Abstract
Microbiota and its contribution to brain function and diseases has become a hot topic in neuroscience. We discuss the emerging role of commensal bacteria in the course of stroke. Further, we review potential pitfalls in microbiota research and their impact on how we interpret the available evidence, emerging results, and on how we design future studies.
Keywords: Brain ischemia, brain trauma, immunology, physiology, regeneration and recovery
All disease begins in the gut
—attributed to Hippocrates. (ca. 400 BC)
Throughout the last decade, we have witnessed remarkable and unexpected new insights into the complex interplay of the immune system and the brain after acute brain damage, including stroke. While many aspects of this interaction remain to be elucidated, a role of innate and adaptive immunity in tissue damage and perhaps endogenous protection, as well as functional recovery after stroke, is firmly established. Partially building on this knowledge, inspired by research on the role of gut microbiota in disease pathophysiology of other organs, and enabled by major advances in sequencing technologies and bioinformatics, stroke researchers have begun to ask whether gut microbial communities impact outcome after stroke. Meanwhile, fecal transplants have been proposed for use in fighting diseases from obesity1 to autism2 whence some scientists have started to criticize the hype surrounding microbiome research and have called for a healthy dose of skepticism.3 In this opinion article, we would like to provide a speculative framework for brain–gut microbiota interaction after focal cerebral ischemia, and at the same time raise some critical issues to discourage stroke researchers from reporting spectacular findings that eventually may not withstand the test of time.
Microbiota and the brain
Humans and bacteria coevolved throughout millions of years and have established, alongside host-pathogen relation, a tight commensal alliance. Human host niches are colonized by several bacterial populations – microbiota, with a huge collection of genes – themicrobiome. Each body site has its specific microbiota composition and interindividual differences between the same body habitat are smaller than intraindividual variance, between different body sites.4 The largest and most complex bacterial community resides within the gastrointestinal system, particularly the large intestine, and comprises trillions of bacteria.5 Gut microbiota is protective for the organism through competition with pathogens. By participating in formation of the intestinal barrier and development of the immune system, it implements structural functions. Finally, it is crucially involved in metabolism, fermenting indigested food, producing vitamins, and deactivating xenobiotics.6
Recent evidence from different fields of medicine suggests that microbiota plays an important role in many diseases, including those of the nervous system.7 The microbial population of the gut has been found to engage in an intense signaling with the central nervous system (CNS) via neural, immunological, and direct humoral signaling pathways and hence has been incorporated into the concept of bidirectional brain–gut signaling.8 Indeed, CNS alters the intestinal microenvironment by regulating gut motility and secretion, as well as mucosal immune responses via the enteric nervous system and the neuronal-glial-epithelial unit.9–11 Bacteria react to catecholamines or host hormones and produce neurotransmitters and neuromodulators or their precursors.12–14 Moreover, gut microorganisms influence the enteric nervous system and might stimulate afferent signaling from the gut to the brain via the vagus nerve.15 In a recent study, gut microbiota was identified as a regulator of host serotonin production.16 If not tightly controlled by the host, microbiota and bacterial components (e.g. LPS) threaten to harm or even kill the host. Not surprisingly then, there is a very close interaction of gut microbiota and the host immune system, with important consequences for systemic immunity.17–19 Consequently, mice raised and kept germ free (GF) appear not to develop a normal immune system,20 which needs to be kept in mind when working with such mice in animal models of disease (see below).
Studies in GF mice have shown that gut microbiota is important for the development and proper function of the host CNS. This in turn has important consequences for modeling brain diseases in GF mice (see below). For example, GF animals have a leaky blood brain barrier (BBB),21 as well as an impaired microglia structure and function. The latter findings were replicated in animals after depletion of microbiota with antibiotics.22 Further, gut microbiota possibly influences brain chemistry and behavior, culminating in the claim that psychological phenotypes (e.g. anxiety) could be transferred between individuals via fecal transplantion.23–28
Animal and human studies have suggested involvement of microbiota in the pathogenesis of autism,29,30 experimental autoimmune encephalomyelitis,31,32 neuromyelitis optica,33 Guillain-Barré syndrome,34 and Parkinson.35 Neuropsychiatric diseases related with alterations in the brain–gut axis include depression,34,36 anxiety,36,37 and pain disorders.38,39 In the context of stroke, it is also important to mention that gut microbiota is linked to metabolic profiles of symptomatic atherosclerosis.40–44
Microbiota and acute stroke
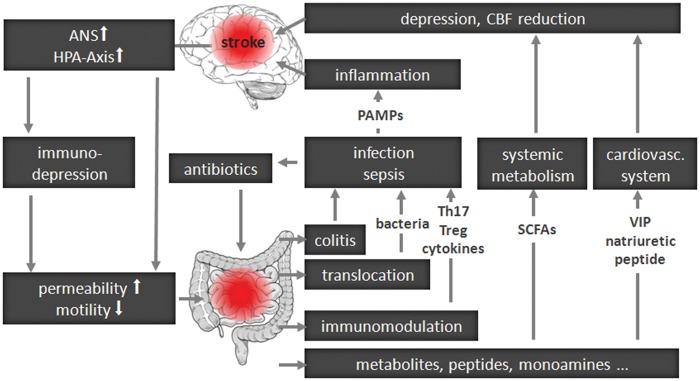
In Figure 1, we summarize a speculative framework for brain–gut communication after acute brain lesion, such as a stroke. We synthesize in it recent insights into the interplay between a brain lesion and the immune and autonomic nervous system (ANS), as well as accumulating knowledge about the interaction of the gut associated lymphatic system (GALT) with the gut microbiota. Briefly, via the ANS and/or the hypothalamus-pituitary-adrenal glands axis (HPA) a brain lesion can affect intestinal function, including immunity. In experimental stroke, T and B cell numbers are significantly reduced in Peyer's patches already 24 h after stroke onset.45 Increased intestinal permeability may promote bacterial translocation to extraintestinal organs, blood stream, or lymphatic fluid, engaging local and systemic immunity and potentially leading to organ infection (e.g. pneumonia) or even sepsis.46
Figure 1.
General concept of brain–gut microbiota interactions after central nervous system lesion.
ANS: autonomic nervous system; HPA axis: hypothalamus-pituitary-adrenal glands axis; CBF: cerebral blood flow; PAMPs: pathogen associated molecular patterns; SCFAs: short chain fatty acids; VIP: vasoactive intestinal peptide.
Brain function can be modulated by metabolic signaling via short chain fatty acids (SCFAs), products of bacterial fermentation in the intestine, or intestinal hormones such as ghrelin or leptin reaching the CNS. Further, an active interplay of commensal bacteria and GALT can modulate systemic immunity, impacting the outcome after brain lesion. Injury to the BBB following stroke allows brain-infiltration of immune cells, reactive to CNS antigens. Concurrent with changes in BBB integrity, microbiota may bias the immune system by inducing, instructing, and shifting immune cell populations (such as regulatory T-cells or IL-17 producing Th17 cells).20,47,48 As post-stroke infections are common complications,49 stroke patients are often treated with a potent combination of antibiotics,50 which might lead to a dramatic alteration in the composition of gut microbiota,51 adding a further level of complexity.
Following in the footsteps of researchers in other fields, many groups worldwide have now started to study the impact of gut microbiota on stroke outcome in experimental models, and to characterize microbiota composition changes after stroke in humans. Surprisingly, the existing knowledge, based on brain–gut interaction after stroke, is rather limited. Tascillar et al. reported bacterial translocation to extraintestinal structures in a mouse middle cerebral artery occlusion model (MCAo).52 Caso et al. found that in rats, stress before MCAo fostered bacterial translocation to diverse organs.53 Swidsinski et al. observed transient colonic inflammation and abrupt disappearance of several bacterial groups after stroke in humans and animals after acute brain injury.54
In summary, it is plausible that gut microbiota plays a role in shaping the outcome after acute cerebral ischemia. However, as we anticipate a certain “hype” regarding the role of gut-brain interactions after stroke, we would like to draw attention to some of the issues that have led to an overselling of the microbiome or microbiomania.55
Potential pitfalls in microbiota studies
Our understanding of stroke pathophysiology has greatly benefited from the availability of rodent models of focal cerebral ischemia, in which the middle cerebral artery is occluded, either transiently or permanently. Experimental gut microbiome research, on the other hand, is based to a large extent on rodent models of primary abiosis or depletion of microbiota with antibiotics, as well as on the transfer of microbiota among animals. Stroke microbiome research is now witnessing a fusion of those models, often combined with bone marrow chimerism and/or adoptive transfer of immune cells. This experimental toolkit, together with sequencing and bioinformatic analysis of whole metagenomes, provides powerful approaches to study the functional role of the gut and its microbiota in acute stroke. However, a number of issues preclude a straightforward interpretation of the results of such modeling. Researchers in a number of fields, including gastroenterology, obesity research, and diabetology have been working with GF mice, metagenomics, and fecal transplants for more than a decade now. Brain research can benefit from their experience and potentially avoid some of the over-interpretations and distortions which threaten to discredit microbiome science.3 Box 1 summarizes important caveats, which we explore briefly below.
Box 1.
Key issues in investigating the role of gut microbiome for stroke outcome.
| • Differences in the anatomy of the rodent and human gastrointestinal tracts56 |
| • Different composition of human and rodent microbiota in health and disease56,57 |
| • Differences in immunology between rodents and humans48,58,59 |
| • Developmental disturbances in primary abiotic (germ free) animals21–24,26–28,60–62 |
| • Effect of antibiotics used to deplete microbiota on nervous and immune system63–68 |
| • Influencing effect of gut virome69–74 |
| • Effect of housing conditions and animal husbandry75–78 |
| • Impact of genetic heterogeneity between humans79 |
| • Impact of (human) diets and their changes under stroke care80–82 |
| • Impact of (human) comorbidities and their treatments83–86 |
| • Sample preparation and bioinformatics of gut metagenomics87–93 |
| • Correlation vs. causation94–96 |
Mouse vs. man
Despite similarities, there are several important differences between rodents and humans in gastrointestinal anatomy and function, many of them driven by differences in diet. For example, the caecum of mice plays a very important role in fermentation of digested food, whereas in humans, it has no known function. Goblet and Paneth cells are distributed differently in rodent and man, and rodents do not have haustra in the colon.56 These differences are not substantial, but may have important implications for the region of the gut from which feces are sampled.
Since rodents are coprophagic and in the laboratory live on a specialized diet, it is not surprising that microbiota composition differs substantially from that of humans. Coprophagy and cohousing may lead to a homogenization of microbiota between individual animals.56 Housing conditions,75 familial transmission,76 caging,77 vendor, as well as genotype77,78 strongly influence the composition of intestinal microbiota in rodents. Although both in mice and man the most abundant phyla are Firmicutes and Bacteroidetes, at the genus level more than 80% of the bacterial sequences found in mice are not found in humans.57
The intestinal immune system maintains the balance between combating pathogens and tolerance for commensal microbiota and food antigens. Dendritic cells (DC) are centrally involved in the generation of gut homing and differentiation of T cells, and thereby in determination of immunogenic or tolerogenic T-cell responses.58,59 Available data suggest similar properties for murine and human intestinal immunity. However, information from humans is scarce, and marked differences between the human and the mouse intestinal immune system have already been described. For example, γδ T cells are found significantly less frequently in the intraepithelial compartment of humans than in that of mice.48,58,59 Moreover, mouse, but not human B cells express TLR4. Therefore, mice, but not humans demonstrate polyclonal B cell responses for antibacterial defense, independently of T-cells.48 This might have relevance for the secretion of immunoglobulin A (IgA), which is known to protect the gut epithelium from luminal antigens and contributes to host-microbe symbiosis.97
Sampling, DNA extraction, and metagenomics
The spatial organization of microbiota along the gut and across the gut lumen complicates microbiome analysis. All this makes sampling location and technique (e.g. aerobic vs. anaerobic; feces vs. biopsy) critical contributors to the results. In addition to the variance introduced by sampling, discrepancies between studies resulting from different DNA extraction methods have to be considered.87,88 The standardization in microbiome research, as proposed for animals89 and for humans,90 is urgently needed.
Largely advances in metagenomics drove the recent surge in interest and knowledge on the role of commensal microorganisms in health and disease. Bacterial 16S rRNA sequencing or, more recently, shotgun sequencing techniques have enabled the quantification of known and unknown microorganisms at species-level resolution and revealed a tremendous microbial diversity that was not captured previously by cultivation-based methods. Many studies find metagenomic differences between healthy or diseased individuals (including experimental animals), or cohorts at risk. However, interpretation of switches in the taxonomic and functional composition of the microbiome is often limited by low sample numbers and missing replications, missing normalization to genome size, as well as heterogeneous sample processing, variability in amplification efficiency and bias by copy number variation, among others.91–93
Additionally, the gut contains not only bacteria but also viruses such as plant-derived viruses, giant viruses, and bacteriophages.69 Bacteriophages have a high host specificity and impact microbial activity. The human gut phageome is estimated at 1015 bacteriophages. The role of the gut virome for health and disease remains opaque.70 Bacteriophages are considered to shape the bacterial community in the gut71 and to modify host immune responses, e.g. by changing bacterial PAMPs and supporting the mucosal barrier of the host.71,72 Thus, phage-viral-bacterial host dynamics in the gut also need to be considered in human health and diseases.73 Very recent data suggest that even the gut virome is altered in patients suffering from ischemic bowel disease, showing disease-specific patterns in ulcerative colitis as compared with Crohn's disease.74 The role of the gut virome in human stroke or animal models has not been investigated so far.
Rodent models to study functional role of microbiota
Mouse models using animals raised in germ-free isolators – “GF mice” – are the workhorse of experimental microbiome research. When interpreting data from GF animals, however, one has to consider that they may deviate from normal physiology in several important ways.60,61
GF mice have underdeveloped immune structures (Peyer’s patches, mesenteric lymph nodes, and splenic white pulp) and differ from conventionally colonized mice in the abundance of several immune cell populations, such as proinflammatory invariant NK cells (iNK),20,62 IgA-producing plasma cells, and lamina propria CD4+ cells. Serum of GF mice contains fewer immunoglobulins, in particular IgG.60
The morphology and physiology of the gastrointestinal tract is altered in GF animals. They have an enlarged caecum, a reduced overall intestinal surface area, longer and thinner intestinal villi and an increased gastrointestinal transit time.62 Interestingly, absence of gut microbiota influences the development not only of the gastrointestinal system but also of other organs. GF mice have higher bone mineral density than conventionally colonized mice and a different metabolic status.62 As discussed above, recent data show that the development and function of the CNS of GF animals is also affected by the absence of microbiota. Besides a “leaky” BBB21 and altered microglia morphology and function,22 they differ from conventionally colonized mice in levels of neurotransmitters, synaptogenesis,26 and behavioral phenotypes.23,24,26–28
To circumvent the developmental deficiencies that result from raising a rodent in an environment lacking viral, bacterial, or parasitic agents, severe reductions of microbial diversity and abundance can be induced by combination therapies with antibiotics.63–65,98,99 However, antibiotics may have modulatory effects on the immune system66,67 and protective as well as detrimental effects on the CNS.67,68 In addition, antibiotic treatment starting in early adolescence may lead to disorders found in GF animals, such as behavioral changes (reduced anxiety and memory deficits) or altered levels of neuromodulators.65
Other models include selective colonization with specific strains or colonization with human microbiota. Selective colonization, however, still remains an artificial system, not taking into account the effects of complex community interactions within the microbiome. In the case of “humanized” mice, it is still not possible to fully mimic the host-microbiota interplay due to differences in the composition of normal mouse and human microbiota and differences in the immune system.89
Human studies
The ultimate goals in microbiota research are studies in humans linking disease susceptibility or pathology to changes in the composition of commensal bacteria. Not surprisingly, human microbiota research also suffers from idiosyncratic limitations. Substantial genetic variation between individuals, quite diverse living conditions and nutritional habits result in major heterogeneity of microbiota composition. Further, microbiota is affected by co-morbidities and their treatments, and depends on the age of the host.100 Different drugs taken, and in particular administration of antibiotics affect human bacterial communities.83–86 For example, Dethlefsen et al. reported restoration of the microbiome composition only 4 weeks after the end of ciprofloxacin treatment. However, shifts in several taxa were detected even 6 months after antibiotic therapy.51 The individual adult human microbiome seems to be relatively stable,101 but nonetheless there are studies reporting high variability over time.102 An important consideration, in particular with respect to studies in stroke patients, is our own experience that patients who are existentially threatened by a CNS disease may object to microbial sampling in observational studies (GUTSTROKE study, NCT02008604). In addition, no information on pre-stroke microbiota composition is available; the majority of patients experience a change of nutrition and receive antibiotics or other drugs in acute stroke care, which impact the immune system and microbial communities. As the effects of stress, life style change, and a potential influence of hospital bacterial strains have to be taken into account, complex combinations of controls are necessary (such as patients with non-CNS emergencies, TIA patients, healthy individuals who have lived with the patient, etc.). Box 1 summarizes critical issues when investigating the role of gut microbiome for stroke outcome.
In conclusion, the recent availability of powerful and inexpensive sequencing methods that allow the characterization of individual metagenomes and the use of GF animals and experimental fecal transplant approaches have led to a surge in insights into the role of gut microbiota in health and disease. In neuroscience, commensal microbiota has been incorporated into the concept of brain–gut signaling. Several groups have provided experimental evidence that intestinal microbiota is involved in the development of neurological diseases. Although as yet, little is known about the role of gut microbiota in stroke, it is highly plausible that microbiota affect outcome after stroke, and a number of groups worldwide are pursuing research into this relevant topic. However, we should learn from other fields of biomedicine, in which microbiota research is more advanced, to avoid overconfidence and overenthusiasm in interpreting our results. Cognizant of the many pitfalls and limitations of the emerging field of gut microbiome research, we will successfully elucidate the highly complex interactions of the trillions of foreign organisms we harbor in our guts with our immune and nervous systems and drawing from this knowledge we will be able to develop novel therapeutic strategies against CNS disorders.
Acknowledgments
The authors would like to thank Ms. Catharine Aubel for proofreading the manuscript.
Funding
The authors received financial support from the German Research Foundation (Exc 257) and the Federal Ministry of Education and Research (01 EO 08 01). KW received PhD-scholarship from the International Max Planck Research School for Infectious Diseases and Immunology (IMPRS-IDI) and the Sonnenfeld-Stiftung.
Declaration of conflicting interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Ridaura VK, Faith JJ, Rey FE, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 2013; 341: 1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu MQ, Cao HL, Wang WQ, et al. Fecal microbiota transplantation broadening its application beyond intestinal disorders. World J Gastroenterol 2015; 21: 102–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hanage WP. Microbiology: Microbiome science needs a healthy dose of scepticism. Nature 2014; 512: 247–248. [DOI] [PubMed] [Google Scholar]
- 4.Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science 2009; 326: 1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lepage P, Leclerc MC, Joossens M, et al. A metagenomic insight into our gut's microbiome. Gut 2013; 62: 146–158. [DOI] [PubMed] [Google Scholar]
- 6.O'Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Rep 2006; 7: 688–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sekirov I, Russell SL, Antunes LC, et al. Gut microbiota in health and disease. Physiol Rev 2010; 90: 859–904. [DOI] [PubMed] [Google Scholar]
- 8.Collins SM, Surette M, Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol 2012; 10: 735–742. [DOI] [PubMed] [Google Scholar]
- 9.Neunlist M, Van Landeghem L, Mahe MM, et al. The digestive neuronal-glial-epithelial unit: A new actor in gut health and disease. Nat Rev Gastroenterol Hepatol 2013; 10: 90–100. [DOI] [PubMed] [Google Scholar]
- 10.Furness JB. The enteric nervous system and neurogastroenterology. Nat Rev Gastroenterol Hepatol 2012; 9: 286–294. [DOI] [PubMed] [Google Scholar]
- 11.Matteoli G, Boeckxstaens GE. The vagal innervation of the gut and immune homeostasis. Gut 2013; 62: 1214–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett E, Ross RP, O'Toole PW, et al. Gamma-aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol 2012; 113: 411–417. [DOI] [PubMed] [Google Scholar]
- 13.Lyte M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol 2004; 12: 14–20. [DOI] [PubMed] [Google Scholar]
- 14.Freestone P. Communication between bacteria and their hosts. Scientifica (Cairo) 2013; 2013: 361073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Forsythe P, Kunze WA. Voices from within: Gut microbes and the cns. Cell Mol Life Sci 2013; 70: 55–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yano JM, Yu K, Donaldson GP, et al. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015; 161: 264–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhee SH, Pothoulakis C, Mayer EA. Principles and clinical implications of the brain-gut-enteric microbiota axis. Nat Rev Gastroenterol Hepatol 2009; 6: 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maynard CL, Elson CO, Hatton RD, et al. Reciprocal interactions of the intestinal microbiota and immune system. Nature 2012; 489: 231–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe 2012; 12: 496–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 2009; 9: 313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braniste V, Al-Asmakh M, Kowal C, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med 2014; 6: 263ra158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Erny D, Hrabe de Angelis AL, Jaitin D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci 2015; 18: 965–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neufeld KM, Kang N, Bienenstock J, et al. Reduced anxiety-like behavior and central neurochemical change in germ-free mice. Neurogastroenterol Motil 2011; 23: 255–264, e119. [DOI] [PubMed] [Google Scholar]
- 24.Bercik P, Denou E, Collins J, et al. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology 2011; 141: 599–609. [DOI] [PubMed] [Google Scholar]
- 25.Bravo JA, Forsythe P, Chew MV, et al. Ingestion of lactobacillus strain regulates emotional behavior and central gaba receptor expression in a mouse via the vagus nerve. Proc Natl Acad Sci USA 2011; 108: 16050–16055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heijtz RD, Wang S, Anuar F, et al. Normal gut microbiota modulates brain development and behavior. Proc Natl Acad Sci USA 2011; 108: 3047–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clarke G, Grenham S, Scully P, et al. The microbiome-gut-brain axis during early life regulates the hippocampal serotonergic system in a sex-dependent manner. Mol Psychiatry 2012; 18: 666-73: 666–673. [DOI] [PubMed] [Google Scholar]
- 28.Sudo N, Chida Y, Aiba Y, et al. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J Physiol 2004; 558: 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsiao EY, McBride SW, Hsien S, et al. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell 2013; 155: 1451–1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang DW, Park JG, Ilhan ZE, et al. Reduced incidence of prevotella and other fermenters in intestinal microflora of autistic children. PLoS One 2013; 8: e68322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee ST, Chu K, Jung KH, et al. Cholinergic anti–inflammatory pathway in intracerebral hemorrhage. Brain Res 2010; 1309: 164–171. [DOI] [PubMed] [Google Scholar]
- 32.Berer K, Mues M, Koutrolos M, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature 2011; 479: 538–541. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun 2014; 38: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochoa-Reparaz J, Mielcarz DW, Begum-Haque S, et al. Gut, bugs, and brain: Role of commensal bacteria in the control of central nervous system disease. Ann Neurol 2011; 69: 240–247. [DOI] [PubMed] [Google Scholar]
- 35.Dobbs SM, Dobbs RJ, Weller C, et al. Peripheral aetiopathogenic drivers and mediators of parkinson's disease and co-morbidities: Role of gastrointestinal microbiota. J Neurovirol 2016; 22: 22–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster JA, McVey Neufeld KA. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci 2013; 36: 305–312. [DOI] [PubMed] [Google Scholar]
- 37.Forsythe P, Sudo N, Dinan T, et al. Mood and gut feelings. Brain Behav Immun 2010; 24: 9–16. [DOI] [PubMed] [Google Scholar]
- 38.Mayer EA, Tillisch K, Gupta A. Gut/brain axis and the microbiota. J Clin Invest 2015; 125: 926–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer EA, Knight R, Mazmanian SK, et al. Gut microbes and the brain: Paradigm shift in neuroscience. J Neurosci 2014; 34: 15490–15496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koren O, Spor A, Felin J, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci USA 2010; 108(Suppl 1): 4592–4598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karlsson FH, Fak F, Nookaew I, et al. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat Commun 2012; 3: 1245. DOI: 10.1038/ncomms2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Z, Klipfell E, Bennett BJ, et al. Gut flora metabolism of phosphatidylcholine promotes cardiovascular disease. Nature 2011; 472: 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang WH, Wang Z, Levison BS, et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N Engl J Med 2013; 368: 1575–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koeth RA, Wang Z, Levison BS, et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 2013; 19: 576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schulte-Herbruggen O, Quarcoo D, Meisel A, et al. Differential affection of intestinal immune cell populations after cerebral ischemia in mice. Neuroimmunomodulation 2009; 16: 213–218. [DOI] [PubMed] [Google Scholar]
- 46.Hagiwara S, Yoshida A, Omata Y, et al. Desulfovibrio desulfuricans bacteremia in a patient hospitalized with acute cerebral infarction: Case report and review. J Infect Chemother 2014; 20: 274–277. [DOI] [PubMed] [Google Scholar]
- 47.Lee YK, Menezes JS, Umesaki Y, et al. Proinflammatory t-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA 2011; 108(Suppl 1): 4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gibbons DL, Spencer J. Mouse and human intestinal immunity: Same ballpark, different players; different rules, same score. Mucosal Immunol 2011; 4: 148–157. [DOI] [PubMed] [Google Scholar]
- 49.Westendorp WF, Nederkoorn PJ, Vermeij JD, et al. Post-stroke infection: A systematic review and meta-analysis. BMC Neurol 2011; 11: 110. DOI: 10.1186/1471-2377-11-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harms H, Hoffmann S, Malzahn U, et al. Decision-making in the diagnosis and treatment of stroke-associated pneumonia. J Neurol Neurosurg Psychiatry 2012; 83: 1225–1230. [DOI] [PubMed] [Google Scholar]
- 51.Dethlefsen L, Huse S, Sogin ML, et al. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16s rRNA sequencing. PLoS Biol 2008; 6: e280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tascilar N, Irkorucu O, Tascilar O, et al. Bacterial translocation in experimental stroke: What happens to the gut barrier? Bratisl Lek Listy 2010; 111: 194–199. [PubMed] [Google Scholar]
- 53.Caso JR, Hurtado O, Pereira MP, et al. Colonic bacterial translocation as a possible factor in stress-worsening experimental stroke outcome. Am J Physiol Regul Integr Comp Physiol 2009; 296: R979–R985. [DOI] [PubMed] [Google Scholar]
- 54.Swidsinski A, Loening-Baucke V, Krüger M, et al. Central nervous system and the colonic bioreactor: Analysis of colonic microbiota in patients with stroke unravels unknown mechanisms of the host defense after brain injury. Intest Res 2012; 10: 332–342. [Google Scholar]
- 55.Eisen J. The tree of life. Microbiomania, http://phylogenomics.blogspot.de/p/blog-page.html (accessed 26 October 2015).
- 56.Nguyen TL, Vieira-Silva S, Liston A, et al. How informative is the mouse for human gut microbiota research? Dis Model Mech 2015; 8: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ley RE, Backhed F, Turnbaugh P, et al. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA 2005; 102: 11070–11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mann ER, Landy JD, Bernardo D, et al. Intestinal dendritic cells: Their role in intestinal inflammation, manipulation by the gut microbiota and differences between mice and men. Immunol Lett 2013; 150: 30–40. [DOI] [PubMed] [Google Scholar]
- 59.Bekiaris V, Persson EK, Agace WW. Intestinal dendritic cells in the regulation of mucosal immunity. Immunol Rev 2014; 260: 86–101. [DOI] [PubMed] [Google Scholar]
- 60.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nat Rev Immunol 2004; 4: 478–485. [DOI] [PubMed] [Google Scholar]
- 61.Gordon HA, Pesti L. The gnotobiotic animal as a tool in the study of host microbial relationships. Bacteriol Rev 1971; 35: 390–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sommer F, Backhed F. The gut microbiota–masters of host development and physiology. Nat Rev Microbiol 2013; 11: 227–238. [DOI] [PubMed] [Google Scholar]
- 63.Membrez M, Blancher F, Jaquet M, et al. Gut microbiota modulation with norfloxacin and ampicillin enhances glucose tolerance in mice. Faseb J 2008; 22: 2416–2426. [DOI] [PubMed] [Google Scholar]
- 64.Reikvam DH, Erofeev A, Sandvik A, et al. Depletion of murine intestinal microbiota: Effects on gut mucosa and epithelial gene expression. PLoS One 2011; 6: e17996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Desbonnet L, Clarke G, Traplin A, et al. Gut microbiota depletion from early adolescence in mice: Implications for brain and behaviour. Brain Behav Immun 2015; 48: 165–173. [DOI] [PubMed] [Google Scholar]
- 66.Tauber SC, Nau R. Immunomodulatory properties of antibiotics. Curr Mol Pharmacol 2008; 1: 68–79. [PubMed] [Google Scholar]
- 67.Pasquale TR, Tan JS. Nonantimicrobial effects of antibacterial agents. Clin Infect Dis 2005; 40: 127–135. [DOI] [PubMed] [Google Scholar]
- 68.Grill MF, Maganti RK. Neurotoxic effects associated with antibiotic use: Management considerations. Br J Clin Pharmacol 2011; 72: 381–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Scarpellini E, Ianiro G, Attili F, et al. The human gut microbiota and virome: Potential therapeutic implications. Dig Liver Dis 2015; 47: 1007–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dalmasso M, Hill C, Ross RP. Exploiting gut bacteriophages for human health. Trends Microbiol 2014; 22: 399–405. [DOI] [PubMed] [Google Scholar]
- 71.De Paepe M, Leclerc M, Tinsley CR, et al. Bacteriophages: An underestimated role in human and animal health? Front Cell Infect Microbiol 2014; 4: 39. DOI: 10.3389/fcimb.2014.00039. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ogilvie LA, Jones BV. The human gut virome: A multifaceted majority. Front Microbiol 2015; 6: 918. DOI: 10.3389/fmicb.2015.00918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reyes A, Wu M, McNulty NP, et al. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci USA 2013; 110: 20236–20241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015; 160: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ma BW, Bokulich NA, Castillo PA, et al. Routine habitat change: A source of unrecognized transient alteration of intestinal microbiota in laboratory mice. PLoS One 2012; 7: e47416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ubeda C, Lipuma L, Gobourne A, et al. Familial transmission rather than defective innate immunity shapes the distinct intestinal microbiota of tlr-deficient mice. J Exp Med 2012; 209: 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hildebrand F, Nguyen TL, Brinkman B, et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol 2013; 14: R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ericsson AC, Davis JW, Spollen W, et al. Effects of vendor and genetic background on the composition of the fecal microbiota of inbred mice. PLoS One 2015; 10: e0116704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blekhman R, Goodrich JK, Huang K, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol 2015; 16: 191. DOI: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Turnbaugh PJ, Ridaura VK, Faith JJ, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med 2009; 1: 6ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.David LA, Maurice CF, Carmody RN, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014; 505: 559–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Carmody RN, Gerber GK, Luevano JM, Jr, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe 2015; 17: 72–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Willing BP, Russell SL, Finlay BB. Shifting the balance: Antibiotic effects on host-microbiota mutualism. Nat Rev Microbiol 2011; 9: 233–243. [DOI] [PubMed] [Google Scholar]
- 84.Wlodarska M, Finlay BB. Host immune response to antibiotic perturbation of the microbiota. Mucosal Immunol 2010; 3: 100–103. [DOI] [PubMed] [Google Scholar]
- 85.Seto CT, Jeraldo P, Orenstein R, et al. Prolonged use of a proton pump inhibitor reduces microbial diversity: Implications for clostridium difficile susceptibility. Microbiome 2014; 2: 42. DOI: 10.1186/2049-2618-2-42. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shin NR, Lee JC, Lee HY, et al. An increase in the akkermansia spp. Population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 2014; 63: 727–735. [DOI] [PubMed] [Google Scholar]
- 87.Kennedy NA, Walker AW, Berry SH, et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16s rrna gene sequencing. PLoS One 2014; 9: e88982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wesolowska-Andersen A, Bahl MI, Carvalho V, et al. Choice of bacterial DNA extraction method from fecal material influences community structure as evaluated by metagenomic analysis. Microbiome 2014; 2: 19. DOI: 10.1186/2049-2618-2-19. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Laukens D, Brinkman BM, Raes J, et al. Heterogeneity of the gut microbiome in mice: Guidelines for optimizing experimental design. FEMS Microbiol Rev 2016; 40: 117–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.International Human Microbiome Standards, http://www.microbiome-standards.org/ (accessed 27 October 2015).
- 91.Sunagawa S, Mende DR, Zeller G, et al. Metagenomic species profiling using universal phylogenetic marker genes. Nat Methods 2013; 10: 1196–1199. [DOI] [PubMed] [Google Scholar]
- 92.Salter SJ, Cox MJ, Turek EM, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol 2014; 12: 87. DOI: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Filippidou S, Junier T, Wunderlin T, et al. Under-detection of endospore-forming firmicutes in metagenomic data. Comput Struct Biotechnol J 2015; 13: 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Harley IT, Karp CL. Obesity and the gut microbiome: Striving for causality. Mol Metab 2012; 1: 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao L. The gut microbiota and obesity: From correlation to causality. Nat Rev Microbiol 2013; 11: 639–647. [DOI] [PubMed] [Google Scholar]
- 96.de Vos WM, de Vos EA. Role of the intestinal microbiome in health and disease: From correlation to causation. Nutr Rev 2012; 70(Suppl 1): S45–S56. [DOI] [PubMed] [Google Scholar]
- 97.Kawamoto S, Tran TH, Maruya M, et al. The inhibitory receptor pd-1 regulates iga selection and bacterial composition in the gut. Science 2012; 336: 485–489. [DOI] [PubMed] [Google Scholar]
- 98.Heimesaat MM, Bereswill S, Fischer A, et al. Gram-negative bacteria aggravate murine small intestinal th1-type immunopathology following oral infection with toxoplasma gondii. J Immunol 2006; 177: 8785–8795. [DOI] [PubMed] [Google Scholar]
- 99.Elinav E, Strowig T, Kau AL, et al. Nlrp6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell 2011; 145: 745–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Claesson MJ, Cusack S, O'Sullivan O, et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc Natl Acad Sci USA 2011; 108(Suppl 1): 4586–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Faith JJ, Guruge JL, Charbonneau M, et al. The long-term stability of the human gut microbiota. Science 2013; 341: 1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome Biol 2011; 12: R50. [DOI] [PMC free article] [PubMed] [Google Scholar]