Abstract
We report the cloning and molecular analysis of Drosophila mitochondrial DNA helicase (d-mtDNA helicase) homologous to human TWINKLE, which encodes one of the genes responsible for autosomal dominant progressive external ophthalmoplegia. An RNA interference construct was designed that reduces expression of d-mtDNA helicase to an undetectable level in Schneider cells. RNA interference knockdown of d-mtDNA helicase decreases the copy number of mitochondrial DNA (mtDNA) ~5-fold. In a corollary manner, overexpression of d-mtDNA helicase increases mtDNA levels 1.4-fold. Overexpression of helicase active site mutants K388A and D483A results in a severe depletion of mtDNA and a dominant negative lethal phenotype. Overexpression of mutants analogous to human autosomal dominant progressive external ophthalmoplegia mutations shows differential effects. Overexpression of I334T and A442P mutants yields a dominant negative effect as for the active site mutants. In contrast, overexpression of A326T, R341Q, and W441C mutants results in increased mtDNA copy number, as observed with wild-type overexpression. Our dominant negative analysis of d-mtDNA helicase in cultured cells provides a tractable model for understanding human autosomal dominant progressive external ophthalmoplegia mutations.
One of the important functions of mitochondria is the production of ATP by the oxidative phosphorylation pathway. Animal mitochondrial DNA (mtDNA)2 encodes 13 polypeptides involved in oxidative phosphorylation, whereas all of the factors essential for mtDNA replication are encoded in the nuclear genome (1). In general, mutations in these factors result in both loss of mtDNA integrity via base substitution mutations, duplications, and deletions and mtDNA depletion (2).
Autosomal dominant progressive external ophthalmoplegia (adPEO) is a human mitochondrial disorder associated with the presence of multiple deletions of mtDNA (2–5). The disease is manifest in adulthood, typically at 20–40 years of age; symptoms include muscle weakness, wasting, exercise intolerance, ataxia, hearing loss, cardiomyopathy, and peripheral neuropathy (2). Most of the adPEO families carry heterozygous mutations in one of three genes: ANT1 (the adenine nucleotide translocator 1), POLG (mitochondrial DNA polymerase), and TWINKLE (mtDNA helicase) (2, 6–8).
TWINKLE protein shares high homology with the bacteriophage T7 gene 4 protein (T7 gp4), which contains both helicase and primase catalytic activities located in its carboxyl- and amino-terminal halves, respectively (7). Like other DNA helicases of the DnaB family, T7 gp4 functions as a hexameric ring (9–11). The amino acid sequence of the helicase domain of T7 gp4 is well conserved in TWINKLE, whereas that of the primase domain is not. TWINKLE exhibits 5′–3′ DNA helicase activity on double-stranded DNA substrates and functions in reconstituted mtDNA replication forks in vitro (12, 13). It co-localizes with mtDNA in structures designated as mitochondrial nucleoids in vivo (7). Recently, TWINKLE was demonstrated to modulate mtDNA copy number in vivo (14), and a transgenic mouse expressing a TWINKLE variant with an in-frame duplication of amino acids 353–365 that is analogous to a human adPEO mutation was shown to harbor multiple mtDNA deletions and to exhibit mitochondrial dysfunction (15). Here, we have studied the effects of overexpression in Drosophila cell culture of d-mtDNA helicase and a number of variants carrying mutations analogous to those in adPEO patients, and characterize their dominant negative effects.
EXPERIMENTAL PROCEDURES
Identification and Sequence Analysis of d-mtDNA Helicase cDNA
The amino acid sequence of TWINKLE was used to search the Berkeley Drosophila Genome Project Data base. One sequence (CG5924) was identified that has a high level of homology to TWINKLE (7). Full-length cDNA was prepared using the SMART RACE cDNA amplification kit (Clontech), total RNA from Drosophila Schneider S2 cells, and the following primers: 5′-TGACTGGCATCGTAGTGCAAC-3′ and 5′-gggctcgagGCTCTTTGAAGCGTTCCAAGG-3′ for 5′-rapid amplification of cDNA ends (RACE) and 5′-ACGCATTGCGTTGTATGCCTGC-3′ and 5′-gggtctgagTACGAACTGAAGACTCTACCAC-3′ for 3′-RACE. RACE products were purified from an agarose gel and sequenced, and sequence analysis was performed using MacDNASIS version 3.7 (Hitachi Software). Similarity searches against the nonredundant GenBankTM data base were performed using BLAST (16). The deduced amino acid sequence was checked for targeting signal peptides using iPSORT (17) and MITPROT (18). Multiple sequence alignments were performed with ClustalW (19).
Bacterial Overexpression of d-mtDNA Helicase and Preparation of d-mtDNA Helicase Antibody
To express d-mtDNA helicase in Escherichia coli, a PCR fragment of d-mtDNA helicase cDNA encoding amino acid residues Glu252 to Leu586 was cloned into pET-28a (Novagen) cleaved with BamHI and EcoRI. Bacterial cells harboring the plasmid were grown in LB medium containing 100 μg/ml kanamycin, and expression was induced with 1 mM isopropylthiogalactoside. The N-terminal hexahistidine tag was utilized for Ni2+-nitrilotriacetic acid affinity purification of recombinant d-mtDNA helicase according to the manufacturer’s instructions (Qiagen). Rabbits were immunized by injection of column-purified d-mtDNA helicase in Freund’s complete adjuvant, followed by secondary immunization in Freund’s incomplete adjuvant. To prepare affinity-purified antiserum, purified d-mtDNA helicase (100 μg) was bound to a polyvinylidene difluoride membrane, and the filter was preincubated for 1 h with 5% skim milk in phosphate-buffered saline (PBS), incubated for 1 h with crude antiserum (1 ml in 5 ml of PBS containing 0.1% Tween 20), washed four times with PBS containing 0.1% Tween 20, and eluted with 0.2 M glycine-HCl, pH 2.4 (~5 ml). The eluted antibody was neutralized with 1 M Tris base.
Glycerol Gradient Sedimentation of d-mtDNA Helicase
An N-terminally hexahistidine-tagged d-mtDNA helicase as above was constructed for expression in the baculovirus system and was purified by Ni2+-nitrilotriacetic acid chromatography according to the manufacturer’s instructions (Qiagen). The protein was layered onto preformed 12–30% glycerol gradients containing 35 mM Tris-HCl, pH 7.5, 330 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium metabisulfite, 2 μg/ml leupeptin, and 10 mM 2-mercaptoethanol. Centrifugation was at 37,000 rpm for 30 h at 4 °C in a Beckman SW 41 rotor. Fractions were analyzed by SDS-PAGE and immunoblotting.
Immunoblotting
Total cellular protein (20 μg/lane) was fractionated by 10.5% SDS-PAGE and transferred to nitrocellulose filters. Filters were preincubated for 1 h with 5% skim milk in PBS, followed by incubation for 1 h with d-mtDNA helicase antibody (1:20 ml in PBS containing 0.1% Tween 20). Filters were washed four times with PBS containing 0.1% Tween 20, incubated for 1 h with horseradish peroxidase-conjugated anti-rabbit IgG (Bio-Rad), and washed with PBS containing 0.1% Tween 20. Protein bands were visualized using ECL Western blotting reagents (Amersham Biosciences). Rabbit polyclonal antibody against Drosophila ATPase β subunit was provided by Rafael Garesse (Consejo Superior de Investigaciones Científicas-Universidad Autónoma de Madrid, Madrid, Spain).
Preparation of an Inducible Plasmid Expressing d-mtDNA Helicase-targeted RNAi
The insert in pMt/invHeli/Hy was generated from two PCR-amplified fragments of d-mtDNA helicase cDNA. One fragment has terminal XhoI and EcoRI sites and was prepared using the following pair of primers: 5′-gcgcctcgagagatcttgGACACACGTGCTTGCAGCTGGAATG-3′ (forward) and 5′-gcgcgaattcgggatcgatCGTAACTTAAAGTGGAGCAGCTG-3′(reverse). A second fragment has terminal BglII and EcoRI sites and was prepared using the primers 5′-gcgcctcgagagatcttgGACACACGTGCTTGCAGCTGGAATG-3′ (forward) and 5′-gcgcgaattcaaaaagcttCGTAACTTAAAGTGGAGCAGCTG-3′ (reverse). The two PCR products were ligated and cloned into the pMt/Hy vector cleaved with XhoI and BamHI.
Generation and Induction of Stable Cell Lines
Drosophila Schneider S2 cells were cultured at 25 °C in Drosophila Schneider medium (Invitrogen) supplemented with 10% fetal bovine serum. Cells were subcultured to 5 × 106 cells/ml every third day. Cells were transfected using Effecten (Qiagen). Hygromycin-resistant cells were selected with 200 μg/ml hygromycin. Cells were passaged at least five times in hygromycin-containing medium and then cultured in standard medium. The cell lines were grown to a density of 3 × 106/ml and then treated with 0.2 or 0.4 mM CuSO4 as indicated to induce high level expression from the metallothionein promoter (20). Overexpression of d-mtDNA helicase was effected by growth of the cell culture in the presence of 0.2 or 0.4 mM CuSO4 as indicated.
Subcellular Fractionation and Preparation of Mitochondrial Extracts for Velocity Sedimentation
Subcellular fractionation was performed, and mitochondrial extracts were prepared from Schneider cells as follows. Schneider cells (250 ml at ~5 × 109 cells/ml) that had been induced with copper for 7 days were suspended in Buffer A (10 mM Tris-HCl, pH 7.4, 100 mM KCl, 0.5 mM EDTA, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium metabisulfite, 2 μg/ml leupeptin, and 10 mM 2-mercaptoethanol) and incubated for 15 min at 4°C. Homogenization was carried out in a Dounce homogenizer, and the homogenate was centrifuged twice at 1000 × g for 10 min. The supernatant was then centrifuged at 7500 × g for 10 min, and after removal of the cytosolic fraction, the mitochondrial pellet was washed once with Buffer A and centrifuged at 7500 ×g for 10 min and then resuspended in 25 mM HEPES, pH 8.0, 0.3 M NaCl, 10% glycerol, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, 10 mM sodium metabisulfite, 2 μg/ml leupeptin, and 10 mM 2-mercaptoethanol. Sodium cholate was added to a final concentration of 2%, and the suspension was incubated on ice for 30 min with mixing by inversion at 5-min intervals. The resulting extract was centrifuged at 140,000 × g for 30 min at 4 °C. The supernatant fluid was recovered as the mitochondrial extract. To analyze the oligomeric state of the wild type and each of the mutant forms of d-mtDNA helicase, 200–500 μg of mitochondrial protein was sedimented in glycerol gradients as described above. The wild type and all of the mutant mtDNA helicases used in this study were found to have sedimentation properties similar to the recombinant form and thus to exist as hexamers. Immunoblot analyses to examine subcellular localization were performed with each of the d-mtDNA helicase-overexpressing cell lines and confirmed that >90% of the overexpressed protein in whole cell extracts was localized in the mitochondrial fraction. A similar value was obtained upon subcellular fractionation of control cells that contained only the endogenous d-mtDNA helicase.
Northern and Southern Blotting
Total cellular RNA was extracted using TRIzol reagent (Invitrogen). RNA (10 μg/lane) was fractionated in a 1.2% agarose/formaldehyde gel, blotted onto Hybond-N+ nylon membrane (Amersham Biosciences), and hybridized to 32P-labeled probes for each of the following four genes: RP49 (ribosomal protein 49), Cytb (cytochrome b), ND4 (NADH dehydrogenase subunit 4), or 12 S rRNA. Hybridization was carried out for 16 h at 42 °C in 5× SSPE (150 mM NaCl, 10 mM sodium phosphate, pH 7.4, 1 mM EDTA), 0.5% SDS, 5× Denhardt’s solution, and 50% formamide. The membrane was washed twice at room temperature with 2× SSC containing 0.1% SDS and twice with 0.1× SSC containing 0.1% SDS for 30 min at 65 °C and then analyzed with a Phosphor-Imager (Amersham Biosciences). The signal for RP49 was used to normalize mitochondrial transcripts. Genomic DNA was purified from Drosophila Schneider S2 cells by standard methods. DNA (10 μg/lane) was cleaved with XhoI, fractionated in 0.8% agarose gel/TBE, and transferred to nylon membrane. Hybridization was performed as above. Filters were washed three times for 10 min at room temperature with 2× SSC containing 0.1% SDS, once for 30 min at 65 °C with 0.2× SSC containing 0.1% SDS, and then analyzed with a PhosphorImager. Blots were probed with radiolabeled DNAs for the mitochondrial gene Cytb and the nuclear histone gene cluster. The ratio of the signals for these two genes was used to determine the relative copy number of mtDNA.
The Northern and Southern blot experiments shown in Figs. 1, 3, 4, and 6 were performed twice with each of the two independent cell lines carrying each plasmid construct, including the control (no plasmid), vector only, wild type, and each of the mutant d-mtDNA helicases. The data presented represent one such experiment, and quantitation is provided for the duplicate experiments from one of the two cell lines. All of the comparable data for each construct vary by <15%.
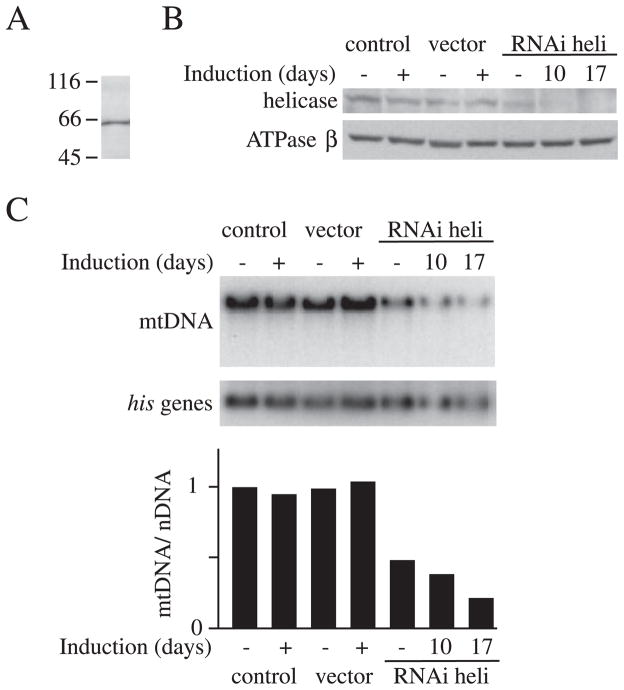
FIGURE 1. Expression of d-mtDNA helicase-targeted RNAi in Schneider cells.
A, immunoblot analysis of d-mtDNA helicase. Schneider cell extract (20 μg) was probed with affinity-purified rabbit antiserum against d-mtDNA helicase. B, Schneider cells with no plasmid (control) or carrying pMt/Hy (vector) or pMt/ invHeli/Hy (RNAi heli) were cultured for 10 days (RNAi heli) or 17 days (control, vector, and RNAi heli) in the presence or absence of 0.4 mM CuSO4. Protein extracts (20 μg) were fractionated by 10.5% SDS-PAGE, transferred to nitrocellulose filters, and probed with affinity-purified rabbit antiserum against d-mtDNA helicase or antiserum against d-ATPase β as indicated. C, effect of RNAi on the copy number of mtDNA. Upper panel, total DNA (10 μg) was extracted from Schneider cells (control) or Schneider cells carrying pMt/Hy (vector) or pMt/invHeli/Hy (RNAi heli) after 10 days or 17 days of culture in the presence or absence of 0.4 mM CuSO4. DNA was digested with XhoI, fractionated in a 0.7% agarose/TBE gel, and blotted to nylon membrane. The membrane was hybridized with radiolabeled probe for the histone gene cluster (his genes) and then stripped and rehybridized with radiolabeled probe for Cytb. Lower panel, relative mtDNA copy number relative was quantitated as described under “Experimental Procedures.”
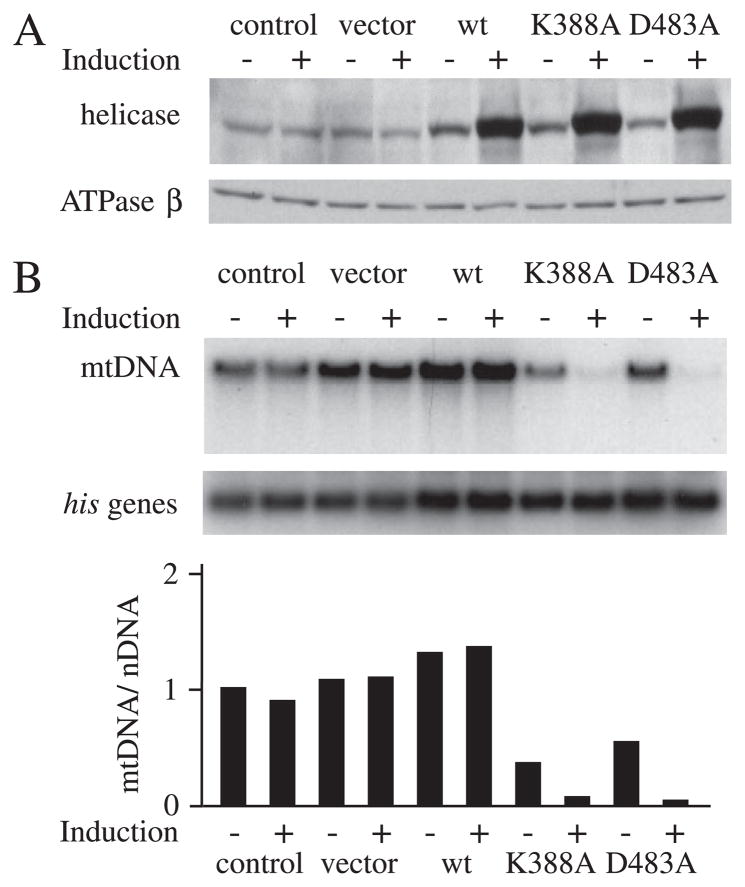
FIGURE 3. Expression of d-mtDNA helicase and active site variants in Schneider cells.
A, Schneider cells with no plasmid (control) or carrying pMt/Hy (vector), pMt/WT/Hy (WT), pMt/K388A/Hy (K388A), or pMt/D483A/Hy (D483A) were cultured for 14 days in the presence or absence of 0.2 mM CuSO4. A, immunoblot analysis of d-mtDNA helicase and d-ATPase β was carried out as described in the legend to Fig. 1. B, Southern blots were probed with radiolabeled mtDNA or control probes, and mtDNA abundance was determined as described in the legend to Fig. 1.
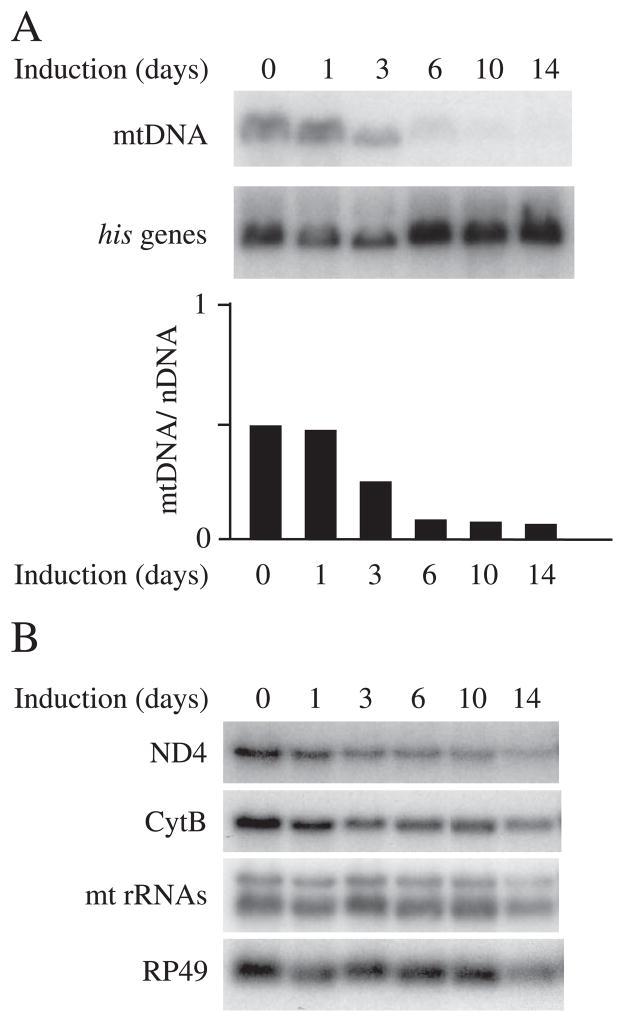
FIGURE 4. Differential changes in mtDNA copy number and mitochondrial transcript levels in cells expressing mutant d-mtDNA helicase.
Total DNA (10 μg) or RNA (10 μg) was extracted from Schneider cells expressing pMt/D483A/Hy at each time point. A, relative mtDNA copy number was estimated by Southern blot. B, RNA was fractionated in a 1.2% agarose/formaldehyde gel, blotted to nylon membrane, and hybridized with radiolabeled probes for the mitochondrial transcripts 12 S rRNA, ND4, and Cytb and the nuclear transcript RP49.
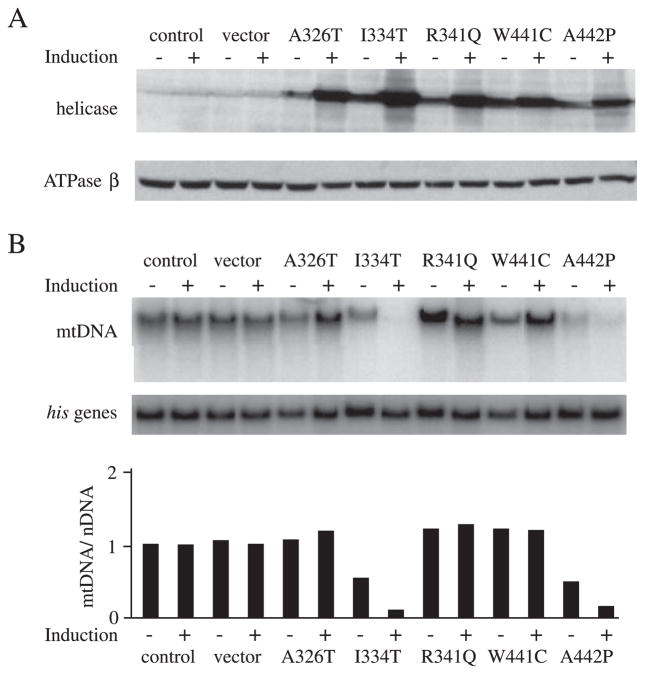
FIGURE 6. Expression of d-mtDNA helicase mutants analogous to human adPEO mutations in Schneider cells.
Schneider cells with no plasmid (control) or carrying pMt/Hy (vector), pMt/A326T/Hy (A326T), pMt/I334T/Hy (I334T), pMt/R341Q/Hy (R341Q), pMt/W441C/Hy (W441C), or pMt/A442P/Hy (A442P) were cultured for 14 days in the presence or absence of 0.2 mM CuSO4. A, immunoblot analysis of d-mtDNA helicase and d-ATPase β was carried out as described in the legend to Fig. 1. B, mtDNA abundance was determined as described in the legend to Fig. 1.
Preparation of an Inducible Plasmid Expressing d-mtDNA Helicase and Its Variants
The plasmid pMt/WT/Hy, in which d-mtDNA helicase cDNA is regulated by the metallothionein promoter, was constructed as follows: a fragment of d-mtDNA helicase cDNA was amplified by PCR using as 5′-primer 5′-gggctcgagAATCTTAAGAAATGGAAAATGAGACGCGCCGGTTTAATC-3′ and as 3′-primer 5′-gcgcactagtTCAGTTCTCGGATGGCGTCTTTTCC-3′. The PCR fragment was cleaved by XhoI and SpeI and subcloned. The expression vectors carrying mutated d-mtDNA helicase were prepared by QuikChange mutagenesis with Pfu DNA polymerase. A typical PCR was carried out in 50 μl of reaction mixture with 50 ng of pMt/WT/Hy and 2 units of Pfu DNA polymerase. A specific primer pair was used for each mutant as follows: A326T, 5′-GCACCGGTGTCGtTTTGGCCAGGATG-3′ and 5′-CATCCTGGCCAAAaCGACACCGGTGC-3′; I334T, 5′-GCACAAGGCCAcCACAACATTCG-3′ and 5′-CGAATGTTGTGgTGGCCTTGTGC-3′; R341Q, 5′-CGGAGCAATGCaAAATGACATTC-3′ and 5′-GAATGTCATTTtGCATTGCTCCG-3′; K388A, 5′-ACGGGTAGTGGAgcGACAACCTTTATGAG-3′ and 5′-CTCATAAAGGTTGTCgcTCCACTACCCGT-3′; W441C, 5′-GAGTTTAACCACTGtGCAGCGGAGTTTGAG-3′ and 5′-CTCAAACTCCGCTGCaCAGTGGTTAAACTC-3′; A442P, 5′-GTTTAACCACTGGcCAGCGGAGTTTG-3′ and 5′-CAAACTCCGCTGgCCAGTGGTTAAAC-3′; D483A, 5′-CACGTGATCATCGcCAATCTGCAATTTATG-3′ and 5′-CATAAATTGCAGATTGgCGATGATCACGTG-3′.
PCR Analysis
PCRs were performed to detect deletions in mtDNA in Schneider cells. Reactions contained 100 ng of total DNA, 50 mM KCl, 10 mM Tris-HCl, pH 7.9, 1.5 mM MgCl, 30 pmol of 5′-primer, 30 pmol of 3′ primer, and 2 units of TaqDNA polymerase (Invitrogen). PCR amplification (94 °C for 30 s, 57 °C for 30 s, 72 °C for 4 min (40 cycles), with an initial step of 94 °C for 1 min and a final step of 72 °C for 4 min) was performed to amplify mtDNA spanning the indicated sequences with the indicated primers as follows: nucleotides 3,014 –5,348, 5′-GGGCTCGAGTAATATGGCAGATTAGTGC-3′ and 5′-CCGATTAATACATGAATTCCGTG-3′; nucleotides 11,315–13,246, 5′-GTAACACCTGCCCATATTCAACCAGAATG-3′ and 5′-AAGGCTGGAATGAATGGTTGGAC-3′; nucleotides 8,493–11,272, 5′-GGAGGAGCTGCTATATTAGCTGATC-3′ and 5′-CAGGTGTTACTAAAGGAGTTGCTGG-3′. The amplified DNA was separated in a 0.7% agarose gel and stained with ethidium bromide.
RESULTS
RNAi-dependent Knockdown of d-mtDNA Helicase Induces mtDNA Depletion in Schneider Cells
The sequence of a putative d-mtDNA helicase was used to identify and clone a full-length cDNA by RACE. The cDNA clone encodes a predicted polypeptide of 613 amino acids. Rabbit antiserum was raised against a recombinant, truncated form of d-mtDNA helicase (Glu252–Leu586). The antibody detects a single polypeptide in mitochondrial extracts from Drosophila Schneider cells with an electrophoretic mobility corresponding to an approximate molecular mass of 65 kDa (Fig. 1A). In particular, no additional species arising from variation of splice site selection were detected. Immunoblot analysis of subcellular fractions of Drosophila Schneider cells demonstrated that d-mtDNA helicase is localized to the mitochondria (data not shown).
The abundance of d-mtDNA helicase was reduced by expressing a metallothionein-inducible d-mtDNA helicase-targeted RNAi species (21–23) from the plasmid pMt/invHeli/Hy. The RNA species produced forms a double-stranded RNA hairpin homologous to d-mtDNA helicase. Our previous studies indicate that double-stranded RNA hairpins are efficient RNAi inhibitors of mitochondrial proteins in Schneider cells (24–26).
Schneider cells stably expressing pMt/invHeli/Hy were cultured for 10 and 17 days in the absence or presence of 0.4 mM CuSO4. Immunoblot analysis showed that basal, uninduced expression of pMt/invHeli/Hy resulted in a 3-fold suppression of d-mtDNA helicase as compared with that in the control cells, most likely due to leaky expression from the metallothionein promoter (Fig. 1B). After 10 days of culture in the presence of copper, cells carrying pMt/invHeli/Hy expressed 8-fold less d-mtDNA helicase than the control cells, and after 17 days, it was undetectable. In contrast, expression of the mitochondrial ATP synthase β subunit, used as a control, was unchanged. Copper-treated cells carrying pMt/invHeli/Hy showed moderate growth retardation, in contrast with the control cells, which showed no adverse growth effects (data not shown).
The effects of reduction of d-mtDNA helicase on mtDNA maintenance were examined in cells expressing d-mtDNA helicase-targeted RNAi. To determine mtDNA copy number, total cellular DNA was isolated from cells carrying no plasmid, pMt/Hy, or pMt/invHeli/Hy, cleaved with XhoI, and analyzed by Southern blot. Blots were hybridized sequentially with probes for the nuclear histone gene cluster and for the mitochondrial gene Cytb (Fig. 1C). Relative mtDNA copy number was estimated from the ratio of Cytb hybridization to histone gene cluster hybridization. The basal level of d-mtDNA helicase-targeted RNAi reduced mtDNA copy number to 48% as compared with that in the control cells. In cells induced with copper for 10 and 17 days, relative mtDNA copy number was reduced further to 38 and 21%, respectively. These results argue that d-mtDNA helicase is essential for mtDNA maintenance.
Overexpression of Active Site Variants of d-mtDNA Helicase Inhibits mtDNA Replication in Schneider Cells
We constructed metallothionein-inducible plasmids expressing wild type d-mtDNA helicase and two variants carrying either a K388A or D483A substitution. These amino acids were selected because they are analogous to Lys318 in the Walker A motif and Asp424 in the Walker B motif in the helicase domain of T7 gp4, respectively (Fig. 2) (27, 28). In T7 gp4, Lys318 and Asp424 have been demonstrated to be essential for helicase activity but not for hexamer formation (27, 28). The constructs were transfected into Schneider cells, and we established two independent stable cell lines for each. After 14 days of incubation in the absence and presence of 0.2 mM CuSO4, immunoblot analysis indicated increases in the range of 2–2.5- and 15–20-fold in the levels of the various d-mtDNA helicases, respectively, relative to the control cell lines, whereas there was no significant change in the level of ATP synthase β subunit (Fig. 3A). At the same time, reverse transcription-PCR analysis showed that the level of transcripts of the endogenous d-mtDNA helicase was unchanged by overexpression of either the wild type or variant d-mtDNA helicases (data not shown). In the absence of copper, we observed no effects on cell growth or viability in any of the cell lines. In contrast, whereas copper treatment had no adverse effects on the growth of cells carrying either no plasmid, empty vector, or pMt/WT/Hy, copper induction of cells carrying pMt/K388A/Hy and pMt/D483A/Hy grew very poorly and produced a lethal phenotype within 4–6 weeks (data not shown).
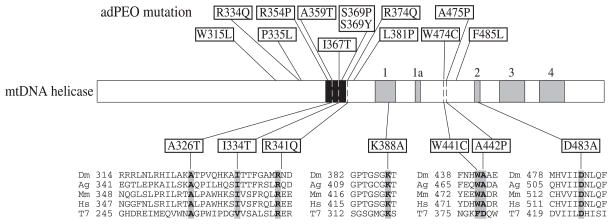
FIGURE 2. Sequence alignment and location of amino acid substitutions in mtDNA helicase.
Schematic diagram of the sequence organization of mtDNA helicase; five amino acid sequence motifs common to ring helicases are indicated in gray, and the bacteriophage T7 gp4 linker-like region is indicated in black. Mutations in the human mtDNA helicase protein (TWINKLE) found in adPEO are shown above the schematic (7, 34–37). The positions of mutations in d-mtDNA helicase used in this study are shown below. Sequence alignment of the regions containing altered amino acids is shown in the lower panel. Dm, fly; Ag, mosquito; Mm, mouse; Hs, man; T7, bacteriophage T7.
Next, we evaluated the effects on mtDNA maintenance in cells overexpressing the wild type d-mtDNA helicase and its variants. Total cellular DNA was isolated from cells grown for 14 days in the presence or absence of copper, cleaved with XhoI, and analyzed by Southern blot (Fig. 3B). Upon 15-fold overexpression of the wild type d-mtDNA helicase, relative mtDNA copy number was somewhat increased over that in control cells. However, upon only 1.5-fold expression of the K388A mutant under uninduced conditions relative to endogenous levels of d-mtDNA helicase, we observed a 2.8-fold decrease in mtDNA copy number. Furthermore, under induced conditions that yielded a 20-fold overexpression of K388A, there was a correspondingly dramatic mtDNA depletion of 14-fold. Similarly, in cells overexpressing the D483A mutant, the relative mtDNA copy number was reduced to 54 and 4% of control cells, correlating with the 2- and 20-fold overexpression of D483A in the absence and presence of induction, respectively. After 4 weeks of induction with copper, Southern blot analysis showed an undetectable level of mtDNA in both of the mutant cell lines (data not shown).
Finally, we investigated the physiological effects on mtDNA transcription concomitant with the development of mtDNA depletion. Fig. 4 shows a time course of mtDNA depletion and mtDNA transcript levels in cells expressing the D483A mutant. Northern blots were probed for transcripts from the mitochondrial ND4, Cytb, and 12 S rRNA genes and for nuclear RP49 as a control. Prior to copper induction, the mtDNA copy number in the D483A-expressing cells was reduced to 50% of that in the control cells (Fig. 4A), without an apparent decrease in the levels of mitochondrial transcripts (Fig. 4B). However, after 6 days of induction with copper, when the relative mtDNA copy number in the D483A-expressing cells was decreased to 10% of that in the control cells, the levels of mitochondrial transcripts in the D483A-expressing cells decreased to 50%. After 14 days of induction, when relative mtDNA copy number in the D483A-expressing cells remained at a level of about 10% of that in the control cells, the levels of mitochondrial transcripts were decreased further to 20% of those in control cells. Thus, mtDNA depletion due to the dominant negative effect of over-expression of active site mutants of d-mtDNA helicase leads to loss of mitochondrial transcription, most likely as an indirect effect of reduction in mtDNA copy number. In contrast, the levels of mitochondrial transcripts were unchanged over the entire time course in cells overexpressing the wild type d-mtDNA helicase (data not shown).
Drosophila mtDNA Helicase Forms a Hexamer
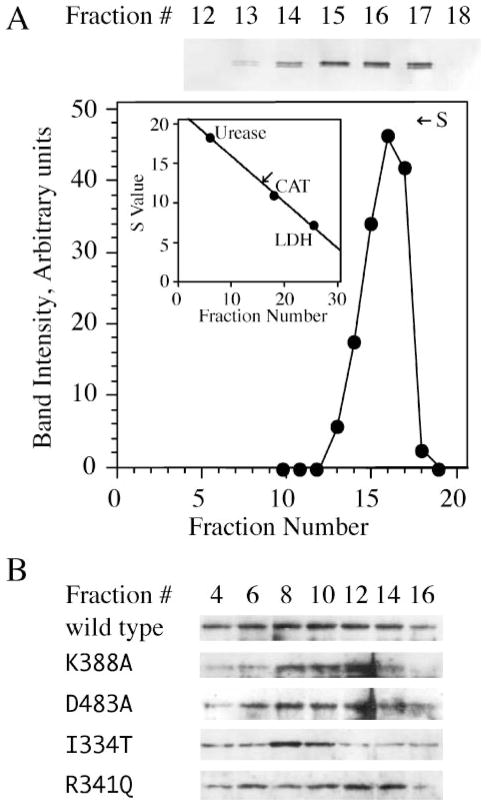
d-mtDNA helicase is highly homologous to the bacteriophage T7 gp4. Crystal structure analysis demonstrates that T7 gp4 assembles into ring-shaped hexamers/heptamers (10, 29). To explain the dominant-negative effects we observed in Schneider cells over-expressing the active site mutants of d-mtDNA helicase, we constructed and purified a recombinant form and examined its native conformation by sedimentation analysis as described under “Experimental Procedures.” We determined the sedimentation coefficient of the wild type d-mtDNA helicase to be 12.4, consistent with the assembly of a hexamer of 70-kDa protomers (Fig. 5A).
FIGURE 5. Drosophila mtDNA helicase forms a hexamer.
Purified, recombinant d-mtDNA helicase (A) and mitochondrial extracts from Schneider cells overexpressing d-mtDNA helicases as indicated (B) were sedimented in 12–30% glycerol gradients as described under “Experimental Procedures.” Protein markers run in parallel gradients were as follows: jack bean urease, 18.6 S; bovine liver catalase (CAT), 11.3 S; and rabbit muscle L-lactate dehydrogenase (LDH), 7.3 S. Fractions were analyzed by SDS-PAGE and immunoblotting. The data in A were quantitated and plotted to obtain a sedimentation coefficient. B, representative data for the mitochondrial forms of wild type and mutant mtDNA helicases from Schneider cells; all of the mutant mtDNA helicases used in this study were found to have sedimentation properties similar to the recombinant form.
Our active site mutations K388A and D483A are analogous to positions K318 and D424 in the helicase domain of T7 gp4, respectively, and the latter are not essential for hexamer formation (28). We analyzed the sedimentation properties of each of the mutant and wild-type mtDNA helicases used in this study as described above after overexpression in Schneider cells and preparation of mitochondrial extracts, and found them to have sedimentation coefficients similar to the purified recombinant form; representative data are shown in Fig. 5B. Thus, we conclude that the mtDNA depletion and lethal cellular phenotype in Drosophila Schneider cells expressing K388A and D483A are due to the formation of mixed oligomers carrying helicase-deficient protomers.
Overexpression of d-mtDNA Helicase Variants Analogous to Mutant Human mtDNA Helicases in adPEO Patients Induces Differential Phenotypes in Schneider Cells
Genomic analysis has documented a number of mutations associated with adPEO in the human mtDNA helicase, TWINKLE (7). Ten of the 12 mutated amino acids are conserved between flies and humans, and the remaining two represent conservative substitutions in the fly protein (Fig. 2). To compare the effects of the human adPEO mutations with those we observed with active site mutants in Schneider cells, we constructed metallothionein-inducible plasmids expressing five d-mtDNA helicase variants carrying A326T, I334T, R341Q, W441C, and A442P substitutions, which are analogous to the A359T, I367T, R374Q, W474C, and A475P mutations found in adPEO patients, respectively, and map within the linker region and helicase domain (Fig. 2). The constructs were transfected into Schneider cells, and we established two independent stable cell lines for each. After 14 days of incubation in the absence and presence of 0.2 mM CuSO4, immunoblot analysis indicated increases in the range of 2.5–3.5- and 15–25-fold in the levels of the various d-mtDNA helicases, respectively, relative to the control cell lines, whereas there was no significant change in the level of ATP synthase β subunit (Fig. 6A).
The effect of overexpression of the d-mtDNA helicase variants on mtDNA maintenance was evaluated by Southern blot analysis as described above. After 14 days of induction, we found a 7–11-fold decrease in relative mtDNA copy number in cells overexpressing I334T and A442P as compared with the control cells (Fig. 6B). After 4 weeks of induction, mtDNA decreased to an undetectable level in both mutant cell lines (data not shown). These data are comparable with data, which we obtained upon overexpression of the active site mutants. In contrast, in cells overexpressing A326T, R341Q, and W441C, the relative mtDNA copy number was increased to 118 –127% of the control upon induction; these values are comparable with that which we observed upon overexpression of the wild type d-mtDNA helicase. Correspondingly, whereas copper treatment had no adverse effects on the growth of cells expressing A326T, R341Q, and W441C, copper-induced cells expressing I334T and A442P grew very poorly and produced a lethal phenotype within 4 –7 weeks (data not shown). Furthermore, we found that in cell lines expressing I334T and A442P, the levels of mitochondrial transcripts were reduced to 30% of that in the control cells after induction (data not shown).
Multiple mtDNA deletions represent the molecular hallmark of adPEO mutations. To detect such multiple deletions, we probed Southern blots of total genomic DNA from all of the cell lines (control, vector only, wild type, and seven mutant d-mtDNA helicases) with the mitochondrial genes encoding Cytb, ND4, 12 S rRNA, and COXI, but we failed to detect any deletions around the Drosophila mtDNA genome (data not shown). Furthermore, we used a sensitive PCR-based approach to detect multiple deletions, amplifying 40 cycles of mtDNA spanning nucleotide positions 3,014–5,348, 8,493–11,272, and 11,315–13,246 from total genomic DNA of all of the above cell lines cultured in the presence and absence of copper induction. Again, we failed to detect any mtDNA deletions in any of the cell lines (data not shown).
DISCUSSION
In Drosophila Schneider cells treated with RNAi, the expression of d-mtDNA helicase is suppressed to an undetectable level, inducing cellular phenotypes of slow growth, reduced viability, and a 5-fold reduction in mtDNA copy number. Overexpression of d-mtDNA helicase increases modestly mtDNA copy number. At the same time, overexpression of the active site mutants K388A and D483A results in a dose-dependent mtDNA depletion. The dominant negative mutants show a more severe effect on mtDNA copy number than is observed by RNAi. Furthermore, overexpression of the mutants I334T and A442P that are analogous to those found in human adPEO patients reduces mtDNA copy number dramatically, but overexpression of the mutants A326T, R341Q, and W441C does not. Unlike the results reported in adPEO patients (3), no mtDNA deletions were found in any of the Drosophila cell lines.
In cloning the Drosophila mtDNA helicase analogous to human TWINKLE, we did not identify any splice variants like human TWINKY by either RACE or immunoblot analyses. Human TWINKLE comprises 684 amino acids, whereas TWINKY is a spliced product of 582 amino acids that lacks residues 579–684 and terminates with four unique amino acids (7). The open reading frame of human TWINKLE consists of five exons, and a 43-bp extension in exon 4 generates TWINKY. In contrast, the open reading frame of d-mtDNA helicase consists of only three exons and lacks introns in the helicase domain. Our data demonstrate that there is not a TWINKY-like splice variant in Schneider cells. Although no data have been reported regarding the physiological significance of TWINKY, our data argue that its functional role, if any, must not be essential in animals.
We found that mtDNA copy number varied with d-mtDNA helicase levels, which is in agreement with data reported for a transgenic mouse model (14). A minor difference was that the increase of mtDNA was 1.4-fold upon overexpression of d-mtDNA helicase in Schneider cells, as compared with 3-fold in the transgenic mice. Regardless, the composite data show clearly that mtDNA helicase is essential for mtDNA maintenance in animals and is modestly limiting for mtDNA replication.
High level overexpression of the d-mtDNA helicase mutants K388A, D483A, I334T, and A442P in Schneider cells causes a lethal phenotype and severe mtDNA depletion. However, low level expression maintains normal growth despite a 2-fold reduction in mtDNA copy number. These expression conditions mimic those in human adPEO patients, in which the gene dosage is 1:1 in terms of wild-type versus mutant DNA. Interestingly, we found that the mitochondrial transcript level is unchanged until the mtDNA content is reduced more than 2-fold. After induction of the mutant D483A, mtDNA copy number was reduced rapidly, but decreases in the levels of mitochondrial transcripts occurred more slowly. Likewise, the increase in mtDNA copy number by overexpression of wild-type d-mtDNA helicase does not increase mitochondrial transcript level. These results indicate that mtDNA is in excess of that needed for normal mitochondrial transcription in Schneider cells. Similar results were obtained upon either suppression or overexpression of the mitochondrial transcription factor A (25, 30, 31). These data contrast with our finding that mitochondrial transcription factor B2 regulates directly both mtDNA copy number and transcription (25), whereas mitochondrial transcription factor B1 affects neither mtDNA nor transcript levels (26).
In adPEO patients, many of the mutations map within a small region, which corresponds to the linker region of bacteriophage T7 gp4. The linker region separates the primase from the helicase domain and is important for hexamer formation (32). Thus, mtDNA helicase mutations that lie in this region (shown in black shading in Fig. 2) might affect subunit interactions. We found that the I334T and A442P mutants display a dominant negative defect as for the helicase active site mutants K388A and D483A. Lys388 and Asp483 are analogous to Lys318 and Asp424 in T7 gp4 and are not essential for T7 gp4 hexamer formation (27, 28). Notably, human TWINKLE variants carrying a duplication of amino acids 352–364 in the linker region form multimers in vivo (7). Here, we demonstrate directly that the wild type and mutant mtDNA helicases form hexamers and that the overexpression of mutants K388A, D483A, I334T, and A442P reduces mtDNA copy number in a dose-dependent manner. Typically, under induced conditions, mtDNA copy number in these mutants is <10% of wild type, and the molar ratio of endogenous to exogenous mtDNA helicase polypeptides is 1:19. If the mutants form hexamers with the same efficiency as the wild-type polypeptides, >75% of the hexamers should comprise only mutant protomers, and <25% should contain one wild type and five mutant protomers. The severe mtDNA depletion that results indicates clearly that one wild-type subunit is insufficient for helicase activity in vivo. In contrast, under uninduced conditions, mtDNA copy number is maintained at ~50% of wild type. Here, the molar ratio of endogenous to exogenous polypeptides is 1:1.5, and only 0.4% of the hexamers should comprise six wild-type protomers; hexamers containing five wild-type and one mutant and containing four wild-type and two mutant protomers comprise only 3.7 and 13.8%, respectively. This argues that these heterohexamers retain substantial helicase activity. In any case, that the dominant negative phenotypes of the active site mutants and the I334T and A442P mutants are highly similar under several conditions suggests that the latter mutants exhibit defects in helicase function per se. These residues lie in predicted α helices that correspond to the A and D3 helices in the linker and helicase domains, respectively, of T7 gp4. In the T7 gp4, the A helix of one protomer participates in the subunit interface with the D2 and D3 helices of the adjacent protomer (10), and amino acid substitutions near Ile334 (Ala359 in T7 gp4) show specific defects in translocation on single-stranded DNA and in helix unwinding (33).
We found that wild-type overexpression increases mtDNA copy number, indicating that d-mtDNA helicase is modestly limiting for DNA replication under physiological conditions. Thus, it seems that the maintenance of ~50% of the mtDNA in cells that overexpress the mutant forms I334T and A442P best fits a model in which only a few wild-type protomers per hexamer are essential for mtDNA replication. Of course, the physiological outcome we observed may be mitigated by other factors. One intriguing possibility to explain our findings might be a cellular response that alters steady-state levels of mtDNA in these mutants by reducing either mtDNA or helicase turnover or both.
In our study, two Drosophila mutants analogous to human adPEO mutations showed dominant-negative phenotypes, whereas three others did not (A326T, R341Q, and W441C). All of these mutations lead to mtDNA deletions in humans. Notably however, transgenic mice expressing the linker region duplication of amino acids 353–365 showed a 2-fold mtDNA depletion in the brain but unchanged levels of mtDNA in muscle and heart (15). Moreover, it is not clear that depletion and deletion of mtDNA are related mechanistically, and in the human adPEO patients examined to date, mtDNA copy number has not been evaluated systematically. Our results would suggest that patients carrying the I334T and A442P mutations may exhibit moderate reduction of mtDNA copy number not only in muscle, heart, and brain but also in other tissues. In the case of transgenic mice expressing a mutation analogous to A326T, a modest phenotype was observed in terms of the evaluation of COX negative muscle fibers by a histochemical analysis as compared with transgenic mice expressing the linker region duplication. A326 is well conserved between mtDNA helicase and bacteriophage T7 gp4, where the analogous residue is Ala257. Although mutants at position Ala257 in bacteriophage T7 gp4 (A257L, A257P, and A257V) show defects in helicase activity in vitro, they fail to show dominant negative effects either in vivo or in vitro in the presence of the wild-type protein (33). These results corroborate our findings with the Drosophila A326T mutant. Interestingly, the A257V and A257L mutants in bacteriophage T7 gp4 form oligomers under conditions in which the wild-type protein does not readily oligomerize, implying that they might preferentially form homo-oligomers, such that a very small population of heterohexamers may exhibit only limited defects in DNA replication in the presence of a nearly normal level of wild-type hexamers. Likewise, the amino acid positions analogous to Drosophila R341Q and W441C are in A helix and D3 helix in bacteriophage T7 gp4, respectively. These helices are involved in subunit interaction, suggesting that the corresponding Drosophila mutants may also exhibit altered efficiencies of oligomerization.
Multiple deletions are regarded to be the molecular hallmark of the adPEO diagnosis. We have cultured each of our Drosophila cell lines carrying the wild type and seven mutant d-mtDNA helicase constructs for over a year in the absence of induction (where the endogenous to exogenous mtDNA helicase ratio is 1:1.0–1.5) but have not detected any mtDNA deletions. One possible explanation is that the efficiency of generating deleted mtDNA might be very low in Schneider cells, just as the proportion of deleted mtDNAs varies widely in different tissues in adPEO patients (3, 5). In general, the accumulation of deleted mtDNA in adPEO patients occurs in tissues that comprise mainly nondividing cell types, such as heart, muscle, and brain, with the first symptoms arising at 20–40 years of age (3, 5). Similarly, in transgenic mice expressing the adPEO duplication, mtDNA deletions were detected by PCR at 18 months of age (15). Thus, it seems likely that accumulation of deleted mtDNAs in actively dividing cultured cells might be very slow. Furthermore, the turnover of deleted mtDNAs may differ. In any case, the mechanism by which a dominant mutation in mtDNA helicase generates multiple deletions is unclear, and more detailed genetic, physiological, and biochemical studies are needed to understand functional differences among these mutants in mtDNA replication. Our dominant negative analysis of d-mtDNA helicase in cultured cells provides a working model for extension of these studies to the whole animal, where we may see both the development of the human disease-like phenotypes, and any generational effects.
Acknowledgments
We thank Carol Farr for performing the glycerol gradient analysis of d-mtDNA helicase and for help with the figures.
Footnotes
This work was supported by National Institutes of Health Grant GM45295.
The abbreviations used are: mtDNA, mitochondrial DNA; d-mtDNA helicase, Drosophila mitochondrial DNA helicase; adPEO, autosomal dominant progressive external ophthalmoplegia; RNAi, RNA interference; T7 gp4, bacteriophage T7 gene 4 protein; RACE, rapid amplification of cDNA ends; PBS, phosphate-buffered saline.
References
- 1.Kaguni LS. Annu Rev Biochem. 2004;73:293–320. doi: 10.1146/annurev.biochem.72.121801.161455. [DOI] [PubMed] [Google Scholar]
- 2.Spinazzola A, Zeviani M. Gene (Amst) 2005;354:162–168. doi: 10.1016/j.gene.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 3.Suomalainen A, Majander A, Haltia M, Somer H, Lonnqvist J, Savontaus ML, Peltonen L. J Clin Invest. 1992;90:61–66. doi: 10.1172/JCI115856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeviani M, Servidei S, Gellera C, Bertini E, DiMauro S, DiDonato S. Nature. 1989;339:309–311. doi: 10.1038/339309a0. [DOI] [PubMed] [Google Scholar]
- 5.Suomalainen A, Majander A, Wallin M, Setala K, Kontula K, Leinonen H, Salmi T, Paetau A, Haltia M, Valanne L, Lonnqvist J, Peltonen L, Somer H. Neurology. 1997;48:1244–1253. doi: 10.1212/wnl.48.5.1244. [DOI] [PubMed] [Google Scholar]
- 6.Van Goethem G, Dermaut B, Lofgren A, Martin JJ, Van Broeckhoven C. Nat Genet. 2001;28:211–212. doi: 10.1038/90034. [DOI] [PubMed] [Google Scholar]
- 7.Spelbrink JN, Li FY, Tiranti V, Nikali K, Yuan QP, Tariq M, Wanrooij S, Garrido N, Comi G, Morandi L, Santoro L, Toscano A, Fabrizi GM, Somer H, Croxen R, Beeson D, Poulton J, Suomalainen A, Jacobs HT, Zeviani M, Larsson C. Nat Genet. 2001;28:223–231. doi: 10.1038/90058. [DOI] [PubMed] [Google Scholar]
- 8.Kaukonen J, Juselius JK, Tiranti V, Kyttala A, Zeviani M, Comi GP, Keranen S, Peltonen L, Suomalainen A. Science. 2000;289:782–785. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- 9.Sawaya MR, Guo S, Tabor S, Richardson CC, Ellenberger T. Cell. 1999;99:167–177. doi: 10.1016/s0092-8674(00)81648-7. [DOI] [PubMed] [Google Scholar]
- 10.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 11.Patel SS, Picha KM. Annu Rev Biochem. 2000;69:651–697. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 12.Korhonen JA, Gaspari M, Falkenberg M. J Biol Chem. 2003;278:48627–48632. doi: 10.1074/jbc.M306981200. [DOI] [PubMed] [Google Scholar]
- 13.Korhonen JA, Pham XH, Pellegrini M, Falkenberg M. EMBO J. 2004;23:2423–2429. doi: 10.1038/sj.emboj.7600257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tyynismaa H, Sembongi H, Bokori-Brown M, Granycome C, Ashley N, Poulton J, Jalanko A, Spelbrink JN, Holt IJ, Suomalainen A. Hum Mol Genet. 2004;13:3219–3227. doi: 10.1093/hmg/ddh342. [DOI] [PubMed] [Google Scholar]
- 15.Tyynismaa H, Mjosund KP, Wanrooij S, Lappalainen I, Ylikallio E, Jalanko A, Spelbrink JN, Paetau A, Suomalainen A. Proc Natl Acad Sci U S A. 2005;102:17687–17692. doi: 10.1073/pnas.0505551102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 17.Horton P, Nakai K. Proc Int Conf Intell Syst Mol Biol. 1997;5:147–152. [PubMed] [Google Scholar]
- 18.Claros MG, Vincens P. Eur J Biochem. 1996;241:779–786. doi: 10.1111/j.1432-1033.1996.00779.x. [DOI] [PubMed] [Google Scholar]
- 19.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bunch TA, Grinblat Y, Goldstein LS. Nucleic Acids Res. 1988;16:1043–1061. doi: 10.1093/nar/16.3.1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammond SM, Bernstein E, Beach D, Hannon GJ. Nature. 2000;404:293–296. doi: 10.1038/35005107. [DOI] [PubMed] [Google Scholar]
- 22.Baulcombe DC. Curr Biol. 1999;9:R599–601. doi: 10.1016/s0960-9822(99)80383-2. [DOI] [PubMed] [Google Scholar]
- 23.Kennerdell JR, Carthew RW. Cell. 1998;95:1017–1026. doi: 10.1016/s0092-8674(00)81725-0. [DOI] [PubMed] [Google Scholar]
- 24.Farr CL, Matsushima Y, Lagina AT, III, Luo N, Kaguni LS. J Biol Chem. 2004;279:17047–17053. doi: 10.1074/jbc.M400283200. [DOI] [PubMed] [Google Scholar]
- 25.Matsushima Y, Garesse R, Kaguni LS. J Biol Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- 26.Matsushima Y, Adan C, Garesse R, Kaguni LS. J Biol Chem. 2005;280:16815–16820. doi: 10.1074/jbc.M500569200. [DOI] [PubMed] [Google Scholar]
- 27.Patel SS, Hingorani MM, Ng WM. Biochemistry. 1994;33:7857–7868. doi: 10.1021/bi00191a013. [DOI] [PubMed] [Google Scholar]
- 28.Washington MT, Rosenberg AH, Griffin K, Studier FW, Patel SS. J Biol Chem. 1996;271:26825–26834. doi: 10.1074/jbc.271.43.26825. [DOI] [PubMed] [Google Scholar]
- 29.Toth EA, Li Y, Sawaya MR, Cheng Y, Ellenberger T. Mol Cell. 2003;12:1113–1123. doi: 10.1016/s1097-2765(03)00442-8. [DOI] [PubMed] [Google Scholar]
- 30.Goto A, Matsushima Y, Kadowaki T, Kitagawa Y. Biochem J. 2001;354:243–248. doi: 10.1042/0264-6021:3540243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsushima Y, Matsumura K, Ishii S, Inagaki H, Suzuki T, Matsuda Y, Beck K, Kitagawa Y. J Biol Chem. 2003;278:31149–31158. doi: 10.1074/jbc.M303842200. [DOI] [PubMed] [Google Scholar]
- 32.Guo S, Tabor S, Richardson CC. J Biol Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 33.Lee SJ, Richardson CC. J Biol Chem. 2004;279:23384–23393. doi: 10.1074/jbc.M400857200. [DOI] [PubMed] [Google Scholar]
- 34.Kiechl S, Horvath R, Luoma P, Kiechl-Kohlendorfer U, Wallacher-Scholz B, Stucka R, Thaler C, Wanschitz J, Suomalainen A, Jaksch M, Willeit J. J Neurol Neurosurg Psychiatry. 2004;75:1125–1128. doi: 10.1136/jnnp.2003.025890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Goethem G, Lofgren A, Dermaut B, Ceuterick C, Martin JJ, Van Broeckhoven C. Hum Mutat. 2003;22:175–176. doi: 10.1002/humu.10246. [DOI] [PubMed] [Google Scholar]
- 36.Arenas J, Briem E, Dahl H, Hutchison W, Lewis S, Martin MA, Spelbrink H, Tiranti V, Jacobs H, Zeviani M. Ann Neurol. 2003;53:278. doi: 10.1002/ana.10430. [DOI] [PubMed] [Google Scholar]
- 37.Lewis S, Hutchison W, Thyagarajan D, Dahl HH. J Neurol Sci. 2002;201:39–44. doi: 10.1016/s0022-510x(02)00190-9. [DOI] [PubMed] [Google Scholar]