Abstract
AIM: To investigate the role of miR-125b in regulating monocyte immune responses induced by hepatitis C virus (HCV) core protein.
METHODS: Monocytic THP-1 cells were treated with various concentrations of recombinant HCV core protein, and cytokines and miR-125b expression in these cells were analyzed. The requirement of Toll-like receptor 2 (TLR2) or MyD88 gene for HCV core protein-induced immune responses was determined by the transfection of THP-1 cells with gene knockdown vectors expressing either TLR2 siRNA or MyD88 siRNA. The effect of miR-125b overexpression on TLR2/MyD88 signaling was examined by transfecting THP-1 cells with miR-125b mimic RNA oligos.
RESULTS: In response to HCV core protein stimulation, cytokine production was up-regulated and miR-125b expression was down-regulated in THP-1 cells. The modulatory effect of HCV core protein on cellular events was dose-dependent and required functional TLR2 or MyD88 gene. Forced miR-125b expression abolished the HCV core protein-induced enhancement of tumor necrosis factor-α, interleukin (IL)-6, and IL-10 expression by 66%, 54%, and 66%, respectively (P < 0.001), by inhibiting MyD88-mediated signaling, including phosphorylation of NF-κBp65, ERK, and P38.
CONCLUSION: The inverse correlation between miR-125b and cytokine expression after HCV core challenge suggests that miR-125b may negatively regulate HCV-induced immune responses by targeting TLR2/MyD88 signaling in monocytes.
Keywords: miR-125b, Hepatitis virus C, Toll like receptor 2, Monocytes, Innate immunity
Core tip: Increasingly many studies have shown that microRNAs are critical regulators of the innate immune response. Many anti-pathogen pathways, including the pattern recognition Toll-like receptor (TLR)-mediated signaling pathway, are known to be regulated by a network of microRNAs. Here we investigated the possible role of miR-125b in regulating the monocyte immune responses induced by HCV core protein through TLR2/MyD88 signaling. Our findings indicated that miR-125b may function as a negative regulator of HCV-induced cellular events, which may provide insight into the role of miR-125b in the innate immune responses of monocytes to HCV.
INTRODUCTION
Hepatitis C virus (HCV), one of the primary causes of chronic hepatitis, end-stage cirrhosis, and hepatocellular carcinoma (HCC), contributes to an estimated 200 million chronic infections worldwide and 3-4 million or more each year[1]. HCV is a small enveloped RNA virus belonging to the Flavivridae family. The HCV genome encodes a polyprotein precursor of approximately 3000 amino acids that is processed by host and viral proteases into at least 10 different proteins. The viral core protein, a major cleaved product of HCV polyprotein by host peptidases, is known for its role in mediating HCV-induced pathogenesis. The RNA-binding HCV core protein is involved not only in the formation of the viral nucleocapsid but is capable of directly interacting with various host cell components, resulting in cellular dysfunction and carcinogenesis[2].
HCV not only infects hepatocytes, but it replicates in other immune cells, including B cells, T cells, NK cells and monocytes/macrophages[3,4]. It has been postulated that HCC is a consequence of prolonged liver injuries induced by chronic and persistent HCV infections. Numerous reports have indicated that defective host innate and adaptive immune systems are the major culprits facilitating the inability of the host to eradicate viruses[5]. While there is clear evidence that the synthesis of anti-viral cytokines, interferon (IFN)-α and IFN-γ were defective in chronic HCV infected patients[6], various others cytokines, such as tumor necrosis factor (TNF)-α, interleukin(IL)-6 and IL-10, were excessively expressed in patients with HCV viremia[7]. These skewed cytokine productions have been suggested to play a key role in liver injury, viral persistence and the progression to chronic HCV infection[8].
Monocytes/macrophages function as both innate immune cells by killing pathogens directly, and producing anti-pathogen cytokines and as adaptive immune cells by presenting antigens to other immune cells to produces antibodies. Although monocytes/macrophages dysfunction has been documented in patients with chronic HCV infection, particularly defective response to TLR ligand stimulation[9], the precise mechanism as how these cells are involved in the pathogenesis remains unclear.
MicroRNAs are highly conserved endogenous non-coding small RNAs that act as translational repressors to regulate a wide range of biological processes[10]. MicroRNAs have also been implicated in the pathogenesis, diagnosis and therapeutic aspects of HCV infections[11]. Recent studies have shown that several microRNAs, including miR-21, miR-146a and miR-155, were involved in regulation of virus-host interactions and may play important roles in the pathogenesis of HCV infection[12-14].
Since miR-125b is highly expressed in peripheral blood mononuclear cells, particularly in monocytes/macrophages[15], and its expression was found to be inversely associated with better treatment outcome of chronic HCV infection[16], we explored the role of miR-125b in modulating HCV-induced events in monocytic THP-1 cells. Here we show that miR-125b expression was negatively correlated with HCV core-induced cytokines expression in a TLR2/MyD88-dependent manner.
MATERIALS AND METHODS
Cell culture and reagents
The THP-1 human monocytic leukemia cell line was obtained from the Institute of Biochemistry and Cell Biology at the Shanghai Institute for Biological Sciences and cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, penicillin (100 IU/mL), and streptomycin (100 mg/mL). Bacterial HCV core recombinant protein was obtained from Immune Technology Co., China. Stability-enhanced miR-125b RNA oligonucleotide, miR-125b mimic (miR-125bM), and the control non-targeting RNA oligonucleotide were purchased from GenePharma.
RNA extraction and real-time quantitative reverse transcription-polymerase chain reaction analyses of cellular and miRNA genes
Total RNA (cellular and miRNA) was extracted from 1-3 × 106 THP-1 cells using TRIzol reagent (Life Technologies). For cellular gene expression, first strand cDNA and real-time quantitative polymerase chain reaction (RT-qPCR) analyses of TLR2 and MyD88 were performed according to the manufacturer’s instructions (Kakara). The following PCR primers were used in this study: TLR2 (F: 5’-TGGCATGTGCTGTGCTCTGTT-3’; R: 5’-AGCTTTCCTGGGCTTCCTTTT-3’); MyD88 (F: 5’-CTCCTCCACATCCTCCCTTC-3’; R: 5’-GCTTGTGTCTCCAGTTGCC-3’); internal control β-actin (F: 5’-CACGATGGAGGGGCCGGACTCATC-3’; R: 5’-TAAAGACCTCTATGCCAACACAGT-3’). The reaction conditions for PCR were 95 °C for 60 s, 53 °C for 60 s, and 72 °C for 60 s for 40 cycles, followed by an extension at 72 °C for 8 min. The relative gene expression of TLR2 and MyD88 was calculated using 2-ddCt, where ddCt = (Ct[gene]-Ct[β-actin]) and Ct is the crossing threshold value returned by the PCR instrument for each gene amplification.
To measure miRNA expression, total RNA was reversely transcribed at 16 °C for 30 min, 42 °C for 30 min and 85 °C for 5 min using miRNA gene specific primer (has-miR-125-5p and U6 snRNA NR_004394; Applied Biosystems). MicroRNA expression was determined by quantitative PCR using TaqMan Universal PCR System (Life Technologies) in 20 μL reactions, containing 1 μL TaqMan probes. The reaction was performed at 95 °C for 10 min, followed by 95 °C for 15 s and 60 °C at 60 s for 40 cycles. Expression of target genes was calculated as relative to that of the internal U6 snRNA.
Short interfering RNA knockdown vectors and transfections
TLR2-short interfering RNA (TLR2-siRNA) and MyD88-siRNA were constructed as previously described[12,13]. In brief, double-stranded oligonucleotides corresponding to the 164-182 position of TLR2 gene (NM_003264) or the 880-898 position of the MyD88 gene (NM_001172566) sequences were selected according to BLOCK-iT™ RNA Designer (Life Technologies) and cloned into Bsa I/Sac I sites of the pBSilence1.1 plasmid (Sirui Biologicl Co.). The recombinant plasmids were verified by sequencing from both ends. Silencing of TLR2 or MyD88 in THP-1 cells was performed using Lipofectamine 2000 (Life Technologies) and 50 nmol/L siRNA vector DNA according to the manufacturer’s protocol. Cells transfected with empty pBSilence1.1 vector was used as a transfection control. Twenty-four hours after transfection, the cells were analyzed for RNA (by RT-qPCR) or protein (by Western blot) expression.
Transfection of miR-125b mimic or control RNA oligos was similarly performed using Lipofectamine 2000. Six hours post transfections, the cells were treated with 5 μg/mL HCV core protein and incubated for an additional 6 h at 37 °C prior to further assays.
Enzyme-linked immunosorbent assay
The levels of TNF-α, IL-6 and IL-10 in the supernatants were measured using a human ELISA kit (elabscience) according to the manufacturer’s instructions. The detection ranges of TNF-α, IL-6 and IL-10 are > 8 pg/mL, > 4 pg/mL, and 7.813-500 pg/mL, respectively.
Western blot analysis
Cell lysates were prepared in a sodium dodecyl sulfate (SDS) sample buffer [62.5 mmol/L Tris-HCl (pH 6.8), 2% SDS, 10% glycerol, 50 mmol/L 1,4-dithiothreitol, and 0.1% bromophenol blue] containing a mixture of protease and phosphatase inhibitors. Lysates (25 μg) were separated on 12% acrylamide gels and transferred to a nitrocellulose membrane in Tris-glycine buffer containing 20% methanol. The membranes were then blocked with 5% milk in 1 × Tris-buffered saline and 0.1% Tween-20 for 1 h at room temperature and probed with various diluted primary monoclonal antibodies overnight at 4 °C with constant rocking. After extensive washing, the membranes were incubated with horseradish peroxidase-conjugated anti-mouse immunoglobulin G (Boster Biological Tech) at room temperature for 2 h. Protein expression was determined using a chemiluminesence method (Thermo Scientific). Antibodies recognizing TLR2, total NF-κBp65, phosphor-NF-κBp65 (Abcam), MyD88(Bioworld), phospho-ERK (Cell Signaling), and phospho-P38 (Santa Cruz Biotech) were used to probe the membranes. To control protein loading, the blots were stripped and re-probed with an anti-glyceraldehyde 3-phosphate dehydrogenase antibody (Xianzhi Lifescience).
Statistical analysis
Data are shown as mean ± SEM. Unpaired two-tailed Student’s t-test was used to compare two independent groups. Graphpad 5.0 software (San Diego, CA) was used for all statistical analyses, and P-values < 0.05 were considered statistically significant.
RESULTS
HCV core protein up-regulates TNF-α, IL-6 and IL-10 expression and down-regulates miR-125b expression in THP-1 cells
We first tested whether treatment with HCV core protein regulates miRNA expression. THP-1 was incubated with various concentrations (0.5, 1, 5 μg/mL) of HCV core protein for different time periods. Expression of miR-125b, miR-146 and miR-155, along with selected cytokine genes (TNF-α, IL-6 and IL-10) was examined by RT-qPCR analyses. Consistent with those of Yao et al[17], our results (Figure 1) showed that HCV core protein could directly stimulate THP-1 cells to express TNF-α, IL-6 and IL-10. The induction was dose-dependent; HCV core protein at 0.5 μg/mL significantly induced these cytokines more than two folds. The induction was also rapid; it peaked at 6 h after HCV core protein stimulation and slowly leveled off through the study period (24 h).
Figure 1.
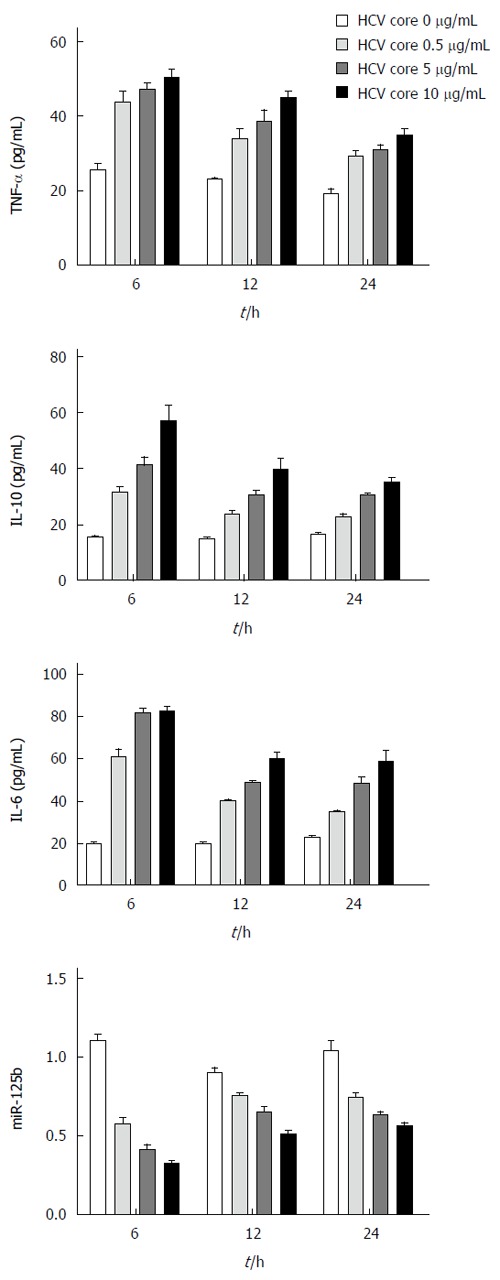
Increased cytokine expression and decreased miR-125b expression in hepatitis C virus core protein treated THP-1 cells. THP-1 cells were treated with HCV core recombinant protein at a final concentration of 0.5, 1.0 or 5.0 μg/mL and incubated for 6, 12 or 24 h. The levels of tumor necrosis factor (TNF)-α, interleukin (IL)-10, and IL-6 in culture supernatants were analyzed by enzyme-linked immunosorbent assay. The cellular miR-125b levels were determined by real-time quantitative polymerase chain reaction. The data are representative of three experiments and shown as mean ± SEM.
In parallel to the cytokine induction, miR-146 and miR-155 expression was also induced (data not shown)[12]. In contrast, miR-125 expression was down-regulated by HCV core stimulation in a dose-dependent manner. The miR-125b expression reached the lowest level (decreased by 68%) at 6 h with 5.0 μg/mL HCV core protein challenge. An inverse correlation between miR-125b expression and that of cytokines suggests that miR-125b may play a regulatory role in HCV-induced cellular responses.
TLR2 or MyD88 genes knockdown abolishes the suppressive effect of HCV core protein on miR-125b expression
TLR2 and its downstream signaling protein Myd88 play a prominent role in HCV core-induced inflammatory responses[18]. We employed a gene knockdown strategy to test if TLR2 or Myd88 could be directly involved in the repression of miR-125b. THP-1 cells were transfected with siRNA plasmids that could silence TLR2, MyD88 or the control plasmid for 24 h. The transfected cells were challenged with HCV core protein at 5 μg/mL for an additional 6 h. The miR-125b level was detected by RT-qPCR analysis. As shown in Figure 2A and 2B, we found that TLR2 and MyD88 mRNA and protein were reduced more than 2.5-fold when cells were transfected with these siRNA plasmids. When these transfected cells were exposed to HCV core protein, miR-125b expression was significantly restored compared to that of control-transfected cells. (P < 0.05, n = 3, Figure 2C). This result suggests that the suppression of miR-125b expression by HCV core treatment depends at least in part on the TLR2/MyD88 signaling.
Figure 2.
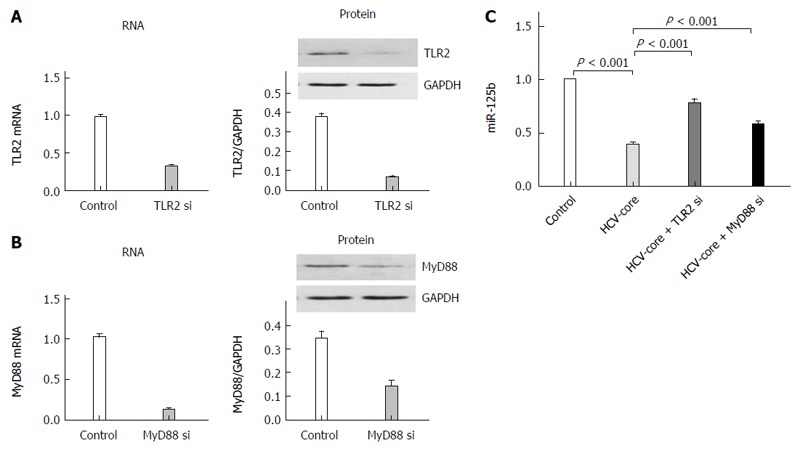
TLR2-MyD88 pathway dependent HCV core protein-induced miR-125b repression. TLR2 (A) or MyD88 (B) mRNA was analyzed by real-time quantitative polymerase chain reaction (RT-qPCR) and protein lysates were analyzed by Western blot at 6 h post transfection. The transfected cells were infected with HCV core protein at 5 μg/mL for 6 h and miR-125b expression was detected by RT-qPCR analysis (C). Cell data are representative of three experiments and are shown as mean ± SEM (P < 0.001 by unpaired two-tailed Student’s t-test).
Upregulation of cytokines by HCV core protein is abrogated by the miR-125b mimic
The inverse correlation noted between cytokine and miR-125b in HCV core protein-treated cells prompts us to examine whether miR-125b can directly participate in regulation of these HCV core protein-induced events. We transfected THP-1 cells with a chemically modified miR-125bM (miR-125b mimic) RNA oligos or the control RNA oligos for 24 h, followed by HCV core stimulation at 5.0 μg/mL for 6 h and examined the expression of TNF-α, IL-6 and IL-10. The effectiveness of miR-125b mimic transfection is shown in Figure 3A. In the experiment in which cells were treated with HCV core protein (Figure 3B), we found that transfection with control miRNA had no effect on the enhancement of cytokine expression by HCV core stimulation but that transfection with miR-125b mimic abrogated upregulated expression of TNF-α, IL-6 and IL-10 by 66%, 54% and 66%, respectively, compared with the control. These results indicate that overexpression of miR-125b down-regulates HCV-mediated cytokine induction.
Figure 3.
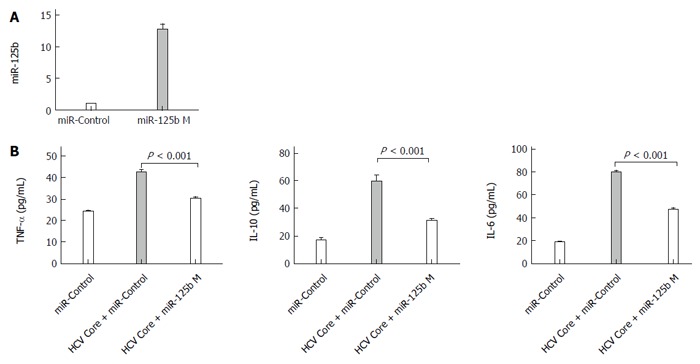
The miR-125b mimic abrogats hepatitis C virus core protein-induced upregulation of cytokines. A: Real-time quantitative polymerase chain reaction analysis of miR-125b levels in THP-1 cells transfected with 0.4 nmol/L miRNA negative control (miR-control) or miR-125b mimic (miR-125b M), respectively; B: Twenty four hours after transfection, cells were treated with 5.0 μg/mL HCV core protein for an additional 6 h. The concentrations of tumor necrosis factor (TNF)-α, interleukin (IL)-10 and IL-6 in culture supernatants were analyzed by enzyme-linked immunosorbent assay. These data are representative of three experiments and shown as mean ± SEM (P < 0.001 by unpaired two-tailed Student’s t-test).
MiR-125b mimic suppresses TLR2/MyD88 signaling
To understand the mechanism of miR-125b in regulating cytokine expression in HCV core protein-treated cells, we performed immunoblot analyses to determine the expression levels of the signaling molecules possibly involved in the TLR signaling pathways. THP-1 cells were transfected with or without miR-125b mimic and incubated with 5 μg/mL HCV core protein. The expression of various TLR signaling proteins was analyzed. As shown in Figure 4A, expression of TLR2 and MyD88 and the phosphorylation of their downstream signaling proteins ERK1/2, NF-κBp5 and p38 were up-regulated by HCV core treatments. The transfection of cells with the miR-125b mimic did not change the expression of TLR2 or NF-κBp65 but slightly down-regulated that of MyD88. In contrast, the phosphorylation of NF-κBp65, p38 and ERK1/2 was significantly down-regulated. Quantitation of three independent experiments showed two-fold down-regulation (Figure 4B).
Figure 4.
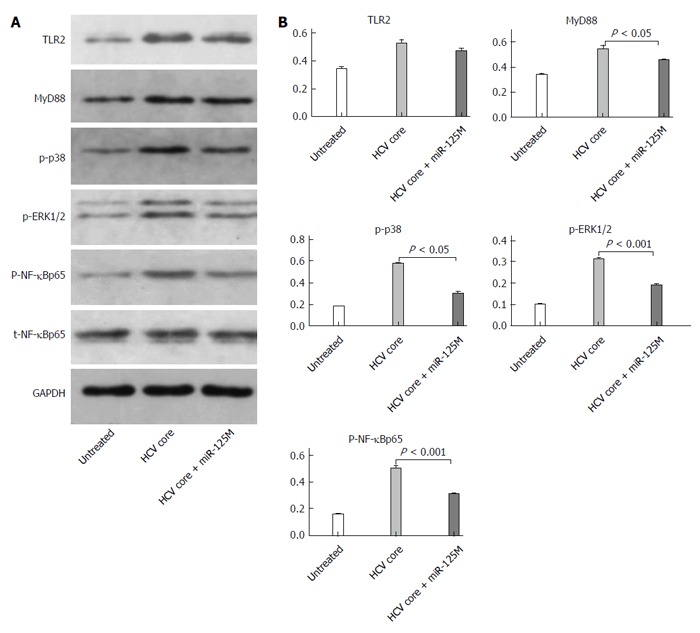
MiR-125b suppresses phosphorylation of NF-κBp65, ERK1/2 and p38. A: THP-1 cells were transfected with control or miR-125b mimic (miR-125bM) for 24 h, followed by incubation with hepatitis C virus (HCV) core protein at 5.0 μg/mL for 6 h. Western blot analysis of Toll-like receptor 2 (TLR2), MyD88, NF-κB, phosphorylated NF-κB, phosphorylated ERK1/2 and phosphorylated P38 in the collected cells; B: Densitometric quantification of the western blot data was performed using Quantity One Software. These data are representative of three experiments and shown as mean ± SEM (P < 0.05, P < 0.001 by unpaired two-tailed Student’s t-test).
DISCUSSION
In this study, we used recombinant HCV core protein-treated THP-1 cells as a model to investigate the interaction between HCV and monocytes. HCV core protein has been shown to be capable of directly interacting with various cellular proteins including TLR2 and resulting in activation of TLR2-MyD88 signaling cascade in monocytes[18]. In agreement with these findings, our current findings indicate that HCV core protein can induce a similar profile of cytokine production in THP-1 cells. We further extended the study by demonstrating for the first time that treatment with HCV core protein suppressed the expression of miR-125b in a dose- and time-dependent manner (Figure 1). In addition, we demonstrated that this suppression was dependent on the TLR2/MyD88 pathway (Figure 2). However, the mechanism by which miR-125b expression is inhibited by HCV core protein treatment remains unknown.
MiR-125b has gained special interest in the field of cancer research because of its dysregulation in a broad variety of tumors[19]. Studing its role in tumorigenesis suggests that miR-125b could have the opposite effect depending on cellular context; miR-125b is upregulated in some tumors such as colon cancer and hematopoietic tumors, suggesting its oncogenic potential capability of facilitating cell proliferation and blocking apoptotic pathways[20,21]. On the other hand, miR-125b is down-regulated in other types of tumors, such as mammary tumors and HCC. The overexpression of miR-125b inhibits HCC cell proliferation via promoting apoptosis and other anti-tumor mechanisms[22].
Our finding that forced expression of miR-125b suppressed HCV core protein-induced cytokine production (Figure 3) and phosphorylation of the TLR/MyD88 signaling cascade (Figure 4) supports the notion that miR-125b suppresses HCV core protein-mediated events by inhibition of TLR2/MyD88 signaling. Although many receptors, signaling molecules, and transcriptional factors of the TLR signaling pathways have been verified to be regulated by various miRNAs[23], the molecules targeted by miR-125b in the TLR2/MyD88 signaling pathway remain unknown. Like other microRNAs, it is assumed that miR-125b exerts its biological function by directly targeting the 3′ untranslated (3′UTR) region of genes. Target site predictions using web-accessible miRNA database search programs (http://www.microrna.org,http://www.miRBase.org and http://www.targetscan.org) identified a miR-125b target site within the 1.3 kB 3′UTR of murine MyD88 mRNA. A study by Wang et al[24] indicated that the treatment of mouse macrophages with a miR-125b inhibitor induced MyD88 expression and that cellular transfection with a murine miR-125b mimic down-regulated the reporter gene activities mediated by 3′UTR MyD88 construct. These results provided clear evidence that murine miR-125b suppresses TLR/MyD88 by directly targeting MyD88 mRNA. We are currently testing this hypothesis by transfecting HCV core protein-treated THP-1 cells with the 3′UTR of MyD88 reporter construct.
In summary, our findings indicated that miR-125b may function as a negative regulator for HCV core protein-induced cellular events, which may provide insight into the role of miR-125b in the innate immune responses against HCV in monocytes. Our study may lay some groundwork for the development of a novel therapeutic approach for treating HCV core protein-induced inflammation and immune activation.
COMMENTS
Background
Macrophages/monocytes have been shown to be important immune cells mediating both innate and adaptive immunity in hepatitis C virus (HCV)-infected patients. Many anti-pathogen pathways, including pattern recognition Toll-like receptors (TLRs) mediating signaling, are known to be regulated by a network of microRNAs. MiR-125b is highly expressed in peripheral blood mononuclear cells, particularly monocytes/macrophages; however, its role in regulating monocyte immune responses induced by HCV has not yet been investigated.
Research frontiers
Recent studies have shown that several microRNAs, including miR-21, miR-146a and miR-155, are involved in regulation of virus-host interactions and may play important roles in the pathogenesis of HCV infection.
Innovations and breakthroughs
This is the first study to report that miR-125b may negatively regulate HCV core protein-induced host immune responses by targeting TLR2/MyD88 signaling in monocytes.
Applications
The present findings indicated that miR-125b may function as a negative regulator for HCV induced cellular events, which may provide insight into the role of miR-125b in the innate immune responses against HCV in monocytes. This study may lay some groundwork for the development of a novel therapeutic approach for treating HCV-induced inflammation and immune activation.
Terminology
MicroRNAs are highly conserved endogenous non-coding small RNAs that act as translational repressors to regulate a wide range of biological processes. They have also been implicated in the pathogenesis, diagnosis, and therapeutic aspects of HCV infections. miR-125b is highly expressed in peripheral blood mononuclear cells, particularly in monocytes/macrophages, and its expression is inversely associated with better treatment outcomes for chronic HCV infection.
Peer-review
The authors investigated the possible role of miR-125b in regulating monocyte immune responses induced by HCV core protein. This study found that cytokine production was up-regulated and miR-125b expression was down-regulated by HCV core protein treatment through TLR2/MyD88 signaling in THP-1 cells. Overexpression of miR125b resulted in suppressed MAPK and NF-κB activity and cytokine production. This research is novel and manuscript presentation and readability are good. Additionally, the manuscript is well organized and its results are sound with respect to the integrity of the paper.
Footnotes
Supported by National Natural Science Foundation of China, No. 81202321 and No. 81461130019; Transregio-SFB (TRR) of the Deutsche Forschungsgemeinschaft (DFG), No. TRR60.
Institutional review board statement: This study was approved by the Institutional Review Board of Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, China.
Conflict-of-interest statement: We declare that we have no financial or personal relationships with other people or organizations that can inappropriately influence our work, and there is no professional or other personal interest of any nature or kind in any product, service and company. The authors declare that there is no conflict of interests regarding the publication of this paper.
Data sharing statement: Technical appendix, statistical code, and dataset available from the corresponding author at dlyang@hust.edu.cn. Participants gave informed consent for data sharing. No additional data are available.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Peer-review started: December 31, 2015
First decision: January 28, 2016
Article in press: March 14, 2016
P- Reviewer: Celikbilek M, Lee WH, Sunbul M S- Editor: Yu J L- Editor: Wang TQ E- Editor: Liu XM
References
- 1.Mohd Hanafiah K, Groeger J, Flaxman AD, Wiersma ST. Global epidemiology of hepatitis C virus infection: new estimates of age-specific antibody to HCV seroprevalence. Hepatology. 2013;57:1333–1342. doi: 10.1002/hep.26141. [DOI] [PubMed] [Google Scholar]
- 2.Li HC, Ma HC, Yang CH, Lo SY. Production and pathogenicity of hepatitis C virus core gene products. World J Gastroenterol. 2014;20:7104–7122. doi: 10.3748/wjg.v20.i23.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bouffard P, Hayashi PH, Acevedo R, Levy N, Zeldis JB. Hepatitis C virus is detected in a monocyte/macrophage subpopulation of peripheral blood mononuclear cells of infected patients. J Infect Dis. 1992;166:1276–1280. doi: 10.1093/infdis/166.6.1276. [DOI] [PubMed] [Google Scholar]
- 4.Lerat H, Berby F, Trabaud MA, Vidalin O, Major M, Trépo C, Inchauspé G. Specific detection of hepatitis C virus minus strand RNA in hematopoietic cells. J Clin Invest. 1996;97:845–851. doi: 10.1172/JCI118485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heim MH, Thimme R. Innate and adaptive immune responses in HCV infections. J Hepatol. 2014;61:S14–S25. doi: 10.1016/j.jhep.2014.06.035. [DOI] [PubMed] [Google Scholar]
- 6.Lechmann M, Woitas RP, Langhans B, Kaiser R, Ihlenfeldt HG, Jung G, Sauerbruch T, Spengler U. Decreased frequency of HCV core-specific peripheral blood mononuclear cells with type 1 cytokine secretion in chronic hepatitis C. J Hepatol. 1999;31:971–978. doi: 10.1016/s0168-8278(99)80307-9. [DOI] [PubMed] [Google Scholar]
- 7.Woitas RP, Petersen U, Moshage D, Brackmann HH, Matz B, Sauerbruch T, Spengler U. HCV-specific cytokine induction in monocytes of patients with different outcomes of hepatitis C. World J Gastroenterol. 2002;8:562–566. doi: 10.3748/wjg.v8.i3.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zampino R, Marrone A, Restivo L, Guerrera B, Sellitto A, Rinaldi L, Romano C, Adinolfi LE. Chronic HCV infection and inflammation: Clinical impact on hepatic and extra-hepatic manifestations. World J Hepatol. 2013;5:528–540. doi: 10.4254/wjh.v5.i10.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Villacres MC, Literat O, DeGiacomo M, Du W, Frederick T, Kovacs A. Defective response to Toll-like receptor 3 and 4 ligands by activated monocytes in chronic hepatitis C virus infection. J Viral Hepat. 2008;15:137–144. doi: 10.1111/j.1365-2893.2007.00904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ambros V, Chen X. The regulation of genes and genomes by small RNAs. Development. 2007;134:1635–1641. doi: 10.1242/dev.002006. [DOI] [PubMed] [Google Scholar]
- 11.Gragnani L, Piluso A, Fognani E, Zignego AL. MicroRNA expression in hepatitis C virus-related malignancies: A brief review. World J Gastroenterol. 2015;21:8562–8568. doi: 10.3748/wjg.v21.i28.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Y, Chen J, Wang H, Shi J, Wu K, Liu S, Liu Y, Wu J. HCV-induced miR-21 contributes to evasion of host immune system by targeting MyD88 and IRAK1. PLoS Pathog. 2013;9:e1003248. doi: 10.1371/journal.ppat.1003248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang WJ, Wang H, Tong QX, Jie SH, Yang DL, Peng C. Involvement of TLR2-MyD88 in abnormal expression of miR-146a in peripheral blood monocytes of patients with chronic hepatitis C. J Huazhong Univ Sci Technolog Med Sci. 2015;35:219–224. doi: 10.1007/s11596-015-1414-5. [DOI] [PubMed] [Google Scholar]
- 14.Bala S, Tilahun Y, Taha O, Alao H, Kodys K, Catalano D, Szabo G. Increased microRNA-155 expression in the serum and peripheral monocytes in chronic HCV infection. J Transl Med. 2012;10:151. doi: 10.1186/1479-5876-10-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chaudhuri AA, So AY, Sinha N, Gibson WS, Taganov KD, O’Connell RM, Baltimore D. MicroRNA-125b potentiates macrophage activation. J Immunol. 2011;187:5062–5068. doi: 10.4049/jimmunol.1102001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hsi E, Huang CF, Dai CY, Juo SH, Chou WW, Huang JF, Yeh ML, Lin ZY, Chen SC, Wang LY, et al. Peripheral blood mononuclear cells microRNA predicts treatment outcome of hepatitis C virus genotype 1 infection. Antiviral Res. 2014;105:135–142. doi: 10.1016/j.antiviral.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Yao Z, Song X, Cao S, Liang W, Lu W, Yang L, Zhang Z, Wei L. Role of the exogenous HCV core protein in the interaction of human hepatocyte proliferation and macrophage sub-populations. PLoS One. 2014;9:e108278. doi: 10.1371/journal.pone.0108278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung H, Watanabe T, Kudo M, Chiba T. Hepatitis C virus core protein induces homotolerance and cross-tolerance to Toll-like receptor ligands by activation of Toll-like receptor 2. J Infect Dis. 2010;202:853–861. doi: 10.1086/655812. [DOI] [PubMed] [Google Scholar]
- 19.Banzhaf-Strathmann J, Edbauer D. Good guy or bad guy: the opposing roles of microRNA 125b in cancer. Cell Commun Signal. 2014;12:30. doi: 10.1186/1478-811X-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willimott S, Wagner SD. miR-125b and miR-155 contribute to BCL2 repression and proliferation in response to CD40 ligand (CD154) in human leukemic B-cells. J Biol Chem. 2012;287:2608–2617. doi: 10.1074/jbc.M111.285718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishida N, Yokobori T, Mimori K, Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y, Kuwano H, Mori M. MicroRNA miR-125b is a prognostic marker in human colorectal cancer. Int J Oncol. 2011;38:1437–1443. doi: 10.3892/ijo.2011.969. [DOI] [PubMed] [Google Scholar]
- 22.Alpini G, Glaser SS, Zhang JP, Francis H, Han Y, Gong J, Stokes A, Francis T, Hughart N, Hubble L, et al. Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. J Hepatol. 2011;55:1339–1345. doi: 10.1016/j.jhep.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Neill LA, Sheedy FJ, McCoy CE. MicroRNAs: the fine-tuners of Toll-like receptor signalling. Nat Rev Immunol. 2011;11:163–175. doi: 10.1038/nri2957. [DOI] [PubMed] [Google Scholar]
- 24.Wang Z, Filgueiras LR, Wang S, Serezani AP, Peters-Golden M, Jancar S, Serezani CH. Leukotriene B4 enhances the generation of proinflammatory microRNAs to promote MyD88-dependent macrophage activation. J Immunol. 2014;192:2349–2356. doi: 10.4049/jimmunol.1302982. [DOI] [PMC free article] [PubMed] [Google Scholar]


