Abstract
Regionally variable rates of aerobic glycolysis in brain networks identified by resting-state functional magnetic resonance imaging (R-fMRI) imply regionally variable adenosine triphosphate (ATP) regeneration. When regional glucose utilization is not matched to oxygen delivery, affected regions have correspondingly variable rates of ATP and lactate production. We tested the extent to which aerobic glycolysis and oxidative phosphorylation power R-fMRI networks by measuring quantitative differences between the oxygen to glucose index (OGI) and the oxygen extraction fraction (OEF) as measured by positron emission tomography (PET) in normal human brain (resting awake, eyes closed). Regionally uniform and correlated OEF and OGI estimates prevailed, with network values that matched the gray matter means, regardless of size, location, and origin. The spatial agreement between oxygen delivery (OEF≈0.4) and glucose oxidation (OGI ≈ 5.3) suggests that no specific regions have preferentially high aerobic glycolysis and low oxidative phosphorylation rates, with globally optimal maximum ATP turnover rates (VATP ≈ 9.4 µmol/g/min), in good agreement with 31P and 13C magnetic resonance spectroscopy measurements. These results imply that the intrinsic network activity in healthy human brain powers the entire gray matter with ubiquitously high rates of glucose oxidation. Reports of departures from normal brain-wide homogeny of oxygen extraction fraction and oxygen to glucose index may be due to normalization artefacts from relative PET measurements.
Keywords: Default mode network, energy metabolism, fMRI, glutamate, neurophysiology, PET
Introduction
Functional magnetic resonance imaging (fMRI) detects large-scale network activity in the human brain.1 The fMRI data analysis of blood oxygenation level-dependent (BOLD) signal yields statistically correlated activity in gray matter with both task-based (T-fMRI, stimuli-correlated signals) and resting-state (R-fMRI, region correlated signals) paradigms, from which estimates are assigned to neuronal networks. R-fMRI, in contrast to T-fMRI, reflects the correlated activity present when subjects are not stimulated nor are asked to execute a specific mental task. Instead, the temporal correlations of R-fMRI signals signify correspondingly small and spontaneous fluctuations of the BOLD signal.
The neurophysiological basis of the spontaneous BOLD signal fluctuations has not been fully revealed, but the fluctuations are held to reflect variations of underlying neuronal activity.2 The fluctuations of the correlated BOLD signal ride on top of a much larger background of neuronal activity present throughout the brain at rest.3 It is unknown whether the fluctuations represent large changes in activity of a small number of neurons or small fluctuations of the activity of the entire ensemble. Also, it is unknown whether specifically correlated networks are of functional significance to mental processes at rest. For example, the default mode network (DMN) is a term assigned to an assembly of regions with correlated fluctuations of activity during introversive awareness, and specific functional assignments have been made to other R-fMRI-derived networks.4 As the neurophysiological basis of these networks is elusive,5 other properties have been used to differentiate between the networks. A recent functional differentiation of DMN from other brain regions and networks was proposed on the basis of certain measures regarding the magnitude of aerobic glycolysis.6
In the brain, the substantial oxygen consumption7 serves to support of Na+, K+-adenosine triphosphate (ATP)ase activity needed for the maintenance of constant ion concentrations in different tissue compartments in support of membrane depolarization.8 Glucose is the primary energy substrate in the adult healthy brain, with an ATP yield that is about 16–18 times higher during full oxidation than during exclusive glycolysis.9 However, it is unknown how the energy yields are distributed between oxidative phosphorylation and aerobic glycolysis, as they power the large-scale network activity in the human brain captured by resting-state network activity. To assess regionally variable ATP regeneration rates, the degree of glucose oxidation must be known, as reflected in the oxygen to glucose index (OGI) definition
| (1) |
which is determined from direct measurements of cerebral metabolic rates of oxygen (CMRO2) and glucose (CMRglc) consumption or as the ratio of the corresponding arteriovenous deficits in oxygen vs. glucose. An OGI of less than 6 indicates a degree of aerobic glycolysis that terminates in the generation of lactate or other metabolites that remain in the tissue without yielding carbon dioxide and water through metabolism.
When glucose utilization is mismatched in relation to the oxygen delivery to specific regions, the regions in question regenerate relatively less ATP per mole of glucose. Note that networks derived by R-fMRI does not reflect glucose utilization per se, but rather the oxygen extracted by the tissue because the BOLD signal depends on the oxygen delivery as reflected in the microvascular deoxyhemoglobin content and the oxygen extraction fraction (OEF), according to Fick’s principle9
| (2) |
which is obtained by relating the CMRO2 in relation to the arterial delivery of oxygen, as given by multiplying cerebral blood flow (CBF) and arterial oxygen concentration (Ca).
As the metabolic basis of networks revealed by R-fMRI remains uncertain, we tested the recent claim that a specific R-fMRI-derived network, the DMN, has a higher fraction of aerobic glycolysis compared to other networks,6 which inherently implies regional variation of ATP rephosphorylation as well as substantial generation of lactate or other metabolites, subject either to export from the brain or to permanent deposit in the tissue. If lactate or other metabolites were not exported from the brain, then the metabolites would add to brain weight in proportion to the magnitude of aerobic glycolysis. According to this claim, DMN is comparatively more active than other networks when a person is at wakeful rest and not focused on a specific task.4 The benefit of a higher fraction of aerobic glycolysis remains uncertain, although explanations such as a relationship to increased synaptogenesis, and by inference biomass, have been proposed.6
To determine OGI and OEF in tomographic voxels of brain tissue with quantitative positron emission tomography (PET), we measured, in the same young and healthy subjects, CMRglc, CMRO2, and CBF in the resting awake, eyes-closed condition used as the common baseline state of many R-fMRI studies. The quantitative PET measurements allowed distributions of the total ATP regeneration rate (VATP) to be determined according to,
| (3) |
where the first and second terms on the right-hand side, respectively, represent ATP generated from oxidative phosphorylation (ϕ = 6 ATP/oxygen) and aerobic glycolysis (γ = 2 ATP/glucose), assuming dedication of all oxygen consumed to oxidative phosphorylation of ATP in the absence of reduced ATP gain by mitochondrial uncoupling (i.e. ϕ < 6 ATP/oxygen).
From equation (3), it is important to note that regional variations of VATP must be determined from quantitative CMRglc and CMRO2 data. Here, we found VATP calculated from quantitative PET measurements in young healthy brain to be in good agreement with absolute VATP measured by 31P and 13C magnetic resonance spectroscopy (MRS).10–13 Regardless of size, spatial coordinates, or origin of the extended R-fMRI-networks that we examined, the regional distributions revealed remarkably similar OEF and OGI values that were comparable in magnitude to gray matter means. The results are consistent with the notion that aerobic glycolysis is a minor, but consistent, contributor to network energy turnover at rest. Thus, high glucose oxidation rates are maintained over nearly the entire gray matter as behooves the major supplier of energy to resting neuronal activity.
Materials and methods
At Aarhus University Hospital, healthy male volunteers (n = 13) underwent PET and MRI at resting awake and eyes-closed condition. The study was conducted in accordance with the Helsinki Protocol, as approved by the official science ethics committee of the Central Denmark Region and Aarhus University Hospital, Aarhus, Denmark. All participants gave written informed consent.
Subjects and tomography
Male volunteers (n = 13, healthy, right handed) underwent PET and MRI in the resting awake and eyes-closed condition, where the average arterial oxygen content (Ca), age, weight, and height were 9.4 ± 0.5 µmol/mL, 26.1 ± 3.8 years, 77.0 ± 11.5 kg, and 1.82 ± 0.10 m, respectively. All tomographies were performed on the same day, during a single session. The order of tracer administration was: (i) 15O-H2O, (ii) 15O-O2, and (iii) 18F-fluorodeoxyglucose (FDG). Each imaging session was separated by the previous one by at least five half-lives of the 15O tracer (i.e. 10 min), corresponding to less than 3% residual radioactivity. Any effects on quantification of the K1-parameter for CMRO2 are likely to be minute, as it is most sensitive to the initial rapid influx-efflux of inhaled radioactivity (not the baseline).14,15 Subjects fasted overnight before FDG imaging sessions.
Each subject underwent high resolution MRI (GE Signa Excite HDx 3T; 3D IR-fSPGR: TE = 3.0 ms, TI = 450 ms, flip angle = 20°, slices = 120, slice thickness = 1.5 mm, FOV = 240 × 240 mm, matrix = 256 × 256) and PET (CTI/Siemens ECAT EXACT HR 47; 3D mode: slices = 47, slice thickness = 3.1 mm, FOV = 256 × 256 mm, matrix = 128 × 128). PET images were reconstructed using filtered back-projection with a 0.5 cycles−1 ramp filter, followed by application of a 7 mm Gaussian filter. Radial artery blood samples were collected with an automatic sampling device (1 s temporal resolution). Hematocrit and blood gas values were measured in arterial blood samples collected immediately prior to each PET recording. CBF quantification used 500 MBq bolus injection of 15O-H2O delivered intravenously, and CMRO2 quantification used 1000 MBq of 15O-O2 inhaled in single breath. CMRglc quantification was based on 200 MBq bolus of FDG injected intravenously. Both CBF and CMRO2 data were acquired in 21 frames (12 × 5 s, 6 × 10 s, 3 × 20 s) over 3 min and modeled using single tissue compartment with added vascular space, according to established procedures.14,15 CMRglc data were acquired in 27 frames (6 × 0.5 min, 7 × 1 min, 5 × 2 min, 4 × 5 min, 5 × 10 min) over 45 min and modeled using two tissue compartments and irreversible binding with a lumped constant of 0.8 according to established procedure,16 with the assumption that the lumped constant is invariable across the healthy young group of male subjects (see above). While the values of injected activity were standard amounts used in numerous past studies, the issue of the lower injectates has to be weighed against the benefit of doing the quantitative studies in the same subjects.
Subjects lay quiet and still for PET scanning in a dark quiet room with their eyes closed. Parameter estimation was performed in each subjects’ native PET space. A transformation between native PET and MRI spaces was calculated for each subject using a six-parameter rigid-body transformation with a mutual information cost function.17 MRI volumes were finally mapped to a common reference space (MNI 1 mm) with linear and non-linear registrations in BioImage Suite (www.bioimagesuite.com). In addition, MRI images were skull-stripped and segmented into gray/white/cerebral spinal fluid (CSF) space using FSL BET, and FAST (fsl.fmrib.ox.ac.uk). Once in common space, averaged PET maps were computed for different parameters in Matlab (MathWorks, Natick, MA) using custom scripts. In addition, volume of interest analysis of PET data was conducted using Brodmann’s areas to define regions and networks (see Tables S1 and S2).
Statistical analysis
Quantitative PET maps from all 13 healthy subjects were co-registered into common space via their own MRI, and CSF space was excluded. Analyses of PET data were conducted on a voxel-by-voxel basis, with voxels identified as gray or white matter based on MRI contrast, and then parcellated into gray matter regions (Table S1; Figure S1(a)) and networks (Table S2; Figure S1(b)). Here, a region was defined as a well-defined gray matter area based on cytoarchitecture (e.g. Brodmann’s areas), whereas a network was defined as a group of regions that function together.1 The cerebellum was excluded from analysis because of unequal coverage in all subjects.
Since each subject underwent CMRglc, CMRO2, and CBF measurements, there were no group comparisons across subjects. Absolute CMRglc, CMRO2, and CBF maps were compared and from which OGI, OEF, and VATP were calculated on a voxel-by-voxel basis using equations (1) to (3), respectively. Using Student’s t-test and analysis of variance (ANOVA, two-way), we assessed statistical differences between gray vs. white matter and across regions and networks (Figure S1). The CMRglc, CMRO2, and CBF data were also put through global mean normalization. In each case, global mean normalization was achieved by dividing each voxel by the mean of some brain region (i.e. excluding voxels outside the brain and inside the ventricles).18 We used either gray or white matter of the cerebrum for the global mean normalization signal. The mean globally normalized values, in each case for OGI and OEF, were then obtained with equations (1) and (2).
The conventional approach to visualizing relations among CMRglc, CMRO2, and CBF estimates is a 2D plot. However, a 2D plot provides no information of when and how well the three parameters are coupled in the present case. We completed 3D regression analysis to illustrate the degree of the 3D correlation reflecting the overall coupling across the independent CMRglc, CMRO2, and CBF estimates. The best-fit of the 3D data was obtained using the fit_3d_data function in Matlab,19 which is an orthogonal linear regression in 3D space based on principal component analysis. The fit_3d_data treats CMRglc, CMRO2, and CBF as state variables. For each parameter, the function generates three values: a normal vector (indicating the 3D space relation of a given parameter to the other two parameters), a data centroid (indicating the global mean of a given parameter), and an error of approximation (indicating the sum of orthogonal distances of data points from the best-fit 3D regression line).
Results
Quantitative CMRglc, CMRO2, and CBF estimates
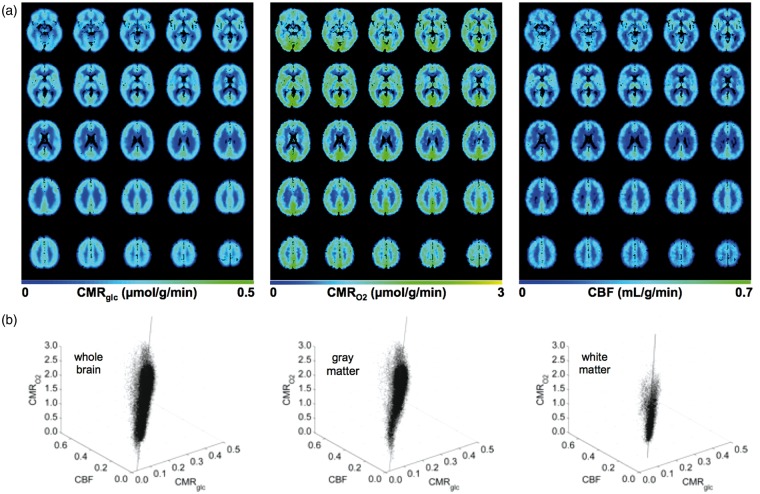
The mean CMRglc, CMRO2, and CBF maps had mutually similar spatial relations (Figure 1(a)), with minor (<15%) variability across subjects as reflected by the standard deviation (SD) maps (Figure S2(a)). Gray and white matter differences were significant, even at the whole brain level (i.e. P < 0.0008; Table S3). In regions with the least partial volume, the gray matter values exceeded the white matter values by about three-fold, in agreement with prior studies.20–24
Figure 1.
Absolute CMRglc, CMRO2, and CBF in normal human brain. (a) Mean maps from 25 representative slices, where CSF was masked out. See Table S3 for statistical analysis, where the mean ± SD values (excluding the cerebellum) for whole brain, gray matter, white matter for CMRglc were 0.26 ± 0.07, 0.28 ± 0.07, and 0.22 ± 0.05 µmol/g/min, for CMRO2 were 1.36 ± 0.37, 1.44 ± 0.35, and 1.04 ± 0.29 µmol/g/min, and for CBF were 0.37 ± 0.11, 0.39 ± 0.10, and 0.29 ± 0.09 mL/g/min. See Figure S2A for SD maps. (b) Global coupling between absolute CMRglc, CMRO2, and CBF with 3D voxel-wise analysis. Black dots and lines, respectively, indicate all voxels and the best-fit 3D regressions (see statistical analysis section). The fit_3d_data function in Matlab treats CMRglc, CMRO2, and CBF as state variables and for each parameter the function generates three values: a normal vector defining the 2D plane on which the data varies maximally, a data centroid, and an error of approximation. See Table S4 for the 3D regression analysis results and Figures S2(b) and S2(c) for the 2D voxel-wise analysis of CMRglc, CMRO2, and CBF data. Units for CMRglc and CMRO2 are µmol/g/min, for CBF the unit is mL/g/min.
We examined the coupling among the CMRglc, CMRO2, and CBF estimates on a voxel-by-voxel basis by 3D regression. The vectors from 3D regression of voxels in whole brain, gray matter, and white matter were nearly identical (Figure 1(b)), suggesting excellent spatial similarity between CMRglc, CMRO2, and CBF maps (Table S4). The centroids from 3D regression for whole brain and gray matter were comparable and higher than the centroids for white matter (Table S4). Voxels in whole brain, gray matter, and white matter were well clustered in 3D, and the goodness-of-fit estimates for regressions were high (i.e. R values higher than 0.95; Table S4). Similarly, the slopes from 2D regressions were analogous for whole brain, gray matter, and white matter (i.e. R values ranging from 0.86 to 0.91; Figure S2(b) and (c)), where the 95% confidence interval (CI) encompassed 97.3% of all voxels in the brain (Figure S2(b) and (c)).
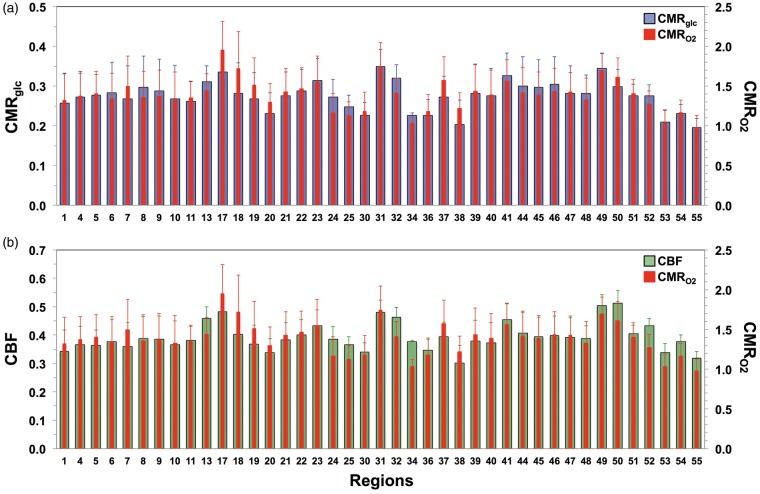
We also examined the coupling among the CMRglc, CMRO2, and CBF estimates across gray matter regions. Specifically, we assessed how the pattern of variations in CMRO2 was related to the pattern of variations in CMRglc and CBF, because they respectively relate to OGI and OEF (equations (1) and (2), respectively). The patterns of CMRO2 and CMRglc (for OGI) were remarkably similar (Figure 2(a)), in agreement with results from a previous meta-analysis of quantitative PET data.24 The patterns of CMRO2 and CBF (for OEF) were similarly consistent (Figure 2(b)), in general agreement with trends from previous quantitative PET data.21,22 Two-way ANOVA suggested that the pattern of variations in CMRglc, CMRO2, and CBF were similar (P ≫ 0.05). For example, in almost every region where CMRO2 was slightly higher than CMRglc, CMRO2 also exceeded CBF (e.g. in regions 7, 17, 37, and 38; see Table S1).
Figure 2.
Regional coupling between CMRglc, CMRO2, and CBF examined in non-overlapping gray matter regions. Absolute variations across regions for (a) CMRO2 (red) vs. CMRglc (blue) and (b) CMRO2 (red) vs. CBF (green). Values shown are mean and SD of each region, where all regions together comprise the entire gray matter (Table S1; Figure S1(a)). Two-way ANOVA tests suggest that the pattern of variations in CMRO2 was identical to pattern of variations in (a) CMRglc and (b) CBF (P≫0.05). Units for CMRglc and CMRO2 are µmol/g/min, for CBF the unit is mL/g/min.
Absolute OGI and OEF in the brain
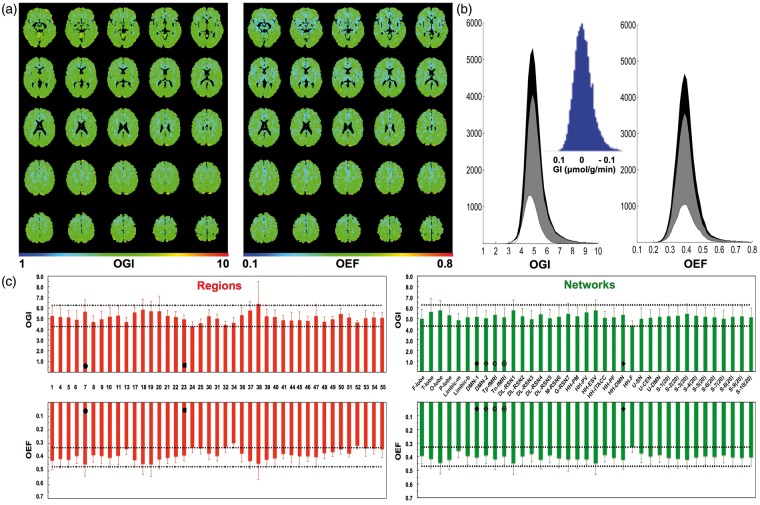
We calculated maps of OGI and OEF with equations (1) and (2), using the 95% CI portions for the CMRglc, CMRO2, and CBF data (Figure 3(a)). Using all voxels generated very similar results (Table S3), but the voxels outside the 95% CI represented less than 2.7% of all voxels in the brain and based on their location were not considered accurate due to large vessel artifacts that distort CMRO2 data (to very high values) in the single tissue compartment model used here.15 The gray and white matter differences were significant for both OGI and OEF (i.e. P < 0.0143; Table S3), where the OGI and OEF histograms were bell-shaped with high kurtosis (Figure 3(b)). The OGI and OEF values of whole brain were clustered (i.e. R values ranging from 0.77 to 0.81; Figure S3(a)) with excellent spatial correlation between OGI and OEF values in nearly 90% of all voxels (Figure S3(b)). At the current spatial resolution of the PET study, analysis of the CMRglc, CMRO2, and CBF maps on a voxel-by voxel basis (Figure 1), and Brodmann’s regions (Figure 2) showed similar global/regional OGI and OEF trends (Figure 3).
Figure 3.
Absolute OGI and OEF in normal human brain derived from equations (1) and (2), respectively, using the data in Figure 1. (a) Maps and (b) histograms, where the voxels outside the 95% CI (representing less than 2.7% of voxels in the whole brain) were replaced by regional averages. The mean ± SD values (excluding the cerebellum) for whole brain (i.e. gray + white), gray matter, white matter, respectively, for OGI were 5.17 ± 0.95, 5.26 ± 0.99, and 4.83 ± 0.60 and for OEF were 0.40 ± 0.07, 0.40 ± 0.0.07, and 0.39 ± 0.06. In (b) histograms shaded in black, gray, and white represent whole brain, gray matter, white matter, respectively. Inset of (b) shows the GI, glycolytic index, calculated by equation (4), where the slope s of ∼0.19 is inversely proportional to an OGI of ∼5.3. See Figure S3 for voxel-wise comparison between OGI and OEF. See Table S3 for statistical analysis between whole brain, gray matter, and white matter histograms. (c) OGI and OEF across regions (left) and networks (right) in normal human brain. OGI (top) and OEF (bottom) across 41 regions and 37 networks in relation to gray matter means, which are indicated by the black dotted horizontal lines representing the ± SD around the means (i.e. OGI = 5.26 ± 0.99 and OEF = 0.40 ± 0.07). Values shown are mean and SD of each region, where all regions together comprise the entire gray matter (Table S1; Figure S1(a)). Two-way ANOVA tests suggest that the pattern of variations in OGI was identical to pattern of variations in OEF, both for regions and networks (P≫0.05). For regions, the symbols • and ▪ are two key regions associated with the DMN (i.e. regions 7 (somatosensory association cortex or precuneus) and 24 (ventral anterior cingulate cortex), respectively; see Table S1 and Figure S1(a) for description and size of regions). For networks, the symbols  ,
,  ,
,  ,
,  and
and  represent networks related to DMN (i.e. DMN-1, DMN-2, Tp-fMRI, Tn-fMRI, HH-DMN, respectively; see Table S2 for and Figure S1(b) description of networks).
represent networks related to DMN (i.e. DMN-1, DMN-2, Tp-fMRI, Tn-fMRI, HH-DMN, respectively; see Table S2 for and Figure S1(b) description of networks).
The quantitative OGI data (Figure 3(a), left) had small (<10%) regional differences throughout gray matter, quite similar to homogenous patterns of absolute OEF data (Figure 3(a), right). Variations of OGI and OEF were calculated for 41 regions and across 37 networks (Figure 3(c)). None of the regions within the DMN were more glycolytic than the average of other regions (Figure 3(c), left), and the DMN, along with other networks related to it, had similarly high OGI values as the remainder of gray matter (Figure 3(c), right), as did patterns of OGI and OEF investigated for random networks in gray matter space (Figure S4). Hence, the patterns of OGI were remarkably similar to the patterns of OEF for both regions and networks.
The spatial relationship between CMRglc and CMRO2, reflected by OGI in equation (2), was also analyzed with the glycolytic index (GI), a recently introduced parameter by Vaishnavi et al.6
| (4) |
where the slope s from a plot of CMRglc vs. CMRO2 (i.e. 0.19 for gray matter with R value of 0.88; Figure S2(b), middle) is equivalent to 1/OGI. Mismatch between the first and second terms on the right-hand side of equation (4) reflects the GI in absolute units (i.e. µmol/g/min). The GI histogram was also bell-shaped with similarly high kurtosis as the OGI histogram (Figure 3(b), left inset), where the OGI determined from equations (1) and (4) were identical for gray matter (i.e. ∼5.3). GI deviations from the zero peak of the histogram showed nearly symmetric contributions from higher and lower OGI values (respectively indicated by negative and positive GI values). Given the 16–18-fold lower ATP yield of glycolysis compared to oxidative phosphorylation, the symmetric shape and relatively small deviation of the GI distribution about the mean OGI of 5.3 for gray matter confirms that oxidative phosphorylation is the major contributor to total ATP production (see below).
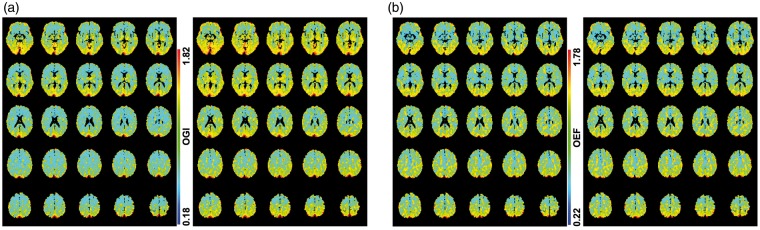
Given the regional and global homogeny of quantitative CMRO2, CMRglc, and CBF estimates within gray matter (Figures 1 and 2), we found uniform distributions of OGI and OEF using the absolute data (Figure 3). However, it is well known that global mean normalization (i.e. using mean of specific region(s) to normalize) generates apparent regions of hypo- and hyper-activity that artefactually accentuate relative differences across regions, at the cost of distorting global/regional differences in the absolute scale.25 Given the limited information about the normalizing region in the Vaishnavi et al.6 study and the fact that they also used conjunction analysis between their relative OGI maps and fMRI correlation maps (of different networks), we simply tested the degree of distortion introduced by global mean normalization as a direct contrast to analysis of the absolute data presented above. The global mean normalization, conducted with gray and white matter means, revealed artefactual increased and decreased areas of activity for both OGI and OEF maps (Figure 4). The large difference between the two normalized OGI maps (Figure 4(a), left and right) was due to absolute OGI difference between gray and white matter (see above), whereas the negligible difference between the two normalized OEF maps (Figure 4(b), left and right) was attributed to minimal OEF difference between gray and white matter (see above). These data reveal the potential for regional biases in OGI and OEF data when using mean globally normalized data, instead of absolute PET data.18,25,26
Figure 4.
Two types of global mean normalization. (a) Mean globally normalized OGI maps produced by equation (1) with mean globally normalized CMRO2 and CMRglc data, where gray matter (left) and white matter (right) were normalizing regions. The relative OGI scale corresponds to the same scale as the absolute OGI data shown in Figure 3. Note the stark high/low regions in these normalized OGI data compared to the homogeneity in the absolute OGI maps. The difference between the left and right normalized OGI maps is due to absolute OGI difference between gray and white matter. (b) Mean globally normalized OEF maps produced by equation (2) using mean globally normalized CMRO2 and CBF data, where gray matter (left) and white matter (right) were normalizing regions. The relative OEF scale corresponds to the same scale as the absolute OEF data in Figure 3. Note the stark high/low regions in these normalized OEF data compared to the homogeneity in the absolute OEF maps. The negligible difference between the left and right normalized OEF maps is due to minimal OEF difference between gray and white matter.
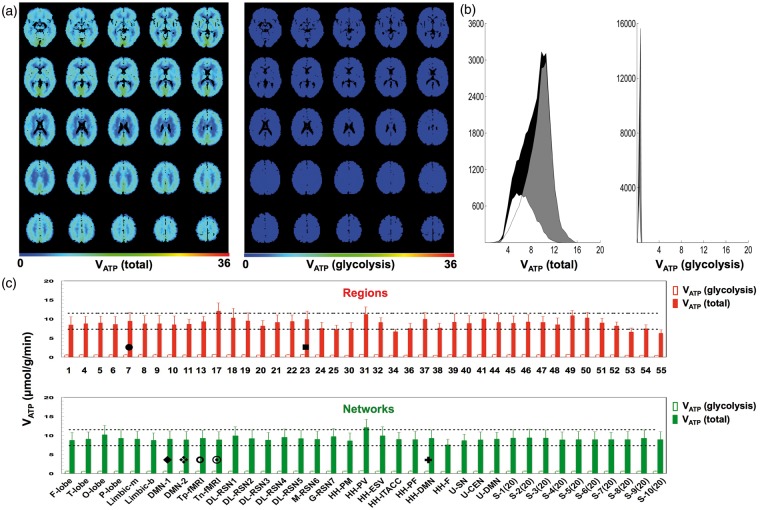
Absolute VATP in the brain
The contributions of oxidative phosphorylation and glycolysis to total ATP (re)generation rate (VATP) were estimated by equation (3), assuming the value of ϕ to be equal to 6. The averaged VATP maps distinguished significantly between gray and white matter (Figure 5(a) and (b), left), i.e. P < 0.0023 (Table S3). The whole brain, gray matter, and white matter estimates of VATP averaged 8.7 ± 2.4, 9.4 ± 2.1, and 6.7 ± 1.8 µmol/g/min, respectively. The ATP production from glycolysis (i.e. the second term of equation (3)) in all of gray matter averaged 0.5 ± 0.3 µmol/g/min, which represented only ∼5% of the gray matter mean for VATP (Figure 5(a) and (b), right). This glycolytic ATP fraction would likely be even smaller if the VATP calculation included the voxels outside the 95% CI that represented about 2.7% of all voxels in the brain. Regardless of this consideration, expansion of the second term on the right-hand side of equation (3) shows that the glycolytic source of ATP production has oxidative (A) and non-oxidative (B) contributions,
| (5) |
where A and B are given by OGI/6 and (1 − (OGI/6)), respectively. Using measured OGI values in equation (5) shows that A and B are 0.88 and 0.12, respectively. Thus, for all of the glucose completing glycolysis in gray matter, 12% terminates as lactate that is not subsequently oxidized, indicating that the non-oxidative glycolytic contribution to VATP was less than 1% of total ATP regenerated (i.e. (0.12 × 0.5 µmol/g/min)/9.4 µmol/g/min). Thus, the VATP maps predominantly represent oxidative phosphorylation. The total ATP (re)generated across all regions (Figure 5(c), top) and networks (Figure 5(c), bottom) yielded mean VATP values of 9.2 ± 1.2 and 9.5 ± 0.7 µmol/g/min, respectively, that are quite comparable to the gray matter mean of 9.4 µmol/g/min.
Figure 5.
Absolute VATP in normal human brain derived from equation (3) using the data in Figure 2. (a) Maps of total ATP regenerated (left) and ATP derived from glycolysis (right). The mean ± SD values of VATP (excluding the cerebellum) for whole brain (i.e. gray + white), gray matter, white matter averages, respectively, were 8.7 ± 2.4, 9.4 ± 2.1, and 6.7 ± 1.9 µmol/g/min. (b) Total ATP regenerated (left) and ATP derived from glycolysis (right). Histograms shaded in black, gray, and white represent whole brain, gray matter, white matter, respectively. Units of maps and histograms are in µmol/g/min. The ATP production from glycolysis (right) in all of gray matter averaged to 0.5 ± 0.3 µmol/g/min, which represented only 5% of the gray matter mean for total ATP regenerated (left). (c) Variations of VATP (solid bars) and ATP derived from glycolysis (open bars) across 41 regions (top) and 37 networks (bottom). The mean ± SD values of VATP for regions (9.2 ± 1.2 µmol/g/min) and networks (9.5 ± 0.7 µmol/g/min) were within 95% CI of the gray matter VATP mean (9.4 ± 2.1 µmol/g/min). The black horizontal lines indicate the mean ± SD of VATP of all gray matter (9.4 ± 2.1 µmol/g/min). See Tables S1 and S2 for description of regions and networks (Figure S1). For regions, the symbols • and ▪ are two key regions associated with the DMN (i.e. regions 7 (somatosensory association cortex or precuneus) and 24 (ventral anterior cingulate cortex), respectively; see Table S1 and Figure S1(a) for description and size of regions). For networks, the symbols  ,
,  ,
,  ,
,  and
and  represent networks related to DMN (i.e. DMN-1, DMN-2, Tp-fMRI, Tn-fMRI, HH-DMN, respectively; see Table S2 for and Figure S1(b) description of networks).
represent networks related to DMN (i.e. DMN-1, DMN-2, Tp-fMRI, Tn-fMRI, HH-DMN, respectively; see Table S2 for and Figure S1(b) description of networks).
Discussion
Glucose (1–3 mM) and oxygen (50–100 μM) levels in brain differ by at least an order of magnitude.27 While some extra glucose can be stored transiently as glycogen or used for biosynthesis,28 excess oxygen is highly reactive as free radicals.29 While glucose delivery is not rate-limiting for energy metabolism under normal conditions, spatially mismatched oxygen delivery and glucose metabolism may be pathogenic when ATP production is impaired (e.g. because of oxidative stress). Using quantitative CMRglc, CMRO2, and CBF data from normal human brain, we tested whether there are significant regional variations of ATP production rate, and if so, what the source is (i.e. elevated rate of oxidative phosphorylation or aerobic glycolysis).
Regional coupling between quantitative CMRglc, CMRO2, and CBF values
Using quantitative PET data, we found excellent coupling among CMRglc, CMRO2, and CBF (Figures 1 and 2), consistent with brain-wide homogeneity of OGI and OEF estimates (Figure 3(a) and (b)) where the variations across regions and networks remained within 5% of the global gray matter means (Figure 3(c)). Whole-brain cortical OGI averaged about 5.3, with OEF values of 0.4 in large swathes of the brain, indicating uniform fractions of glucose oxidation across different brain regions. The gray and white matter values for CMRglc, CMRO2, and CBF as well as the OGI and OEF calculated from these values (equations (1) and (2), respectively) are in agreement with prior quantitative PET results.20–24 In contrast, the relative OGI and OEF maps demonstrated regions of higher and lower activity compared to the global mean (Figure 4). The quantitative PET data revealed a calculated VATP (equation (3)) value of 9.4 µmol/g/min for gray matter, assuming minimal mitochondrial uncoupling (Figure 5(a), left), which is in good agreement with directly measured VATP using 31P MRS 10,11 and similar to VATP estimates derived from 13C MRS measurements of the rate of tricarboxylic acid (TCA) cycle, assuming a stoichiometric relationship between ATP production and CMRO2 12,13
About 95% of the VATP estimate was derived from glucose oxidation (Figure 5(a) and (b)). While there is little dispute regarding γ in equation (3) (the glycolytic contribution to VATP), it is evident that ϕ in equation (3) (the oxidative phosphorylation rate of VATP) arguably can be as low as 3 (i.e. 18 ATP per 1 mole of glucose or per 6 moles of oxygen), depending on the degree of mitochondrial uncoupling. If ϕ were reduced to 3 (instead of 6) in the presence of unchanged glucose consumption, VATP would decrease by about half for gray matter (i.e. ∼4.6 µmol/g/min), but the total glycolytic ATP contribution would increase to only 6% of VATP. This suggests that 1% additional ATP generated by glycolysis comes at a price of 50% reduction of total ATP produced, which is unlikely because the VATP from the present PET measurements is in good agreement with previous 31P and 13C MRS measurements in the awake resting human brain.10–13 The agreement between PET and MRS measured values of VATP suggests that there is negligible mitochondrial uncoupling in young healthy brain, but which may be altered during the course of healthy aging process.30
Because the gray matter thickness of gyri and sulci is less than optimal for the spatial resolution of the PET voxels,31 the CMRglc, CMRO2, and CBF values of gray and white matter are contingent on partial volume effects of a voxel, as well as on thresholds used in the MRI data to classify tissue as white or gray matter. Improved PET and MRI spatial resolution would reduce the white matter fraction of voxels assigned to gray matter (and vice versa) and improve the accuracy of these measurements. However, in regions with minimal partial volume effects, approximately three-fold higher rates of CMRglc, CMRO2, and CBF were measured in gray matter than in white matter. Based on the bell-shaped distributions of OGI and OEF in the current study (Figure 3(b)), we expect that higher PET resolution data for the normal brain will raise the kurtosis of these distributions.32
Minimal non-oxidative glycolytic ATP contributions in DMN and other networks
We found that nearly all networks, including DMN, have similar aerobic glycolysis and oxygen extraction indices (Figures 2 and 3). Vaishnavi et al.6 observed similar correlations between relative CMRO2 and CMRglc measures (i.e. Figures S1 in Vaishnavi et al.6 vs. Figure S2(b) here), but nonetheless concluded that the DMN differed from other regions with respect to a substantially greater contribution from aerobic glycolysis. To determine GI across regions, Vaishnavi et al.6 applied equation (4) to relative CMRO2 and CMRglc measures. While the relative GI can be calculated with equation (4), the slope s of the relative CMRO2 and CMRglc measures that is close to unity for gray matter is not indicative of the magnitude of quantitative OGI measurements and hence the GI remains a relative parameterized index. For the null-hypothesis test of equation (4), Vaishnavi et al.6 reasoned that any voxel with a higher relative CMRglc measure than indicated by the regression against the relative CMRO2 measure would serve as statistically significant evidence of GI being higher or OGI being lower. As the null-hypothesis test of equation (4) with relative data ignores the large majority of voxels in the brain, the relative GI parameterization by them did not reveal the quantitative extent of ATP regeneration from oxidative phosphorylation vs. aerobic glycolysis when very few voxels reside along the regression line.
We assessed the spatial relations of quantitative CMRglc and CMRO2 measurements to reveal the highly clustered absolute OGI estimates with symmetric distributions (Figure 3(b), left), made possible because of the small inter-subject variability (Figure S2(a)). The absolute OGI histogram was nearly identical to the absolute GI histogram (Figure 3(b), left and inset) because the value of the slope s in equation (4) is related quantitatively to the measured OGI (see Results section). The absolute GI shows that the extent of ATP contributions from glycolytic source is within 0.5 µmol/g/min for the entire brain (Figure 3(b), left and inset). This glycolytic ATP yield is quite small compared to the total ATP turnover (i.e. 9.4 µmol/g/min).
With quantitative PET data, we showed that VATP is uniformly high with at least 95% contribution from glucose oxidation (Figure 5(a) and (b)) and with a maximum of 5% contribution from aerobic glycolysis to the total ATP production. Thus, we conclude that all regions and networks had similar but low ATP contributions from aerobic glycolysis (Figure 5(c)). Yet, the non-oxidative glycolytic contribution to VATP is even smaller, because equation (5) shows that non-oxidative glycolysis contributes less than 1% of the total ATP regenerated. These types of distinctions between generating sources for ATP cannot be made with relative CMRO2 and CMRglc measures.
Oxidative phosphorylation supports R-fMRI networks
We found that the majority of R-fMRI networks had VATP variations within 5% of the global gray matter mean (Figure 5(c), bottom), regardless of size, spatial location, or definition. These small VATP variations across these networks, that represent at most a ±0.4 µmol/g/min deviation of ATP demand from the gray matter mean value of 9.4 µmol/g/min, arose from purely oxidative sources. Since the OGI measurements suggest that 10–12% of glucose undergoing metabolism is not subsequently oxidized as indicated by the non-oxidative glycolysis term in equation (5), the less than 1% regional variations in VATP due to non-oxidative glycolysis would vary no more than ±0.05 µmol/g/min from the observed regional VATP variations. In the future, it is possible to use R-fMRI to examine how these regions are correlated, whereas 13C MRS can investigate neuronal and glial glucose oxidations in relation to the relative contributions of glutamatergic and GABAergic neuronal activities.33 In turn, 31P MRS can measure VATP directly10 while 13C MRS can determine if VATP variations across regions are due to alternate substrate oxidation (e.g. ketones34 or lactate35).
Since R-fMRI networks are based on inter-regional correlations of spontaneous BOLD signal variations and not on the actual neuronal activity, an important question is how much ATP demand the fluctuations of neuronal activity actually represent, estimates of which can be obtained from calibrated fMRI which includes BOLD and CBF measurements to calculate CMRO2.36 In the awake human gray matter, approximately ±0.5% fluctuations of the BOLD signal at 3T correspond to at most ±5% variations in CMRO2,5,37,38 which is close to ±5% variability of VATP. Together these estimates imply that variations in ATP demand across regions and networks, spatially (∼0.4 µmol/g/min) and dynamically (∼0.5 µmol/g/min), are of the same order of magnitude. The brain-wide homogeny of oxidative phosphorylation, with similar levels of variation across the spatiotemporal landscape of gray matter, suggests that there are no large discrepancies of energy turnover across regions and networks of the brain (Figure 5(c)).
Regional homogeneity between absolute OGI and OEF
The possibility of regionally specialized glucose metabolism was tested further by examining the spatial differences between normalized OGI and OEF estimates. For most of the brain, we found that OEF and OGI estimates were well matched (Figure S3(b)), implying that the normal brain has the metabolic capacity necessary for high ATP yield by glucose oxidation. However, two small regions potentially could be vulnerable to brain disorders: dorsal white matter where more oxygen was extracted than glucose oxidized, and ventral gray matter areas where higher oxidation was observed, possibly indicating additional substrate utilization. Excessive metabolic activity in medial limbic regions has been implicated in paraneoplastic limbic encephalitis,39 whereas dorsal white matter can be linked to oxidative stress in multiple sclerosis.40 We found several R-fMRI networks including the DMN to have highly oxidative glucose metabolism with OGI ≥ 5.2, but these networks also had relatively higher OEF (Figure 3(c)), consistent with a constant relation between values of CMRglc and CBF.41,42 The relation suggests that conditions of high-yield ATP production prevailed in all these networks (Figure 5(c)). Future studies can be designed to test if brain-wide homogeny between OGI and OEF is lost in different pathophysiological states of the brain, indicative of a disturbed role of glucose metabolism in the regulation of blood flow. Given the regionalized nature of OGI and OEF changes in task studies, we hypothesize that regional homogeny between OGI and OEF would also be lost in pathophysiological states compared to normal brain during T-fMRI studies.
Although only a small fraction of total ATP production stems from aerobic glycolysis, this pathway potentially may still serve as an important energetic role, particularly when subpopulations of neurons transiently have very high activity. A potential explanation is provided by the glycogen shunt. Vesicular glutamate release into the synaptic cleft is rapidly cleared by Na+-coupled glial uptake, where it is converted to glutamine. This process requires 2 moles of ATP per mole of glutamate cycled between neurons and astrocytes. If 1 mole of glucose produces 2 moles of ATP in glia via glycolysis, then two equivalents of lactate generated in glia may be completely oxidized at an OGI of 6. Alternatively, to generate 2 moles of ATP in the glia via the glycogen shunt,43 2 moles of glucose (i.e. twice as much of glucose) have to be used for 4 moles of lactate to be produced. Thus, extra lactate is produced and exported to the circulation when the OGI drops below 6 in the absence of other nutrient sources. When the rate of glial energy production via glycolysis is not rapid enough for higher neuronal firing rates, the glycogen shunt could be activated to restore Na+ gradient and glutamine synthesis in glia such that the glutamate release by the neuron remains matched. Thus, the glycogen shunt is energetically less efficient than aerobic glycolysis, but when OGI is less than 6 with brain activation, more lactate is produced via the glycogen shunt.44 While the metabolites of the glucose that undergoes glycolysis were not measured in this study, lactate (measurable by 1H MRS) is the obvious candidate with a known mechanism of efflux from the brain.41,45 To what degree the glycogen shunt contributes to the support of network function in different physiological and even pathophysiological situations remains an important goal of future studies.
While the present evidence suggests that oxidative phosphorylation is the dominant target of energy support for synaptic transmission, the OGI measurements suggest that 10–12% of glucose may yield metabolites that are not subsequently oxidized. A recent report claims that this entire fraction of glycolysis is devoted to synaptogenesis.6 However, the claim fails to consider the availability of other sources of carbon such as lipids, ketone bodies, and lactate which can directly support biosynthesis.9 An interesting feature of alternative carbon sources is their direct entry into the TCA cycle, bypassing the generation of metabolites of glucose (ATP and lactate). Regionally differential utilization of glucose, lactate, and ketone bodies could mean that lactate potentially would vary regionally. It is clear that future quantitative PET and MRS studies investigating the role of alternative substrates for the brain would be needed to reveal to what extent synaptogenesis is supported and how the support varies regionally in relation to the functional energy demands of synaptic transmission.
Quantitative vs. relative PET imaging
Recently, there has been a growing interest in using relative CMRglc mapping with PET to help interpret R-fMRI-based networks,46,47 because merging of PET and R-fMRI studies can lead to multimodal information about both functional and metabolic brain networks.48 In the present study, no attempt was made to compute standardized uptake values (SUVs) of tracers. By normalizing the tissue concentration to the administered dose or the subject’s weight, it is thought that SUVs improve the precision of PET quantification. However, SUV computation is time-dependent, and it is affected by different plasma clearance rates between subjects and study conditions (e.g. competition for tracer by different organs). Since these differences on a per subject basis is an accurate reflection of the subject’s metabolic state, we obtained quantitative CMRglc, CMRO2, and CBF maps by including plasma tracer kinetics with tissue concentration on a per subject basis. We believe that the small inter-subject variability in the quantitative PET data (Figure S2(a)) indicates the homogeneity of the group of young healthy subjects that we studied. However, kinetic modeling of PET data remains hampered by “noise” in the input function derived from plasma tracer kinetics. Given the invasiveness of blood sampling and the fact that such approaches cannot be utilized in all scenarios, alternatives such as image-derived input functions are being sought to improve the precision of model parameter fitting.49
In this study, the CMRglc, CMRO2, and CBF maps also underwent global mean normalization as a test of the distortion introduced by this procedure (Figure 4). In many PET studies, it is assumed that the average metabolic rate across gray matter or the entire brain (or some other region such as the cerebellum) can be used to normalize the PET data to smooth, or remove experimental or physiological nuisance components from, the data. However, the normalization resulted in expression of the images in units of fractional deviation from the global mean. This normalization took only relative regional differences of brain metabolism into account and hence obscured the real quantitative metabolic differences among the different measures that determine the OGI and OEF indices.25 But normalization may be needed in some cases, as pointed out by Horwitz et al.,20 e.g. if inter-individual metabolic differences are large compared to intra-individual metabolic differences, which can mask correlations between regions in a multisubject analysis.
Conclusion
In healthy human brain, we found that oxidative phosphorylation, not glycolysis, powers resting-state fMRI networks in normal human brain, given the observable ranges and spatial distributions of CMRO2 and CMRglc. No R-fMRI network was found to have a substantially larger fraction of glycolysis than other networks, including the DMN that was found to have a slightly higher OGI than the average. Although not a major source of ATP, increased aerobic glycolysis may reflect rapid ATP production from glycogen,43 which is uniquely placed for glial Na+, K+-ATPase activity associated with glutamate uptake for neurotransmission.33 Alternatively, glycolytic ATP regeneration may reflect biosynthetic needs,6 but proof of this proposal needs testing of regional utilization of alternative fuels34,35 and as well as the contribution to biomass changes of brain weight over the natural aging process.
The current results indicate that oxygen delivery (OEF) is well matched to glucose oxidation (OGI) in the majority of gray matter voxels in the normal human brain. Ubiquitously high ATP yields from glucose oxidation are needed to support normal conscious human behavior,50 and significant changes in glucose oxidation are reproducibly measured subjects experiencing a wide range of stimuli.24 Since somatic mutations of mitochondrial DNA are associated with various pathophysiological states (e.g. healthy aging, neurodegeneration, cancer, Leber hereditary optic neuropathy) that limit the ability of brain cells to generate high-yield ATP via oxidative energy metabolism,51 combining R-fMRI with quantitative PET studies may reveal the metabolic condition of different networks of individual patients.52,53 The approach has advantages over methods that normalize across gray matter and probably miss global changes in VATP. We propose that clinical R-fMRI studies can be combined with quantitative PET measures of metabolic profiles to evaluate patients with brain disorders.
Supplementary Material
Acknowledgements
The authors thank Xenophon Papademetris and Kevin L. Behar, respectively, for inputs on image analysis software and ATP yield differences between oxidative phosphorylation and aerobic glycolysis.
Funding
This study was supported by NIH grants (R01 MH-067528, R01 AG-034953, R01 NS-087568, P30 NS-052519).
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Authors’ contributions
FH, CJB, AM, AG designed and performed research. FH, PH, CJB, RG, RKF, DLR, AG analyzed data. FH, PH, CJB, DLR, AG wrote paper.
Supplementary material
Supplementary material for this paper can be found at http://jcbfm.sagepub.com/content/by/supplemental-data
References
- 1.Smith SM, Miller KL, Moeller S, et al. Temporally-independent functional modes of spontaneous brain activity. Proc Natl Acad Sci U S A 2012; 109: 3131–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scholvinck ML, Maier A, Ye FQ, et al. Neural basis of global resting-state fMRI activity. Proc Natl Acad Sci U S A 2010; 107: 10238–10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maandag NJ, Coman D, Sanganahalli BG, et al. Energetics of neuronal signaling and fMRI activity. Proc Natl Acad Sci U S A 2007; 104: 20546–20551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckner RL, Andrews-Hanna JR, Schacter DL. The brain’s default network: anatomy, function, and relevance to disease. Ann N Y Acad Sci 2008; 1124: 1–38. [DOI] [PubMed] [Google Scholar]
- 5.Hyder F, Rothman DL. Neuronal correlate of BOLD signal fluctuations at rest: err on the side of the baseline. Proc Natl Acad Sci U S A 2010; 107: 10773–10774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vaishnavi SN, Vlassenko AG, Rundle MM, et al. Regional aerobic glycolysis in the human brain. Proc Natl Acad Sci U S A 2010; 107: 17757–17762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aiello LC, Wheeler P. The expensive-tissue hypothesis – the brain and the digestive-system in human and primate evolution. Curr Anthropol 1995; 36: 199–221. [Google Scholar]
- 8.Hyder F, Rothman DL, Bennett MR. Cortical energy demands of signaling and nonsignaling components in brain are conserved across mammalian species and activity levels. Proc Natl Acad Sci U S A 2013; 110: 3549–3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siesjo BK. Brain energy metabolism, New York, NY: Wiley and Sons, Ltd., 1978. [Google Scholar]
- 10.Zhu XH, Qiao H, Du F, et al. Quantitative imaging of energy expenditure in human brain. Neuroimage 2012; 60: 2107–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin AL, Fox PT, Hardies J, et al. Nonlinear coupling between cerebral blood flow, oxygen consumption, and ATP production in human visual cortex. Proc Natl Acad Sci U S A 2010; 107: 8446–8451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rothman DL, De Feyter HM, de Graaf RA, et al. 13C MRS studies of neuroenergetics and neurotransmitter cycling in humans. NMR Biomed 2011; 24: 943–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Graaf RA, Rothman DL, Behar KL. State of the art direct 13C and indirect 1H-[13C] NMR spectroscopy in vivo. A practical guide. NMR Biomed 2011; 24: 958–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohta S, Meyer E, Thompson CJ, et al. Oxygen consumption of the living human brain measured after a single inhalation of positron emitting oxygen. J Cereb Blood Flow Metab 1992; 12: 179–192. [DOI] [PubMed] [Google Scholar]
- 15.Ohta S, Meyer E, Fujita H, et al. Cerebral [15O]water clearance in humans determined by PET: I. Theory and normal values. J Cereb Blood Flow Metab 1996; 16: 765–780. [DOI] [PubMed] [Google Scholar]
- 16.Kuwabara H, Gjedde A. Measurements of glucose phosphorylation with FDG and PET are not reduced by dephosphorylation of FDG-6-phosphate. J Nucl Med 1991; 32: 692–698. [PubMed] [Google Scholar]
- 17.Collins DL, Neelin P, Peters TM, et al. Automatic 3D intersubject registration of MR volumetric data in standardized Talairach space. J Comput Assist Tomogr 1994; 18: 192–205. [PubMed] [Google Scholar]
- 18.Borghammer P, Aanerud J, Gjedde A. Data-driven intensity normalization of PET group comparison studies is superior to global mean normalization. Neuroimage 2009; 46: 981–988. [DOI] [PubMed] [Google Scholar]
- 19.Petras I, Podlubny I. State space description of national economies: the V4 countries. Comput Stat Data Anal 2007; 52: 1223–1233. [Google Scholar]
- 20.Horwitz B, Duara R, Rapoport SI. Intercorrelations of glucose metabolic rates between brain regions: application to healthy males in a state of reduced sensory input. J Cereb Blood Flow Metab 1984; 4: 484–499. [DOI] [PubMed] [Google Scholar]
- 21.Roland PE, Eriksson L, Stone-Elander S, et al. Does mental activity change the oxidative metabolism of the brain? J Neurosci 1987; 7: 2373–2389. [PMC free article] [PubMed] [Google Scholar]
- 22.Hatazawa J, Fujita H, Kanno I, et al. Regional cerebral blood flow, blood volume, oxygen extraction fraction, and oxygen utilization rate in normal volunteers measured by the autoradiographic technique and the single breath inhalation method. Ann Nucl Med 1995; 9: 15–21. [DOI] [PubMed] [Google Scholar]
- 23.Rumsey JM, Duara R, Grady C, et al. Brain metabolism in autism. Resting cerebral glucose utilization rates as measured with positron emission tomography. Arch Gen Psychiatry 1985; 42: 448–455. [DOI] [PubMed] [Google Scholar]
- 24.Hyder F, Fulbright RK, Shulman RG, et al. Glutamatergic function in the resting awake human brain is supported by uniformly high oxidative energy. J Cereb Blood Flow Metab 2013; 26: 865–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borghammer P, Jonsdottir KY, Cumming P, et al. Normalization in PET group comparison studies – the importance of a valid reference region. Neuroimage 2008; 40: 529–540. [DOI] [PubMed] [Google Scholar]
- 26.Borghammer P, Cumming P, Aanerud J, et al. Subcortical elevation of metabolism in Parkinson’s disease – a critical reappraisal in the context of global mean normalization. Neuroimage 2009; 47: 1514–1521. [DOI] [PubMed] [Google Scholar]
- 27.Hyder F. Dynamic imaging of brain function. Methods Mol Biol 2009; 489: 3–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Riera JJ, Schousboe A, Waagepetersen HS, et al. The micro-architecture of the cerebral cortex: functional neuroimaging models and metabolism. Neuroimage 2008; 40: 1436–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jesberger JA, Richardson JS. Oxygen free radicals and brain dysfunction. Int J Neurosci 1991; 57: 1–17. [DOI] [PubMed] [Google Scholar]
- 30.Boumezbeur F, Mason GF, de Graaf RA, et al. Altered brain mitochondrial metabolism in healthy aging as assessed by in vivo magnetic resonance spectroscopy. J Cereb Blood Flow Metab 2010; 30: 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.la Fougere C, Grant S, Kostikov A, et al. Where in-vivo imaging meets cytoarchitectonics: the relationship between cortical thickness and neuronal density measured with high-resolution [18F]flumazenil-PET. Neuroimage 2011; 56: 951–960. [DOI] [PubMed] [Google Scholar]
- 32.Huisman MC, van Golen LW, Hoetjes NJ, et al. Cerebral blood flow and glucose metabolism in healthy volunteers measured using a high-resolution PET scanner. EJNMMI Res 2012; 2: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hyder F, Patel AB, Gjedde A, et al. Neuronal-glial glucose oxidation and glutamatergic-GABAergic function. J Cereb Blood Flow Metab 2006; 26: 865–877. [DOI] [PubMed] [Google Scholar]
- 34.Nugent S, Tremblay S, Chen KW, et al. Brain glucose and acetoacetate metabolism: a comparison of young and older adults. Neurobiol Aging 2014; 35: 1386–1395. [DOI] [PubMed] [Google Scholar]
- 35.Herzog RI, Jiang L, Herman P, et al. Lactate preserves neuronal metabolism and function following antecedent recurrent hypoglycemia. J Clin Invest 2013; 123: 1988–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hyder F, Sanganahalli BG, Herman P, et al. Neurovascular and neurometabolic couplings in dynamic calibrated fMRI: transient oxidative neuroenergetics for block-design and event-related paradigms. Front Neuroenergetics 2010; 2: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gu H, Stein EA, Yang Y. Nonlinear responses of cerebral blood volume, blood flow and blood oxygenation signals during visual stimulation. Magn Reson Imaging 2005; 23: 921–928. [DOI] [PubMed] [Google Scholar]
- 38.Wu CW, Gu H, Lu H, et al. Mapping functional connectivity based on synchronized CMRO2 fluctuations during the resting state. Neuroimage 2009; 45: 694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scheid R, Lincke T, Voltz R, et al. Serial 18F-fluoro-2-deoxy-D-glucose positron emission tomography and magnetic resonance imaging of paraneoplastic limbic encephalitis. Arch Neurol 2004; 61: 1785–1789. [DOI] [PubMed] [Google Scholar]
- 40.Witherick J, Wilkins A, Scolding N, et al. Mechanisms of oxidative damage in multiple sclerosis and a cell therapy approach to treatment. Autoimmune Dis 2010; 2011: 164608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bergersen LH, Gjedde A. Is lactate a volume transmitter of metabolic states of the brain? Front Neuroenergetics 2012; 4: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vafaee MS, Gjedde A. Spatially dissociated flow-metabolism coupling in brain activation. Neuroimage 2004; 21: 507–515. [DOI] [PubMed] [Google Scholar]
- 43.Shulman RG, Hyder F, Rothman DL. Cerebral energetics and the glycogen shunt: neurochemical basis of functional imaging. Proc Natl Acad Sci U S A 2001; 98: 6417–6422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prichard J, Rothman D, Novotny E, et al. Lactate rise detected by 1H NMR in human visual cortex during physiologic stimulation. Proc Natl Acad Sci U S A 1991; 88: 5829–5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shulman RG, Hyder F, Rothman DL. Lactate efflux and the neuroenergetic basis of brain function. NMR Biomed 2001; 14: 389–396. [DOI] [PubMed] [Google Scholar]
- 46.Wehrl HF, Hossain M, Lankes K, et al. Simultaneous PET-MRI reveals brain function in activated and resting state on metabolic, hemodynamic and multiple temporal scales. Nat Med 2013; 19: 1184–1189. [DOI] [PubMed] [Google Scholar]
- 47.Di X, Biswal BB. Alzheimer’s Disease Neuroimaging Initiative. Metabolic brain covariant networks as revealed by FDG-PET with reference to resting-state fMRI networks. Brain Connect 2012; 2: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wehrl HF, Wiehr S, Divine MR, et al. Preclinical and translational PET/MR imaging. J Nucl Med 2014; 55: 11S–18S. [DOI] [PubMed] [Google Scholar]
- 49.Alf MF, Wyss MT, Buck A, et al. Quantification of brain glucose metabolism by 18F-FDG PET with real-time arterial and image-derived input function in mice. J Nucl Med 2013; 54: 132–138. [DOI] [PubMed] [Google Scholar]
- 50.Shulman RG, Hyder F, Rothman DL. Baseline brain energy supports the state of consciousness. Proc Natl Acad Sci U S A 2009; 106: 11096–11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schon EA, DiMauro S, Hirano M. Human mitochondrial DNA: roles of inherited and somatic mutations. Nat Rev Genet 2012; 13: 878–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riedl V, Bienkowska K, Strobel C, et al. Local activity determines functional connectivity in the resting human brain: a simultaneous FDG-PET/fMRI study. J Neurosci 2014; 34: 6260–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Spetsieris PG, Ko JH, Tang CC, et al. Metabolic resting-state brain networks in health and disease. Proc Natl Acad Sci U S A 2015; 112: 2563–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.