Abstract
Human mitochondrial diseases are associated with a wide range of clinical symptoms, and those that result from mutations in mitochondrial DNA affect at least 1 in 8500 individuals. The development of animal models that reproduce the variety of symptoms associated with this group of complex human disorders is a major focus of current research. Drosophila represents an attractive model, in large part because of its short life cycle, the availability of a number of powerful techniques to alter gene structure and regulation, and the presence of orthologs of many human disease genes. We describe here Drosophila models of mitochondrial DNA depletion, deafness, encephalopathy, Freidreich’s ataxia, and diseases due to mitochondrial DNA mutations. We also describe several genetic approaches for gene manipulation in flies, including the recently developed method of targeted mutagenesis by recombinational knock-in.
Keywords: Drosophila, Mitochondria, Mitochondrial diseases, Gene targeting, Oxidative phosphorylation
1. Introduction
The fruit fly Drosophila has played a critical role in the origin and development of modern biology, particularly in the field of genetics. In 1910, T. H. Morgan’s experiments demonstrating the chromosome theory of heredity initiated Drosophila as a tool to investigate the basis of genetics and development [1]. Features that made Drosophila useful as an animal model in the first half of the past century are still attractive today [1,2]. These include (i) a short life cycle, (ii) the facility to feed and maintain stocks and populations without specialized instrumentation or infrastructure, (iii) the availability of non-recombinogenic balancer chromosomes, (iv) physical mapping of genes on polytene chromosomes, (v) the straightforward use of X-rays and other mutagenic agents to generate large collections of mutant stocks and, (vi) the physical accessibility of the mechanosensory apparatus for experimentation. During the remainder of the century, new technologies were developed to manipulate and understand the Drosophila genome, leading to remarkable discoveries such as the genetic control of embryonic development by E. Lewis, C. Nüsslein-Volhard and E. Wieschaus [3]. Advances in molecular biology have allowed the generation of transgenic flies using P-element transposons [4], the development of the yeast UAS-GAL4-based gene overexpression system [5] and more recently, technologies based on site specific recombination to knock-in and knock-out specific genes, and RNA interference (RNAi) to knock-down gene expression (for a review, see [6]). At the same time, Drosophila melanogaster was one of the first organisms to be sequenced in its entirety. The first annotated sequence was published in March 2000, and it has been updated regularly (http://flybase.net/annot/dmel-release4-notes.html). The Drosophila genome contains a large number of human orthologs, demonstrating its potential as a model of human disease [7].
2. Mitochondrial biogenesis in Drosophila
Mitochondrial biogenesis is an essential process in cell proliferation and differentiation. It depends on the co-ordinated expression of two genomes located in different subcellular compartments, the nucleus and the mitochondrion [8]. Mitochondrial DNA (mtDNA) is a circular double-stranded DNA molecule with an extremely compact organization, and a gene content that is well conserved in the animal kingdom. It encodes 22 tRNAs, 2 rRNAs and 13 polypeptides, all of which are components of the oxidative phosphorylation system (OXPHOS). The rest of the subunits of the OXPHOS system (about 90) and all of the proteins involved in the maintenance and expression of mtDNA, and all other mitochondrial functions, are encoded in nuclear DNA [8].
In the last few years, several mammalian transcriptional regulators have been characterized that control the expression of both nuclear and mitochondrial genes, and therefore play a critical role in intergenomic communication [9]. Extracellular signals induce expression of PGC-1 family coactivators, which in cooperation with specific transcription factors (mainly Nuclear Respiratory Factors, NRF-1 and NRF-2), have been found to regulate the expression of many genes involved in mitochondrial biogenesis. These include structural subunits of the OXPHOS system, enzymes involved in intermediary metabolism, and several factors involved in mtDNA maintenance and transcription (reviewed in [10]). A similar framework to explain the integrated control of Drosophila mitochondrial gene expression has not been identified. The ortholog of nrf-1 in Drosophila, erect wing, is essential for neurogenesis and myogenesis [11], but it has not been linked directly to regulation of mitochondrial gene expression. Thus, it is presently unknown if the circuitry that coordinates mitochondrial biogenesis at the transcriptional level in mammals is conserved in Drosophila.
Our laboratories and others have carried out a systematic characterization of the promoter regions of Drosophila genes encoding factors involved in mtDNA replication, maintenance and transcription [reviewed in [12]). To date, these include the genes encoding the two subunits of DNA polymerase γ (α and β) [13], mtSSB [14], mtDNA helicase (L.S. Kaguni, unpublished data), mtRNA polymerase (R. Garesse, unpublished data), TFAM [15],mtTFB1 [16],mtTFB2 [17] and DmTTF [18] (Table 1). We have found that the expression of most of these genes is regulated by DREF, a transcription factor that also regulates the expression of genes involved in nuclear DNA replication and cell cycle control, including the catalytic subunit of DNA polymerase α, proliferating cell nuclear antigen, cyclin A and E2F [12]. In addition, a recently identified conserved sequence element (RTTAYRTAAY), designated as the Nuclear Respiratory Gene element (NRG), is located downstream of the transcriptional initiation site of a high proportion of nuclear genes encoding mitochondrial proteins [19]. Although there are no functional studies and the transcription factors that recognize the NRG site have yet to be identified, this element represents a good candidate for involvement in the coordinate response of genes involved in energy production in Drosophila, perhaps in combination with DREF. In support of this hypothesis, several of the genes encoding factors of the Drosophila mtDNA replication and transcription machineries contain both DRE and NRG sites.
Table 1.
DNA sequence elements involved in the regulated expression of essential factors for mtDNA metabolism in Drosophila
| Gene | DNA sequence elements |
||
|---|---|---|---|
| DRE | NRG | EWG | |
| pol γ-α | − | − | + |
| pol γ-β | + | + | − |
| mtSSB | + | + | − |
| mtDNA helicase | + | − | − |
| mtRNA pol | − | + | − |
| TFAM | + | + | − |
| mtTFB1 | + | − | − |
| mtTFB2 | + | − | − |
| mtTTF | + | − | − |
The proximal promoter (2 Kb) and 3′-upstream gene regions of several genes encoding essential factors for Drosophila mtDNA metabolism have been analyzed in silico, to search for potential binding sites for DREF, NRG and EWG. The functional role of all DREF binding sites except that in the gene for mtTFB1 has been demonstrated by EMSA and cell transfection analyses. [14 (mtSSB), 65 (pol γ-β), 66 (Tfam) and unpublished data from our laboratories].
To begin to understand the regulation of mitochondrial biogenesis during embryonic development, we have studied by in situ hybridization and immunostaining the spatio-temporal expression pattern of genes encoding mtDNA replication factors as markers of mitochondrial proliferation, and those encoding structural components of the OXPHOS system as markers of mitochondrial differentiation. We have found that genes involved in mtDNA metabolism are highly expressed in the gut, and in some cases in additional cellular domains of the embryo. OXPHOS genes are also highly expressed in the gut, but are also expressed at high levels in other tissues including the mesoderm (M.A. Fernández-Moreno and R. Garesse, unpublished data). These results indicate that mitochondrial proliferation and differentiation are not strictly coupled during development.
Interestingly, the molecular characterization of bonsai mutants has also revealed that the gut is extremely active in mitochondrial biogenesis during Drosophila development [20]. The Bonsai protein is highly expressed in the gut, localizes to mitochondria and shows strong homology to prokaryotic ribosomal protein S15, suggesting a role in mitochondrial translation. However, the precise role of Bonsai in mitochondrial function remains to be determined.
3. Drosophila as an animal model to study human mitochondrial diseases
Drosophila provides a useful model for studying several complex biological processes because it is ideally tractable at the genetic, biochemical, molecular and physiological levels. It has served as a model in studies of development and differentiation [3], aging [21], cell cycle [22], transcriptional and translaional control [23], signaling pathways [24], response to hypoxia [25], sensorial perception [26], and circadian rhythms [27]. In addition, the presence in Drosophila of orthologs of genes involved in human diseases has spawned interest in using it to understand the molecular basis of disease, and to discover new therapies. Representative research using Drosophila has been focused on neurodegenerative diseases [7,28], cancer [29], cardiac pathologies [30], age-associated dysfunction [31], sensitivity to pollutants [32], and more recently on mitochondrial diseases (see below).
Mitochondrial diseases, mainly due to the alteration of OXPHOS functions, are associated with a wide range of clinical symptoms, especially those in neurodegenerative disorders, such as blindness, deafness, dementia, movement disorders, ataxia, corea, and encephalopathies [33,34]. Mitochondrial defects are also associated with muscular weakness, cardiac failure, diabetes, renal dysfunction and hepatic disease. The clinical and genetic diversity of mitochondrial pathologies makes it difficult to estimate their incidence in the population, which in composite, is considered relatively high. The alteration of OXPHOS function caused by known mutations in mtDNA affects at least 1 in 8500 people. One of the main objectives in understanding the physiopathology of mitochondrial diseases is to create amenable animal models that reproduce the variety of symptoms associated with this group of complex human disorders. Although a great effort is currently focused on developing mammalian models of human mitochondrial pathologies, Drosophila emerges as an attractive, tractable alternative.
3.1. mtDNA depletion
mtDNA depletion syndrome (MDS) is caused by a defect in intergenomic communication that results in decreased mtDNA content (i.e., a low mtDNA/ nDNA ratio). Children usually present with hypotonia, lactic acidosis and elevated serum creatine kinase. Some also have severe hepatopathy or renal involvement mimicking de Toni–Fanconi syndrome. The symptoms can also involve multiple systems affecting the muscle. Mutations in the genes encoding the mitochondrial deoxyguanosine and deoxythymidine kinases are associated with the hepatocerebral and the myopathic forms of MDS [35]. In addition, preliminary studies have also shown a deficient activity of DNA polymerase γ in tissues of patients suffering MDS [36].
Our laboratories have used the UAS-GAL4 system to alter the level of mtDNA in the fly. Overexpression of a wild-type version of the catalytic subunit of mitochondrial DNA polymerase (pol γ-α) interferes with the process of mtDNA replication and produces a significant decrease in the amount of mtDNA [37]. We have analyzed the consequences of the mtDNA depletion in the whole animal, and specifically in two tissues affected in MDS patients, muscle and nervous system. mtDNA depletion resulting from constitutive overexpression is lethal at the pupal stage, a phenotype that is also observed upon specific overexpression in the muscle. Interestingly, the induction of mtDNA depletion in the nervous system is not lethal. The main phenotype detected in adults is an increase in the population mortality rate, and a moderate but significant increase in apoptosis in the larval brain, which likely contributes to the phenotype. This model presents the opportunity to characterize the molecular and metabolic responses of various tissues to mtDNA depletion, and to develop methods that alleviate it. At the same time, recognizing that overexpression of the wild-type catalytic subunit may generate secondary effects not related directly to the mtDNA depletion, we are complementing these studies with the introduction of specific mtDNA mutations in the pol γ-α gene by homologous recombination (see below).
3.2. Mitochondrial deafness
The Drosophila mutant technical knock-out, tko, carries a point mutation affecting a phylogenetically conserved residue in the nuclear gene encoding the mitochondrial ribosomal protein S12 (MRPS12). This behavioural mutant exhibits developmental retardation at the larval stage, bang sensitivity, defective response to sound, quantitatively reduced mitochondrial translational capacity and impaired male courtship [38]. The primary biochemical defect is a decrease in OXPHOS and ATP synthase activities in mitochondria. tko mutants possess various features of mitochondrial disorders in humans, and these phenotypes are reverted by transgenic expression of a wild-type copy of the tko gene. The transgenic reversion analysis revealed critical steps and cell types that were responsible for the disease-like phenotype [39]. Furthermore, inbreeding of mutant lines resulted in a systematic improvement of the phenotypic outcome. These features have made the tko mutant a suitable model for many forms of human mitochondrial disease that are characterized by a reduction in the mitochondrial respiratory capacity and a generalized OXPHOS deficiency and in particular, for sensorineural deafness.
3.3. Mitochondrial encephalopathy
Mutations in mtDNA and in nuclear-encoded mitochondrial proteins cause a group of devastating encephalomyopathies with complex clinical features [40]. Leigh syndrome (LS) is the most common infantile mitochondrial encephalopathy and is characterized by symmetric necrotic lesions in the brainstem, diencephalon and basal ganglia. Symptoms usually include nystagmus, ataxia, dystonia, hypotonia and optic atrophy. Although the cause of the disease is genetically heterogeneous, mutations in the surf-1 gene are the single most prevalent cause of LS [41]. The surf-1 gene is highly conserved from prokaryotes to humans, and is expressed ubiquitously in human tissues, albeit at higher levels in aerobic tissues. It encodes a 31 kDa protein located in the inner mitochondrial membrane. Previous studies in human cells [42], mice [43] and yeast [44] showed that Surf-1 is involved in COX complex assembly. Nonetheless, the molecular mechanism leading to the pathogenic course of LS is not fully understood.
Zordan and co-workers performed a functional knock-down of Surf-1 in Drosophila using an RNAi strategy [45]. Post-transcriptional gene silencing was induced by GAL4-mediated expression of an UAS inverted-repeat transgene. Ubiquitous silencing results in severely impaired development. Larvae are smaller and exhibit profound alterations in spontaneous and light-induced locomotion. Most individuals die before entering pupariation. Notably, behavioural and electrophysiological abnormalities were found not to be due to structural defects, but were rather caused by a deficient energy supply. When surf-1 was silenced exclusively in the central nervous system (CNS), individuals developed to adulthood and exhibited altered mitochondria in larval muscles, and decreased COX activity in cephalic sections. Because both development and metamorphosis are high energy-requiring processes, a defect in respiration and ATP synthesis may be responsible for the developmental delay and the late larval lethality observed in Surf-1-deficient larvae. The results obtained in the Drosophila model suggest important functions for Surf-1 in COX activity, and establish a critical role for mitochondrial energy pathways in organogenesis, development and CNS function that are likely to be similar in humans.
3.4. Friedreich ataxia
Friedreich ataxia (FA) is an inherited recessive neurodegen-erative disorder associated with a genetic insufficiency of frataxin in humans [46]. Frataxin is encoded by the FRDA gene and has a predicted size of 210 amino acids [47]. It is a mitochondrial iron chaperone that plays a critical role in iron storage, cellular iron homeostasis and biogenesis of Fe–S clusters. As an iron binding protein, frataxin prevents Fe+2 from generating toxic hydroxyl radicals inside the mitochondrial matrix.
Most FA patients are homozygous for a GAA triplet repeat expansion in the first intron of the FRDA gene. While normal chromosomes contain up to 40 repeats, FA chromosomes harbor between 90 and 1000 triplets. Due to the meiotic and mitotic instability of the expanded alleles, FA patients have insufficient frataxin levels, resulting in iron accumulation in mitochondria and dysfunctional iron-containing proteins, specifically aconitase and the OXPHOS complexes. The peripheral nervous system (PNS) and heart are among the most severely affected tissues. Hence, FA is characterized by progressive neurological disability and heart abnormalities. The age of onset may vary from infancy to adulthood, but FA usually appears during childhood. Variations in symptoms and age of onset suggest that other factors in addition to the degree of triplet expansion influence disease progression, and there is currently no effective therapy for this devastating disease.
Frataxin is highly conserved throughout evolution. Orthologs have been identified in mammals, invertebrates, yeast and plants, and several models have already been used to gain insight into the functions of frataxin and its related disease. Previous studies in yeast revealed that deletion of YFH1, the frataxin homolog, produces iron accumulation in mitochondria and a progressive loss of respiratory competence and mtDNA. As a result, ΔYFH1 strains exhibit permanent mitochondrial damage and dysfunction [48]. Cell differentiation experiments performed in mouse cell lines demonstrated that frataxin is essential for development of neural lineages [49]. Although mouse models for FA showed early embryonic lethality [50] conditional inactivation of the mouse FA gene led to cardiomyopathy, sensory nerve defects and Fe–S enzyme deficiency [51].
A RNAi strategy was used in Drosophila to suppress production of the frataxin homolog (DFH) in a global or tissue-specific manner [52]. The main advantage in the Drosophila system as compared to other eukaryotic models is that inverted repeat transgenes used in RNAi reduce rather than totally eliminate the corresponding protein. Thus, the Drosophila model mimics more accurately the genetic origin of FA, which arises from a decrease rather than a complete loss of frataxin. Furthermore, the fly model also circumvents the early embryonic lethality reported in frataxin-null mice. Systemic suppression of the dfh gene led to large, long-lived larvae that showed diminished iron cofactor-dependent enzyme activity, and increased susceptibility to iron toxicity. Because prolonged larval development and failure to initiate and complete metamorphosis are common features of Drosophila mutants defective in mtDNA maintenance, observations from this model prompted the hypothesis that DFH depletion may lead to increased mtDNA damage. Whereas silencing of the dfh gene in motor neurons had no deleterious effect, silencing in the PNS resulted in normal larval development but a reduced adult life span. Over-expression of primary ROS-metabolizing enzymes, such as SOD1, SOD2 and catalase in combination with silencing of the dfh gene did not improve the phenotype, suggesting that oxidative stress is not a major contributor.
The Drosophila model of FA is further validated by the fact that the vulnerability of the PNS to frataxin depletion is conserved across the animal kingdom. Because a DFH deficiency produces a robust phenotype in Drosophila, the fly model may aid in the identification of factors that alleviate FA symptoms, and the development of novel treatment strategies.
3.5. Mitochondrial diseases caused by mtDNA mutations
Point mutations in mtDNA are associated with a wide range of severe mitochondrial diseases, including a number of encephalomyopathies with complex clinical features [33]. Although some of the mutations have been characterized in cybrid cell lines, and demonstrated to produce a clear mitochondrial impairment, it has not been possible to study the consequences of the mutations in animal model systems because of the currently inability to transform mitochondria with exogenous DNA. Very recently, the first pathogenic mtDNA mutation in an intact animal was reported in Drosophila, a G–A transition that affects an evolutionarily conserved amino acid residue in the ATP6 gene [53]. Point mutations in the human ATP6 gene cause neuropathy, ataxia, and retinitis pigmentosa (NARP), maternally inherited Leigh’s syndrome (MILS) and familial bilateral striatal necrosis (FBSN) [33]. Notably, the G–A transition in the ATP6 gene was found in virtually homoplasmic levels in a Drosophila sesB stock that carries a mutation in the ANT1 gene, which is associated with autosomal dominant progressive external ophtalmoplegia (adPEO) in humans [54]. Patients with PEO are predisposed to secondary mtDNA mutations, and it is thus possible that a similar mutator effect has occurred in Drosophila. The ATP6 mutation inhibits significantly the activity of ATP synthase, but surprisingly does not affect the respiration rate, and therefore the mitochondria are uncoupled. Most interestingly, Drosophila ATP6 mutants have a short life span, locomotion impairment and myodegeneration that are strictly progressive, a situation similar to human mitochon-drial encephalomyopathy of mitochondrial origin. Although the mutation per se does not induce neurodegeneration, neurological dysfunction was apparent.
4. New perspectives for gene manipulation in Drosophila
Until recently, Drosophila models used in the study of mitochondrial dysfunction were based on the analysis of mutants generated by random mutagenic screens, or by gene silencing by RNAi that is based on P-element transgenesis. One of the main advantages of the fly system is the UAS– GAL4 binary system, which can be exploited to knockdown a target gene under the control of alternative promoters, thus restricting silencing to a desired spatio-temporal pattern. More recently, new technologies for gene manipulation in Drosophila have been expanded to include new genome integration methods based on bacteriophage ΦC31 or piggybacks that show much less insertional specificity than P-elements [6]. Despite these advances, the study of specific mutations associated with human diseases has been hampered by the lack of gene targeting techniques, one of the main limitations in working with Drosophila. Golic and co-workers have recently circumvented this drawback with the development of efficient homologous recombination-based methods [55]. Any endogenous locus can be replaced or deleted by the expression of a site-specific recombinase and a site-specific nuclease, taking advantage of the fact that double-strand breaks (DSB) in Drosophila DNA are recombinogenic. There are currently two techniques that use homologous recombination for gene replacement, “Ends-in” and “Ends-out”. These systems have similar efficiencies but present different advantages and disadvantages.
4.1. Ends-in
The Ends-in approach first introduces a version of the gene containing the mutation of interest in tandem with the endogenous allele, and then removes one of the copies leaving only the mutated or wild-type allele in the chromosome. The main advantage of this approach is that intact alleles can be recovered that harbor the desired mutation.
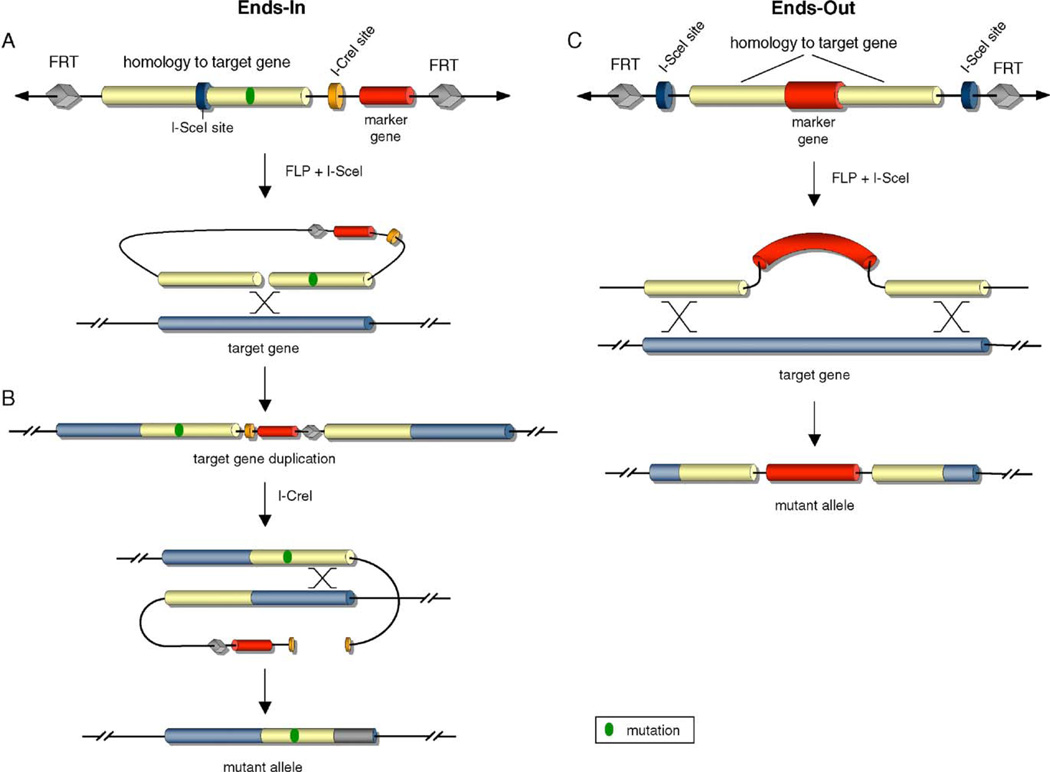
The Ends-in targeting technique is executed in three steps (Fig. 1A and B). First, a P-element construct is generated that incorporates several specific features: a DNA region homologous to the target gene containing the specific mutation(s), two FLP Recombination Target (FRT) sequences, an I-SceI endonuclease recognition site, an I-CreI endonuclease recognition site, and a marker gene, usually white+. P-element transformation is performed by standard methods [4]. The range of total local homology that has led to successful targeting is between 2 kb and 9 kb, with >4 kb utilized in most cases [56]. Second, a series of crosses between the transgenic flies carrying the donor element with fly stocks harboring FLP and I-SceI transgenes allow the expression of the enzymes by heat shock induction, thus generating the recombinogenic donor, and inducing targeted homologous recombination. Homologous recombination generates a typical structure in which the endogenous and the mutant copies of the gene flank the marker gene at the target locus. The wild-type–white+-mutant gene structure is not expected to manifest a phenotype until reduction. Finally, a second homologous recombination event resulting from expression of the endonuclease I-CreI removes one copy of the gene, the wild-type or the mutant, and the white marker. This results in the replacement of the endogenous copy with a version containing the specific mutation without any other alterations in the genome. To date, this novel method has been used to generate mutant alleles of several endogenous Drosophila genes [57].
Fig. 1.
Gene targeting in Drosophila. A, Ends-in technique and the elements of the donor construct. After transgenesis the combined action of FLP and endonuclease I-SceI generates the recombinogenic template, which produces the duplication at the target gene locus by homologous recombination. B, reduction of target gene repeats. The double-strand break caused by endonuclease I-CreI induces the second recombination. Ends-in targeting introduces a point mutation (green dot) in one of the alleles of the target gene. C, Ends-out technique. FLP and I-SceI generate a linear DNA molecule that undergoes homologous recombination with the target gene.
4.2. Ends-out
Ends-out targeting is the most frequently used method in mice and yeast. This technique starts with a different donor construct structure, and also differs from Ends-in in the position of the specific-site endonuclease cleavage sites (Fig. 1C). The double-stranded breakage occurs at the outer ends of donor DNA. The process of Ends-out targeting is similar to Ends-in targeting; through the action of FLP and I-SceI, a linear DNA extrachromosomal donor fragment is generated, and it is targeted to the endogenous allele by homologous recombination. Two of the advantages of the Ends-out method are that the target gene can be replaced without the involvement of a tandem duplication, thus generating a knock-in or knock-out in one step. In addition, the donor construct is easier to make than in Ends-in. The disadvantage of this technique is that the end product leaves exogenous sequence at the altered locus, usually the marker gene [58]. Ends-out gene targeting has been used successfully to target yellow, the myocardin-related transcription factor DMRTF, and the odorant-receptor gene, Or83b (reviewed in [6]).
5. Current models of mitochondrial diseases in flies
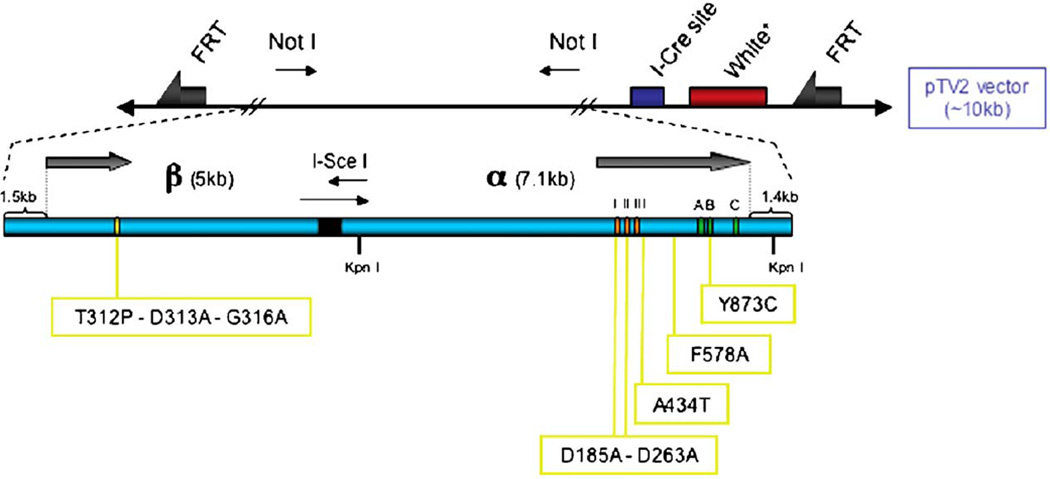
In our laboratories, we are developing a Drosophila model of mitochondrial pathology associated with pol γ mutations using the knock-in technology. Even though we have been successful in circumventing by low-level expression the deleterious phenotypes engendered with high-level overexpression of wild-type pol γ-α we recognize that potential problems may result from developmental and spatial mis-expression of a transgene. Thus, we have altered our strategy for mutagenesis in Drosophila to use targeted gene replacement by homologous recombination (Fig. 2). Our objective has been to introduce in Drosophila versions of the pol γ-α gene that are associated with PEO in humans, and to examine the effects of several mutations in the Drosophila pol γ-α an dpol γ-β genes for which disease phenotypes have not yet been reported in humans. Of the 30 different mutations that have been identified in the human pol γ-α gene [59], approximately half are in strictly conserved positions in the orthologous gene in Drosophila. We have selected two, A467T and Y955C (A434T and Y873C in Drosophila), which map within the conserved γ1 element in the spacer region and the pol domain, respectively, and for which we and others have already evaluated the biochemical defects upon recombinant expression in the baculovirus system [60–63]. We have also selected the Drosophila γ3 spacer-region mutation F578A, for which we have defined the biochemical defects [64]. In addition, we have introduced a double mutation (D185A and D263A) within two active site aspartate residues in the exonuclease domain, which we have shown to cause developmental defects, delays and arrest, and to limit life span when it is expressed at a low level using the UAS-GAL4 system in transgenic animals. Finally, we took advantage of our prior discovery that the two subunits of Drosophila pol γ map within a compact gene cluster [13] to generate a fragment containing ~ 12 kb donor homology that carries both the pol γ-α and -β genes, with the I-SceI site introduced between them. This allowed us to introduce specific amino acid substitutions in both pol γ-α and -β in a single recombinant donor construct. After homologous recombination, the expected product is a tandem partial duplication of the target genome sequence, with one copy containing the desired pol γ-α mutation and the other copy containing the desired pol γ-β mutation. As a result, we have also introduced three mutations in the C-terminal region of the Drosophila accessory subunit gene, which we have shown to be deleterious in our biochemical analysis of human pol γ-β.
Fig. 2.
Pol γ knock-in in Drosophila. Approximately 12 kb of DNA from the D. melanogaster pol γ cluster genomic region [60] was amplified by PCR and cloned into the vector pBluescript. For simplicity, only the pol γ-α and pol γ-β genes are shown schematically (arrows). The I-SceI recognition site was introduced between the genes, and this construct was used to generate specific mutations (boxed in yellow) in the pol γ-α or pol γ-β coding regions. The conserved exonuclease (I, II and III), and polymerase (A, B and C) active site motifs are indicated by boxes. Each of the modified genomic DNAs was cloned into the NotI site of the pTV2 vector, and the orientation of the inserted DNA was confirmed by KpnI digestion. The main features of the pTV2 vector are indicated as follows: FRTs (arrows), white + marker gene (red box), I-CreI recognition site (blue box), NotI recognition sites (thin arrows).
Our immediate goals will be to evaluate these models using established protocols in Drosophila to analyze mtDNA content and integrity, respiratory chain complex activity, mitochondrial distribution and function, apoptotic signaling and importantly, larval and adult behaviour. We expect to establish an experimental system of mtDNA replication failure that is analogous to that associated with human pathologies and in doing so, we will establish a direct correlation between the defect(s) that the mutants exhibit in our biochemical assays and their phenotypic effects in vivo. We anticipate that this will set the stage for us to use the power of Drosophila molecular genetics to search for factors that modulate the phenotype, either augmenting or ameliorating it.
6. Concluding remarks
Although tissues requiring high oxidative metabolism are affected most severely in mitochondrial disorders, specific tissue involvement varies considerably. In order to develop therapies for these diseases, it is necessary to determine both the cellular processes and the tissues that are altered by insufficient energy supply. Drosophila models can help us to understand common aspects of these pathologies because they enable the identification of critical times, cell types and tissues that account for the disease-like phenotypes. Global transcriptional analysis of these models has already revealed many sets of genes, including those related to stress responses and metabolism, which are systematically up- or down-regulated according to the degree of the mitochondrial dysfunction. The ultimate aim of such transcriptomic approaches is to identify potential pharmacological targets in mitochondrial disease. Because the recent development of efficient gene targeting methods has paved the way for the introduction of specific mutations responsible for human mitochondrial diseases, Drosophila offers substantial promise for understanding the pathogenic mechanisms of this devastating group of diseases.
Acknowledgments
We thank Erin Wakeling for critical reading and comments on the manuscript. This work was supported by European Union Project QLG1-CT-2001-00966, Ministerio de Ciencia y Tecnologia, Spain (Grants BMC01-1525 and BFU2004-04591) and Instituto de Salud Carlos III, Redes de centros RCMN (C03/08) and Temáticas (G03/011 (to R.G.) and National Institutes of Health Grant GM45295 (to L.S.K.).
Abbreviations
- mtDNA
mitochondrial DNA
- OXPHOS
oxidative phosphorylation
- NRG
Nuclear respiratory gene
- COX
cytochrome oxidase
- MDS
mtDNA depletion syndrome
- pol γ-α
DNA polymerase γ catalytic subunit
- LS
Leigh syndrome
- CNS
central nervous system
- FA
Friedreich’s ataxia
- adPEO
autosomal dominant progressive external ophthalmoplegia
- RNAi
RNA interference
- PNS
peripheral nervous system
- DSB
double-strand break
References
- 1.Rubin GM, Lewis BE. A brief history of Drosophila’s contributions to genome research. Science. 2000;287:2216–2218. doi: 10.1126/science.287.5461.2216. [DOI] [PubMed] [Google Scholar]
- 2.Fernández-Moreno MA, Farr CL, Kaguni LS, Garesse R. Drosophila melanogaster as a Model System to Study Mitochondrial Function, in Methods in Molecular Biology. Totowa, NJ: Humana Press Inc.; (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bate M, Martínez-Arias A. The Development of Drosophila melanoga-ster. New York: Cold Spring Harbor Laboratory Press; 1993. [Google Scholar]
- 4.Rubin GM, Spradling CA. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–353. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 5.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 6.Venken KJ, Bellen HJ. Emerging technologies for gene manipulation in Drosophila melanogaster. Nat. Rev. Genet. 2005;6:167–178. doi: 10.1038/nrg1553. Erratum in: Nat. Rev. Genet. 6 (2005)340) [DOI] [PubMed] [Google Scholar]
- 7.Bilen J, Bonini NM. Drosophila as a model for human neurodegener-ative disease. Annu. Rev. Genet. 2005;39:153–171. doi: 10.1146/annurev.genet.39.110304.095804. [DOI] [PubMed] [Google Scholar]
- 8.Garesse R, Vallejo CG. Animal mitochondrial biogenesis and function: a regulatory cross-talk between two genomes. Gene. 2001;263:1–16. doi: 10.1016/s0378-1119(00)00582-5. [DOI] [PubMed] [Google Scholar]
- 9.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim. Biophys. Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 10.Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J. Cell. Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- 11.DeSimone SM, White K. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol. Cell. Biol. 1993;13:3641–3649. doi: 10.1128/mcb.13.6.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garesse R, Kaguni LS. A Drosophila model of mitochondrial DNA replication: proteins, genes and regulation. IUBMB Life. 2005;57:555–561. doi: 10.1080/15216540500215572. [DOI] [PubMed] [Google Scholar]
- 13.Lefai E, Fernandez-Moreno MA, Kaguni LS, Garesse R. The highly compact structure of the mitochondrial DNA polymerase genomic region of Drosophila melanogaster: functional and evolutionary implications. Insect Mol. Biol. 2000;9:315–322. doi: 10.1046/j.1365-2583.2000.00191.x. [DOI] [PubMed] [Google Scholar]
- 14.Ruiz de Mena I, Lefai E, Garesse R, Kaguni LS. Regulation of mitochondrial single-stranded DNA-binding protein gene expression links nuclear and mitochondrial DNA replication in Drosophila. J. Biol. Chem. 2000;275:13628–13636. doi: 10.1074/jbc.275.18.13628. [DOI] [PubMed] [Google Scholar]
- 15.Takata K, Yoshida H, Hirose F, Yamaguchi M, Kai M, Oshige M, Sakimoto I, Koiwai O, Sahaguchi K. Drosophila mitochondrial transcription factor A: characterization of its cDNA and expression pattern during development. Biochem. Biophys. Res. Commun. 2001;287:474–483. doi: 10.1006/bbrc.2001.5528. [DOI] [PubMed] [Google Scholar]
- 16.Matsushima Y, Adán C, Garesse R, Kaguni LS. Drosophila transcription factor B1 modulates mitochondrial translation but not transcription or DNA copy number in Schneider cells. J. Biol. Chem. 2005;280:16815–16820. doi: 10.1074/jbc.M500569200. [DOI] [PubMed] [Google Scholar]
- 17.Matsushima Y, Garesse R, Kaguni LS. Drosophila transcription factor B2 regulates mitochondrial DNA copy number and transcription in Schneider cells. J. Biol. Chem. 2004;279:26900–26905. doi: 10.1074/jbc.M401643200. [DOI] [PubMed] [Google Scholar]
- 18.Roberti M, Fernández-Silva P, Polosa PL, Fernández-Viana E, Bruni F, Deceglie S, Montoya J, Gadaleta MN, Cantatore P. In vivo transcription termination activity of the Drosophila mitochondrial DNA-binding protein DmTTF. Biochem. Biophys. Res. Commun. 2005;331:357–362. doi: 10.1016/j.bbrc.2005.03.173. [DOI] [PubMed] [Google Scholar]
- 19.Sardiello M, Tripoli G, Romito A, Minervini C, Viggiano L, Caggese C, Pesole G. Energy biogenesis: one key for coordinating two genomes. Trends Genet. 2005;21:12–16. doi: 10.1016/j.tig.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Galloni M, Bonsai a ribosomal protein S15 homolog, involved in gut mitochondrial activity and systemic growth. Dev. Biol. 2003;264:482–494. doi: 10.1016/j.ydbio.2003.08.021. [DOI] [PubMed] [Google Scholar]
- 21.Helfand SL, Rogina B. Genetics of aging in the fruit fly, Drosophila melanogaster. Annu. Rev. Genet. 2003;37:329–348. doi: 10.1146/annurev.genet.37.040103.095211. [DOI] [PubMed] [Google Scholar]
- 22.Song YH. Drosophila melanogaster: a model for the study of DNA damage checkpoint response. Mol. Cell. 2005;19:67–179. [PubMed] [Google Scholar]
- 23.Wilhelm JE, Smibert CA. Mechanisms of translational regulation in Drosophila. Biol. Cell. 2005;97:235–252. doi: 10.1042/BC20040097. [DOI] [PubMed] [Google Scholar]
- 24.Davies SA. Signalling via cGMP: Lessons from Drosophila. Cell. Signal. 2006;18:409–421. doi: 10.1016/j.cellsig.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 25.Farahani R, Haddad CG. Understanding the molecular responses to hypoxia using Drosophila as a genetic model. Respir. Physiol. Neurobiol. 2003;135:221–229. doi: 10.1016/s1569-9048(03)00049-1. [DOI] [PubMed] [Google Scholar]
- 26.Davis RL. Olfactory memory formation in Drosophila: from molecular to systems neuroscience. Annu. Rev. Neurosci. 2005;28:275–302. doi: 10.1146/annurev.neuro.28.061604.135651. [DOI] [PubMed] [Google Scholar]
- 27.Hardin EP. The circadian timekeeping system of Drosophila. Curr. Biol. 2005;15:714–722. doi: 10.1016/j.cub.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 28.Chen L, Feany MB. Alpha-synuclein phosphorylation controls neuro-toxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat. Neurosci. 2005;8:657–663. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- 29.Woodhouse EC, Liotta LA. Drosophila invasive tumors: a model for understanding metastasis. Cell Cycle. 2004;3:38–40. [PubMed] [Google Scholar]
- 30.Wolf MJ, Amrein H, Izatt JA, Choma MA, Reedy MC, Rockman HA. Drosophila as a model for the identification of genes causing adult human heart disease. Proc. Natl. Acad. Sci. U.S.A. 2006;103:1394–1399. doi: 10.1073/pnas.0507359103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saitoe M, Horiuchi J, Tamura T, Ito N. Drosophila as a novel animal model for studying the genetics of age-related memory impairment. Rev, Neuroscience. 2005;16:137–149. doi: 10.1515/revneuro.2005.16.2.137. [DOI] [PubMed] [Google Scholar]
- 32.Wilson M, Widdicombe JH, Gohil K, Burtis KC, Reznick AZ, Cross CE, Elserich JP. Are Drosophila a useful model for understanding the toxicity of inhaled oxidative pollutants: a review. Inhal. Toxicol. 2005;17:765–774. doi: 10.1080/08958370500225141. [DOI] [PubMed] [Google Scholar]
- 33.McFarland R, Taylor RW, Turnbull DM. The neurology of mitochon-drial DNA disease. Lancet Neurol. 2002;1:343–351. doi: 10.1016/s1474-4422(02)00159-x. [DOI] [PubMed] [Google Scholar]
- 34.Smeitink JA, Zeviani M, Turnbull DM, Jacobs HT. Mitochondrial medicine: a metabolic perspective on the pathology of oxidative phosphorylation disorders. Cell Metab. 2006;3:9–13. doi: 10.1016/j.cmet.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 35.Spinazzola A, Zeviani M. Disorders of nuclear-mitochondrial inter-genomic signalling. Gene. 2005;354:16–18. doi: 10.1016/j.gene.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 36.Naviaux RK, Nyhan WL, Barshop BA, Poulton J, Markusic D, Karpinski NC, Haas RH. Mitochondrial DNA polymerase γ deficiency and mtDNA depletion in a child with Alpers’ syndrome. Ann. Neurol. 1999;45:54–58. doi: 10.1002/1531-8249(199901)45:1<54::aid-art10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 37.Lefai E, Calleja M, Ruiz de Mena I, Lagina AT, III, Kaguni LS, Garesse R. Overexpression of the catalytic subunit of DNA polymerase γ results in depletion of mitochondrial DNA in Drosophila melanogaster. Mol. Gen. Genet. 2000;264:37–46. doi: 10.1007/s004380000301. [DOI] [PubMed] [Google Scholar]
- 38.Toivonen JM, O’Dell KM, Petit N, Irvine SC, Knight GK, Lehtonen M, Longmuir M, Luoto K, Touraille S, Wang Z, Alziari S, Shah ZH, Jacobs HT. Technical knockout, a Drosophila model of mitochondrial deafness. Genetics. 2001;159:241–254. doi: 10.1093/genetics/159.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobs HT, Fernandez-Ayala J, Manjiry S, Kemppainene E, Toivonen JM, O’Dell KM. Mitochondrial disease in flies. Biochim. Biophys. Acta. 2004;1659:190–196. doi: 10.1016/j.bbabio.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 40.DiMauro S, Schon EA. Mitochondrial respiratory chain diseases. N. Engl. J. Med. 2003;348:2656–2658. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- 41.Bohm M, Pronicka E, Karcmarewicz E, Pronicki M, Piekutowska-Abramczuk D, Sykut-Cegielska J, Mierzewska H, Hansikova H, Vesela K, Tesarova M, Houstkova H, Houstek J, Zeman J. Restrospective, multicentric study of 180 children with cytochrome c oxidase deficiency. Pediatr. Res. 2006;59:21–26. doi: 10.1203/01.pdr.0000190572.68191.13. [DOI] [PubMed] [Google Scholar]
- 42.Tiranti V, Galimberti C, Nijtmans L, Bovolenta S, Perini MP, Zeviani M. Characterization of SURF-1 expression and Surf-1p function in normal and disease conditions. Hum. Mol. Genet. 1999;8:2533–2540. doi: 10.1093/hmg/8.13.2533. [DOI] [PubMed] [Google Scholar]
- 43.Agostino A, Invernizzi F, Tiveron C, Fagiolari G, Prelle A, Lamantea E, Giavazzi A, Battaglia G, Tatangelo L, Tiranti V, Zeviani M. Constitutive knockout of Surf1 is associated with high embryonic lethality, mitochondrial disease and cytochrome c oxidase deficiency in mice. Hum. Mol. Genet. 2003;12:399–413. doi: 10.1093/hmg/ddg038. [DOI] [PubMed] [Google Scholar]
- 44.Nijtmans LG, Artal-Sanz M, Bucko M, Farhoud MH, Feenstra M, Hakkaart GA, Zeviani M, Grivell LA. Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 2001;498:46–51. doi: 10.1016/s0014-5793(01)02447-4. [DOI] [PubMed] [Google Scholar]
- 45.Zordan MA, Cisotto P, Benna C, Agostino A, Rizzo G, Piccin A, Pegoraro M, Sandrelli F, Perini G, Tognon G, De Caro R, Peron S, Kronnie TT, Megighian A, Reggiani C, Zeviani M, Costa R. Post transcriptional silencing and functional characterization of the Drosophila melanogaster homolog of HumanSurf1. Genetics. 2006;172:229–241. doi: 10.1534/genetics.105.049072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campuzano V, Montermini L, Molto MD, Pianese L, Cossee M, Cavalcanti F, Monros E, Rodius F, Duclos F, Monticelli A, Zara F, Canizares J, Koutnikova H, Bidichandani SI, Gellera C, Brice A, Trouillas P, De Michele G, Filla A, De Frutos R, Palau F, Patel PI, Di Donato S, Mandel JL, Cocozza S, Koenig M, Pandolfo M. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 47.Pandolfo M. Frataxin deficiency and mitochondrial dysfunction. Mitochondrion. 2002;2:87–93. doi: 10.1016/s1567-7249(02)00039-9. [DOI] [PubMed] [Google Scholar]
- 48.Babcock M, de Silva D, Oaks R, Davis-Kaplan S, Jiralerspong S, Montermini L, Pandolfo M, Kaplan J. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276:1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 49.McBurney M, Jones-Villeneuve WEM, Edwards MK, Anderson PJ. Control of muscle and neuronal differentiation in a cultured embryonal carcinoma cell line. Nature. 1982;299:165–167. doi: 10.1038/299165a0. [DOI] [PubMed] [Google Scholar]
- 50.Cossee M, Puccio H, Gansmuller A, Koutnikova H, Dierich A, LeMeur M, Fischbeck K, Dolle P, Koenig M. Inactivation of the Friedreich ataxia mouse gene leads to early embryonic lethality without iron accumulation. Hum. Mol. Genet. 2000;9:1219–1226. doi: 10.1093/hmg/9.8.1219. [DOI] [PubMed] [Google Scholar]
- 51.Puccio H, Simon D, Cossee M, Criqui-Filipe P, Tiziano F, Melki J, Hindelang C, Matyas R, Rustin P, Koenig M. Mouse models for Friedreich ataxia exhibit cardiomyopathy, sensory nerve defect and Fe-S enzyme deficiency followed by intramitochondrial iron deposits. Nat. Genet. 2001;27:181–186. doi: 10.1038/84818. [DOI] [PubMed] [Google Scholar]
- 52.Anderson PR, Kirby K, Hilliker AJ, Phillips JP. RNAi-mediated suppression of the mitochondrial iron chaperone, frataxin, in Drosophila. Hum. Mol. Genet. 2005;14:3397–3405. doi: 10.1093/hmg/ddi367. [DOI] [PubMed] [Google Scholar]
- 53.Celotto AM, Frank AC, McGrath SW, Fergestad T, Van Voorhies WA, Buttle KF, Mannella CA, Palladino MJ. Mitochondrial encephalomyopathy in Drosophila. J. Neurosci. 2006;26:810–820. doi: 10.1523/JNEUROSCI.4162-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaukonen J, Juselius JK, Tiranti V, Kittala A, Zeviani M, Comi GP, Keranene S, Peltonene L, Suomalainen A. Role of adenine nucleotide translocator 1 in mtDNA maintenance. Science. 2000;289:782–785. doi: 10.1126/science.289.5480.782. [DOI] [PubMed] [Google Scholar]
- 55.Rong YS, Golic KG. Gene targeting by homologous recombination in Drosophila. Science. 2000;288:2013–2018. doi: 10.1126/science.288.5473.2013. [DOI] [PubMed] [Google Scholar]
- 56.Bi X, Rong YS. Genome manipulation by homologous recombination in Drosophila. Briefings in Functional Genomics and Proteomics. 2003;2:142–146. doi: 10.1093/bfgp/2.2.142. [DOI] [PubMed] [Google Scholar]
- 57.Rong YS, Titen SW, Xie HB, Golic MM, Bastiani M, Bandyo-padhyay P, Olivera BM, Brodsky M, Rubin GM, Golic KG. Targeted mutagenesis by homologous recombination in D melanogaster. Genes Dev. 2002;16:1568–1581. doi: 10.1101/gad.986602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gong WJ, Golic KG. Ends-out, or replacement, gene targeting in Drosophila. Proc. Natl. Acad. Sci. U.S.A. 2003;100:2556–2561. doi: 10.1073/pnas.0535280100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Di Fonzo A, Bordoni A, Crimi M, Sara G, Del Bo R, Bresolin N, Comi GP. POLG mutations in sporadic mitochondrial disorders with multiple mtDNA deletions. Hum. Mutat. 2003;22:498–499. doi: 10.1002/humu.9203. [DOI] [PubMed] [Google Scholar]
- 60.Luoma PT, Luo N, Loscher WN, Farr CL, Horvath R, Wanschitz J, Kiechl S, Kaguni LS, Suomalainen A. Functional defects due to spacer-region mutations of human mitochondrial DNA polymerase in a family with an ataxia-myopathy syndrome. Hum. Mol. Genet. 2005;14:1907–1920. doi: 10.1093/hmg/ddi196. [DOI] [PubMed] [Google Scholar]
- 61.Chan SS, Longley MJ, Copeland WC. The common A467T mutation in the human mitochondrial DNA polymerase (POLG) compromises catalytic efficiency and interaction with the accessory subunit. J. Biol. Chem. 2005;280:31341–31346. doi: 10.1074/jbc.M506762200. [DOI] [PubMed] [Google Scholar]
- 62.Ponamarev MV, Longley MJ, Nguyen D, Kunkel TA, Copeland WC. Active site mutation in DNA polymerase gamma associated with progressive external ophthalmoplegia causes error-prone DNA synthesis. J. Biol. Chem. 2002;277:15225–15228. doi: 10.1074/jbc.C200100200. [DOI] [PubMed] [Google Scholar]
- 63.Graziewicz MA, Longley MJ, Bienstock RJ, Zeviani M, Copeland WC. Structure-function defects of human mitochondrial DNA polymerase in autosomal dominant progressive external ophthalmoplegia. Nat. Struct. Mol. Biol. 2004;11:770–776. doi: 10.1038/nsmb805. [DOI] [PubMed] [Google Scholar]
- 64.Luo N, Kaguni LS. Mutations in the spacer region of Drosophila mitochondrial DNA polymerase affect DNA binding, processivity, and the balance between Pol and Exo function. J. Biol. Chem. 2005;280:2491–2497. doi: 10.1074/jbc.M411447200. [DOI] [PubMed] [Google Scholar]
- 65.Lefai E, Fernández-Moreno MA, Alahari A, Kaguni LS, Garesse R. The expression of the catalytic and accessory subunits of the mitochondrial DNA polymerase are differentially regulated in Drosophila melanogaster. J. Biol. Chem. 2000;275:33123–33133. doi: 10.1074/jbc.M003024200. [DOI] [PubMed] [Google Scholar]
- 66.Takata K, Inoue YH, Hirose F, Murakami S, Shimanouchi K, Sakimoto I, Sakaguchi K. Spatio-temporal expression of Drosophila mitochon-drial transcription factor A during development. Cell Biol. Int. 2003;27:361–374. doi: 10.1016/s1065-6995(02)00355-4. [DOI] [PubMed] [Google Scholar]