Lancet. 1989;2:588–591. doi: 10.1016/s0140-6736(89)90713-7. [DOI] [PubMed] [Google Scholar]
- 75.Lee H, Kim H, Lee SA, Won YS, Kim HI, Inn KS, Kim BJ. Upregulation of endoplasmic reticulum stress and reactive oxygen species by naturally occurring mutations in hepatitis B virus core antigen. J Gen Virol. 2015;96:1850–1854. doi: 10.1099/vir.0.000134. [DOI] [PubMed] [Google Scholar]
- 76.Ono Y, Onda H, Sasada R, Igarashi K, Sugino Y, Nishioka K. The complete nucleotide sequences of the cloned hepatitis B virus DNA; subtype adr and adw. Nucleic Acids Res. 1983;11:1747–1757. doi: 10.1093/nar/11.6.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saha D, Pal A, Biswas A, Panigrahi R, Sarkar N, Das D, Sarkar J, Guha SK, Saha B, Chakrabarti S, et al. Molecular characterization of HBV strains circulating among the treatment-naive HIV/HBV co-infected patients of eastern India. PLoS One. 2014;9:e90432. doi: 10.1371/journal.pone.0090432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Honda T, Ishigami M, Luo F, Ishizu Y, Kuzuya T, Hayashi K, Itoh A, Hirooka Y, Ishikawa T, Nakano I, et al. Hepatitis B e antigen and hepatitis B surface antigen seroclearance with the emergence of lamivudine-associated and core mutations following CD4 elevation in a patient with hepatitis B and HIV. Intern Med. 2015;54:585–590. doi: 10.2169/internalmedicine.54.2038. [DOI] [PubMed] [Google Scholar]
- 79.Liu Z, Luo K, He H, Hou J. Hot-spot mutations in hepatitis B virus core gene: eliciting or evading immune clearance? J Viral Hepat. 2005;12:146–153. doi: 10.1111/j.1365-2893.2005.00552.x. [DOI] [PubMed] [Google Scholar]
- 80.Akarca US, Lok AS. Naturally occurring hepatitis B virus core gene mutations. Hepatology. 1995;22:50–60. [PubMed] [Google Scholar]
- 81.Saikia N, Talukdar R, Mazumder S, Khanna S, Tandon R. Management of patients with HBeAg-negative chronic hepatitis B. Postgrad Med J. 2007;83:32–39. doi: 10.1136/pgmj.2006.044826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hui CK, Lau GK. Treatment of Hepatitis B e Antigen-negative Patients. Curr Treat Options Gastroenterol. 2007;10:474–482. doi: 10.1007/s11938-007-0047-6. [DOI] [PubMed] [Google Scholar]
- 83.Yeh ML, Peng CY, Dai CY, Lai HC, Huang CF, Hsieh MY, Huang JF, Chen SC, Lin ZY, Yu ML, et al. Pegylated-interferon alpha therapy for treatment-experienced chronic hepatitis B patients. PLoS One. 2015;10:e0122259. doi: 10.1371/journal.pone.0122259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang D, Ma S, Zhang X, Zhao H, Ding H, Zeng C. Prevalent HBV point mutations and mutation combinations at BCP/preC region and their association with liver disease progression. BMC Infect Dis. 2010;10:271. doi: 10.1186/1471-2334-10-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inoue J, Ueno Y, Wakui Y, Fukushima K, Kondo Y, Kakazu E, Ninomiya M, Niitsuma H, Shimosegawa T. Enhanced replication of hepatitis B virus with frameshift in the precore region found in fulminant hepatitis patients. J Infect Dis. 2011;204:1017–1025. doi: 10.1093/infdis/jir485. [DOI] [PubMed] [Google Scholar]
- 86.Wang Z, Tanaka Y, Huang Y, Kurbanov F, Chen J, Zeng G, Zhou B, Mizokami M, Hou J. Clinical and virological characteristics of hepatitis B virus subgenotypes Ba, C1, and C2 in China. J Clin Microbiol. 2007;45:1491–1496. doi: 10.1128/JCM.02157-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Makondo E, Bell TG, Kramvis A. Genotyping and molecular characterization of hepatitis B virus from human immunodeficiency virus-infected individuals in southern Africa. PLoS One. 2012;7:e46345. doi: 10.1371/journal.pone.0046345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guarnieri M, Kim KH, Bang G, Li J, Zhou Y, Tang X, Wands J, Tong S. Point mutations upstream of hepatitis B virus core gene affect DNA replication at the step of core protein expression. J Virol. 2006;80:587–595. doi: 10.1128/JVI.80.2.587-595.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang HI, Lu SN, Liaw YF, You SL, Sun CA, Wang LY, Hsiao CK, Chen PJ, Chen DS, Chen CJ. Hepatitis B e antigen and the risk of hepatocellular carcinoma. N Engl J Med. 2002;347:168–174. doi: 10.1056/NEJMoa013215. [DOI] [PubMed] [Google Scholar]
- 90.Wu JF, Hsu HY, Ni YH, Chen HL, Wu TC, Chang MH. Suppression of furin by interferon-γ and the impact on hepatitis B virus antigen biosynthesis in human hepatocytes. Am J Pathol. 2012;181:19–25. doi: 10.1016/j.ajpath.2012.03.036. [DOI] [PubMed] [Google Scholar]
- 91.Wynne SA, Crowther RA, Leslie AG. The crystal structure of the human hepatitis B virus capsid. Mol Cell. 1999;3:771–780. doi: 10.1016/s1097-2765(01)80009-5. [DOI] [PubMed] [Google Scholar]
- 92.Locarnini S, Shaw T, Dean J, Colledge D, Thompson A, Li K, Lemon SM, Lau GG, Beard MR. Cellular response to conditional expression of the hepatitis B virus precore and core proteins in cultured hepatoma (Huh-7) cells. J Clin Virol. 2005;32:113–121. doi: 10.1016/j.jcv.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 93.Preikschat P, Günther S, Reinhold S, Will H, Budde K, Neumayer HH, Krüger DH, Meisel H. Complex HBV populations with mutations in core promoter, C gene, and pre-S region are associated with development of cirrhosis in long-term renal transplant recipients. Hepatology. 2002;35:466–477. doi: 10.1053/jhep.2002.30698. [DOI] [PubMed] [Google Scholar]
- 94.Ehata T, Omata M, Chuang WL, Yokosuka O, Ito Y, Hosoda K, Ohto M. Mutations in core nucleotide sequence of hepatitis B virus correlate with fulminant and severe hepatitis. J Clin Invest. 1993;91:1206–1213. doi: 10.1172/JCI116281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bock CT, Buerke B, Tillmann HL, Tacke F, Kliem V, Manns MP, Trautwein C. Relevance of hepatitis B core gene deletions in patients after kidney transplantation. Gastroenterology. 2003;124:1809–1820. doi: 10.1016/s0016-5085(03)00396-2. [DOI] [PubMed] [Google Scholar]
- 96.Okumura A, Takayanagi M, Aiyama T, Iwata K, Wakita T, Ishikawa T, Yoshioka K, Kakumu S. Serial analysis of hepatitis B virus core nucleotide sequence of patients with acute exacerbation during chronic infection. J Med Virol. 1996;49:103–109. doi: 10.1002/(SICI)1096-9071(199606)49:2<103::AID-JMV6>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 97.Ferrari C, Bertoletti A, Penna A, Cavalli A, Valli A, Missale G, Pilli M, Fowler P, Giuberti T, Chisari FV. Identification of immunodominant T cell epitopes of the hepatitis B virus nucleocapsid antigen. J Clin Invest. 1991;88:214–222. doi: 10.1172/JCI115280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ceres P, Stray SJ, Zlotnick A. Hepatitis B virus capsid assembly is enhanced by naturally occurring mutation F97L. J Virol. 2004;78:9538–9543. doi: 10.1128/JVI.78.17.9538-9543.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ning B, Shih C. Nucleolar localization of human hepatitis B virus capsid protein. J Virol. 2004;78:13653–13668. doi: 10.1128/JVI.78.24.13653-13668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lin YJ, Huang LR, Yang HC, Tzeng HT, Hsu PN, Wu HL, Chen PJ, Chen DS. Hepatitis B virus core antigen determines viral persistence in a C57BL/6 mouse model. Proc Natl Acad Sci USA. 2010;107:9340–9345. doi: 10.1073/pnas.1004762107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lin YJ, Wu HL, Chen DS, Chen PJ. Hepatitis B virus nucleocapsid but not free core antigen controls viral clearance in mice. J Virol. 2012;86:9266–9273. doi: 10.1128/JVI.00608-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Suppiah J, Mohd Zain R, Bahari N, Haji Nawi S, Saat Z. G1896A Precore Mutation and Association With HBeAg Status, Genotype and Clinical Status in Patients With Chronic Hepatitis B. Hepat Mon. 2015;15:e31490. doi: 10.5812/hepatmon.31490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tong MJ, Blatt LM, Kao JH, Cheng JT, Corey WG. Basal core promoter T1762/A1764 and precore A1896 gene mutations in hepatitis B surface antigen-positive hepatocellular carcinoma: a comparison with chronic carriers. Liver Int. 2007;27:1356–1363. doi: 10.1111/j.1478-3231.2007.01585.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sun L, Zhang Y, Zhao B, Deng M, Liu J, Li X, Hou J, Gui M, Zhang S, Li X, et al. A new unconventional HLA-A2-restricted epitope from HBV core protein elicits antiviral cytotoxic T lymphocytes. Protein Cell. 2014;5:317–327. doi: 10.1007/s13238-014-0041-4. [DOI] [PMC free article] [PubMed] [Google Scholar]


