Abstract
Human-mediated dispersal interplays with natural processes and complicates understanding of the biogeographical history of species. This is exemplified by two invasive tunicates, Ciona robusta (formerly Ciona intestinalis type A) and C. intestinalis (formerly Ciona intestinalis type B), globally distributed and sympatric in Europe. By gathering new mitochondrial sequences that were merged with published datasets, we analysed genetic patterns in different regions, with a focus on 1) their sympatric range and 2) allopatric populations in N and S America and southern Europe. In the sympatric range, the two species display contrasting genetic diversity patterns, with low polymorphism in C. robusta supporting the prevalent view of its recent introduction. In the E Pacific, several genetic traits support the non-native status of C. robusta. However, in the NE Pacific, this appraisal requires a complex scenario of introduction and should be further examined supported by extensive sampling efforts in the NW Pacific (putative native range). For C. intestinalis, Bayesian analysis suggested a natural amphi-North Atlantic distribution, casting doubt on its non-native status in the NW Atlantic. This study shows that both natural and human-mediated dispersal have influenced genetic patterns at broad scales; this interaction lessens our ability to confidently ascertain native vs. non-native status of populations, particularly of those species that are globally distributed.
For centuries, species have been intentionally or accidentally transported beyond their native range by human activities (i.e. biological introduction1). Human-mediated dispersal has radically altered species’ distributions, sometimes leading to global distributions (i.e. with occupation of several biogeographic regions2). When introduction of a given taxon commenced in the relatively distant past, the identification of its native range can be difficult or even impossible, conferring cryptogenic status (uncertainty regarding native or non-native status3) over part or all of the species’ range. Several invertebrate species in the NW Atlantic that correspond to this category4,5,6 have been the subject of extensive debate in the scientific community7.
These issues are exemplified by the class Ascidiacea, in which more than 1600 species (out of 2815 valid species) were described only after 1950 and several have a worldwide distribution8. For instance, molecular and morphological studies have only recently confirmed Ciona robusta Hoshino and Tokioka, 1967 and Ciona intestinalis (Linnaeus, 1767) as distinct species. The two species were synonymized in 1985 under the name Ciona intestinalis9. However, in the early 2000s10,11,12, molecular studies reported four major evolutionarily divergent lineages in this species, among which were two types that were named C. intestinalis type A and type B. The taxonomic status of these two taxa has been recently re-evaluated: C. intestinalis type A was assigned to C. robusta described by Hoshino and Tokioka in 196713 and C. intestinalis type B to C. intestinalis (Linnaeus, 1767), sensu Millar14. Several lines of evidence supported this re-classification: 1) morphological evidence showed distinctive features between the two taxa15,16,17, 2) the two taxa were first described in distinct biogeographic regions and oceans, in the North Pacific and North Atlantic for C. robusta and C. intestinalis, respectively, and 3) genetic and genomic studies showed their strong evolutionary divergence, estimated to have occurred ca. 3–5 My BP12,18,19.
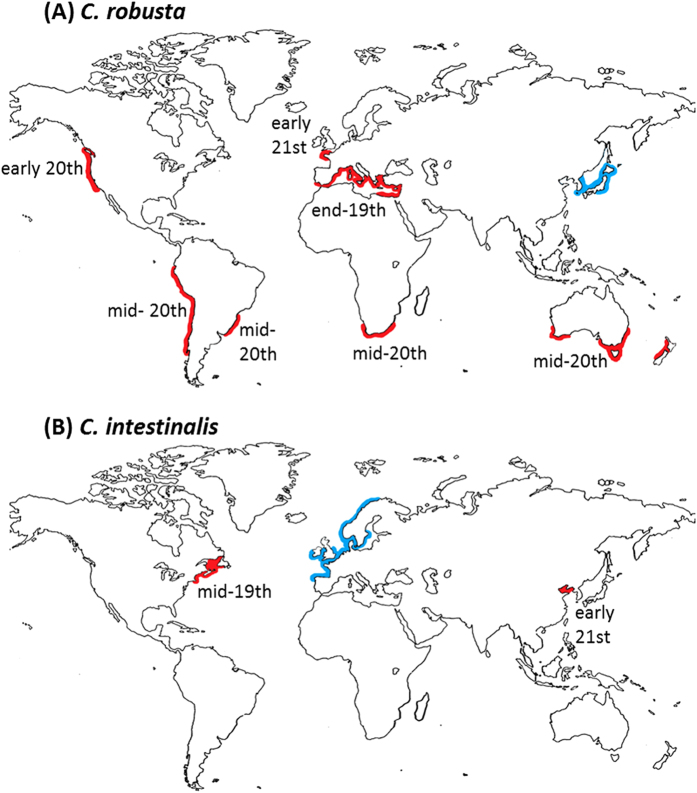
The two species are distributed along temperate and warm-temperate coasts20,21 (Fig. 1). The larger range of C. robusta is, at least partly, explained by its tolerance to a larger temperature range compared to C. intestinalis22. The species are well established in artificial habitats including marinas and harbours, are important members of fouling communities and are both recognized as introduced/invasive species in parts of the world23,24.
Figure 1.
Worldwide distribution of Ciona robusta (A) and C. intestinalis (B) based on literature records. Areas where the species have been regarded as native or as non-native or cryptogenic (i.e. of undefined status) are in blue and red respectively. For each allopatric geographical region, the approximate time is given of the first report in the literature of Ciona intestinalis sensu lato (i.e. including both C. intestinalis and C. robusta before the distinction was appreciated); for the EC (sympatric region), the time shown for C. robusta is of its first documented presence as distinct from C. intestinalis; details in Supplementary Note. Map was obtained from http://www.pedagogie.ac-aix-marseille.fr/jcms/c_67064/fr/cartotheque under the permission of the creator (Daniel Dalet©Académie Aix-Marseille).
C. robusta is generally assumed to be native to the NW Pacific, where it was described, and has been reported as an introduced species in the northern and southern hemisphere: in the Atlantic, Mediterranean Sea, Oceania, and North and South Pacific oceans (Fig. 1a, see the Supplementary Note for details). C. intestinalis is generally considered native to the NE Atlantic but non-native or cryptogenic in the NW Atlantic (e.g.25,26) and also occurs in the Bohai and Yellow Seas, China21 (Fig. 1b, Supplementary Note). The western English Channel and south of Brittany (hereafter named EC), is the only area confirmed so far where the two species live in sympatry, thought to result from the introduction of C. robusta into the native range of C. intestinalis22,27, although the Bohai and Yellow Seas are a second possible area21, as yet unconfirmed (Fig. 1b, Supplementary Note). In the EC contact zone, only extremely rare interspecific gene flow is occurring in the wild despite the two species living there in syntopy22,27: only a few hybrids were observed and all of them were shown to be first-generation (F1) hybrids; no other types of recent hybrid (e.g. backcrosses, F2 individuals) were observed so far22,28. Shared polymorphism was however documented and explained by gene flow during previous secondary contact between the two species19. According to Roux et al.19, this secondary contact occurred at a time estimated between 4–57 KY ago in an unknown location, before human activities substantially altered species’ natural ranges. Signs of this historical gene flow have indeed been reported in several allopatric regions in the Atlantic and Pacific19,28 suggesting a spread at a worldwide scale after the secondary contact, by natural expansion and/or human-mediated introduction.
Considering the re-classification of the two species, the recent report of new introductions (i.e. C. robusta in EC), the uncertainty of the native vs. native status in some regions (i.e. cryptogenic status of C. intestinalis in NW America) and their status as model species in evolutionary and developmental biology20, it is timely to describe global genetic patterns to better examine their histories and understand their contemporary distribution patterns and future on a global scale29.
Several studies have considered phylogenetic relationships within the genus Ciona or the C. intestinalis species complex, (e.g.12,18,21), or investigated genetic diversity and connectivity of local populations (e.g. in North America25, in South Africa30, and in Mediterranean Sea31). None of these examined in detail the global genetic patterns of the two species comparing different regions of introduction and notably the single sympatric area described so far. Here, we carried out genetic studies using sequence data obtained with two mitochondrial markers. Our data were merged with published information of Zhan et al.25 to produce population data at a global scale for the two species.
Besides providing a global picture, we had two more specific objectives. The first was to compare genetic patterns of the two species, which provide an almost perfectly matched set of biological properties (i.e. with shared phylogeny and life-history traits and very similar environmental requirements). They are both sessile as adults, with a short life cycle involving broadcast spawning for external fertilization, and with a non-feeding larva providing a short planktonic phase20,22,32. It has been argued33,34,35,36,37 that species’ basic biology will profoundly affect patterns of natural dispersal and thus population-genetic structuring and the extent of natural geographical ranges and rates of speciation. By this token, the two Ciona species should show very similar population-genetic properties. The extent to which they actually fulfil this expectation offers insight into the relative strength of biological characteristics vs. stochastic influences on dispersal and population demography, potentially including the intervention of anthropogenic vectors and major environmental events such as climatic fluctuations, which might by chance affect the species differently. Particularly close comparison of the species is possible in their contact zone in the EC, where the two species live side by side at many sites22.
Our second objective was to use the additional genetic information to assess the native vs. non-native status of the study species in the global regions sampled. The current understanding of the biogeographical status of the various populations is detailed, with references, in the Supplementary Note. In summary, C. robusta is presumed to be native to the NW Pacific, but a recent introduction in the English Channel (in the 2000s) and an older (19th to mid-20th century) introduction in the NE Pacific and Mediterranean Sea, while the age of the establishment in the SE Pacific is unclear. C. intestinalis is considered native to the NE Atlantic, while its status in the NW Atlantic is debated. Our analyses are based on patterns of genetic diversity and population structuring, with the assumption that both natural expansion and human-mediated contemporary dispersal could have influenced the observed patterns of genetic diversity. In the case of C. intestinalis, Bayesian analysis with an Isolation with Migration Model (IMM) is used to contrast alternative scenarios for the expansion of C. intestinalis in the North Atlantic.
Results
By merging our dataset with the published dataset of Zhan et al.25 on Pacific and Atlantic coasts of North America, totals of 714 C. robusta and 1140 C. intestinalis individuals were examined on COX3-ND1 sequences of 580 and 529 bp respectively. A subset of 501 individuals of C. robusta and 683 individuals of C. intestinalis sampled for this study (details about sampling are provided in Fig. 2 and Supplementary Table S1) were sequenced on an additional mtDNA fragment (COI). Concatenating CO1 and COX3-ND1 fragments allowed statistical analyses using a long fragment with a total of 1404 base pairs (bp) for C. robusta and 1313 bp for C. intestinalis. In the following text, for clarity, the results will be detailed for the largest sampling analyzed, thus with COX3-ND1; the results obtained with concatenated sequences are presented in the Supplementary Material and used in the main text only when relevant (e.g. to reinforce findings or point out contradictory results). Haplotype frequencies per population and sequences of haplotypes are deposited in DRYAD (DOI: 10.5061/dryad.7g555).
Figure 2.
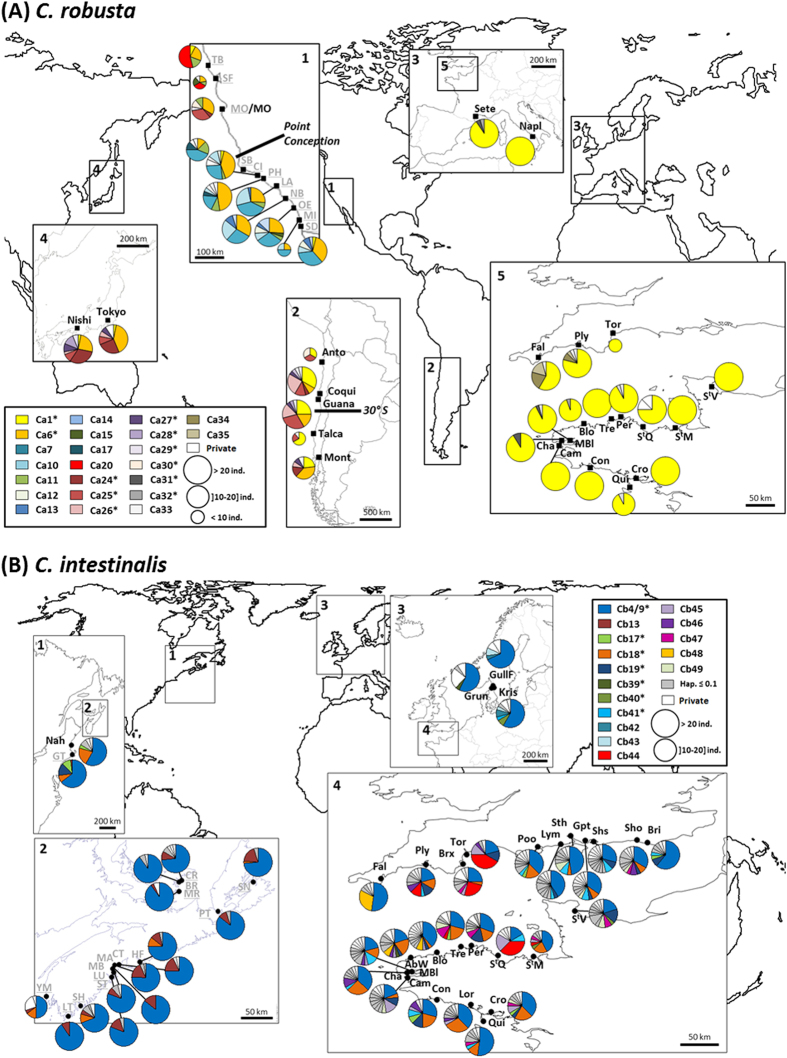
Location of the study populations and haplotype frequencies (COX3-ND1) at the population level for C. robusta (A) and C. intestinalis (B). Sites of Ciona robusta (A) and of Ciona intestinalis (B) sampled for this study are indicated in black and bold (for population codes and sampling, see Table S1). The localities studied by Zhan and co-authors25 that are included in the present study are in grey and underlined. Each color in pie charts refers to a given haplotype (see haplotype name in the box) as defined in the network in Fig. 3ab for C. robusta and C. intestinalis, respectively. The haplotypes with a name followed by an asterisk are shared between at least two regions. For sake of clarity, only the haplotypes shared between regions or with frequency higher than 0.1 at population level are shown wth a specific color for C. intestinalis. Maps were obtained from http://www.pedagogie.ac-aix-marseille.fr/jcms/c_67064/fr/cartotheque under the permission of the creator (Daniel Dalet©Académie Aix-Marseille).
Diversity analyses across species and populations
Genetic diversity indices obtained using our COX3-ND1 dataset with additional data from Zhan et al.25 are summarized for each region in Table 1 and detailed for each population in Supplementary Table S1. Haplotypic frequencies per population are illustrated in Fig. 2a,b for C. robusta and C. intestinalis, respectively. Very high polymorphism was observed over the two species but C. robusta was clearly less polymorphic than C. intestinalis, with 45 haplotypes (39 segregating sites) and 147 haplotypes (114 segregating sites) for C. robusta (N = 714) and C. intestinalis (N = 1140) respectively. As expected, the divergence was clearly higher between (14.4%) than within species (C. robusta: 0.71%, C. intestinalis: 1.96%).
Table 1. Regional genetic diversity indices of Ciona robusta and C. intestinalis based on COX3-ND1 sequences (source: this study and Zhan et al. 25).
| Mean per population (±SD) |
Total per region |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Code | Npop | Nind | Nh | Rh | Npr | Rpr | Hd | π (102) | Nind | Nh | Rh | Npr | Rpr | Hd | π (102) | |
| C. robusta | ||||||||||||||||
| North Western Pacific | NWP | 2 | 32.00 (0.00) | 6.50 (0.71) | 6.06 (0.48) | 0.50 (0.71) | 0.73 (0.22) | 0.790 (0.045) | 0.301 (0.057) | 64 | 9 | 8.98 | 1 | 1.18 | 0.792 | 0.301 |
| North Eastern Pacific | NEP | 10 | 20.50 (6.70) | 6.20 (1.48) | 5.33 (0.88) | 0.80 (1.14) | 0. 82 (0.84) | 0.766 (0.050) | 0.402 (0.098) | 213 | 22 | 15.48 | 16 | 4.46 | 0.814 | 0.419 |
| -North NEP | nNEP | 3 | 13.33 (4.51) | 5.00 (1.00) | 4.77 (0.78) | 0.33 (0.58) | 0.84 (0.73) | 0.777 (0.092) | 0.448 (0.162) | 40 | 16 | 16.00 | 2 | 3.81 | 0.837 | 0.457 |
| -South NEP | sNEP | 7 | 23.57 (4.89) | 6.71 (1.38) | 5.56 (0.87) | 1.00 (1.29) | 0.81 (0.94) | 0.775 (0.032) | 0.382 (0.065) | 173 | 9 | 5.51 | 12 | 3.35 | 0.775 | 0.393 |
| South Eastern Pacific | SEP | 4 | 17.50 (7.68) | 5.25 (2.06) | 5.36 (1.66) | 1.25 (0.50) | 1.05 (0.28) | 0.703 (0.111) | 0.253 (0.024) | 73 | 14 | 12.86 | 6 | 5.20 | 0.749 | 0.259 |
| -North SEP | nSEP | 2 | 24.00 (0.00) | 6.50 (2.12) | 6.35 (0.76) | 1.50 (0.71) | 0.97 (0.42) | 0.725 (0.056) | 0.266 (0.031) | 51 | 12 | 5.8 | 3 | 2.30 | 0.724 | 0.385 |
| -South SEP | sSEP | 2 | 11.00 (2.83) | 4.00 (1.41) | 4.37 (1.94) | 1.00 (0.00) | 1.13 (0.19) | 0.682 (0.178) | 0.241 (0.013) | 22 | 8 | 8.0 | 2 | 2.00 | 0.771 | 0.246 |
| English Channel | EC | 14 | 22.79 (3.31) | 1.93 (1.00) | 1.87 (0.97) | 0.50 (0.76) | 0.45 (0.59) | 0.129 (0.153) | 0.023 (0.028) | 320 | 13 | 6.41 | 10 | 1.67 | 0.138 | 0.025 |
| Mediterranean Sea | MedS | 2 | 22.00 (1.41) | 2.00 (1.41) | 1.68 (0.96) | 0.00 (0.00) | 0.10 (0.14) | 0.093 (0.132) | 0.017 (0.023) | 44 | 3 | 3 | 0 | 1.15 | 0.090 | 0.016 |
| Total | 714 | 45 | 0.703 | 0.334 | ||||||||||||
| C. intestinalis | ||||||||||||||||
| North Eastern Atlantic | NEA | 28 | 23.54 (1.69) | 10.68 (3.04) | 10.40 (2.94) | 2.89 (1.52) | 2.81 (1.42) | 0.815 (0.107) | 0.732 (0.253) | 659 | 126 | 44.45 | 119 | 21.19 | 0.854 | 0.815 |
| -English Channel | EC | 25 | 23.68 (1.68) | 11.24 (2.67) | 10.93 (2.59) | 3.00 (1.58) | 2.90 (1.48) | 0.839 (0.081) | 0.793 (0.189) | 592 | 117 | 43.36 | 110 | 20.79 | 0.870 | |
| -North Sea | NS | 3 | 22.33 (1.53) | 6.00 (1.73) | 5.93 (1.65) | 2.00 (0.00) | 2.04 (0.00) | 0.608 (0.067) | 0.223 (0.009) | 67 | 13 | 13.00 | 9 | 6.19 | 0.619 | 0.232 |
| North Western Atlantic | NWA | 16 | 30.06 (8.87) | 4.81 (1.83) | 4.14 (1.31) | 0.88 (0.96) | 0.83 (0.86) | 0.482 (0.188) | 0.241 (0.228) | 481 | 25 | 13.47 | 21 | 9.81 | 0.498 | 0.324 |
| Total | 1140 | 147 | 0.737 | 0.635 | ||||||||||||
For each region, per-population means and overall values for the region are given. Npop: number of populations; Nind: number of individuals; Nh: number of haplotypes; Rh: haplotypic richness (number of haplotypes corrected for sampling size); Npr: number of private haplotypes; Rpr: number of private haplotypes corrected for sampling size; Hd: haplotype diversity; π: nucleotide diversity. Genetic diversity indices per population are detailed in Supplementary Table S1. Study localities are shown in Fig. 2a for C. robusta and Fig. 2b for C. intestinalis.
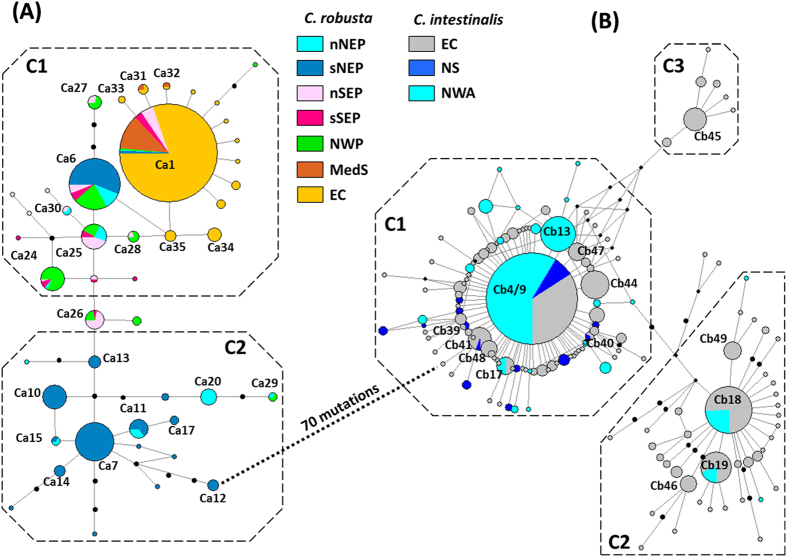
Regarding genetic diversity indices for C. robusta at regional level, all Pacific areas sampled were similar (Hd = 0.792 for NW, Hd = 0.814 for NE and Hd = 0.749 for SE Pacific; Table 1). The same holds when considering mean values per population, for each region (Table 1). These values contrast with the very low genetic diversity found in EC at the regional (Hd = 0.138 for EC) and population (Hd(mean) = 0.129 ± 0.153) level. These differences of population genetic diversity are significant between NE Pacific and EC (P < 0.001, Mann-Whitney rank sum test), SE Pacific and EC (P = 0.003) but not between the NE and SE Pacific ranges (P = 0.289). Similar statistical tests could not be reliably carried out with the NW Pacific as two populations only were examined in this region. Note that large differences were observed between the two Mediterranean populations studied (Hd(mean) = 0.093 ± 0.132) indicating the need for further sampling in this region to investigate possible regional trends. The haplotypic richness, which makes correction for sampling effort, is also higher in Pacific ranges (9.0, 15.5 and 12.9 for NW, NE and SE Pacific, respectively) compared to EC (6.41) (Table 1). The low haplotypic diversity in European populations as compared to all other populations studied is well illustrated by the map in Fig. 2a; some populations were monomorphic (e.g. Cam, Con, StM for EC and Napl) despite the extensive polymorphism of the markers used. The nucleotide diversity over all populations of the EC was also very low with a π value 12 times lower than in the NW Pacific, 17 times lower than in the NE Pacific, and 10 times lower than in the SE Pacific. This is illustrated in the network (Fig. 3a) in which all the Europe haplotypes are closely clustered in a star-like feature (1 and 2 mutation steps) around a central dominant haplotype (Ca1). Conversely, Pacific haplotypes are distributed in the network in two clusters: C1 incorporated all individuals from the EC and Mediterranean Sea, 87% of the individuals of the NW Pacific, 88% of the SE Pacific and 37% of the NE Pacific; and C2 included the remaining individuals from the NE Pacific and 3% of the individuals of the NW Pacific (Supplementary Table S2a). Only private haplotype richness distinguished the three Pacific regions, with the highest values (4.46 and 5.20) for NE and SE Pacific whereas NW Pacific displayed a lower value (1.18) that was more similar to EC (1.67) (Table 1).
Figure 3.
Median-joining haplotype networks of Ciona robusta (A) and C. intestinalis (B) based on COX3-ND1 sequences. Data from this study and from Zhan et al.25. Haplotype circles are proportional to haplotype frequency in the whole dataset. Branch lengths are proportional to number of mutational steps between two haplotypes. Missing haplotypes are indicated by small black circles. Colors represent the regions where the individuals possessing the haplotypes were found (regional codes are provided in Table 1).
For C. intestinalis, the NE Atlantic showed almost twice the haplotypic diversity observed in the NW Atlantic (Hd = 0.854 for NE, Hd = 0.498 for NW, Table 1). Conversely to C. robusta, EC area was significantly more genetically diverse (Hd(mean) = 0.839 ± 0.081) than elsewhere: the North Sea (Hd(mean) = 0.608 ± 0.067, P < 0.01, Mann-Whitney rank sum test) and the NW Atlantic (Hd(mean) = 0.482 ± 0.188, P < 0.001). However, Hd was not significantly different between North Sea and the NW Atlantic (P = 0.162). The high molecular diversity of the EC populations is illustrated by the haplotypic frequencies map (Fig. 2b) and the median joining haplotype network (Fig. 3b). EC haplotypes were distributed in three star-like clusters separated by 3–4 mutation steps (C1, C2 and C3, Supplementary Table S2b): Cluster C1 was represented by 67% of the EC individuals, C2 by 28% and C3 by 5%; for the NW Atlantic, haplotypes were represented by 94% of the individuals in C1 and 6% in C2 while haplotypes of the North Sea were all found in C1. Dominant haplotypes of the two major clusters (Cb4/9 for C1 and Cb18 of C2) were both shared by individuals of both Atlantic coasts. It is however important to note that most haplotypes were private to one coast (i.e. 84% and 94% of haplotypes were found only in NW and NE Atlantic respectively, Table 1).
The findings obtained with COX3-ND1 were all confirmed by using the concatenated sequences. In particular, C. robusta was clearly less polymorphic than C. intestinalis, with 48 haplotypes (63 segregating sites over 1404 bp) and 255 haplotypes (227 segregating sites over 1313 bp) for C. robusta and C. intestinalis respectively. Genetic diversity indices (Hd: haplotypic diversity and π: nucleotide diversity) are detailed in Supplementary Table S3. For C. robusta, as shown with COX3-ND1 only, the populations in the EC displayed the lowest haplotype diversity (mean per population, Hd(mean) = 0.265 ± 0.232) compared to populations of other geographical areas, in particular SE Pacific populations (Hd(mean) = 0.897 ± 0.066), where the highest population genetic diversity indices were found. This low molecular diversity in the EC is well illustrated by the haplotype network (Supplementary Figure S1) with a topology similar to the COX3-ND1 network (i.e. a star-like network with few haplotypes all at 1–3 mutation steps around a central dominant haplotype). The differences already observed between the two Mediterranean populations are even more pronounced with the concatenated dataset. For C. intestinalis, as with the COX3-ND1 locus alone, the genetic diversity was high in every population with an opposite trend to C. robusta in the EC: the EC contributed most of the diversity observed in C. intestinalis, as shown by the high haplotype diversity average Hd (0.960 ± 0.028) and explained by a large proportion of private haplotypes in this region. Compared to COX3-ND1, the concatenated dataset displayed more clearly the presence of three clusters (C1, C2 and C3 in Supplementary Figure S1b) with more divergence between them, i.e. separated by 11–12 mutations. Similarly to COX3-ND1, EC haplotypes were distributed in the three clusters, North Sea haplotypes were in only one cluster, NW Atlantic haplotype in the two major clusters (Supplementary Table S4), with for both, the sharing of central haplotypes by the two sides of the Atlantic.
Genetic structure
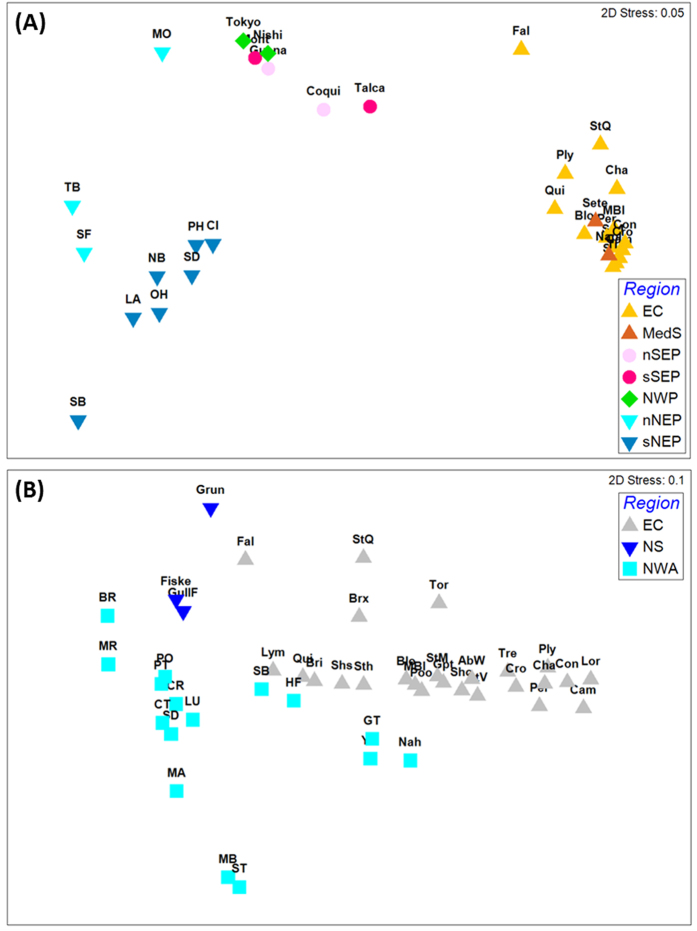
For C. robusta, overall genetic differentiation among all populations at COX3-ND1 locus was high and significant (ϕST = 0.461, P < 10−4). The hierarchical AMOVA showed a large regional effect (ϕCT = 0.497, P < 10−4, Table 2) whereas genetic structure between localities within groups was low although significant (ϕSC = 0.083, P < 10−4, Table 2). Population pairwise ϕST values (detailed in Supplementary Table S5a) are illustrated by the nMDS plot shown in Fig. 4a. The NW and SE Pacific regions appear genetically close. This genetic similarity is confirmed by a hierarchical AMOVA which showed an absence of significant effect of the regional grouping (ϕCT = 0.044, P = 0.075). The other regional groups clearly distinguished by the nMDS plot are also noticeable in the network (Fig. 3a) and the haplotype frequencies map (Fig. 2a). For instance, in the network, a single haplotype (Ca1) is shared by the five geographical areas but at high frequency in the EC and Mediterranean Sea (90% and 95% respectively), moderate frequency in the SE Pacific (33%) and low frequency in the remaining areas, the NW and NE Pacific (3.1 and 1.5% respectively). The NE Pacific, the only region for which haplotypes were found in the two clusters, is also a region with two sub-groups distinguished in the nMDS plot (northern and southern NE Pacific Fig. 4a). Interestingly these two groups are distributed across a well-known biogeographic break in the NE Pacific (i.e. at Point Conception)38. Hierarchical AMOVA carried out by grouping populations according to this biogeographic break showed a significant sub-group effect (ϕCT = 0.10, P < 10−4). A similar biogeographic break has been described in the SE Pacific at 30°S–33°S39, separating in our study the populations Anto, Coqui, Guana from Talca and Mont (Fig. 2a). Conversely to the NE Pacific, AMOVA did not show significant differences between these two groups in the SE Pacific (ϕCT = 0.020, P = 0.198). In Europe, hierarchical AMOVA also did not show genetic differences between EC and Mediterranean Sea (ϕCT = 0.025, P = 0.518).
Table 2. Hierarchical analysis of molecular variance for Ciona robusta and C. intestinalis.
| Fixation index | COX3-ND1 dataset | Concatenated dataset | |||
|---|---|---|---|---|---|
| C. robusta | Groups | NWP, NEP, SEP, EC and MedS | NWP, SEP, EC and MedS | ||
| ϕCT | 0.497 | (P < 0.001) | 0.504 | (P < 0.001) | |
| ϕSC | 0.083 | (P < 0.001) | 0.005 | (P = 0.084) | |
| ϕST | 0.539 | (P < 0.001) | 0.507 | (P < 0.001) | |
| C. intestinalis | Groups | NWA, EC and NS | EC and NS | ||
| ϕCT | 0.154 | (P < 0.001) | 0.125 | (P < 0.001) | |
| ϕSC | 0.035 | (P < 0.001) | 0.071 | (P < 0.001) | |
| ϕST | 0.140 | (P < 0.001) | 0.147 | (P < 0.001) | |
Analyses were done using the COX3-ND1 dataset and the concatenated mtDNA dataset. ϕCT, ϕSC and ϕST refer to the fixation index measuring genetic differences among areas, among populations within areas and among all populations, respectively. Probability values (H0: ϕ = 0) are indicated in brackets.
Figure 4.
Non-metric multidimensional scaling plots constructed using pairwise FST estimates among (A) all populations of C. robusta and (B) all populations of C. intestinalis. The same colour code as in Fig. 3 was used to represent regions.
For C. intestinalis, the overall genetic differentiation was significant among populations (ϕST) and the hierarchical AMOVA showed a significant effect of geographical areas (ϕCT) and of populations within areas (ϕSC) (Table 2). Excluding populations of North Sea from the AMOVA analysis did not change the results, still showing a significant effect of geographical area (ϕCT = 0.115, P < 10−4) and of populations within areas (ϕSC = 0.062, P < 10−4). Altogether, the outcome of the hierarchical genetic variance analyses is consistent with the network analysis showing no particular geographical partitioning of the three clusters (Fig. 3b). Pairwise genetic distances (Supplementary Table S5b) showed that the most differentiated populations in the NE Atlantic were those from the North Sea, as illustrated with the nMDS plot with the Grun population (Fig. 4b). The NW Atlantic populations appear genetically different from each other but with a subset (i.e. SB, HF, Nah, GT and YM, Fig. 4b) more similar to the EC populations than to other NW Atlantic populations.
Using the concatenated dataset on the subset of localities for which COI data were obtained confirm all the findings described with COX3-ND1 only. For example, for C. robusta, the hierarchical AMOVA confirmed a large regional effect (ϕCT) (Table 2). Similarly, genetic similarities were observed between SE Pacific and NW Pacific: 33% of the haplotypes found in the SE and NW Pacific were shared between the two ranges. Also, at the European level, Naples in the Mediterranean Sea and populations of the EC were not genetically different as indicated by pairwise ϕST-values (Supplementary Table S6a). As for C. robusta, the concatenated dataset confirmed the findings detailed above for COX3-ND1 in C. intestinalis (data are detailed in Supplementary Tables S3b and S6b).
Bayesian inference of divergence time for C. intestinalis in the N Atlantic
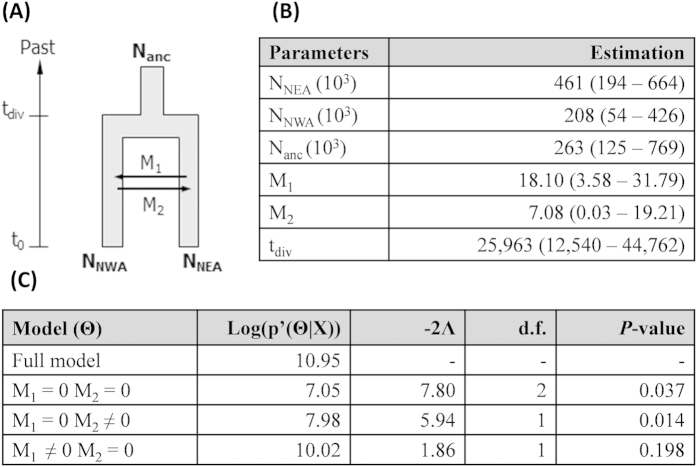
Results of the Isolation with Migration Model (IMM) used for C. intestinalis to evaluate the time of divergence between NW Atlantic and NE Atlantic are provided in Fig. 5, showing the median value and the 95% highest posterior density (95HPD) of each parameter, and in Supplementary Figure S2, showing the marginal posterior distribution of each parameter. Under IMM, the time of divergence between NW Atlantic and NE Atlantic was estimated 25,963 yrs BP (95HPD: 12,540 – 44,762; Fig. 5b). Concerning the migration model, only the model with an asymmetric migration could not be rejected (Fig. 5c), suggesting substantial gene flow from NE to NW Atlantic but not from NW to NE Atlantic.
Figure 5. Demographic and divergence parameters estimated under isolation with migration model for Ciona intestinalis.
(A) Diagram of the population splitting events (see Table 1 for codes of geographical areas). Time axis is not to scale. (B) Estimates of times of divergence and demographic parameters for the two studied regions using COX3-ND1 sequences. Nanc, NNEA, NNWA: effective population size; M1 and M2: number of migrants per generation; tdiv: time since divergence. Median value and the 95% highest posterior density (95 HPD) of each parameter are given. (C) Tests for the migration model; Log(p’(Θ|X): log of the highest posterior density of each model; −2Λ: likelihood ratio; d.f.: degrees of freedom; and P-value.
Discussion
Whatever the dataset, contrasting patterns were observed in Ciona robusta and C. intestinalis. Altogether, C. robusta displayed lower genetic diversity at population and regional levels, but marked spatial genetic structure at these two scales. In contrast, C. intestinalis showed high genetic diversity and a more homogenous genetic structure in the N Atlantic. These contrasting patterns are even more pronounced in the sympatric area (i.e. EC): very low genetic diversity was observed in C. robusta, while C. intestinalis encompassed the whole genetic diversity observed at the species level in our study.
Roux and coauthors19 also recorded lower polymorphism in C. robusta compared to C. intestinalis at a species level. They analyzed few individuals and localities of the two species (i.e. 10 individuals sampled in 3 localities for the two species), but at the genome level (i.e. 852 nuclear loci isolated from full transcriptomes). We here confirm this global pattern with a mitochondrial marker, using a wider geographical coverage and more extensive population sampling, with the addition of three geographic regions: SE Pacific, NW Pacific and EC. mtDNA markers can be sensitive to selective effects40. However, results from our mitochondrial dataset were congruent with those obtained by Roux and coauthors19. In addition, a similar trend is apparent in a microsatellite study25 of C. robusta (type A) in the NE Pacific and C. intestinalis (type B) in the NW Atlantic: Mann-Whitney rank sum tests on data provided in Table 1 of Zhan et al.25 showed significant differences for gene diversity (P = 0.005) and allelic richness (P = 0.018), both lower in C. robusta as compared to C. intestinalis.
What can explain such a pervasive difference between two congeneric species that share common life-history traits and ecological properties? Roux and coauthors19 suggested that after their divergence and isolation (estimated ca. 3–5 My BP during the Pliocene12,19) but before their global spread, demographic expansion of C. intestinalis was more pronounced than in C. robusta, or that a stronger bottleneck occurred in C.robusta than in C. intestinalis. This last hypothesis is potentially realistic given several glacial and interglacial episodes since the divergence of the two species, which severely impacted the distribution ranges of other marine species41,42,43,44. Roux and coauthors19 noted that “Tajima’s D was significantly more negative in C. intestinalis B [C. intestinalis] than in C. intestinalis A [C. robusta]”. They however did not get statistical support for a demographic bottleneck based on various statistics (Tajima’s D, Fu and Li’s D and F), and hypothesized that this result might reflect a lack of statistical power, in particular stemming from insufficient sampling. Dedicated demographic studies, using nuclear markers and including a substantial sampling effort, notably in its Asian range, are needed to investigate in detail the cause of reduced genetic diversity in C. robusta. Decreased polymorphism of C. robusta as compared to C. intestinalis is nevertheless a consistent finding across the few studies carried out so far (this study and19,25) at different scales, in different regions and with various markers.
C. robusta is distributed across biogeographic provinces that are separated by natural barriers to dispersal (Fig. 2a). Our study revealed large differences in the level of genetic diversity across these regions. Based on the COX3-ND1 and concatenated datasets, we could divide the current range of C. robusta into two categories: i) regions of relatively high genetic diversity (all those sampled in the Pacific) and ii) one region of lower genetic diversity (EC). The Mediterranean Sea is not classified as only two populations displaying much contrasting features were available. Pacific populations are genetically distinct from European areas (Figs 2a and 4a). The presence of evolutionarily divergent and private haplotypes suggest long-term residence of C. robusta in Pacific regions4.
However, the NE Pacific shows distinctive features as compared to NW and SE Pacific regions, in particular the largest haplotypic diversity accompanied by the largest nucleotide diversity values (Table 1), thus highlighting the co-existence of particularly diverse and evolutionarily divergent haplotypes as pictured by the network analysis (Fig. 3a). In addition, except for the population MO (Monterey), NE Pacific populations are genetically distinct from the NW and SE Pacific, the two last-named regions being quite similar to each other as pictured by the nMDS plot (Fig. 4a). Finally, as shown by Zhan et al.25, individuals of the NE Pacific are distributed within two genetically differentiated groups located in the north and the south along the coasts (here after refered to nNEP and sNEP)
Can these genetic patterns cast light on the putative native (in the NW Pacific) and non-native or cryptogenic (in the NE and SE Pacific) status of C. robusta in the Pacific? Compared to the native range of a species, some genetic signs are expected in the non-native range, in particular: lower frequency or absence of private (endemic) haplotypes, weaker genetic structure, lower genetic diversity (except if the introduction was accompanied by genetic admixture between genetically differentiated sources) and lack of concordance between gene genealogies and the geographical distribution of the haplotypes. NE Pacific populations, showing high diversity and private haplotypes, do not fit particularly well with these characteristics. In addition an important genetic discontinuity separates populations located north and south of the well-known biogeographic break at Point Conception (Fig. 2a). This biogeographic boundary is a source of genetic differentiation for species showing a short pelagic phase45,46, like ascidians32. Based on these characteristics, we might hypothesize that C. robusta is native to the NE Pacific. However, these arguments are insufficient to dismiss the possible non-native status of C. robusta in this region. High levels of genetic diversity have in fact been reported in most non-native populations of marine invertebrates29, including non-native ascidians47,48. This is explained by propagule pressure due to the existence of multiple vectors (polyvectism sensu Carlton & Ruiz49) from genetically diverse sources29,49. Furthermore, not only the frequent haplotype Ca6 is shared by the two sub-regions nNEP and sNEP but also the less frequent haplotype Ca11 (Fig. 2a), and these two haplotypes belong to opposite clusters (C1 and C2 respectively) of the gene genealogy (Fig. 3a). NE Pacific populations thus display admixture between genetically divergent lineages, a pattern often observed in non-native species due to repeated introductions50. Such genetic mixing has already been described in the introduced range of C. robusta in South Africa30. In the NE Pacific, a non-native status for C. robusta is thus possible but the genetic characteristics observed involve 1) numerous and repeated introduction events from genetically diversified sources and 2) two distinct and independent introduction events, in the northern and southern part of this region, from at least two genetically differentiated sources, and subsequent gene flow between the two regions. This is not an exceptional scenario in marine invertebrates, as illustrated by the introduction of Carcinus maenas in the NW Atlantic which involved the establishment of two lineages from two independent introduction events51. So far, such a scenario is not strongly supported by the data obtained in NW Pacific: the two Japanese populations studied are genetically diverse but similar to each other (Fig. 4a) and only the haplotype Ca29 belongs to the C2 cluster common in the NE Pacific, and particularly in sNEP (Fig. 3a). An extensive sampling effort in the NW Pacific is thus needed to seek places where C2 haplotypes are frequent, and thus potential sources of the introductions to the NE Pacific.
By contrast with the NE Pacific, SE Pacific and NW Pacific populations were found to be poorly differentiated (Fig. 4a) and to share numerous haplotypes (Figs 2a and 3a), a situation involving very poor concordance between gene genealogy and geography. Given the great distance between them and their positions in opposite hemispheres, it is not considered feasible that both regions are within the native range of the species as part of an array of undifferentiated populations. Instead, it is presumed that one of these populations was derived from the other as a result of anthropogenic dispersal, apparently directly rather than through secondary introductions. Interestingly, the Chilean populations studied are spread over a recognized biogeographical boundary at 30–33°S: this boundary is known to be associated with a major phylogeographic break in low dispersers39,52, like C. robusta. And yet no genetic structure was found between populations located on both sides of this boundary (Anto, Coqui, Guana versus Talca, Mont). This finding supports the supposed non-native status of C. robusta in this region (Supplementary Note). Another interesting finding is that Chilean populations share one haplotype at high frequency with NE Atlantic populations (i.e. Ca1 haplotype in Figs 2a and 3a), suggesting that they might share the same origin or even that one region might have been seeded by the other. It is noteworthy that, despite their geographical remoteness, Europe hosts recently introduced species considered native to Chile, e.g. the tunicate Corella eumyota53 and the mollusc Crepipatella dilatata54; Chile and EC also share several non-native species, e.g. the tunicate Asterocarpa humilis, the bryozoan Bugula neritina, the mollusc Mytilus galloprovincialis, the green alga Codium fragile fragile (cited as C. fragile tomentosoides in Castilla et al.55) and the red alga Polysiphonia morrowii56,57,58.
The low diversity of the EC populations compared to Pacific populations appears consistent with a very recent introduction of C. robusta in this region. The EC populations did not display signs of genetic admixture (as might arise over time from multiple introductions) and most of them shared several haplotypes including the dominant Ca1 (Figs 2a and 3a) with the SE, NE and NW Pacific and the Mediterranean Sea. There is no overall concordance between gene genealogies and geography at this broad scale. Some haplotypes or branches of the haplotype network are shared across very distinct biogeographic regions. Such a pattern is incompatible with a hypothesis of natural expansion, particularly considering the poor dispersal ability of C. robusta and the broad geographical scale here considered59,60. The disjunct geographical distribution and weak genetic structure between the Mediterranean Sea and the NE Atlantic in C. robusta are also not expected within a native range59,61. Bottleneck events are rarely observed in marine invertebrates29 except in the very early stages of expansion. This scenario seems probable to be the situation for C. robusta in Europe and explain the low genetic diversity in this region. But besides this “selectively neutral” scenario, we need to consider “adaptive” scenarios for reduction in genetic diversity, for instance selection on standing genetic variation, which is one of the evolutionary outcomes of introductions of genetically diversified individuals40. A selective sweep in EC populations of C. robusta could explain our present findings based on mitochondrial loci40. However, 115 polymorphic SNPs examined in one population in Chile (Guanaqueros in the present study) also displayed higher genetic diversity (He = 0.294) than the corresponding markers in the EC populations (7 populations studied here, range of He = 0.236–0.253)28. It would be informative, as recently undertaken for other marine invaders62,63,64, to examine additional populations with nuclear markers and high genome coverage, not only to confirm patterns of lower genetic diversity in English Channel populations compared to the Pacific but also to examine in more detail the genetic diversity and structure of Pacific populations. Looking for the source of a European introduction was beyond the scope of this study as it would have required much more substantial sampling of the Asian range (e.g. C. robusta has also been reported in eastern Korea65) as well as of other regions (i.e. SW Pacific, S Atlantic) where the species is reported, and presumed to be introduced, representing possible sources for a secondary introduction into Europe.
Whereas our study agrees with the literature to support the non-native status of C. robusta in the NE Atlantic (Supplementary Note), it conversely casts doubt on the commonly assumed non-native status of C. intestinalis in NW Atlantic. In a previous study of patterns of marine species’ distributions in the N Atlantic, Haydar7 categorized C. intestinalis as possessing a disjunct distribution, being present on both E and W coasts of Atlantic but absent from the intervening Arctic coastal regions (i.e. Spitzbergen, Iceland, Greenland and northern Canada). The status of this species was accordingly classified as cryptogenic in NW Atlantic, its presence on both temperate coasts being attributable with certainty to neither natural nor anthropogenic dispersal. As an alternative to anthropogenic dispersal, on-going natural dispersal across the Atlantic seems unlikely for organisms such as C. intestinalis which have a brief planktonic phase32, but a naturally disjunct distribution could arise from recolonization of NW Atlantic coasts from glacial refuges, as has been suggested for the gastropod Littorina littorea5, a species alternatively regarded as a non-native in NW Atlantic6,66. Based on the COX3-ND1 dataset for C. intestinalis, the NW Atlantic population displays similarities to EC populations, with four haplotypes shared by populations on both N Atlantic coasts at medium and high frequencies, one being dominant in all populations (Cb4/9, Figs 2b and 3b). Additionally, weak regional genetic structure was found (Table 2). Disjunct distribution, shared dominant haplotypes and weak genetic structure are arguments in favor of an anthropogenic introduction of C. intestinalis across the Atlantic.
On the other hand, several lines of evidence in our study are consistent with the alternative hypothesis of C. intestinalis being native to both sides of the N Atlantic. A natural disjunct distribution occupying both sides of the N Atlantic has been ascribed to the ascidian Cnemidocarpa mollis7. In Ciona intestinalis, most of the haplotypes are private to the American (84%) or European (94%) region, and private haplotypes are often considered characteristic of long-term population establishment4,5. Also, the Bayesian inferences computed under IMM best supported the hypothesis of a natural divergence from a common ancestor (Fig. 5) within an estimated interval of 12,540 to 44,762 yrs BP. These dates not only exclude anthropogenic vectors but also bracket the Last Glacial Maximum (LGM; ca. 21ky BP) of the Pleistocene Period, a major driver of distribution range shifts59. In C. intestinalis, we observed high genetic diversity on both sides of the N Atlantic that was spread over the same two major haplotypic clusters in both mitochondrial datasets (Figs 4b and 5b). Such a partitioning is commonly explained by phylogeographic scenarios of divergence in allopatry followed by survival of lineages in different refugia and then by secondary contacts and gene flow during post-glacial expansion, as documented in the same region in several marine species59.
The three Ciona intestinalis populations of the North Sea (Skagerrak) showed a different pattern: they exhibited the lowest genetic diversity and were all included in the same haplotypic cluster. Such a pattern has already been described in several marine invertebrates and algae in the NE Atlantic (e.g. Palmaria palmata67; see Provan et al.68 for a review) and can reasonably be explained by a genetic bottleneck during retreat into a glacial refugium that was not balanced afterwards by massive expansion during post-glacial recolonization. Finally, it is noteworthy that C. intestinalis individuals have already been reported in the mid-20th century in the Faeroe Islands47 and were more recently found in Iceland (SB, personal observation and sampling in 2014) suggesting that the species’ distribution is not as interrupted in the most northern areas of the N Atlantic as supposed. The present-day distribution of C. intestinalis might thus originate from natural expansion processes like those described for other coastal species, for instance Littorina saxatilis, characterized by an amphi-Atlantic distribution69 explained by its survival during glaciation period in multiple refugia on both sides of the N Atlantic. In this context, we note that C. intestinalis is largely replaced by two taxa of uncertain taxonomic rank along Arctic coasts, C. gelatinosa Bonnevie, 1896 and C. longissima Hartmeyer, 1899. These were regarded as forms, varieties or subspecies of Ciona intestinalis by several authors, e.g.9,70,71,72,73, but were listed as full species by Sanamyan74. They have not been included in recent molecular clarifications of inter-relationships of Atlantic and global Ciona populations. If C. gelatinosa and C. longissima were found to be infraspecific entities within C. intestinalis, this species would have a continuous amphi-Atlantic distribution (see Fig. 8 in Dybern75), favouring the categorization of populations on both the E and W coasts as native according to the logical framework of Haydar7. Examining specimens preserved in museums and analyzing populations from natural habitats, rather than artificial ones, and from Arctic localities could be helpful to further assess the status of C. intestinalis in the NW Atlantic.
Note that natural isolation between the two populations does not exclude additional contemporary migrations. It is thus possible that both historical population splitting and human-mediated introductions have occurred in Ciona intestinalis, at different times, across the N Atlantic. This scenario is feasible given that some populations of the NW Atlantic are closely genetically related to populations in the NE Atlantic (e.g. Nah, GT, HF, Fig. 4b). This interaction can explain the observed genetic features and the ecological reports documenting a sudden expansion of C. intestinalis in the Maritime Provinces of Canada in recent decades76,77,78. It is also important to note that under IMM, mutation and migration rates are constant through time, an assumption rarely met in nature for migration, particularly for introduced species. More complex models assuming differential migration and mutation rates over time and using nuclear markers should be developed to evaluate more fully the demographic history of C. intestinalis in the N Atlantic. Despite these limitations, our data support a natural amphi-Atlantic distribution with plausible recent local population expansion due to human activities (i.e. human-mediated dispersal and increased coastal urbanization that opened new habitats to be colonized). As for the numerous cryptogenic species presenting similar complex patterns7, our study casts doubt on a single non-native origin for C. intestinalis in the NW Atlantic.
Despite the complexity of reconstructing the eco-evolutionary history of cosmopolitan and cryptogenic species that have possibly been exploiting human-made habitats and vectors for a long time, this study showed that mtDNA-based studies can be helpful in documenting contrasting features between congeners at a global scale. In the EC, although the two congeners display very similar biological features and occupy the same localities, they show contrasting genetic features, including notable differences in genetic diversity; this study highlighted the importance of population demographic events and human-mediated dispersal in determining genetic patterns, apparently over-riding the influence of life-history traits. Comparison between geographical regions revealed an apparent example in C. robusta in the NE Atlantic of reduced genetic diversity associated with recent introduction. This phenomenon is not commonly observed in marine invaders, presumably because the effects of founder events are short lived in the face of repeated introductions and high propagule loads. The low diversity observed in C. robusta in the EC is thus expected to disappear relatively quickly with new cryptic introductions, a hypothesis testable by temporal genetic monitoring. The data presented here also provide new insights regarding the native vs. non-native status of the two study species in various parts of their ranges. Our genetic data support a non-native status of C. robusta in the SE Pacific, particularly regarding the absence of genetic differences across the well-known biogeographic break at 30–33°S. However, our data do question the non-native status of C. robusta in the NE Pacific and of C. intestinalis in the NW Atlantic. Analyses of Arctic populations of Ciona sp. to assess the relationship of C. intestinalis populations on the two sides of the N Atlantic and additional studies in the NW Pacific range of C. robusta are priorities for further advancing the enquiries reported here. This study emphasizes the need for caution in the interpretation of global genetic patterns in species in which natural and human-mediated dispersal have interplayed for some time.
Materials and Methods
Sampling and DNA extraction
Ciona robusta and C. intestinalis populations were sampled in 25 sites of the EC (in most of which the two species were found living in syntopy, Fig. 2 and Supplementary Table S1 for details) and in 14 sites located in regions where only one of the two species has been reported so far (i.e. allopatric areas). In detail, C. robusta individuals were collected in 14 marinas in the EC, five locations in the SE Pacific (Chile), two in the putative native range, the NW Pacific (Japan), one in the NE Pacific and two in the Mediterranean Sea. Note that to our knowledge, this study is the first to sample a substantial number of individuals from the putative native range of C. robusta to carry out population genetic analyses. C. intestinalis individuals were collected in 25 marinas in the EC, three locations in the Skagerrak (North Sea region) and one in NW Atlantic (North America). Sampling was generally realized under floating pontoons, either using SCUBA diving or by collection from the surface from pontoon floats, hanging ropes and the underside of buoys. Around 24 individuals were sampled in each population with few exceptions (Supplementary Table S1 for details). For each individual, a piece of branchial basket was preserved in 100% ethanol for genetic analyses. DNA extraction was performed with a Nucleospin® 96 Tissue Kit (Macherey-Nagel, Germany) according to the manufacturer’s protocol.
Sequencing procedure
Two mitochondrial loci (hereafter named mtDNA), cytochrome oxidase subunit 3 - NADH dehydrogenase subunit I (COX3-ND1) and cytochrome oxidase subunit I (COI), were chosen to allow comparisons with previous datasets21,25. The COX3-ND1 fragment was amplified using TX3F and TN1R primers following the PCR protocol described in Iannelli et al.79. The COI fragment was amplified using primer sequences and protocols obtained from Nydam and Harrison12. Sequencing reactions, run on an Applied BiosystemsTM AB3730XL DNA sequencer after purification on ExoSAP-it®, were performed at Centre National de Sequençage-Genoscope (Evry, France) or the LGC Genomics platform (Berlin, Germany). All PCR products were sequenced in both directions. Sequences obtained were edited using CodonCode Aligner v.4.0.2 (CodonCode Corporation, MA) and aligned using BioEdit v.7.1.980. For COI, a final alignment length of 737 base pairs (bp) was obtained for each species. For COX3-ND1, the final alignment was 667 bp for C. robusta and 576 bp for C. intestinalis. Nucleotide sequences were translated into amino acid sequences using the Ascidian mitochondrial genetic code implemented in DnaSP v.5.10.0181.
Statistical analyses
For a detailed comparison of the structure between Pacific and Atlantic coasts of North America25 and other regions of our sampling, the analyses were first carried out using COX3-ND1 only. We aligned our COX3-ND1 sequences and cut them to the same length as Zhan et al.25 (580 bp for C. robusta and 530 bp for C. intestinalis). Two overlapping peaks at position 434 bp for C. intestinalis were observed on the forward sequence but not on the reverse sequence. As we did not have access to the chromatographs obtained by Zhan and coauthors, we chose to exclude this position from the sequence analyses to avoid adding false haplotypes.
For each species and each population, the number of haplotypes (Nh), the number of polymorphic sites (S), the haplotype diversity (Hd) and the nucleotide diversity (π) were computed using DnaSP81. To compare diversity among populations and regions, haplotype richness and corrected number of private haplotypes were computed using the HP-RARE software82, which corrects for unequal sample sizes using a rarefaction procedure. For each species, median-joining networks were generated to infer the most parsimonious phylogenetic relationships among concatenated mtDNA haplotypes using NETWORK v.4.6 (www.fluxus-engineering.com). Analyses of genetic structure were carried out with ARLEQUIN v.3.583. Population pairwise differentiations (ϕST) were carried out based on 10,000 random permutations. To picture the genetic distances between all study populations, a non-metric multi dimensional scaling (nMDS) was done with pairwise ϕST values, using the PRIMER 6 + software84. To investigate genetic differences among (ϕCT) and within (ϕSC) geographic areas, a hierarchical analysis of molecular variance (AMOVA) was done.
The same analyses were also done using a subset of the sampling obtained for the two mitochondrial loci (i.e. all except the populations from Zhan et al.21,25) to check for the consistency of the results over a longer sequence. Due to the non-independence of the two loci, statistical analyses were carried out on concatenated sequences with a total of 1404 bp for C. robusta and 1313 bp for C. intestinalis. Note that similar results (in terms of genetic diversity and structure) were obtained independently when the two loci were analyzed separately (data not shown). Note that for AMOVA analyses for C. intestinalis, to avoid over-representation of the EC populations, the AMOVA was conducted with a random subsampling of 6 populations and was repeated with several other random subsamples of the populations without changing the results.
Population divergence Bayesian analyses
Considering the cryptogenic status of C. intestinalis7, we investigated the divergence history between populations of this species present both sides of the N Atlantic. An Isolation with Migration Model (IMM) was performed using IMa285. The IMM was applied to the COX3-ND1 dataset only, to include data from the NW Atlantic for C. intestinalis25. Briefly, the IMa2 program explores parameter and genealogy space through Markov chain Monte Carlo algorithms (MCMC) from a gene genealogy compatible with the observed dataset and generates posterior probabilities of several parameters. A Hasegawa-Kishino-Yano mutation model86 was used with a range of mutation rate between 1.31 × 10−5 and 2.67 × 10−5 per generation for the COX3-ND1 locus. These rates were estimated from the number of fixed mutations between C. robusta and C. intestinalis on COX3-ND1 sequences, divided by the lowest and the highest bound of the divergence time interval estimated to be 2.7–5.5 My BP, with one generation per year19. To optimize computation time, a subset of 80 individuals on each side of the N Atlantic was used (population splitting pictured in Fig. 5a). MCMC repetitions were done with 106 steps after 100,000 long burn-in cycles. Finally, to test the influence of migration between areas after divergence, likelihood-ratio tests were performed on the nested model of gene flow according to Hey and Nielsen87. Note that the low sampling coverage of the putative Asian native range of C. robusta and the absence of sampling in the S Atlantic (i.e. South Africa and Brazil) and in SW Pacific (i.e. Australia and New Zealand) prevented us carrying out similar analyses for C. robusta in the Pacific.
Additional Information
How to cite this article: Bouchemousse, S. et al. Contrasting global genetic patterns in two biologically similar, widespread and invasive Ciona species (Tunicata, Ascidiacea). Sci. Rep. 6, 24875; doi: 10.1038/srep24875 (2016).
Supplementary Material
Acknowledgments
We are very grateful to all our colleagues who contributed to the collection of samples all over the world: the divers of the Marine Operations department (Service Mer & Observation) at the Roscoff Biological Station, P. Cirino, S. Krueger-Hadfield, C. Lejeusne, S. Le Cam, B. Lundve, M. Otani, J. Pechenik, C. Wood and A. Yunnie. We are thankful to G. Dubois and C. Roby for assistance with DNA extraction and statistical analyses and to N. Bierne for stimulating discussions on these datasets and advice for Bayesian analyses. We thank A. Zhan for kindly providing us with his raw data. This work was supported by the Bibliothèque du Vivant project (CNRS, INRA, MNHN), the Interreg IVa Marinexus program and the ANR project HYSEA (no. ANR-12-BSV7-0011).
Footnotes
Author Contributions S.B., J.D.D.B. and F.V. conceived the experiment design and contributed to sampling collection. S.B. ran the genetic labwork and prepared figures and tables. S.B., J.D.D.B. and F.V. analyzed the data and wrote the manuscript.
References
- Elton C. S. The ecology of invasions by animals and plants (University of Chicago Press, 1958). [Google Scholar]
- Molnar J. L., Gamboa R. L., Revenga C. & Spalding M. D. Assessing the global threat of invasive species to marine biodiversity. Front. Ecol. Environ. 6, 485–492, doi: 10.1890/070064 (2008). [DOI] [Google Scholar]
- Carlton J. T. Pattern, process, and prediction in marine invasion ecology. Biol. Conserv. 78, 97–106, doi: 10.1016/0006-3207(96)00020-1 (1996). [DOI] [Google Scholar]
- Haydar D., Hoarau G., Olsen J. L., Stam W. T. & Wolff W. J. Introduced or glacial relict? Phylogeography of the cryptogenic tunicate Molgula manhattensis (Ascidiacea, Pleurogona). Divers. Distrib. 17, 68–80, doi: 10.1111/j.1472-4642.2010.00718.x (2011). [DOI] [Google Scholar]
- Wares J. P., Goldwater D. S., Kong B. Y. & Cunningham C. W. Refuting a controversial case of a human-mediated marine species introduction. Ecol. Lett. 5, 577–584, doi: 10.1046/j.1461-0248.2002.00359.x (2002). [DOI] [Google Scholar]
- Chapman J. W., Carlton J. T., Bellinger M. R. & Blakeslee A. M. H. Premature refutation of a human-mediated marine species introduction: the case history of the marine snail Littorina littorea in the northwestern Atlantic. Biol. Invasions 9, 737–750, doi: 10.1007/s10530-006-9073-x (2007). [DOI] [Google Scholar]
- Haydar D. What is natural? The scale of cryptogenesis in the North Atlantic Ocean. Divers. Distrib. 18, doi: 10.1111/j.1472-4642.2011.00863.x (2012). [DOI] [Google Scholar]
- Shenkar N. & Swalla B. J. Global diversity of Ascidiacea. PloS ONE 6, e20657, doi: 10.1371/journal.pone.0020657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino Z.-i. & Nishikawa T. Taxonomic Studies of Ciona intestinalis (L.) and its allies. Publ. Seto Mar. Biol. Lab. 30, 61–79 (1985). [Google Scholar]
- Suzuki M. M., Nishikawa T. & Bird A. Genomic approaches reveal unexpected genetic divergence within Ciona intestinalis. J. Mol. Evol. 61, 627–635 (2005). [DOI] [PubMed] [Google Scholar]
- Caputi L. et al. Cryptic speciation in a model invertebrate chordate. Proc. Natl. Acad. Sci. of USA 104, 9364–9369, doi: 10.1073/pnas.0610158104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nydam M. L. & Harrison R. G. Genealogical relationships within and among shallow-water Ciona species (Ascidiacea). Mar. Biol. 151, 1839–1847, doi: 10.1007/s00227-007-0617-0 (2007). [DOI] [Google Scholar]
- Hoshino Z.-i. & Tokioka T. An unusually robust Ciona from the Northeastern coast of Honsyu Island, Japan. Publ. Seto Mar. Biol. Lab. 15, 275–290 (1967). [Google Scholar]
- Millar R. H. In L.M.B.C. Memoirs of typical British marine palnts and animals, XXXV (ed Colman J. S.) 123 (Liverpool University Press, 1953). [Google Scholar]
- Brunetti R. et al. Morphological evidence that the molecularly determined Ciona intestinalis type A and type B are different species: Ciona robusta and Ciona intestinalis. J. Zoolog. Syst. Evol. Res. 53, 186–193, doi: 10.1111/jzs.12101 (2015). [DOI] [Google Scholar]
- Pennati R. et al. Morphological differences between larvae of the Ciona intestinalis species complex: hints for a valid taxonomic definition of distinct species. PLoS ONE 10, e0122879, doi: 10.1371/journal.pone.0122879 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato A., Satoh N. & Bishop J. D. D. Field identification of ‘types’ A and B of the ascidian Ciona intestinalis in a region of sympatry. Mar. Biol. 159, 1611–1619, doi: 10.1007/s00227-012-1898-5 (2012). [DOI] [Google Scholar]
- Nydam M. L. & Harrison R. G. Polymorphism and divergence within the ascidian genus Ciona. Mol. Phylogenet. Evol. 56, 718–726, doi: 10.1016/j.ympev.2010.03.042 (2010). [DOI] [PubMed] [Google Scholar]
- Roux C., Tsagkogeorga G., Bierne N. & Galtier N. Crossing the species barrier: Genomic hotspots of introgression between two highly divergent Ciona intestinalis species. Mol. Biol. Evol. 30, 1574–1587, doi: 10.1093/molbev/mst066 (2013). [DOI] [PubMed] [Google Scholar]
- Procaccini G., Affinito O., Toscano F. & Sordino P. In Evolutionary Biology: Concepts, Biodiversity, Macroevolution and Genome Evolution (ed Pontarotti P.) Ch. 6, 91–106 (2011). [Google Scholar]
- Zhan A., MacIsaac H. J. & Cristescu M. E. Invasion genetics of the Ciona intestinalis species complex: from regional endemism to global homogeneity. Mol. Ecol. 19, 4678–4694 (2010). [DOI] [PubMed] [Google Scholar]
- Bouchemousse S., Lévêque L., Dubois G. & Viard F. Co-occurrence and reproductive synchrony do not ensure hybridization between an alien tunicate and its interfertile native congener. Evol. Ecol. 30, 69–87, doi: 10.1007/s10682-015-9788-1 (2016). [DOI] [Google Scholar]
- Lambert C. C. & Lambert G. Non-indigenous ascidians in southern California harbors and marinas. Mar. Biol. 130, 675–688 (1998). [Google Scholar]
- Therriault T. W. & Herborg L.-M. Predicting the potential distribution of the vase tunicate Ciona intestinalis in Canadian waters: informing a risk assessment. Ices J. Mar. Sci. 65, 788–794, doi: 10.1093/icesjms/fsn054 (2008). [DOI] [Google Scholar]
- Zhan A. et al. Complex genetic patterns in closely related colonizing invasive species. Ecol. Evol. 2, 1331–1346, doi: 10.1002/ece3.258 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carver C. E., Mallet A. L. & Vercaemer B. Biological synopsis of the solitary tunicate Ciona intestinalis. Can. Man. Rep. Fish. Aquat. Sci. 2746, v+ 55p. (2006). [Google Scholar]
- Nydam M. L. & Harrison R. G. Introgression despite substantial divergence in a broadcast spawing marine invertebrate. Evolution 65, 429–442, doi: 10.1111/j.1558-5646.2010.01153.x (2011). [DOI] [PubMed] [Google Scholar]
- Bouchemousse S., Haag-Liautard C., Bierne N. & Viard F. Past and contemporary introgression between two strongly differentiated Ciona species as revealed by a post-genomic SNPs panel. BioRxiv, doi: 10.1101/030346 (2015). [DOI] [Google Scholar]
- Rius M., Turon X., Bernardi G., Volckaert F. A. M. & Viard F. Marine invasion genetics: from spatio-temporal patterns to evolutionary outcomes. Biol. Invasions 17, 869–885, doi: 10.1007/s10530-014-0792-0 (2015). [DOI] [Google Scholar]
- Rius M. et al. Range expansions across ecoregions: interactions of climate change, physiology and genetic diversity. Global Ecol. Biogeogr. 23, 76–88, doi: 10.1111/geb.12105 (2014). [DOI] [Google Scholar]
- Affinito O. et al. High connectivity and directional gene flow in European Atlantic and Mediterranean populations of Ciona intestinalis sp. A. Mar. Ecol. 36, 1230–1243, doi: 10.1111/maec.12226 (2016). [DOI] [Google Scholar]
- Lambert G. Ecology and natural history of the protochordates. Can. J. Zool. 83, 34–50, doi: 10.1139/z04-156 (2005). [DOI] [Google Scholar]
- Bohonak A. J. Dispersal, gene flow, and population structure. Q. Rev. Biol. 74, 21–45, doi: 10.1086/392950 (1999). [DOI] [PubMed] [Google Scholar]
- Kinlan B. P. & Gaines S. D. Propagule dispersal in marine and terrestrial environments: a community perspective. Ecology 84, 2007–2020 (2003). [Google Scholar]
- Lester S. E., Ruttenberg B. I., Gaines S. D. & Kinlan B. P. The relationship between dispersal ability and geographic range size. Ecol. Lett. 10, 745–758, doi: 10.1111/j.1461-0248.2007.01070.x (2007). [DOI] [PubMed] [Google Scholar]
- Marshall D. J., Krug P. J., Kupriyanova E. K., Byrne M. & Emlet R. B. The biogeography of marine invertebrate life histories. Ann. Rev. Ecol. Evol. Systematics 43, 97–114, doi: 10.1146/annurev-ecolsys-102710-145004 (2012). [DOI] [Google Scholar]
- Selkoe K. A. & Toonen R. J. Marine connectivity: a new look at pelagic larval duration and genetic metrics of dispersal. Mar. Ecol. Prog. Ser. 436, 291–305, doi: 10.3354/meps09238 (2011). [DOI] [Google Scholar]
- Wares J. P., Gaines S. D. & Cunningham C. W. A comparative study of asymetric migration events across a marine biogeographic boundary. Evolution 55, 295–306 (2001). [DOI] [PubMed] [Google Scholar]
- Haye P. A. et al. Phylogeographic Structure in Benthic Marine Invertebrates of the Southeast Pacific Coast of Chile with Differing Dispersal Potential. PLoS ONE 9, e88613, doi: 10.1371/journal.pone.0088613 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N., Nabholz B., Glemin S. & Hurst G. D. D. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 18, 4541–4550, doi: 10.1111/j.1365-294X.2009.04380.x (2009). [DOI] [PubMed] [Google Scholar]
- Wares J. P. & Cunningham C. W. Phylogeography and historical ecology of the North Atlantic intertidal. Evolution 55, 2455–2469 (2001). [DOI] [PubMed] [Google Scholar]
- Vermeij G. J. From Europe to America: Pliocene to recent trans-Atlantic expansion of cold-water North Atlantic molluscs. Proc. R. Soc. B 272, 2545–2550, doi: 10.1098/rspb.2005.3177 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marko P. B. et al. The ‘Expansion-Contraction’ model of Pleistocene biogeography: rocky shores suffer a sea change? Mol. Ecol. 19, 146–169, doi: 10.1111/j.1365-294X.2009.04417.x (2010). [DOI] [PubMed] [Google Scholar]
- Guillemin M.-L. et al. In Seaweed Phylogeography (eds Hu Zi-Min & Ceridwen Fraser) Ch. 10, 251–277 (Springer Netherlands, 2016). [Google Scholar]
- Dawson M. N. Phylogeography in coastal marine animals: a solution from California? J. Biogeogr. 28, 723–736, doi: 10.1046/j.1365-2699.2001.00572.x (2001). [DOI] [Google Scholar]
- Hohenlohe P. A. Limits to gene flow in marine animals with planktonic larvae: models of Littorina species around Point Conception, California. Biol. J. Linn. Soc. 82, 169–187, doi: 10.1111/j.1095-8312.2004.00318.x (2004). [DOI] [Google Scholar]
- Goldstien S. J. et al. Global phylogeography of the widely introduced North West Pacific ascidian Styela clava. PLoS ONE 6, e0016755, doi: 10.1371/journal.pone.0016755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock D. G., MacIsaac H. J. & Cristescu M. E. Multilocus genetic analyses differentiate between widespread and spatially restricted cryptic species in a model ascidian. Proc. R. Soc. B, 279, 2377–2385, doi: 10.1098/rspb.2011.2610 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton J. T. & Ruiz G. M. In Biological invasions in changing ecosystems:Vectors, ecological impacts, management and predictions (ed. Canning-Clode João) 24–36 (De Gruyter Open Ltd, 2015). [Google Scholar]
- Rius M. & Darling J. A. How important is intraspecific genetic admixture to the success of colonising populations? Trends Ecol. Evol. 29, 233–242, doi: 10.1016/j.tree.2014.02.003 (2014). [DOI] [PubMed] [Google Scholar]
- Roman J. Diluting the founder effect: cryptic invasions expand a marine invader’s range. Proc. R. Soc. B 273, 2453–2459, doi: 10.1098/rspb.2006.3597 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brante A., Fernandez M. & Viard F. Phylogeography and biogeography concordance in the marine gastropod Crepipatella dilatata (Calyptraidae) along the South Eastern Pacific coast J. Heredity 103, 630–637, doi: 10.11093/jhered/ess030 (2012). [DOI] [PubMed] [Google Scholar]
- Dupont L., Viard F., David P. & Bishop J. D. D. Combined effects of bottlenecks and selfing in populations of Corella eumyota, a recently introduced sea squirt in the English Channel. Divers. Distrib. 13, 808–817, doi: 10.1111/j.1472-4642.2007.00405.x (2007). [DOI] [Google Scholar]
- Collin R., Farrell P. & Cragg S. Confirmation of the identification and establishment of the South American slipper limpet Crepipatella dilatata (Lamark 1822) (Caenogastropoda: Calyptraeidae) in Northern Spain. Aquat. Invasions 4, 377–380, doi: 10.3391/ai.2009.4.2.13 (2009). [DOI] [Google Scholar]
- Castilla J. C. et al. Down under the southeastern Pacific: marine non-indigenous species in Chile. Biol. Invasions 7, 213–232, doi: 10.1007/s10530-004-0198-5 (2005). [DOI] [Google Scholar]
- Castilla J. C. & Neill P. E. In Biological invasions in marine ecosystems (ed Springer) 439–457 (Heidelberg, 2009). [Google Scholar]
- Geoffroy A., Le Gall L. & Destombe C. Cryptic introduction of the red alga Polysiphonia morrowii Harvey (Rhodomelaceae, Rhodophyta) in the North Atlantic Ocean highlighted by a DNA barcoding approach. Aquat. Bot. 100, 67–71, doi: 10.1016/j.aquabot.2012.03.002 (2012). [DOI] [Google Scholar]
- Bishop J. D. D. et al. The Southern Hemisphere ascidian Asterocarpa humilis is unrecognised but widely established in NW France and Great Britain. Biol. Invasions 15, 253–260, doi: 10.1007/s10530-012-0286-x (2013). [DOI] [Google Scholar]
- Maggs C. A. et al. Evaluating signatures of glacial refugia for North Atlantic benthic marine taxa. Ecology 89, S108–S122, doi: 10.1890/08-0257.1 (2008). [DOI] [PubMed] [Google Scholar]
- Cox L. N., Zaslavskaya N. I. & Marko P. B. Phylogeography and trans-Pacific divergence of the rocky shore gastropod Nucella lima. J. Biogeogr. 41, 615–627, doi: 10.1111/jbi.12217 (2014). [DOI] [Google Scholar]
- Waltari E. & Hickerson M. J. Late Pleistocene species distribution modelling of North Atlantic intertidal invertebrates. J. Biogeogr. 40, 249–260, doi: 10.1111/j.1365-2699.2012.02782.x (2013). [DOI] [Google Scholar]
- Riquet F., Daguin-Thiebaut C., Ballenghien M., Bierne N. & Viard F. Contrasting patterns of genome-wide polymorphism in the native and invasive range of the marine mollusc Crepidula fornicata. Mol. Ecol. 22, 1003–1018, doi: 10.1111/mec.12161 (2013). [DOI] [PubMed] [Google Scholar]
- Rohfritsch A. et al. Population genomics shed light on the demographic and adaptive histories of European invasion in the Pacific oyster, Crassostrea gigas. Evol. Appl. 6, 1064–1078, doi: 10.1111/eva.12086 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tepolt C. K. Adaptation in marine invasion: a genetic perspective. Biol. Invasions 17, 887–903, doi: 10.1007/s10530-014-0825-8 (2015). [DOI] [Google Scholar]
- Lee T. & Shin S. Morphological and molecular identification of an introduced aline sea squirt (Tunicata: Ascidiacea) in Korea. P. Biol. Soc. Wash. 127, 284–297, doi: 10.2988/0006-324X-127.1.284 (2014). [DOI] [Google Scholar]
- Blakeslee A. M. H., Byers J. E. & Lesser M. P. Solving cryptogenic histories using host and parasite molecular genetics: the resolution of Littorina littorea’s North American origin. Mol. Ecol. 17, 3684–3696, doi: 10.1111/j.1365-294X.2008.03865.x (2008). [DOI] [PubMed] [Google Scholar]
- Provan J. I. M., Wattier R. A. & Maggs C. A. Phylogeographic analysis of the red seaweed Palmaria palmata reveals a Pleistocene marine glacial refugium in the English Channel. Mol. Ecol. 14, 793–803, doi: 10.1111/j.1365-294X.2005.02447.x (2005). [DOI] [PubMed] [Google Scholar]
- Provan J. & Bennett K. D. Phylogeographic insights into cryptic glacial refugia. Trends Ecol. Evol. 23, 564–571, doi: 10.1016/j.tree.2008.06.010 (2008). [DOI] [PubMed] [Google Scholar]
- Panova M. et al. Glacial history of the North Atlantic marine snail, Littorina saxatilis, inferred from distribution of mitochondrial DNA lineages. PLoS ONE 6, e17511, doi: 10.1371/journal.pone.0017511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmeyer R. In The Danish Ingolf Expedition. Copenhagen 2(7), 275 (1924). [Google Scholar]
- Van Name W. G. The North and South American ascidians. B. Am. Mus. Nat. Hist. 84, 471–431 (1945). [Google Scholar]
- Berrill N. J. The Tunicata with an account of the Bristish species. (Royal Society of London, 1950). [Google Scholar]
- Millar R. H. Tunicata Ascidiacea. Marine Invertebrates of Scandinavia. Vol. No. 1 (Universitetsforlaget, 1966). [Google Scholar]
- Sanamyan K. Ciona Fleming, 1822. In Shenkar N., Gittenberger A., Lambert G., Rius M., Moreira Da Rocha R., Swalla B. J., Turon X. Ascidiacea World Database. (2014) Accessed through: World Register of Marine Species at http://www.marinespecies.org/aphia.php?p=taxdetails&id=103488, 23th December 2014). [Google Scholar]
- Dybern B. I. The life cycle of Ciona intestinalis (L.) f.typica in relation to the environmental temperature. Oikos 16, 109–131, doi: 10.2307/3564870 (1965). [DOI] [Google Scholar]
- Ramsay A., Davidson J., Bourque D. & Stryhn H. Recruitment patterns and population development of the invasive ascidian Ciona intestinalis in Prince Edward Island, Canada. Aquat. Invasions 4, 169–176, doi: 10.3391/ai.2009.4.1.17 (2009). [DOI] [Google Scholar]
- Ramsay A., Davidson J., Landry T. & Arsenault G. Process of invasiveness among exotic tunicates in Prince Edward Island, Canada. Biol. Invasions 10, 1311–1316, doi: 10.1007/s10530-007-9205-y (2008). [DOI] [Google Scholar]
- Sargent P. S., Wells T., Matheson K., McKenzie C. H. & Deibel D. First record of vase tunicate, Ciona intestinalis (Linnaeus, 1767), in coastal Newfoundland waters. Bioinvasions Rec. 2, 89–98, doi: 10.3391/bir.2013.2.2.01 (2013). [DOI] [Google Scholar]
- Iannelli F., Pesole G., Sordino P. & Gissi C. Mitogenomics reveals two cryptic species in Ciona intestinalis. Trends Genet. 23, 419–422, doi: 10.1016/j.tig.2007.07.001 (2007). [DOI] [PubMed] [Google Scholar]
- Hall T. A. BioEdit: a user friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Sympos. Ser. 41, 95–98 (1999). [Google Scholar]
- Rozas J., Sanchez-DelBarrio J. C., Messeguer X. & Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497, doi: 10.1093/bioinformatics/btg359 (2003). [DOI] [PubMed] [Google Scholar]
- Kalinowski S. T. HP-Rare 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol. Ecol. Res. 5, 187–189, doi: 10.1111/j.1471-8286.2004.00845.x (2005). [DOI] [Google Scholar]
- Excoffier L. & Lischer H. E. L. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Res. 10, 564–567, doi: 10.1111/j.1755-0998.2010.02847.x (2010). [DOI] [PubMed] [Google Scholar]
- Clarke K. R. & Gorley R. N. Primer v.6: user manual/tutorial (2006).
- Hey J. Isolation with migration models for more than two populations. Mol. Biol. Evol. 27, 905–920, doi: 10.1093/molbev/msp296 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa M., Kishino H. & Yano T. A. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 22, 160–174, doi: 10.1007/bf02101694 (1985). [DOI] [PubMed] [Google Scholar]
- Hey J. & Nielsen R. Integration within the Felsenstein equation for improved Markov chain Monte Carlo methods in population genetics. Proc. Natl. Acad. Sci. USA 104, 2785–2790, doi: 10.1073/pnas.0611164104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.