Abstract
Aberrant expression of ROS1, ALK or c-MET (RAM) is implicated in carcinogenesis and cancer drug resistance. We retrospectively evaluated the effect of RAM expression on outcomes for advanced biliary tract cancer patients, who were treated with gemcitabine plus oxaliplatin (GEMOX), with or without cetuximab, in a randomized phase II trial. RAM expression levels on archived tissue sections were scored using immunohistochemistry (IHC). Of 110 tumors with IHC staining for all three markers, 18 were RAMhigh (IHC intensity 3+ for any markers). Ninety-two tumors were RAMlow (IHC intensity <3+ for all markers). All RAMhigh tumors were intra-hepatic cholangiocarcinomas (IHCC). Of the patients with IHCC (n = 80), median overall survival (OS) of RAMhigh group was inferior to that of the RAMlow group (5.7 vs. 11.7 months, p = 0.021). In multivariate analysis RAMhigh remained an independently adverse prognostic factor, with a hazard ratio of 2.01 (p = 0.039). In the RAMlow group, GEMOX treatment with cetuximab significantly improved the disease control rate (68% vs. 41%, p = 0.044), median progression-free survival (7.3 vs. 4.9 months, p = 0.026), and marginally prolonged median OS (14.1 vs 9.6 months, p = 0.056), compared to GEMOX treatment alone. Future trials of anti-EGFR inhibitors for IHCC may consider RAM expression as a patient stratification factor.
Systemic chemotherapy with gemcitabine plus platinum is recognized as the standard of care for patients with advanced or metastatic biliary tract cancer (ABTC)1. However, treatment outcomes are largely unsatisfactory with a median overall survival (OS) ranging from 7 to 11 months2,3. Combination of inhibitors targeting the epidermal growth factor receptor (EGFR) pathway with chemotherapy has been extensively studied to improve the efficacy of chemotherapy4,5,6,7. Although trends of improved objective response rate and progression-free survival (PFS) had been observed in certain trials, none of these including “all comers” randomized trials could demonstrate the survival benefit of add-on anti-EGFR inhibitors in ABTC patients receiving gemcitabine plus platinum chemotherapy4,5,7. Identification of predictive and prognostic biomarkers is crucial to better define patients who may benefit from targeted molecular therapies such as anti-EGFR inhibitors.
Various biomarkers predictive for anti-EGFR inhibitor efficacy have been explored for different cancer types. For example, RAS/BRAF mutations predicted poor response to cetuximab in patients with metastatic colorectal cancers8. In another study, high EGFR expression predicted incremental benefit from cetuximab combine with chemotherapy in patients with advanced non-small cell lung cancer (NSCLC)9. However, two previous randomized studies with either pre-planned or post-hoc analyses failed to demonstrate an association between the status of RAS/BRAF mutations and the therapeutic efficacy of cetuximab in ABTC, or between the latter and EGFR expression4,7. Additional molecular alternations have been identified to account for the resistance to EGFR inhibition in NSCLC and provide opportunities for new drug development. Among them, over-expression of c-MET, which could result in activation of hepatocyte growth factor (HGF)-c-MET signaling correlates with resistance to anti-EGFR therapy in patients with NSCLC10. c-MET overexpression has been detected in 11.7% and 16.2% of intra- and extra-hepatic cholangiocarcinoma (IHCC and EHCC) cases, respectively, and associated with inferior post-resection recurrence-free survival than those without11. Chromosome rearrangements involving either anaplastic lymphoma kinase (ALK) or ROS1 genes have been identified in wild-type EGFR NSCLCs that were refractory to anti-EGFR therapy12,13. ROS1 rearrangement has been detected in 8.7% of IHCC cases and shown to be oncogenic in the BTC mouse model, whereas ALK rearrangement or aberrant protein expression has not been described in ABTC14,15. In NSCLC, ALK or ROS1 rearrangement-harboring tumors exhibited compatible response to ALK inhibitors because of the high concordance of kinase domain sequences16,17. Whether alternations in c-MET, ROS1, and ALK signaling pathways may impact the clinical outcomes of patients undergoing anti-EGFR therapy and provide opportunities for new ABTC drug development, remains unexplored.
The present post-hoc study evaluates predictive and prognostic significances of aberrant ROS1, ALK or c-MET (RAM) expression in a prospective cohort of ABTC patients who received gemcitabine plus oxaliplatin (GEMOX) chemotherapy with or without cetuximab in a randomized phase II study.
Results
RAM protein expression and rearrangement of ROS1 and ALK genes
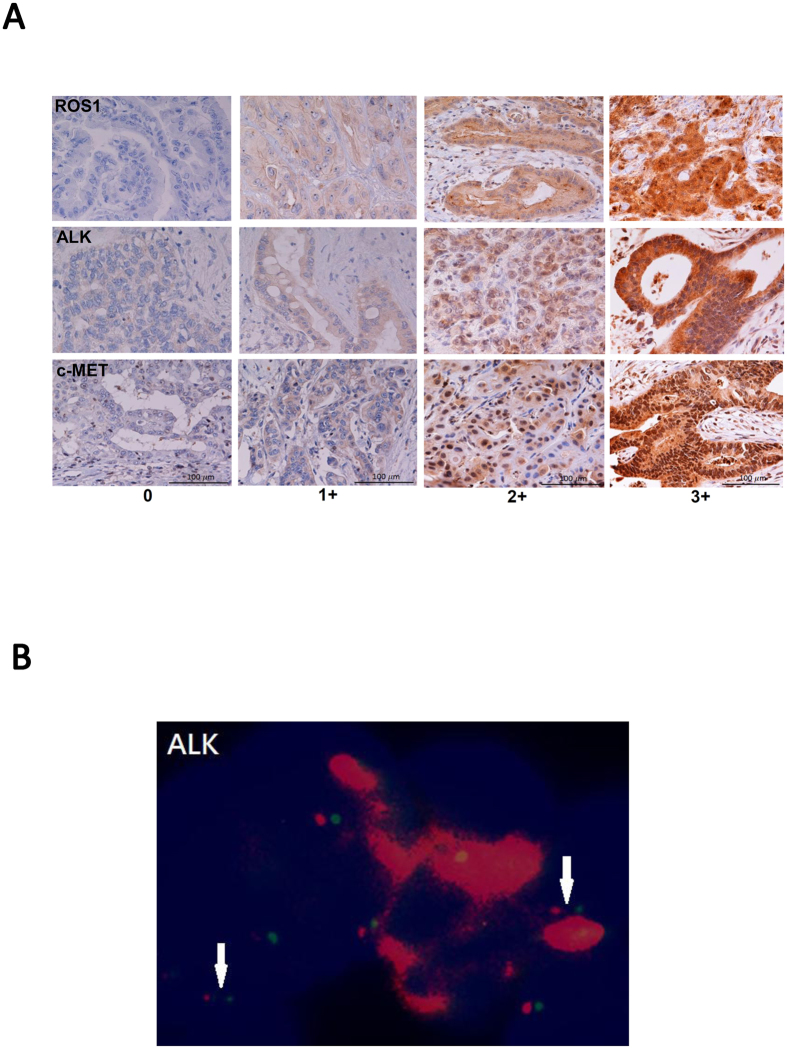
Of the tumor tissue sections from 122 patients who participated in the prospective randomized phase II trial, sections from 110 patients had sufficient tissue for IHC of all three markers. The staining of the RAM markers was mainly localized in the cytoplasm of tumor cells (Fig. 1A). Among them, 18 of cases (16%) were categorized as RAMhigh, with 3+ IHC staining of ROS1 in 9 cases, ALK in 5, and c-MET in 6 (Supplementary Table 1). One tumor overexpressed all three markers. ALK rearrangement was detected in 1 of the 5 tumors presenting ALK overexpression (Fig. 1B), whereas none of the 9 tumors with ROS1 overexpression had detectable ROS1 rearrangement.
Figure 1.
(A) ROS1, ALK and c-MET expression in primary intra-hepatic cholangiocarcinoma (expression score: 0, 1+, 2+ and 3+). ROS1 and ALK are mainly localized in cytoplasm, and c-MET is localized in both cytoplasm and nucleus. (B) Break-apart Fluorescence in situ hybridization for ALK revealed wide-apart of the red and green signals (white arrows), indicating ALK translocation.
Correlations between RAM expression and clinicopathological factors
As of December 31, 2014, the median follow-up was 10.3 months (range 0.9–45.4 months). The demography of the 110 ABTC patients with RAMhigh or RAMlow tumors is listed in Table 1. Compared with RAMlow tumors, RAMhigh tumors were more likely to be IHCC (100% vs. 67.4%, p = 0.015), without prior surgery (0% vs. 48.9%, p < 0.0001), and with concurrent mutant KRAS (55.6% vs. 31.5%, p = 0.063). The trend of differences in the baseline characteristics between RAMhigh and RAMlow groups was similar in the 80 patients with IHCC (Supplementary Table 2).
Table 1. Demographic and clinical characteristic of 110 patients.
| RAMhigh (n = 18) | RAMlow (n = 92) | p | |
|---|---|---|---|
| Age, years | 1.00 | ||
| <60 | 9 (50%) | 47 (51%) | |
| ≥60 | 9 (50%) | 45 (49%) | |
| Sex | 0.61 | ||
| Men | 7 (39%) | 45 (49%) | |
| Women | 11 (61%) | 47 (51%) | |
| ECOG PS | 1.00 | ||
| 0 | 5 (28%) | 29 (32%) | |
| 1 | 13 (72%) | 63 (69%) | |
| Primary site | 0.02 | ||
| Intra-hepatic | 18 (100%) | 62 (67%) | |
| Extra-hepatic/GB | 0 (0%) | 30 (33%) | |
| T stage | 1.00 | ||
| ≤T3 | 14 (78%) | 73 (79%) | |
| T4 | 4 (22%) | 19 (21%) | |
| N stage | 0.39 | ||
| N0 | 3 (17%) | 27 (29%) | |
| N1 | 15 (83%) | 65 (71%) | |
| Disease status at entry | 0.29 | ||
| Locally advanced | 4 (22%) | 34 (37%) | |
| Distant metastasis | 14 (78%) | 58 (63%) | |
| Previous surgery | <0.001 | ||
| No | 18 (100%) | 45 (49%) | |
| Yes | 0 (0%) | 47 (51%) | |
| KRAS mutation | 0.06 | ||
| Negative | 8 (44%) | 63 (69%) | |
| Positive | 10 (56%) | 29 (32%) | |
| EGFR expression | 0.61 | ||
| Negative | 6 (33%) | 37 (41%) | |
| Positive | 12 (67%) | 54 (59%) | |
| HBsAg or anti-HCV Ab | 0.56 | ||
| Negative | 15 (83%) | 69 (75%) | |
| Positive | 3 (17%) | 23 (25%) |
RAM, ROS1/ALK/c-MET; ECOG PS, Eastern Cooperative Oncology Group performance status; GB, gallbladder; EGFR, epidermal growth factor receptor; HBsAg, surface antigen of hepatitis B virus; HCV Ab, antibody of hepatitis C virus.
Comparison of treatment efficacies
Of the 110 patients included in this study, those who received C-GEMOX had a tended to have better ORR, better long-term disease control rates (DCR≥16wk, objective response according to RECIST plus stable disease [SD] ≥ 16 weeks), and longer survival compared to patients who received GEMOX (Supplementary Table 3)2. Because all RAMhigh tumors were IHCCs, further analysis was performed on the IHCC population. Despite similar DCR≥16wk and PFS, IHCC patients with RAMhigh tumors had a significantly inferior median OS than patients with RAMlow tumors (5.7 vs. 11.7 months, p = 0.021, Table 2). In the RAMlow IHCC subgroup, C-GEMOX treatment achieved significantly better DCR≥16wk (68% vs. 41%, p = 0.044) and median PFS (7.3 vs. 4.9 months, p = 0.026) than GEMOX alone did. A trend toward improved OS was also observed (14.1 vs. 9.6 months, p = 0.056, Table 3 and Fig. 2). Both KRASwt and KRASmut patients in the RAMlow IHCC subgroup showed similar trends of improvement in DCR≥16wk and median PFS after C-GEMOX treatment (Supplementary Table 4). This KRAS-independent improvement suggests that KRAS mutation did not interfere with the benefits derived from add-on cetuximab to GEMOX in RAMlow cases. The findings were consistent with our previous report on the entire ABTC intention-to treat population4. However, in the RAMhigh subgroup, the DCR≥16wk, PFS and OS were similar between C-GEMOX and GEMOX-treated patients (Table 3).
Table 2. Therapeutic efficacies stratified by RAM expression in IHCC.
| RAMhigh n = 18 | RAMlow n = 62 | p## | |
|---|---|---|---|
| Objective response rate | 22% (6–48) | 24% (14–37) | 1.000 |
| Disease control rate# | 39% (17–64) | 53% (40–66) | 0.423 |
| Median PFS (months) | 4.5 (1.8–8.5) | 5.9 (4.2‒7.1) | 0.533 |
| Median OS (months) | 5.7 (3.3–9.9) | 11.7 (8.8–14.5) | 0.021 |
RAM, ROS1/ALK/c-MET; IHCC, intrahepatic cholangiocarcinoma; PFS, progression - free survival; OS, overall survival.
#Disease control rate = complete response + partial response + stable disease >16 weeks.
##Objective response rates and disease control rates were compared using Fisher’s exact test, 2-tailed; PFS and OS were compared using log-rank tests.
Table 3. Therapeutic efficacies stratified by treatment arms in IHCC with RAMlow and RAMhigh expression.
| C-GEMOX | GEMOX | p## | |
|---|---|---|---|
| RAMlow | N = 28 | N = 34 | |
| Disease control rate# | 68% (48–84) | 41% (25–59) | 0.044 |
| Median PFS (months) | 7.3 (5.6–10.9) | 4.9 (1.8–6.7) | 0.026 |
| Median OS (months) | 14.1 (10.1–20.0) | 9.6 (6.6–13.8) | 0.056 |
| RAMhigh | N=10 | N = 8 | |
| Disease control rate# | 40% (12–74) | 38% (9–76) | 1.000 |
| Median PFS (months) | 4.7 (1.5–8.6) | 4.5 (1.3–13.8) | 0.610 |
| Median OS (months) | 6.3 (2.6–12.0) | 5.2 (1.4–27.4) | 0.605 |
RAM, ROS1/ALK/c-MET; IHCC, intrahepatic cholangiocarcinoma; C-GEMOX, cetuximab plus gemcitabine and oxaplatin; PFS, progression-free survival; OS, overall survival.
#Disease control rate = complete response+ partial response+ stable disease >16 weeks.
##Disease control rates were compared using Fisher’s exact test, 2-tailed; PFS and OS were compared using log-rank test.
Figure 2.
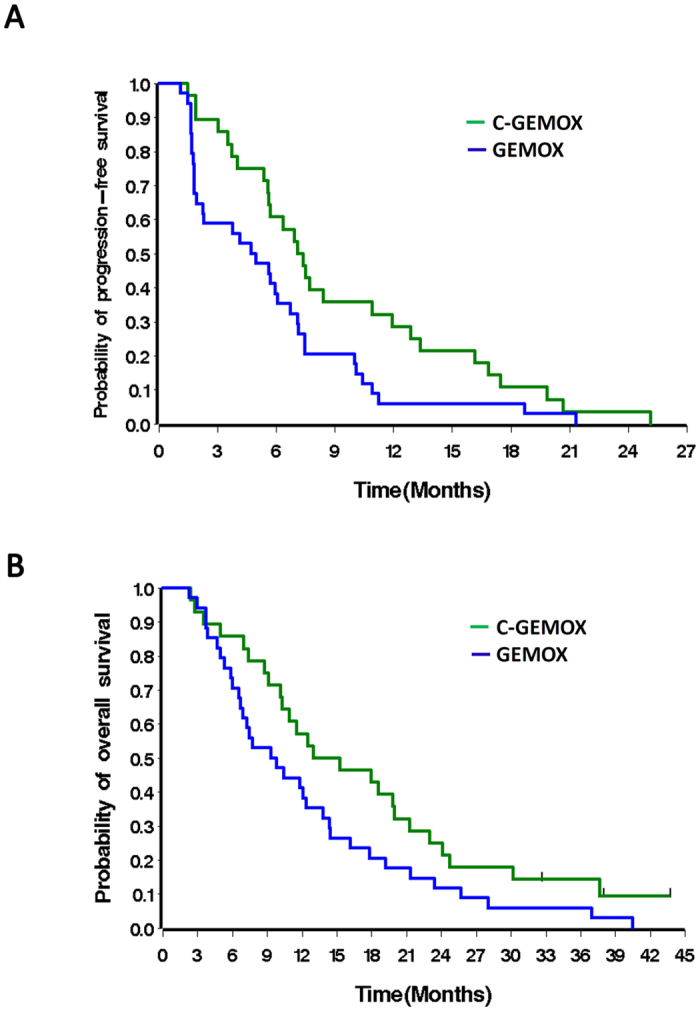
The comparison of PFS (A) and OS (B) in IHCC patients with RAMlow -expressed tumors (n = 62), receiving treatment of C-GEMOX or GEMOX. (C) cetuximab; GEMOX, gemcitabine and oxaliplatin; PFS, progression-free survival; OS, overall survival; IHCC, intrahepatic cholangiocarcinoma; RAM, ROS1/ALK/c-MET.
Multivariate analysis showed that male gender and RAMhigh subgroup were independent adverse prognostic factors for OS in 80 IHCC patients (Table 4). Male gender and GEMOX alone had only a statistically non-significant tendency to prognosticate worse PFS (Supplementary Table 5).
Table 4. Univariate and multivariate Cox regression of prognostic factors for overall survival in patients with IHCC.
| Variables | Univariate |
Multivariate |
||
|---|---|---|---|---|
| Hazard ratio (95% CI) | p | Hazard ratio (95% CI) | p | |
| Age ≥60 (vs. <60) | 1.48 (0.92–2.36) | 0.103 | 1.54 (0.92–2.59) | 0.102 |
| C-GEMOX (vs. GEMOX) | 0.75 (0.48–1.18) | 0.209 | – | – |
| Male (vs. female) | 1.62 (1.03–2.56) | 0.038 | 2.11 (1.27–3.50) | 0.004 |
| ECOG PS 1 (vs. 0) | 1.42 (0.87–2.34) | 0.165 | 1.27 (0.73–2.22) | 0.398 |
| Disease status, Meta (vs. LA) | 1.86 (1.12–3.09) | 0.017 | 1.35 (0.76–2.41) | 0.312 |
| Prior surgery, yes (vs. no) | 0.52 (0.32–0.85) | 0.009 | 0.81 (0.46–1.44) | 0.478 |
| KRAS MT (vs. WT) | 1.62 (1.01–2.59) | 0.045 | 1.48 (0.87–2.52) | 0.153 |
| RAMhigh (vs. RAMlow) | 1.86 (1.09–3.18) | 0.023 | 2.01 (1.04–3.88) | 0.039 |
IHCC, intrahepatic cholangiocarcinoma; C-GEMOX, cetuximab plus gemcitabine and oxaliplatin; ECOG PS, Eastern Cooperative Oncology Group performance status; Meta: metastasis; LA, locally advanced; MT: mutated-type; WT: wild-type; LA: locally advanced RAM, ROS1/ALK/c-MET.
Recommendations for phase III trial design based on phase II trial with biomarkers
In the RAMlow IHCC subgroup, C-GEMOX achieved better median PFS than GEMOX did, with a hazard ratio (HR) of 1.78 at one-sided α = 0.1, and the null hypothesis was rejected, according to Freidlin et al.17. The 2-sided 80% confidence interval (CI) for the HR in the RAMhigh subpopulation was 0.39 to 1.50 (Supplementary Table 6). For RAMhigh cases, this range is consistent with the null hypothesis that the efficacy of C-GEMOX is no different from that of GEMOX. Therefore, future large-scale clinical trials should consider using RAM expression as a patient stratification factor.
Discussion
The present post-hoc analysis is the first report to describe the prognostic significance of aberrant ROS1, ALK, or c-MET expression in ABTC patients undergoing front-line therapy. We found that patients with tumors over-expressing any of the ROS1, ALK, or c-MET proteins (RAMhigh) had inferior OS, regardless of the assigned treatment. Patients with RAMlow tumors, notably of IHCC, were likely to benefit from the addition of cetuximab to GEMOX chemotherapy.
The findings of 18 RAMhigh tumors that were all IHCC cases and without prior surgery at study entry imply that aberrant RAM expression can be site-specific and associated with aggressive tumor biology. Recent comprehensive genomic profiling studies of BTCs also suggested distinct etiologies and molecular characteristics among IHCC, EHCC and gallbladder carcinoma (GBC)18,19,20,21. IDH mutation and FGFR rearrangement are usually specific to IHCC, whereas BAP1 and IDH mutations are more frequently detected in non-parasite related cholangiocarcinoma than in those related to parasite infection. Compared with IHCC, EHCC has a significantly higher frequency of TP53 and KRAS mutations. These BTC subtype-specific genetic alternations indicate that studies of molecular pathogenesis and therapeutic development may be most fruitful on a subtype-specific basis. The observation of higher KRAS mutation rate in RAMhigh tumors indicates that the RAMhigh phenotype might be related to the ‘proliferation’ class of IHCC, which is associated with worse clinical outcomes than other IHCC classes22,23.
Because of the lack of association between the status of KRAS mutation or EGFR positivity and therapeutic benefit of cetuximab as shown in our primary report, we strongly encourage a search for other predictive biomarkers for anti-EGFR therapy in ABTC. ROS1, ALK and c-MET, appear to be reasonable choices because the known crosstalk between EGFR signaling and all three signaling pathways, and their genetic alterations or aberrant expression may confer resistance to EGFR inhibition in NSCLC or other cancer types24,25,26,27,28,29. For example, c-MET activation involves the acquisition of resistance to anti-EGFR therapy via bypass of the EGFR pathway26. EGFR activation can mediate the resistance to ROS1 inhibition in NSCLC25. In addition, ROS1 activation mediates resistance to EGFR inhibition in glioblastoma cells24. ALK kinase is another druggable target in NSCLC patients who do not respond to EGFR inhibitors or have EGFR wild-type tumors; ALK activation may be associated with resistance to EGFR tyrosine kinase inhibitor27,28. Although aberrant ALK signaling has not been reported in BTC, ROS1 rearrangement and c-MET over-expression have been detected in cholangiocarcinoma; the latter observations are associated with aggressive biological behavior11,14,15. For clinically available inhibitors of receptor tyrosine kinase targeting either ALK/ROS1 or ALK/ROS1 and c-MET30,31, we include ALK expression in our analysis. Finally, due to the low respective incidence of aberrant ROS1, ALK, or c-MET expression, a pooled analysis of all 3 molecules was performed to increase the statistical power of our analysis.
Although our previous studies demonstrated a high concordance between ALK rearrangement and protein over-expression in surgically resected NSCLC samples32, FISH detected chromosomal translocation for only 1 of 5 ALK over-expression tumors in the present study (ALK rearrangement, Fig. 1B). However, our present finding of discord between the frequency of ROS1 over-expression and that of ROS1 rearrangement is consistent with results of previous IHCC and NSCLC studies. Seventy-two of 194 surgically resected IHCC tumors (37%) exhibited high expression level of ROS1; none of the 102 samples successfully analyzed using FISH had detectable ROS1 rearrangement33. Analyses of NSCLC tumors found that the frequency of ROS1 over-expression and ROS1 rearrangement was 22% (9 of 399) and 1.7% (18 of 1037), respectively12,34. Lee et al. further showed that a small fraction of NSCLC tumors over-expressing ROS1 exhibited hypo-methylation in the ROS1 promoter region; however, the mechanisms of this dysregulation remain largely un-elucidated34. Our discordant IHC and FISH results could be caused by the technical difficulty posed by FISH analyses of small IHCC biopsy tissue samples. Another possible explanation of the discordance could be that translational or post-translational regulation enhances the expression of ROS1/ALK proteins.
Our data indicate that RAMlow expression predicts therapeutic efficacy of add-on cetuximab in IHCC with GEMOX-based therapy. These results are applicable to this statistical model proposed by Freidlin et al., which aims to clarify the values off biomarkers in designing phase III randomized trials of targeted therapies35. RAM expression status should be considered as a stratification factor (RAMlow vs. RAMhigh subgroups) for future trials of anti-EGFR inhibitors in IHCC. However, the wide 80% CI of the HR estimate (Supplementary Table 6), resulting from the small sample size of patients in the RAMhigh subgroup, possibly misled the conclusion according to Freidlin’s algorithm. Therefore, we strongly recommend adoption of this algorithm at the interim analysis of phase III studies, through monitoring the HR of biomarker-defined subgroups. Such monitoring should be used to determine whether RAM should be used continuously as a stratification factor or shifted to the role of an enrichment factor. Based on the dismal survival outcome of RAMhigh patients and the well-known clinical activity of ALK inhibitors in NSCLC with ALK and/or ROS1 rearrangement, an investigator-initiated phase II trial is ongoing. This trial evaluates the efficacy and safety of ceritinib in patients with IHCC over-expressing ROS1 or ALK (clinicaltrials.gov NCT2374489).
For IHCC patients, ROS1, ALK and c-MET expression levels have prognostic significance on clinical outcomes. Although this finding may require further validation, it has led to proposal of a new stratification or enriched biomarker for future phase III trial of anti-EGFR therapy in IHCC. This example demonstrates the importance of prospective tissue sample collection from well-designed clinical trials for companion exploratory biomarker studies. Such exploratory studies can facilitate further drug development.
Methods
Patients
The current study analyzed archival tumor tissues from patients with ABTC who participated in a Taiwan Cooperative Oncology Group (TCOG)-sponsored randomized phase II trial comparing first-line GEMOX alone versus GEMOX plus cetuximab (C-GEMOX)4. In brief, 122 patients were 1:1 randomized, stratified by tumorous KRAS status, ECOG performance status, and location of primary tumor, and then 1:1 randomized. The patients received GEMOX or C-GEMOX until disease progression or the occurrence of unacceptable toxicity. The primary end point of the study was objective response rate (ORR) by RECIST. Trends toward improvement in ORR (27% vs. 15%, p = 0.12) and median PFS (6.7 vs. 4.1 months, p = 0.05) favoring C-GEMOX were observed. The trends of improvement did not correlate with tumorous KRAS mutation status. However, C-GEMOX did not improve OS (10.6 vs. 9.8 months, p = 0.91) at primary analysis with a cut-off date on April 30, 20134. In the present study, survival follow-up was updated to December 31, 2014. Tumor procurement was approved by the institutional review board of National Health Research Institutes (EC1040427-E). Written informed consent was obtained from all subjects and the study was conducted in accordance with International Conference Statements for Good Clinical Laboratory Practice and the declaration of Helsinki of the World Health Medical Association.
KRAS mutation analysis
Genomic DNA was extracted from tumor tissues using TaKaRa DEXPATTM (TaKaRa Bio Inc, Japan). KRAS mutation status was then determined using direct sequencing of the coding sequences of exons 2 and 3, as well as the TaqMan assay for KRAS codon 12/13 mutations using the LightMix Kit (TIB MOLBIOL GmbH, Germany). Tumors were considered to be KRAS mutation if any test was positive.
Immunohistochemical study
Formalin-fixed, paraffin-embedded (FFPE) tumor tissue sections were subjected to immunohistochemistry (IHC) to determine RAM expression levels. In brief, 4-μm-thick tissue sections were de-paraffinized, followed by antigen retrieval (Cell Conditioning 1, Roche, Rotkreuz, Switzerland) for 10 minutes (for ROS1 staining) or 15 minutes (for ALK and c-MET staining) at 95 °C. The slides were then incubated with primary antibody against ROS1 (Abcam plc., Cambridge, UK, in 1:100 dilution for 1 hour), c-MET (clone C12, Biotechnology, Inc., Santa Cruz, Calif., in 1:200 dilution for 1 hour) or ALK (clone ZAL4, Invitrogen, Life Tech Corp, Carlsbad, Calif., in 1:500 dilution for 16 hours). The detection was performed by the Ventana Discovery XT staining system with ultraView Universal DAB detection kit (Ventana Medical Systems, Inc, Tucson, AZ) according to the manufacturer’s protocol. Two pathologists (YYS and SFH), who were blinded to the clinical information, independently reviewed and scored the IHC staining results. The expression levels of ROS1, ALK and c-MET in tumor tissue was graded as negative (0), weak (1+), moderate (2+), or high (3+), according to the highest staining intensity identified in more than 10% of the tumor tissue. Tumors with high expression level (3+ staining intensity) of any one of the RAM markers were categorized as RAMhigh tumors; those with 0 to 2+ staining intensity for all 3 markers were categorized as RAMlow tumors. EGFR protein expression was determined by immunohistochemical (IHC) staining (primary antibody: DAKO, H11 Cytomation Denmark A/S, Glostrup, Denmark), followed by staining with the ultraView Universal DAB detection kit (Ventana Medical Systems, Inc., Tucson, AZ, USA). EGFR protein expression was considered positive if ≥1% of tumor cells were staining.
Fluorescence in situ hybridization (FISH)
Tumors over-expressing ROS1 or ALK (3+) in IHC were subjected to FISH studies as previously described32. The probes for detecting ALK and ROS1 translocation were Vysis ALK Break Apart FISH Probe (Abbott Molecular Inc. Des Plaines, IL, USA) and ROS1 Split FISH probe (Abnova Biotec Co., Ltd, Taiwan), respectively. At least 100 intact, non-overlapping tumor nuclei were evaluated by using a Leica DMR fluorescence microscope (Leica Microsystems Nussloch GmbH, Nussloch, Germany). Samples subjected to FISH in which more than 15% of tumor nuclei had wide apart of red and green signals for more than 2 nuclei, or an absent green signal, were considered break-apart positive. Positive break-apart samples were considered to have ALK or ROS1 rearrangements.
Decision algorithm for recommendations of phase III trial design
To determine whether the RAM expression status may serve as a patient enrichment or patient stratification factor in future randomized trials of EGFR inhibitors for ABTC, the median PFS of patients who received GEMOX or C-GEMOX was further analyzed using an algorithm proposed by Freidlin B et al. (see supplementary information for detailed methods)35. Briefly, the median PFS was analyzed first to test whether the GEMOX and C-GEMOX treatment groups had the same PFS in the RAMlow subpopulation. If the null hypothesis was rejected, then the 2-sided 80% CI for the HR in the RAMhigh subpopulation was analyzed to determine whether RAM expression was suitable to be used as a patient-enrichment or patient-stratification biomarker in future clinical trials. If the null hypothesis was not rejected, then the 2-sided 80% CI for the HR for all patients was analyzed to determine the value of RAM expression in future clinical trials.
Statistical Analysis
Correlations between the results of IHC and clinicopathological factors were assessed using Fisher’s exact probability test or t-test. The associations between objective response rate stratified by treatment (GEMOX or C-GEMOX) and RAM expression status (RAMhigh vs RAMlow) were evaluated using the Chi-square test or Fisher’s exact test. PFS and OS were calculated using the Kaplan-Meier method and compared using the log-rank test. Univariate and multivariate Cox regression was used to estimate prognostic parameters for PFS and OS. For multivariate analysis of PFS and OS, all predictors with a p value <0.2 in univariate analysis were incorporated. Two-sided p-values less than 0.05 were considered statistically significant. All analysis was performed using the SAS 9.1 software package (SAS Institute, Inc., Cary, NC, USA).
Additional Information
How to cite this article: Chiang, N.-J. et al. Expression levels of ROS1/ALK/c-MET and therapeutic efficacy of cetuximab plus chemotherapy in advanced biliary tract cancer. Sci. Rep. 6, 25369; doi: 10.1038/srep25369 (2016).
Supplementary Material
Acknowledgments
The authors would also like to thank the research nurses of the Taiwan Cooperative Oncology Group (Yung-Hsin Chin, Tsai-Rong Chung, Wei-Lan Yu, Mei-Hua Lee, Ling-Fang Lin, Pei-Chyi Lin, Ya-Ling Wu, Hui-Ling Wang, Li-Ju Lu, Shiang-Yi Chen, Chih-Chu Wu and Te-Chih Wei) for their assistance in collecting clinical data and tumor samples. The authors would also like to thank Mei-Hsing Chuang for her assistance with data management and statistical analysis. Taiwan Cooperative Oncology Group is a government- sponsored organization with budget from National Health Research Institutes (CA-105-PP-19 and CA-105-PP-20) and Cancer Excellence Research Program (CA-105-SP-01), Ministry of Health and Welfare, Executive Yuan, Taiwan. The corresponding author had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Footnotes
J.S.C. received honorariums from Merck Serono, TTY Biopharm, and Sanofi. Y.C. received an honorarium from TTY Biopharm. L.T.C. received honorariums from Merck Serono, TTY Biopharm, Sanofi, and Eli Lilly, and also received study medication from TTY Biopharm for another investigator-initiated trial.
Author Contributions N.J.C., C.H. and L.T.C. wrote the manuscript; N.J.C., H.H.T., Y.Y.S. and S.F.H. performed the experiments and/or analyzed data; J.S.C., M.H.C., Y.C., T.S.Y., Y.S.S. and L.T.C. enrolled patients and collected tissue.
References
- Park J. O. et al. Gemcitabine Plus Cisplatin for Advanced Biliary Tract Cancer: A Systematic Review. Cancer Res Treat 47, 343–361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okusaka T. et al. Gemcitabine alone or in combination with cisplatin in patients with biliary tract cancer: a comparative multicentre study in Japan. Br J Cancer 103, 469–474 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valle J. et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 362, 1273–1281 (2010). [DOI] [PubMed] [Google Scholar]
- Chen J. S. et al. A KRAS mutation status-stratified randomized phase II trial of gemcitabine and oxaliplatin alone or in combination with cetuximab in advanced biliary tract cancer. Ann Oncol 26, 943–949 (2015). [DOI] [PubMed] [Google Scholar]
- Lee J. et al. Gemcitabine and oxaliplatin with or without erlotinib in advanced biliary-tract cancer: a multicentre, open-label, randomised, phase 3 study. Lancet Oncol 13, 181–188 (2012). [DOI] [PubMed] [Google Scholar]
- Leone F. et al. A phase II, open-label, randomized clinical trial of panitumumab plus gemcitabine and oxaliplatin (GEMOX) versus GEMOX alone as first-line treatment in advanced biliary tract cancer: The Vecti-BIL study. ASCO Meeting Abstracts 33, 281 (2015). [Google Scholar]
- Malka D. et al. Gemcitabine and oxaliplatin with or without cetuximab in advanced biliary-tract cancer (BINGO): a randomised, open-label, non-comparative phase 2 trial. Lancet Oncol 15, 819–828 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E. et al. Cetuximab plus irinotecan, fluorouracil, and leucovorin as first-line treatment for metastatic colorectal cancer: updated analysis of overall survival according to tumor KRAS and BRAF mutation status. J Clin Oncol 29, 2011–2019 (2011). [DOI] [PubMed] [Google Scholar]
- Pirker R. et al. EGFR expression as a predictor of survival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: analysis of data from the phase 3 FLEX study. Lancet Oncol 13, 33–42 (2012). [DOI] [PubMed] [Google Scholar]
- Cappuzzo F. et al. MET increased gene copy number and primary resistance to gefitinib therapy in non-small-cell lung cancer patients. Ann Oncol 20, 298–304 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto M. et al. Prognostic significance of overexpression of c-Met oncoprotein in cholangiocarcinoma. Br J Cancer 105, 131–138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergethon K. et al. ROS1 rearrangements define a unique molecular class of lung cancers. J Clin Oncol 30, 863–870 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak E. L. et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med 363, 1693–1703 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu T. L. et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. Plos One 6, e15640 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saborowski A. et al. Mouse model of intrahepatic cholangiocarcinoma validates FIG-ROS as a potent fusion oncogene and therapeutic target. Proc Natl Acad Sci USA 110, 19513–19518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. T. et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med 370, 1189–1197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. T. et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N Engl J Med. 371, 1963–1971 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan-On W. et al. Exome sequencing identifies distinct mutational patterns in liver fluke-related and non-infection-related bile duct cancers. Nat Genet 45, 1474–1478 (2013). [DOI] [PubMed] [Google Scholar]
- Chiu J. W.-Y. et al. Next-generation sequencing: Profiling gallbladder cancer (GBC). ASCO Meeting Abstracts 33, 286 (2015). [Google Scholar]
- Holcombe R. F. et al. Tumor profiling of biliary tract carcinomas to reveal distinct molecular alterations and potential therapeutic targets. ASCO Meeting Abstracts 33, 285 (2015). [Google Scholar]
- Ross J. S. et al. Comprehensive genomic profiling of biliary tract cancers to reveal tumor-specific differences and genomic alterations. ASCO Meeting Abstracts 33, 231 (2015). [Google Scholar]
- Andersen J. B. et al. Genomic and genetic characterization of cholangiocarcinoma identifies therapeutic targets for tyrosine kinase inhibitors. Gastroenterology 142, 1021–1031 e1015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sia D. et al. Integrative molecular analysis of intrahepatic cholangiocarcinoma reveals 2 classes that have different outcomes. Gastroenterology 144, 829–840 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aljohani H. et al. ROS1 amplification mediates resistance to gefitinib in glioblastoma cells. Oncotarget 6, 20388–20395 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies K. D. et al. Resistance to ROS1 inhibition mediated by EGFR pathway activation in non-small cell lung cancer. Plos One 8, e82236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dempke W. C. & Heinemann V. Resistance to EGF-R (erbB-1) and VEGF-R modulating agents. Eur J Cancer 45, 1117–1128 (2009). [DOI] [PubMed] [Google Scholar]
- Miyanaga A. et al. Activity of EGFR-tyrosine kinase and ALK inhibitors for EML4-ALK-rearranged non-small-cell lung cancer harbored coexisting EGFR mutation. BMC Cancer 13, 262 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A. T. et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol 27, 4247–4253 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J. J. et al. Lung cancers with concomitant EGFR mutations and ALK rearrangements: diverse responses to EGFR-TKI and crizotinib in relation to diverse receptors phosphorylation. Clin Cancer Res 20, 1383–1392 (2014). [DOI] [PubMed] [Google Scholar]
- Le X. et al. Detection of Crizotinib-Sensitive Lung Adenocarcinomas With MET, ALK, and ROS1 Genomic Alterations via Comprehensive Genomic Profiling. Clin Lung Cancer 16, e105–109 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raedler L. A. Zykadia (Ceritinib) Approved for Patients with Crizotinib-Resistant ALK -Positive Non-Small-Cell Lung Cancer. Am Health Drug Benefits 8, 163–166 (2015). [PMC free article] [PubMed] [Google Scholar]
- Wu Y. C. et al. Comparison of IHC, FISH and RT-PCR methods for detection of ALK rearrangements in 312 non-small cell lung cancer patients in Taiwan. Plos One 8, e70839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K. H. et al. Clinical and pathological significance of ROS1 expression in intrahepatic cholangiocarcinoma. BMC Cancer 15, 721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H. J. et al. ROS1 receptor tyrosine kinase, a druggable target, is frequently overexpressed in non-small cell lung carcinomas via genetic and epigenetic mechanisms. Ann Surg Oncol. 20, 200–208 (2013). [DOI] [PubMed] [Google Scholar]
- Freidlin B., McShane L. M., Polley M. Y. & Korn E. L. Randomized phase II trial designs with biomarkers. J Clin Oncol 30, 3304–3309 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.