Summary
Lenalidomide is a highly effective treatment for myelodysplastic syndrome (MDS) with deletion of chromosome 5q (del(5q)). Here, we demonstrate that lenalidomide induces the ubiquitination of casein kinase 1A1 (CK1α) by the CRL4CRBN E3 ubiquitin ligase, resulting in CK1α degradation. CK1α is encoded by a gene within the common deleted region for del(5q) MDS and haploinsufficient expression sensitizes cells to lenalidomide therapy, providing a mechanistic basis for lenalidomide's therapeutic window in del(5q) MDS. We found that mouse cells are resistant to lenalidomide but that changing a single amino acid in mouse Crbn to the corresponding human residue enables lenalidomide-dependent degradation of CK1α. We further demonstrate that minor side chain modifications in thalidomide and a novel analogue, CC-122, can modulate the spectrum of substrates targeted by CRL4CRBN. These findings have implications for the clinical activity of lenalidomide and related compounds and demonstrate the therapeutic potential of novel modulators of E3 ubiquitin ligases.
The immunomodulatory (IMiD) agents lenalidomide, thalidomide, and pomalidomide are the first drugs identified that promote the ubiquitination and degradation of specific substrates by an E3 ubiquitin ligase. These compounds bind CRBN1, the substrate adaptor for the CRL4CRBN E3 ubiquitin ligase, and modulate the substrate specificity of the enzyme. Each of these drugs induce degradation of two lymphoid transcription factors—IKZF1 and IKZF3—leading to dramatic clinical efficacy in multiple myeloma and increased interleukin-2 release from T-cells.2-4 However, it has not yet been determined whether degradation of distinct substrates may mediate additional activities and whether all IMiD compounds have the same substrate specificity.
Lenalidomide is also a highly effective treatment for myelodysplastic syndrome (MDS) with deletion of chromosome 5q (del(5q)), inducing cytogenetic remission in more than 50% of patients.5-7 In vitro, lenalidomide selectively induces apoptosis of del(5q) MDS cells.8,9 No biallelic deletions or loss of function mutations on the remaining allele have been detected in any of the genes in the del(5q) common deleted region, implying that MDS with del(5q) is a disease of haploinsufficiency.10,11 We hypothesized that ubiquitination of a distinct CRBN substrate explains the efficacy of lenalidomide in del(5q) MDS.
Lenalidomide induces degradation of CK1α
In order to identify lenalidomide-regulated CRL4CRBN substrates in myeloid cells, we applied stable isotope labeling of amino acids in cell culture (SILAC)-based quantitative mass spectrometry12 to assess global changes in ubiquitination13 and protein levels in the del(5q) myeloid cell line KG-1 (Fig. 1a,b, Extended Data Fig. 1, Extended Data Table 1a,b, Supplementary Table 1). Treatment with 1 μM lenalidomide significantly altered only 7 K-ε-GG sites from 5 proteins out of 13,061 reproducible sites, demonstrating the highly specific effects of this drug. Moreover, lenalidomide significantly altered the protein abundance of 3 out of 5 differentially ubiquitinated proteins. Consistent with previous studies, lenalidomide treatment decreased ubiquitination of CRBN (p=0.026) and increased ubiquitination of IKZF1 (p=7.23 × 10−6 and p=4.97 × 10−4 for two distinct sites), with a reciprocal decrease in IKZF1 protein abundance (p=0.006).2,3
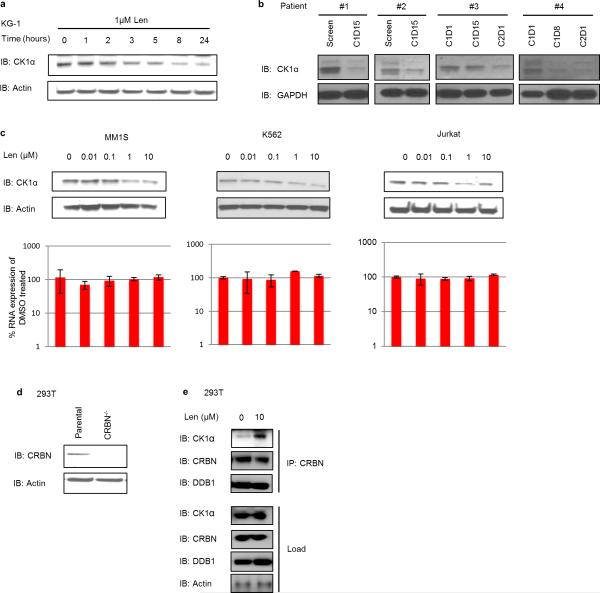
Fig. 1. Lenalidomide-induced changes in ubiquitination and protein levels.
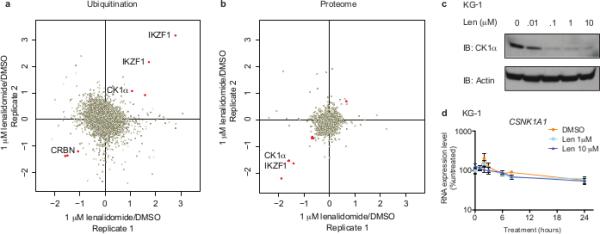
a, Log2 ratios for individual K-ε-GG sites of lenalidomide- (1 μM) versus DMSO-treated KG-1 cells for biological replicates 1 and 2. Each point represents a unique K-ε-GG site. Significantly regulated sites (p<0.05) are red. b, Log2 ratios of protein abundance for lenalidomide- (1 μM) versus DMSO-treated KG-1 cells for biological replicates 1 and 2. Each point represents a unique protein group. Significantly regulated proteins (p<0.05) are red. c, Effects of lenalidomide on endogenous CK1α protein levels in KG-1 cells after 24-hour treatment. Data are representative of 5 independent experiments (n=5). d, CSNK1A1 mRNA levels in KG-1 cells following lenalidomide treatment. Data are mean ± s.d., n=3 biological replicates.
In addition to IKZF1, we detected increased ubiquitination (p=0.04) and decreased protein abundance (p=0.006) of casein kinase 1A1 (CK1α) following treatment with 1 μM lenalidomide (Fig. 1a,b, Extended Data Fig. 1, Extended Data Table 1a,b, Supplementary Table 1). CK1α is encoded by the CSNK1A1 gene, which is located in the del(5q) common deleted region and is expressed at haploinsufficient levels in del(5q) MDS.10,14 CK1α has been implicated in the biology of del(5q) MDS15 and has been shown to be a therapeutic target in myeloid malignancies16, and is therefore an attractive candidate for mediating the effects of lenalidomide in del(5q) MDS.
CK1α is a substrate of CRL4CRBN
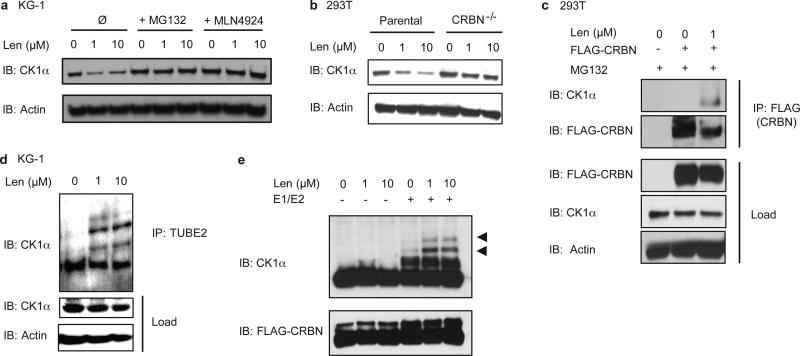
We sought to determine whether CK1α is a lenalidomide-dependent substrate of the CRL4CRBN E3 ubiquitin ligase. We confirmed that lenalidomide treatment decreases CK1α protein levels in multiple human cell lines and in the bone marrow and peripheral blood of AML patients treated in vivo (Fig. 1c, Extended Data Fig. 2, Extended Data Table 2). Lenalidomide treatment resulted in decreased protein levels of both wild-type isoforms of CK1α as well as two somatic CK1α mutations recently identified in del(5q) MDS patients15 (Extended Data Fig. 3). Lenalidomide decreased CK1α protein levels without altering CSNK1A1 mRNA expression (Fig. 1d, Extended Data Fig. 2c), consistent with a post-translational mechanism of regulation. The lenalidomide-dependent decrease in CK1α protein level was abrogated by treatment with the proteasome inhibitor MG132 and the NEDD8-activating enzyme inhibitor MLN4924, which interferes with the activity of cullin-RING E3 ubiquitin ligases, implicating proteasome- and cullin-dependent degradation of CK1α (Fig. 2a). Homozygous genetic inactivation of CRBN by CRISPR-Cas9 genome editing eliminated lenalidomide-dependent degradation of CK1α, demonstrating CRBN-dependent degradation of CK1α (Fig. 2b, Extended Data Fig. 2d).
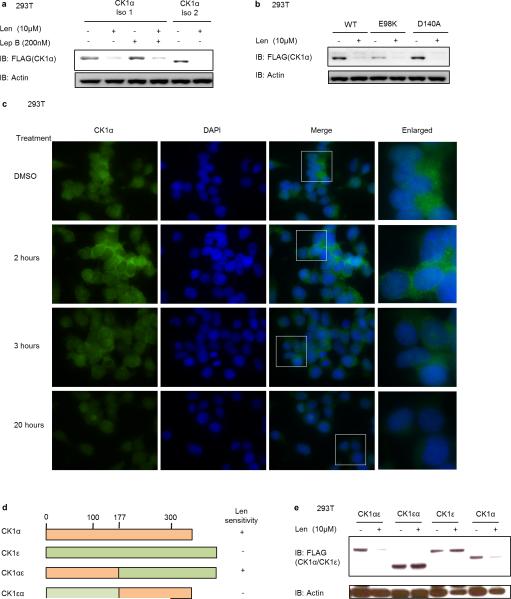
Fig. 2. Lenalidomide induces the ubiquitination of CK1α by CRL4CRBN.
a, CK1α protein levels in KG-1 cells treated with DMSO or lenalidomide alone or in the presence of 10 μM MG132 or 1 μM MLN4924 for 6 hours. b, CK1α protein levels in CRBN knockout 293T cells treated with lenalidomide. c, Immunoprecipitation of FLAG-CRBN in 293T cells treated with DMSO or 1 μM lenalidomide in the presence of 1 μM MG132. d, Ubiquitination of endogenous CK1α in KG-1 cells treated with DMSO or lenalidomide analyzed by TUBE2 pull-down of ubiquitinated proteins followed by staining with a CK1α-specific antibody. Higher molecular weight bands represent ubiquitinated CK1α. e, Ubiquitination of CK1α by CRBN in vitro using lysine-free ubiquitin. Arrowheads indicate ubiquitinated CK1α. Results are representative of two (a, b, d, n=2) or three independent experiments (c, e, n=3). Uncropped blots shown in Supplementary Figure 1.
We next examined whether CK1α binds CRBN and is ubiquitinated by the CRL4CRBN E3 ubiquitin ligase. We observed co-immunoprecipitation of CK1α with endogenous and FLAG-tagged CRBN only in the presence of lenalidomide (Fig. 2c, Extended Data Fig. 2e). Lenalidomide treatment increased the ubiquitination of endogenous CK1α in KG-1 cells (Fig. 2d) and in the presence of CRBN in vitro (Fig. 2e), confirming that CK1α is a direct target of CRL4CRBN. Using a chimeric protein of CK1α and CK1ε, which shares significant homology with CK1α but is not responsive to lenalidomide, we found that the N-terminal half (amino acids 1-177) of CK1α is essential for lenalidomide-induced degradation (Extended Data Fig. 3d,e). Sequence alignment with the previously delineated lenalidomide-responsive degron in IKZF1/3 did not reveal any evident homology, suggesting that CK1α and IKZF1/3 may interact with the CRBN-lenalidomide complex in distinct manners.
Effect of CSNK1A1 expression level
We next explored the biological effects of CK1α depletion. CK1α is a serine/threonine kinase with multiple cellular activities. Most notably, CK1α inhibits p53 through MDM2 and MDMX and negatively regulates Wnt signaling as a component of the β-catenin destruction complex.17-21 In a hematopoietic-specific conditional knockout mouse model, homozygous inactivation of Csnk1a1 induces apoptosis via p53 activation, while heterozygous loss of Csnk1a1 causes β-catenin accumulation and stem cell expansion15. Similarly, cells haploinsufficient for Csnk1a1 preferentially undergo apoptosis in response to the casein kinase 1 inhibitor D447616. Since del(5q) cells express about 50% of normal levels of CSNK1A114, these results led us to hypothesize that del(5q) cells would be more sensitive to the effects of lenalidomide-induced degradation of CK1α as compared to normal cells with two copies of the gene.
To evaluate whether decreased CSNK1A1 expression sensitizes cells to lenalidomide, we transduced primary human CD34+ hematopoietic stem and progenitor cells with GFP-tagged lentiviral vectors expressing CSNK1A1 or control shRNAs. Cells expressing CSNK1A1 shRNAs were depleted in the absence of treatment, demonstrating that knockdown of CSNK1A1 inhibits the growth or survival of hematopoietic cells (Extended Data Fig. 4). Treatment with lenalidomide enhanced the depletion of cells expressing CSNK1A1 shRNAs but had no effect on cells expressing control shRNAs, demonstrating that reduced CSNK1A1 levels sensitize hematopoietic cells to lenalidomide.
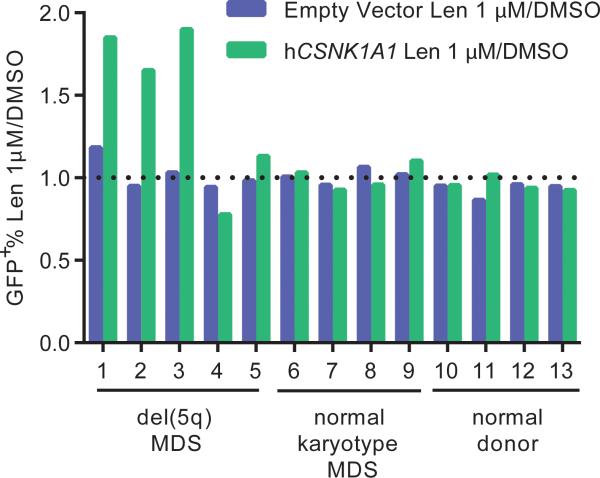
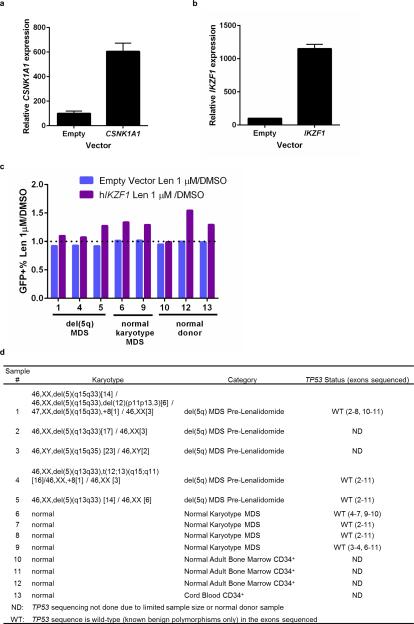
We next evaluated whether overexpression of CSNK1A1 could reduce the lenalidomide sensitivity of del(5q) MDS cells. We obtained bone marrow samples from MDS patients with heterozygous deletions of chromosome 5q, including heterozygous deletion of CSNK1A1, prior to treatment with lenalidomide. We isolated CD34+ cells from these samples and transduced them with a GFP-tagged lentivirus expressing CSNK1A1 cDNA or empty vector (Fig. 3, Extended Data Fig. 5). Overexpression of CSNK1A1 reduced the lenalidomide sensitivity of CD34+ cells from three out of five del(5q) MDS patients, which compares well with the clinically-observed cytogenetic response rate of about 50%.5-7 In contrast, overexpression of CSNK1A1 had no effect on CD34+ cells from patients with normal karyotype MDS or normal donors. Although lenalidomide also induced the degradation of IKZF1 in myeloid cells (Fig. 1a,b), overexpression of IKZF1 had a similar effect on del(5q) MDS, normal karyotype MDS, and normal donor CD34+ cells, suggesting that degradation of IKZF1 does not explain lenalidomide's therapeutic window in del(5q) MDS (Extended Data Fig. 5). These findings demonstrate that increased expression of CSNK1A1 specifically rescues del(5q) cells from lenalidomide treatment.
Fig. 3. Ectopic CSNK1A1 overexpression reduces lenalidomide sensitivity in primary MDS del(5q) cells.
CD34+ cells derived from patient or control bone marrow were transduced with a lentiviral vector overexpressing CSNK1A1 and GFP or an empty control vector and treated with DMSO or 1 μM lenalidomide. Results are reported as a ratio of the percentage of GFP+ cells in the lenalidomide condition to the percentage of GFP+ cells in the DMSO condition after 5 days of treatment. A ratio greater than 1 for the CSNK1A1 vector but not for the empty vector indicates that CSNK1A1 expression reduces lenalidomide sensitivity. Further information about the patients is given in Extended Data Fig. 5d.
Species-specific effects of lenalidomide
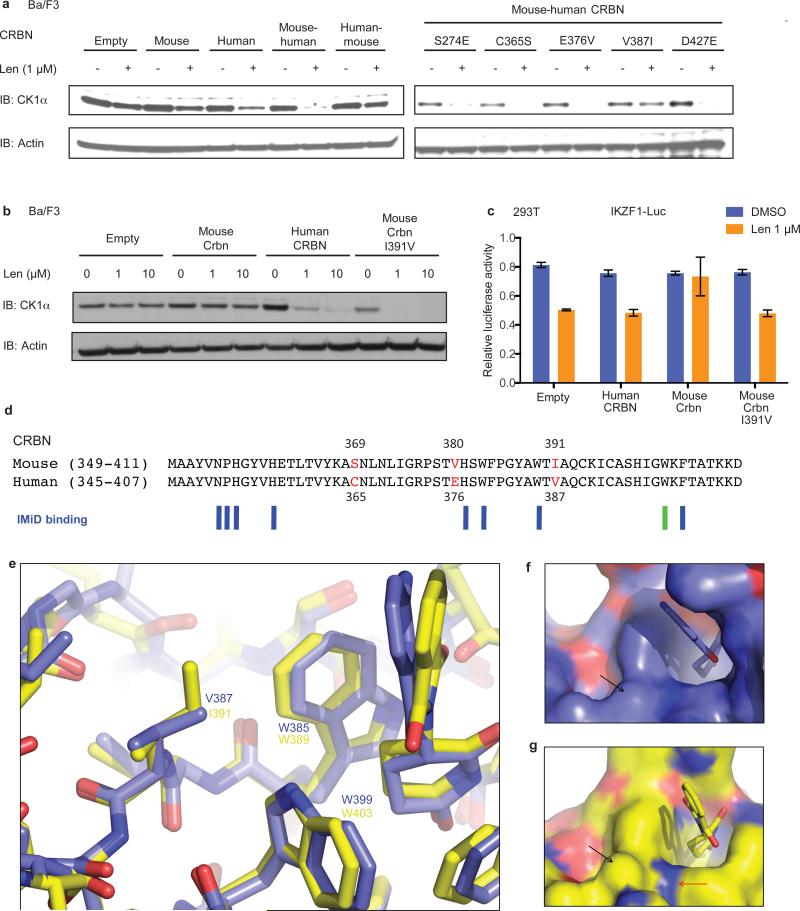
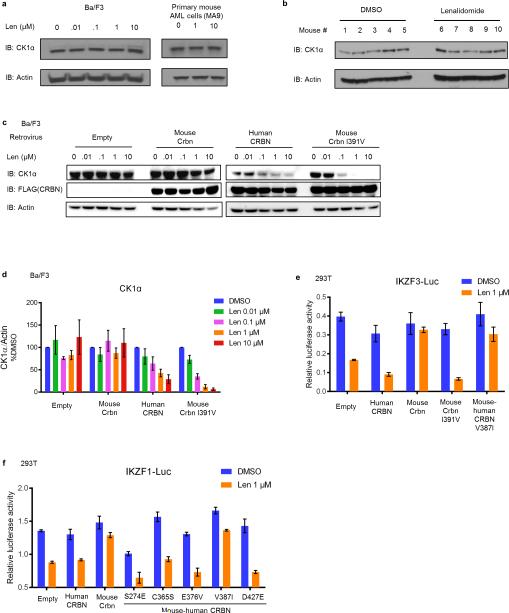
We next sought to use a conditional knockout mouse model to determine whether haploinsufficiency for Csnk1a1 sensitizes cells to lenalidomide treatment.15 In initial experiments, we found that lenalidomide did not decrease CK1α protein levels in mouse Ba/F3 cells, primary murine leukemia cells, or mice treated in vivo (Fig. 4a,b, Extended Data Fig. 6a-d), suggesting that mouse cells are intrinsically resistant to IMiD compounds. Consistent with these findings, mice do not develop the limb malformations observed in human embryos exposed to thalidomide22 and murine multiple myeloma cells do not respond to lenalidomide.23 Since CRBN is the direct protein target of lenalidomide, we examined whether expression of human CRBN could confer lenalidomide sensitivity to mouse cells. Overexpression of human, but not mouse, CRBN in mouse Ba/F3 cells resulted in a lenalidomide-dependent decrease of CK1α protein levels, implying that amino acid differences between mouse CRBN (mCrbn) and human CBRN (hCRBN) are responsible for the species-specific response to lenalidomide (Fig. 4a,b, Extended Data Fig. 6c,d).
Fig. 4. Amino acid changes in CRBN explain species-specific lenalidomide effects.
a, Effect of the expression of human CRBN (h), mouse Crbn (m), chimeras of human and mouse CRBN (m-h and h-m, breakpoint at residue 221 (human) / 225 (mouse)) and variants of the m-h chimera where single amino-acids in the C-terminus were mutated to their corresponding mouse residue on lenalidomide-dependent CK1α degradation in mouse Ba/F3 cells. b, Expression of mouse CrbnI391V restores lenalidomide-dependent CK1α degradation in mouse Ba/F3 cells. See also Extended Data Fig. 6d. c, Effect of hCRBN, mCrbn and mCrbnI391V on lenalidomide sensitivity of an IKZF1-luciferase fusion protein expressed in human 293T cells. Data are mean ± s.e.m. (n=3 biological replicates). d, Alignment of human and mouse CRBN IMiD binding region. Non-conserved amino acids are red. Amino acids involved in IMiD binding24,25 are indicated by blue bars. Mouse W403 is indicated with a green bar. e, Superposition of the IMiD binding domains of human CRBN (blue, PDB accession 4TZ4) and mouse Crbn (yellow, PDB accession 4TZC). Residues are labeled according to human isoform 2 (blue numbers) and mouse isoform 2 (yellow numbers). f, The V387 residue is indicated on the surface of human CRBN with a black arrow. g, The corresponding mouse residue, I391, is indicated on the surface of mouse Crbn with a black arrow. Mouse W403 is indicated by a red arrow. Results are representative of 3 (a, b, c) independent experiments. Uncropped blots shown in Supplementary Figure 1.
To identify the amino acids responsible for this difference, we tested human/mouse CRBN chimeric proteins for their ability to confer lenalidomide-induced CK1α degradation in mouse Ba/F3 cells (Fig. 4a). Lenalidomide sensitivity was determined by the C-terminal half of CRBN, which contains only 5 non-conserved amino acids between human and mouse. When these non-conserved positions in human CRBN were substituted with the corresponding amino acid in mouse Crbn, only one substitution, V387I (human CRBN isoform 2), disrupted the lenalidomide-responsiveness of human CRBN (Fig. 4a). Substitution of the isoleucine at this position in mouse Crbn for the human valine (mCrbnI391V) was sufficient to confer lenalidomide-induced CK1α degradation in mouse cells (Fig. 4b, Extended Data Fig. 6c,d). Similar effects of these two point mutants were observed on lenalidomide-induced degradation of IKZF1 and IKZF3 in human 293T cells (Fig. 4c, Extended Data Fig. 6e,f), suggesting that a single amino acid change in CRBN determines lenalidomide-responsiveness for multiple substrates.
We modeled the effects of this mouse-human amino acid substitution based on recently published crystal structures of the CRBN-DDB1-IMiD drug complex.24,25 V387 of human CRBN (equivalent to I391 of mouse Crbn) is located in the IMiD drug binding region of CRBN, but does not directly interact with lenalidomide (Fig. 4d). To investigate how the substitution of isoleucine for valine in mouse Crbn confers lenalidomide-responsiveness, we superimposed the structures for the mouse and human IMiD-binding regions bound to lenalidomide as solved in Chamberlain et al. (2014).25 No backbone changes are present at the site of the valine-isoleucine species differences (Fig. 4e), but the isoleucine residue is well-defined in the electron density with the long arm of the side chain oriented towards the indole NH moiety of W403 in the mouse structure (Extended Data Fig. 7). The increase in steric bulk of the isoleucine side chain, relative to valine, results in a bulge in the solvent accessible surface of the mouse protein adjacent to both W403 and lenalidomide (Fig. 4f, g). It has been proposed that IMiD binding produces a hotspot for substrate interactions by placement of the hydrophobic phthalimide or isoindolinone ring in an environment of potential hydrogen bond donors and acceptors from the surface of CRBN25. In this case, the larger side chain of the isoleucine residue found in rodents may sterically clash with substrate proteins such as IKZF1 and CK1α, blocking access to key hydrogen bonds from CRBN, such as from the indole NH from tryptophan 403 (mouse numbering). Steric clashes and occlusion of key bonds with substrate proteins thereby provides a potential explanation of why IMiD compounds bind mouse Crbn1 but do not promote degradation of IKZF1 and CK1α.
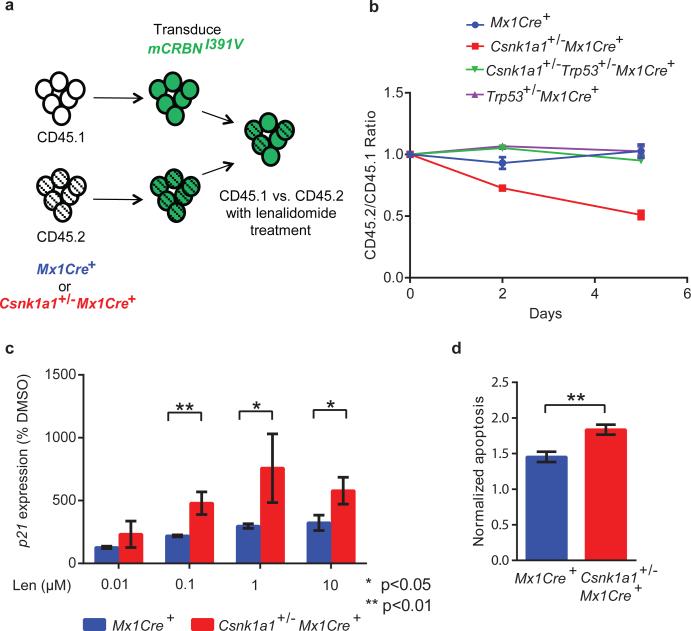
Having determined the mechanism of lenalidomide resistance in mouse cells, we expressed the mCrbnI391V cDNA in hematopoietic cells from Csnk1a1 conditional knockout mice to determine the effects of Csnk1a1 haploinsufficiency on lenalidomide sensitivity. We isolated CD45.2+ c-Kit+ hematopoietic stem and progenitor cells from Csnk1a1+/−Mx1Cre+ and Mx1Cre+ control littermates treated with poly (I:C) to induce gene excision in hematopoietic cells. We transduced these cells with a retroviral vector expressing mCrbnI391V, and cultured them in competition with similarly transduced isogenic CD45.1 c-Kit+ cells in the presence or absence of lenalidomide (Fig. 5a). Lenalidomide had no effect on control cells, but Csnk1a1+/−Mx1Cre+ cells were significantly depleted in the presence of lenalidomide (Fig. 5b). The enhanced sensitivity of Csnk1a1+/−Mx1Cre+ cells to lenalidomide was associated with induction of the p53 target gene p21 (Fig. 5c) and increased levels of apoptosis (Fig. 5d), and was rescued by heterozygous deletion of Trp53 (Fig. 5b), demonstrating a critical downstream role for the p53 pathway. These results are consistent with the clinical observation that TP53 mutations confer lenalidomide resistance in MDS with del(5q).26
Fig. 5. Effects of lenalidomide treatment on Csnk1a1+/− mouse hematopoietic cells.
a, Csnk1a1+/−Mx1Cre+ or Mx1Cre+ c-Kit+ hematopoietic stem and progenitor cells (CD45.2) and competitor cells (CD45.1) were transduced with mCrbnI391V, mixed in equal ratios, and treated with lenalidomide or DMSO. The relative percentage of CD45.1+ and CD45.2+ cells was followed by flow cytometry over 5 days. b, Effects of 0.1 μM lenalidomide on the chimerism of Csnk1a1+/− Mx1Cre+, Csnk1a1+/− Trp53+/− Mx1Cre+, Trp53+/− Mx1Cre+, or Mx1Cre+ cells (CD45.2) transduced with mCrbnI391V in comparison to CD45.1 competitor cells. Data are shown as mean ± s.e.m, n=3 biological replicates. c, Quantitative RT-PCR analysis of p21 expression in Csnk1a1+/− or control cells transduced with mCrbnI391V and treated with DMSO or lenalidomide. Data are normalized to DMSO and shown as mean ± s.d, n=3 biological replicates. d, Ratio of CD45.2+ cells and CD45.1+ cells in late apoptosis (Annexin V+ DAPI+) after transduction with mCrbnI391V and four day treatment with 0.1 μM lenalidomide. CD45.2+ cells are either Csnk1a1+/− Mx1Cre+ or Mx1Cre+. Data are normalized to DMSO treatment. Data are mean ± s.e.m, n=4 biological replicates. Results for b, c, and d are representative of three independent experiments with hCRBN or mCrbnI391V. P-values are from an unpaired two-sided t-test.
Differential substrate specificity
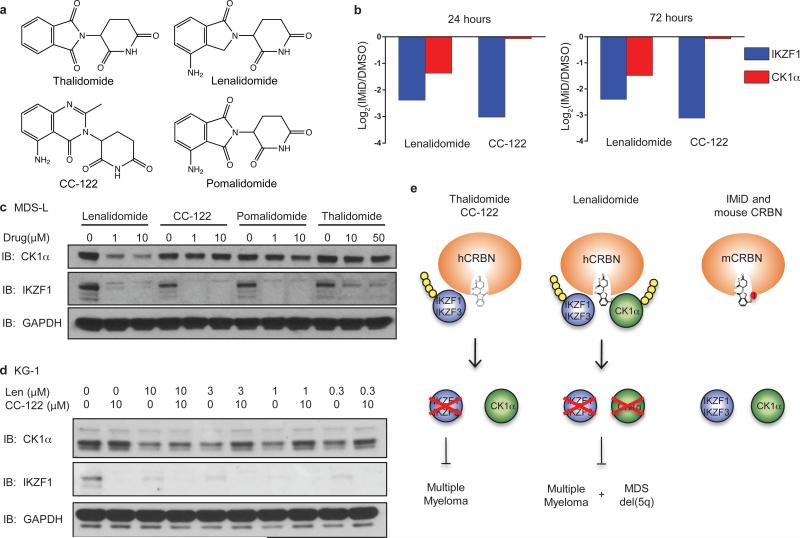
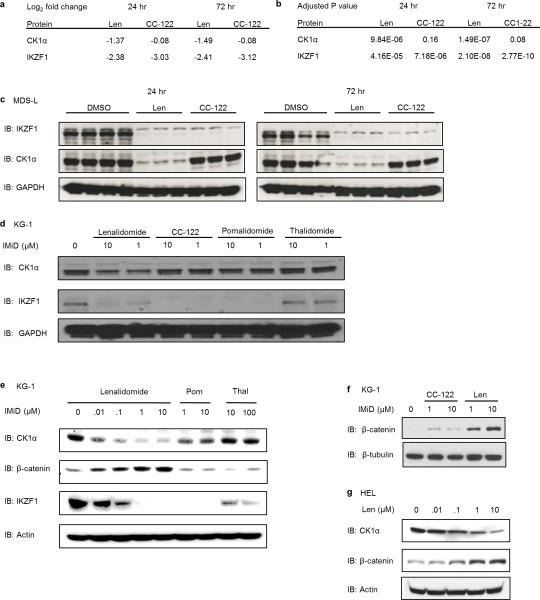
Thalidomide, lenalidomide, and pomalidomide target IKZF1 and IKZF3 for ubiquitination and degradation and are active in multiple myeloma, but only lenalidomide has been shown to be clinically effective in del(5q) MDS.2,5-7 We therefore asked whether different thalidomide analogues induce degradation of the same substrates. We used tandem mass tag (TMT) quantitative proteomics27 in the MDS-L cell line to compare the activities of lenalidomide and CC-122, a novel CRBN-binding agent that shares the glutarimide ring and has recently entered clinical trials (Fig. 6a). As expected, treatment with lenalidomide significantly decreased protein levels of both IKZF1 and CK1α. In striking contrast, treatment with 1 μM CC-122 caused an even greater decrease in IKZF1 (p=2.77 × 10−10, 72 hours) than 10 μM lenalidomide (p=2.10 × 10−8, 72 hours), but had no effect on CK1α protein levels (p>0.05)(Fig. 6b, Extended Data Fig. 8a,b).
Fig. 6. Substrate specificity of thalidomide analogues.
a, Structures of thalidomide, lenalidomide, pomalidomide and CC-122. b, Protein levels of IKZF1 and CK1α assessed by tandem mass tag quantitative proteomics in MDS-L cells treated with DMSO, 10 μM lenalidomide, or 1 μM CC-122 for 24 hours (left panel) or 72 hours (right panel). n=3 (drug treatment) or n=4 (DMSO). c, Western blot analysis of CK1α and IKZF1 protein levels in MDS-L cells treated with DMSO or different concentrations of thalidomide, lenalidomide, pomalidomide, or CC-122. Results are representative of two independent experiments (n=2). d, KG-1 cells were treated with DMSO or lenalidomide in the absence or presence of different concentrations of CC-122. Results are representative of five independent experiments in various cell lines. e, Schematic presentation of the interaction of different thalidomide analogues with CRBN, substrates, and therapeutic indications.
We confirmed the TMT mass spectrometry findings by western blot for IKZF1 and CK1α in MDS-L and KG-1 cells (Fig. 6c, Extended Data Fig. 8c,d). While all compounds induced degradation of IKZF1, thalidomide and CC-122 did not affect CK1α protein levels, even at high concentrations, and pomalidomide had only weak effects on CK1α protein levels. Although CC-122 has a greater potency than lenalidomide for degradation of IKZF1 and IKZF3, it was ineffective in decreasing CK1α protein levels compared to lenalidomide, suggesting that subtle chemical modifications can affect substrate preference (Fig. 6a-c). Furthermore, treatment with excess CC-122 abrogated the lenalidomide-induced degradation of CK1α, demonstrating that lenalidomide and CC-122 compete for the same glutarimide binding site on CRBN (Fig. 6d). Consistent with the role of CK1α as a negative regulator of Wnt signaling, we observed increased levels of β-catenin after treatment with lenalidomide but not CC-122 or thalidomide (Extended Data Fig. 8e-g). These experiments demonstrate that despite structural similarity, the substrate specificities of thalidomide analogues differ. Notably, only lenalidomide has a strong effect on CK1α, suggesting that it may indeed be most appropriate modulator of CRL4CRBN for the treatment of del(5q) MDS (Fig. 6e).
Intriguingly, lenalidomide, but not thalidomide or pomalidomide, has been reported to induce the formation of two β-strands composed of CRBN residues 346-363.25 Although conformational differences are difficult to interpret in the absence of a substrate-bound structure, the formation of these β-strands is expected to make significant changes in the surface of CRBN near the IMiD binding site25 and thus it may contribute to the differential recruitment of IKZF1 and CK1α. The interaction of specific thalidomide analogues with particular substrates may therefore be governed by unique structural determinants, revealing the biological and clinical potential for members of this class of drugs to induce degradation of distinct sets of proteins.
Discussion
We demonstrate that lenalidomide targets CK1α for degradation, and that heterozygous deletion of CSNK1A1 in del(5q) MDS provides a therapeutic window for selective targeting of the malignant cells by lenalidomide. The concept that genes within heterozygous deletions could cause vulnerabilities in cancer cells was first proposed 20 years ago28 and has been more recently demonstrated as CYCLOPS genes (cancer vulnerabilities unveiled by genomic loss) in cell lines.29 Our data demonstrate that in del(5q) MDS, lenalidomide-induced degradation of CK1α below haploinsufficient levels induces p53 activity and growth inhibition, as CK1α is a negative regulator of p53. Deletion of contiguous genes on chromosome 5q, such as RPS14, may further sensitize del(5q) cells to p53 activation.9,30,31 This mechanism of activity is consistent with the acquisition of TP53 mutations in del(5q) MDS patients who develop resistance to lenalidomide. Degradation of CK1α may also contribute to other clinical effects of lenalidomide such as activity in the activated B-cell (ABC) subtype of diffuse large B-cell lymphoma32 and lenalidomide-induced myelosuppression. Further investigation is required to determine the complete biological effects from degradation of each substrate.
Lenalidomide, like thalidomide and pomalidomide, binds CRBN and induces degradation of specific substrates. We found that a single amino acid difference between mouse and human CRBN renders mouse cells insensitive to IMiD compounds. This discovery enabled us to demonstrate, using a genetically engineered mouse model, that Csnk1a1 haploinsufficiency sensitizes cells to lenalidomide. Non-conserved amino acid changes in CRBN may also explain why thalidomide does not cause teratogenicity in mice and was approved for use in pregnant women, leading to the birth of more than 10,000 newborns with limb malformations and other disabilities.
Thalidomide, lenalidomide, and pomalidomide all induce CRL4CRBN-mediated degradation of IKZF1 and IKZF3, but the subtle differences in chemical structure between these molecules cause dramatic changes in potency. We now find that thalidomide and a novel compound, CC-122, induce the degradation of IKZF1 but not CK1α. CC-122 may have a greater therapeutic window for the treatment of B cell malignancies and other diseases that depend on IKZF1 and IKZF3, but would not be predicted to have activity in del(5q) MDS.
CC-122, like thalidomide and its analogues, has a glutarimide ring that anchors the molecule in CRBN, and structural variation in the remainder of the molecule is thought to determine substrate specificity.24,25 These findings provide evidence that thalidomide-related molecules have distinct biological activities, mediated by degradation of distinct sets of substrates, and that these compounds will be the first in a larger class of drugs with therapeutic utility through the targeting of specific proteins for degradation.
Methods
Reagents
Lenalidomide (Toronto Research Chemicals, Selleck Chemicals, and Celgene), Thalidomide (Millipore and Celgene), Pomalidomide (Selleck Chemicals and Celgene), MG-132 (Selleck Chemicals), CC-122 (Celgene), PR619 (Lifesensors), MLN4924 (Active Biochem), and Leptomycin B (Santa Cruz) were dissolved in DMSO at 10 to 100 mM and stored at −20°C for up to 6 months. For cell culture experiments drugs were diluted at least by 1:1000 so that the final DMSO concentration was 0.1% or lower.
Cell lines
KG-1, Ba/F3, K562, MM1S, Jurkat, HEL, and 293T cells were obtained from American Type Culture Collection (ATCC) and their identity was not further authenticated. MDS-L cells were provided by Kaoru Tohyama, Kawasaki Medical School (Japan). Cells were cultured in RPMI 1640 (Mediatech) or DMEM (Mediatech) supplemented with 10-20% heat-inactivated fetal bovine serum (FBS)(Omega Scientific) and 1% penicillin, streptomycin, and L-glutamine (Mediatech). Cells were grown at 37°C in a humidified incubator under 5% CO2. Ba/F3 cells were cultured in the presence of 10 ng/ml mouse IL-3 (Miltenyi) and MDS-L cells were cultured with 10 ng/ml human GM-CSF. 293T cells were transfected using TransIT-LT1 (Mirius Bio) according to the manufacturer's protocol. Cell lines were intermittently tested for mycoplasma.
Cell culture and treatment for K-ε-GG and proteome profiling
KG-1 cells were cultured for 2 weeks (~6 cell doublings) in RPMI depleted of L-arginine and L-lysine (Caisson Labs Inc.) and supplemented with 10% dialyzed FBS (Sigma) and L-arginine (Arg0) and L-lysine (Lys0) (light), 13C614N4-L-arginine (Arg6) and 4,4,5,5-D4-l-lysine (Lys4) (medium) or 13C615N4-l-arginine (Arg10) and 13C615N2-l-Lysine (Lys8) (heavy) to generate light-, medium- and heavy-labeled cells. Media was exchanged every 3rd day. On day 14 cells were treated with 1 μM lenalidomide, 10 μM lenalidomide or DMSO for 4 hours for ubiquitination profiling and 24 hours for protein level assessment. Experiments were performed in two biological replicates with flipped SILAC labeling: Replicate 1: DMSO/light, lenalidomide 1 μM/medium; lenalidomide 10 μM/ heavy; Replicate 2: lenalidomide 1 μM/light; lenalidomide 10 μM / medium; DMSO/heavy.
SILAC based K-ε-GG and proteome profiling of KG-1 cells
Cell lysis and trypsin digestion, basic pH reversed phase fractionation, K-ε-GG enrichment, and LC-MS/MS analysis for KG-1 cells were performed as recently described2 with minor changes. Cell pellets used for K-ε-GG profiling were lysed in 8 M urea, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2 μg/ml aprotinin (Sigma-Aldrich), 10 μg/ml leupeptin (Roche Applied Science), 1 mM phenylmethylsulfonyl fluoride (PMSF), 50 μM PR-619, and 1 mM chloroacetamide at 4°C. Pellets used for proteome profiling were lysed in 8 M urea, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 2 μg/ml aprotinin (Sigma-Aldrich), 10 μg/ml leupeptin (Roche Applied Science), 1 mM phenylmethylsulfonyl fluoride (PMSF), at 4 °C. For this work, 10 mg of protein was input per SILAC state for the ubiquitin workflow. For proteome profiling, 1.5 mg of protein was input per SILAC state. Proteins were reduced with 5 mM dithiothreitol for 45 minutes at room temperature and subsequently carbamidomethylated with 10 mM iodoacetamide for 30 min at RT in the dark. Samples were diluted to 2 M urea with 50 mM Tris-HCl, pH 7.5, and digested with sequencing grade trypsin (Promega) at 25 °C overnight using an enzyme-to-substrate ratio of 1:50. Digested samples were acidified to 1% formic acid (Sigma-Aldrich). Tryptic peptides were centrifuged for 5 min at 3000 × g to remove precipitate. Peptides were desalted exactly as previously described.2
Samples were fractionated by basic pH reversed phase (bRP) fractionation using an Agilent 1100 Series HPLC and Zorbax 300 Å Extend-C18 columns as previously described.13,33 A 9.4 mm × 250 mm column (Agilent, 5 μm bead size) was used to fractionate samples intended for K-ε-GG enrichment, whereas a 4.6 mm × 250 mm column (Agilent, 3.5 μm bead size) was used to fractionate samples intended for proteome analysis.33 For K-ε-GG samples, approximately 15 mg of peptide sample was resuspended in 1.8 ml of basic RP solvent A (2% MeCN, 5 mM ammonium formate, pH 10), separated into 2 HPLC vials and injected with Solvent A at flow rate of 3 ml/min. A 64-min method was used for fractionation exactly as previously described.2,13 A total of 96 2 ml fractions were collected every 0.66 min at a flow rate of 3 ml/min. After separation, bRP fractions were pooled in a serpentine, noncontiguous manner to generate 8 final fractions (final fraction 1 = 1, 9, 17, 25, 33, 41, 49, 57, 65; final fraction 2 = 2, 10, 18, 26, 34, 42, 50, 58, 66; ...). Since 10 mg of protein per SILAC state was used for K-ε-GG samples, and the maximum loading capacity on the 9.4 mm × 250 mm column bRP column is 15 mg, two rounds of fractionation were completed per replicate sample.
Proteome samples were brought up in 0.9 ml of basic RP solvent A and injected with solvent A at a flow rate of 1 ml/min. Peptides were separated using the bRP method previously described for the 4.6 mm × 250 mm column.33 Approximately 5% of each proteome fraction was taken and pooled to generate 24 final fractions (final fraction 1 = 1, 25, 49; final fraction 2 = 2, 26, 50; ...) plus a fraction “A” that contains early eluting peptides. All bRP pooled fractions were dried using a SpeedVac concentrator. K-ε-GG enrichment was completed using the anti-K-ε-GG antibody obtained from the PTMScan® ubiquitin remnant motif (K-ε-GG) kit (Cell Signaling Technology) as previously described.2,13 Briefly, bRP fractions we reconstituted in 1.5 ml of immunoaffinity purification buffer and each fraction was incubated with 31 μg of cross-linked anti-K-ε-GG antibody for 1 hour at 4 °C with end-over-end rotation. Following incubation, samples were spun down and the supernatant was removed. Antibody-bound beads were washed 4x with 1.5 ml of ice cold PBS. Peptides were eluted with 2 × 50 μl of 0.15% TFA. Eluted peptides were desalted using C18 StageTips.
K-ε-GG and proteome fractions were reconstituted in 9 μl and 20 μl of 3% MeCN/1% FA, respectively, and analyzed using a Q Exactive mass spectrometer (Thermo Fisher Scientific) coupled on-line to a Proxeon Easy-nLC 1000 system. For analysis of each fraction, 4 μl and 1 μl of K-ε-GG and global proteome samples was injected, respectively. Samples were injected onto a microcapillary column (360 μm OD × 75 μm ID) packed with 24 cm of ReproSil-Pul C18-AQ 1.9 μm beads (Dr. Maisch GmbH) that was heated to 50 °C and equipped with an integrated electrospray emitter tip (10 μm). For online LC separation, solvent A was 0.1 % FA/3 % MeCN and solvent B was 90% MeCN/0.1% FA. Peptides were eluted into the mass spectrometer using the LC-MS method previously described.2 The Q Exactive instrument was operated in the data-dependent mode acquiring 12 HCD MS/MS scans (R=17,500) after each MS1 scan (R=70,000) using an MS1 ion target of 3 × 106 ions and an MS2 target of 5 × 104 ions. The maximum ion time for the MS/MS scans was set to 120 ms, the collision energy was set to 25, the dynamic exclusion time was set to 20 s, and the peptide match setting was set to on.
The MaxQuant software version 1.3.0.5 was used to analyze MS data. Data was searched against the human Uniprot database as well as a database provided by MaxQuant containing common laboratory contaminants. The search parameters were as follows: enzyme specificity was set to trypsin, maximum number of mixed cleavages set to 2, precursor mass tolerance was set to 20 ppm for the first search, and set to 6 ppm for the main search. Oxidized methionine and N-terminal protein acetylation were searched as variable modifications and carbamidomethylation of cysteine was searched as a fixed modification. Data files from K-ε-GG enriched samples were also searched with Gly-Gly addition to lysine as a variable modification. The minimum peptide length was set to 6, and false discovery rate for peptide, protein, and site identification was set to 1%. Reverse and contaminant hits were removed from datasets. Normalized ratios were utilized for quantification. For proteome data, proteins identified by 2 or more razor/unique peptides and quantified by 2 or more ratio counts in both biological replicates were considered for the final dataset. For the K-ε-GG data, K-ε-GG sites were considered if they were quantified in both biological replicates.
For data analysis, normalized SILAC ratios for the 2 biological replicates were filtered to retain only those deemed reproducible. Reproducibility was based on replicates being confined within the 95% limits of agreement of a Bland-Altman plot34. In the Bland-Altman plot, differences of the replicates are plotted against the average values and the limits of agreement correspond to the prediction confidence interval for a regression line with unit slope. Reproducible replicates were then subjected to a moderated t-test to assess statistical significance35. This statistic is similar to the ordinary t-statistic, with the exception that the standard errors are calculated using an empirical Bayes method utilizing information across all proteins, thereby making inference about each individual protein more robust. The nominal p-values arising from the moderated t-statistic are corrected for multiple testing by controlling the false discovery rate (FDR), as proposed by Benjamini and Hochberg36. Proteins with an FDR adjusted p-value of less than 0.05 were deemed to be reproducibly regulated. Figures containing scatter plots of SILAC data show all points regardless of the reproducibility measure. Statistical significance was assessed using only reproducible data points.
The original mass spectra may be downloaded from MassIVE (http://massive.ucsd.edu) using the identifier: MSV000079014. The data are accessible at ftp://MSV000079014:a@massive.ucsd.edu.
Plasmids and Virus constructs
The following cDNAs were cloned in the pRSF91 retrovirus backbone (kind gift of Christopher Baum, Hanover Medical School) or pEF1a-IRES-GFP lentiviral backbone: CSNK1A1 Isoform 2 (ccsbBroadEN_06055), CSNK1A1 Isoform 1 (kind gift from William G. Kaelin), CSNK1E (ccsbBroadEN_00379), mouse Crbn Isoform 2 (Thermo Scientific), and human CRBN Isoform 2 (ccsbBroadEn_08244). Human IKZF1 isoform 1 was synthesized using gBlocks (IDT) with internal BstXI and BsrGI sites removed using synonymous substitutions. For certain experiments GFP was replaced by GFP-T2A-Puro to allow for drug selection of positively transduced cells. Chimeric cDNAs and point mutations were cloned with overlapping PCR primers. Point mutations in mouse-human CRBN chimeric proteins are annotated according to their position in human CRBN isoform 2. Lentivirus was concentrated by ultracentrifugation for transduction of primary cells.
Lentiviral vectors (TRC005 backbone) expressing shRNAs targeting luciferase (TRCN0000072254: ATGTTTACTACACTCGGATAT) and CSNK1A1 (#1: TRCN0000342505, CATCTATTTGGCGATCAACAT; #2: TRCN0000342507, GCAGAATTTGCGATGTACTTA) were obtained from The RNAi Consortium (TRC) of the Broad Institute. For certain experiments, the puromycin resistance gene was replaced by GFP.
The luciferase reporter plasmid pCMV-IRES-RenillaLUC-IRES-Gateway-FireflyLUC3 was a kind gift from William G. Kaelin (Dana-Farber Cancer Institute). Cloning of cDNAs was performed using Gateway LR reaction (Invitrogen).
CRISPR mediated genetic deletion was performed with the pSgRNA-CAS9-T2A-Puro plasmid37 using CRBN exon 1-specific guide RNAs (CRBN targeting sequence: #1 TCCTGCTGATCTCCTTCGC, #2 AACCACCTGCCGCTCCTGCC). 1 × 105 293T cells were transfected in a 12-well with 500 ng of each plasmid using TransLTI (Mirius). After 24 hours, transfected cells were selected with 2 μg/ml puromycin for 4 days. Then 293T cells were diluted to single cell and plated in 96-well. Colonies were tested by western blot and Sanger sequencing of the endogenous CRBN exon 1 locus for inactivating biallelic out-of-frame mutations.
Western blot and antibodies
Protein lysates were run on Tris-HCl, 1mm Criterion Precast gels (Bio-Rad) or NuPAGE Bis-Tris gels (Novex) gels at a constant voltage. Proteins were transferred onto Immobilon-P transfer membranes (Millipore) at a constant amperage. Before staining with primary antibodies, blots were blocked in 5% non-fat dry milk (Santa Cruz) or 5% BSA in TBS-T 0.1% for 30 minutes.
For protein detection primary antibodies detecting CK1α (C-19, Santa Cruz or Abcam ab108296), β-catenin (Cell Signaling #9587 and #8480), HA (HRP-conjugate, Miltenyi, GG8-1F3.3), FLAG (M2, HRP-conjugate Sigma Aldrich), ubiquitin conjugates (FK2, HRP-conjugate Enzo Life Sciences), Actin (HRP-conjugate, Abcam), β-tubulin (Cell Signaling #2146) and GAPDH (Santa Cruz sc-47724) were used. Secondary antibodies were HRP conjugated Bovine anti-Goat (Jackson ImmunoResearch) and HRP conjugated donkey anti-rabbit (GE Healthcare). SuperSignal (Thermo Scientific) chemi-luminescent substrate was used for detection. For re-probing, blots were stripped in Restore Western Blot Stripping Buffer (Thermo Scientific), activated in methanol, and re-blocked.
Flow cytometry
Flow cytometry was performed on a FACS Canto II (BD Bioscience) using the PE and FITC channels for the detection of dTomato and GFP, respectively. DAPI staining was performed to exclude dead cells. A High-Throughput Sampler (BD) was used for some experiments.
Quantitative RT-PCR
Gene expression was measured by reverse transcription quantitative PCR (RQ-PCR). For RNA isolation and reverse transcription a cDNA Synthesis Kit for MultiMacs (Miltenyi) was used according to the manufacturer's protocol. The following primer-probe sets from Life Technologies were used with TaqMan Gene Expression Master Mix (Life Technologies): human GAPDH (402869), human CSNK1A1 (Hs00793391_m1), human IKZF1 (Hs00958474_m1), mouse GAPDH (Mm99999915_g1), mouse p21 (Mm04205640_g1). Analysis was performed on a 7900HT Fast Real-Time PCR System (Applied Biosystems) in a 384-well plate. Relative expression levels were calculated using the ΔΔCT method.
Immunoblot analysis of patient samples
Frozen viable patient samples from the CC-5013-AML-001 trial (clinicaltrials.gov identifier NCT01358734) were thawed at 37°C, washed with PBS, and cell pellets were frozen at −80°C. Cells were lysed in RIPA buffer containing HALT Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific), quantified with a BCA Protein Assay Kit and 3-5 μg of protein was run on a Bis-Tris gel (Novex). Membranes were stained with anti-CK1α and anti-GAPDH antibodies and detected with chemiluminescence. Informed consent was obtained from all subjects in this trial, including consent to use the collected materials to study the mechanism of lenalidomide and its effects on specific proteins. Samples were collected according to IRB-approved protocols at the 30 sites at which this study was conducted. Due to limited availability of patient samples, this experiment could be performed only once.
Immunoprecipitation of CRBN
For immunoprecipitation of FLAG-CRBN, 3 × 106 293T cells were plated in a 10 cm dish and transfected with 10 μg FLAG-hCRBN or empty vector. Cells were treated with DMSO or 1 μM lenalidomide in the presence of 10 μM MG132 for 3 hours. Cells were lysed in Pierce IP Lysis Buffer and lysates were cleared by centrifugation. FLAG-CRBN was immunoprecipitated overnight using anti-FLAG M2 Affinity Gel (Sigma-Aldrich) in the presence of 10 μM MG132 and DMSO or 1 μM lenalidomide. The beads were washed 3 times with IP lysis buffer (Pierce) and protein was eluted from the affinity gel with 250 μg/ml FLAG peptide (Sigma) after incubation for 30 min at 4°C. Protein lysates were then analyzed as described above.
For immunoprecipitation of endogenous CRBN 5 × 106 293T cells were treated with DMSO or 10 μM lenalidomide and 10 μM MG132 for 4 hours. Protein lysates were incubated overnight at 4 °C with 1 μg of a polyclonal mouse anti-CRBN antibody (abcam) in the presence of lenalidomide or DMSO and MG132, Protein G sepharose beads were added for one hour. The beads were washed once with IP lysis buffer (Pierce) and protein was eluted from the beads by incubation with LDS loading buffer (Life Technologies) at 70 °C for 10 minutes.
In vivo ubiquitination
For assessment of endogenous ubiquitination of CK1α 2 × 107 KG-1 cells were treated with DMSO, 1 or 10 μM lenalidomide for 4 hours and then lysed in IP lysis buffer containing 10 mM NEM and 10 μM MG132. Ubiquitinated proteins were pulled down by Ubiquilin 1 Tandem UBA (TUBE2) Agarose (Boston Biochem) for 4 hours at 4 °C and washed 3x with IP lysis buffer. Protein was eluted by incubation with Laemmli buffer (Biorad) at 95 °C for 5 minutes, separated by SDS-PAGE, transferred to PVDF membrane and probed with anti-CK1α.
In vitro ubiquitination
293T cells were transfected with either HA-CK1α or FLAG-CRBN expressing vectors. After 48 hours, cells were lysed in Pierce IP lysis buffer (Thermo Scientific) and immunoprecipitated overnight with FLAG-Sepharose beads (Anti-FLAG M2 Affinity Gel, Sigma) or HA-Sepharose beads (EZView Red anti-HA affinity gel, Sigma). The beads were washed 3x in IP lysis buffer and 2x in E3 Ligase Reaction buffer (Boston Biochem) and eluted with 250 μg/ml FLAG peptide (Sigma) or 100 μg/ml HA peptide for 30 min at 4 °C. The eluates were mixed in a 1:1 ratio and added to a ubiquitination reaction mixture containing 200 nM E1 (UBE1), 2 μM UbcH5a, 1 μM UbcH5c, 1 μg/μL K0 ubiquitin, 1 μM ubiquitin aldehyde, 1x Mg-ATP, 1x E3 Ligase Reaction Buffer (all Boston Biochem), 10 μM MG132, 100 nM MG101 and 1 μM lenalidomide, 10 μM lenalidomide, or DMSO (1:1000) as appropriate in a total volume of 25 μl. Negative controls did not include E1 and E2 enzymes. After a 90 minute incubation at 30 °C, the reaction was denatured by adding 5x SDS containing loading buffer (Boston Biochem), boiled at 95 °C for 5 minutes, separated by SDS-PAGE and transferred to a PVDF membrane in order to detect HA-CK1α and its ubiquitinated forms with a CK1α-specific antibody. The membrane was then stripped and re-probed with anti-FLAG antibody.
Immunofluorescence
50,000 293T cells were grown on Lab-Tek 8 well chamber slides (Nunc) for 24 hours and then treated with DMSO or 10μM lenalidomide for various durations. At the conclusion of treatment, the media was decanted and the wells were washed 1x with PBS. Cells were fixed in 4% formaldehyde in PBS for 15 minutes, washed 3x 5 min in PBS and blocked for 1 hour at room temperature in PBS with 0.3% Tween-20 and 5% BSA. Primary antibody was anti-CK1α (C-19, Santa Cruz), which was diluted 1:100 in PBS with 0.3% Tween-20 and 1% BSA (antibody dilution buffer) and incubated for 2 hours at room temperature. After 3x 5 min washes in PBS, Alexa Fluor 488 donkey anti-Goat (Life Technologies) was added at 1:200 in antibody dilution buffer and incubated for 1 hour at room temperature. After 3x 5 min washes in PBS slides were coverslipped with Vectashield mounting media with DAPI (Vector Laboratories). Slides were analyzed by fluorescence microscopy at 100x using a Nikon Eclipse 90i and NIS Elements. Channels were merged using ImageJ.
Purification, culture, and lentiviral infection of human CD34+ cells for shRNA experiments
Research cord blood units were obtained from The New York Blood Center according to an Institutional Review Board-approved protocol. Cord blood CD34+ hematopoietic cells were isolated from Ficoll purified PBMCs with an Indirect CD34 MicroBead kit (Miltenyi) and an Auto MACS Pro (Miltenyi) according to the manufacturer's protocol. Cells were cultured in serum free media (SFEM, StemSpan) containing 50 ng/ml recombinant human TPO (Miltenyi), 40 ng/ml human FLT3 ligand (Miltenyi), 25 ng/ml recombinant human SCF (Miltenyi), and 10 ng/ml IL-3 (Miltenyi). For shRNA experiments, CD34+ cells were transduced with a VSV-G pseudotyped TRC pLKO.005 lentiviral vector expressing GFP instead of the puromycin resistance gene. Infection was performed after 24 hours in culture in a 96-well using spinfection in the presence of 2 μg/ml polybrene (hexadimethrine bromide, Sigma). 48 hours after transduction the number of transduced cells was analyzed by flow cytometry and was used as baseline. Then cells were cultured in 1 μM lenalidomide or DMSO and the relative number of infected cells was assessed by flow cytometry for 3 weeks.
Purification, culture, and lentiviral infection of patient samples
Viably frozen bone marrow mononuclear cells were obtained from healthy donors or patients with del(5q) MDS according to IRB approved protocols at the University of Pennsylvania and Roswell Park Cancer Institute. Informed consent was obtained from all subjects. Samples were thawed and CD34+ hematopoietic cells were isolated 20-24 hours later using an Indirect CD34 MicroBead kit (Miltenyi) and an Auto MACS Pro (Miltenyi). Cells were grown in serum free media (SFEM, StemSpan) supplemented with 25 ng/ml SCF, 40 ng/ml FLT3 ligand, 50 ng/ml thrombopoietin, 40 μg/ml lipids, 100 U/ml Pen/Strep and 2 mM glutamine. 6-8 hours after CD34+ isolation, cells were transduced with concentrated VSV-G pseudotyped pEF1a-GFP-IRES-hCSNK1A1, pEF1a-GFP-IRES-hIKZF1 or empty vector control virus via spinfection in the presence of 4 μg/ml polybrene (Sigma, diluted to 2 μg/ml after spinfection). After 3 days, the initial percentage of transduced cells was determined by flow cytometry and remaining cells were split to treatment with either DMSO or 1 μM lenalidomide. The relative abundance of transduced cells in each condition was assessed after 5 days by flow cytometry. Control cord-blood CD34+ cells were isolated as above. Adult bone marrow CD34+ cells were purchased as single-donor lots from AllCells (Alameda, CA). The number of replicates for each patient sample, vector, and treatment was limited by the number of cells available and was as follows: one for samples 1-5, 8, 9 and 12; two for samples 6, 7 and 10; three for sample 11; four for sample 13. Samples were combined from three experiments.
For qPCR validation of CSNK1A1 or IKZF1 mRNA expression, cord blood CD34+ cells were transduced with lentivirus expressing GFP and the cDNA of interest or empty vector. After 3 days, transduced GFP+ cells were FACS sorted and RNA extraction and qPCR was performed as above.
TP53 Sequencing of patient samples
Genomic DNA was extracted from the CD34− fraction of the patient bone marrow samples using a DNA Blood Mini Kit (Qiagen). PCR and sequencing was performed as described in the International Agency for Research on Cancer's Direct Sequencing Protocol (http://p53.iarc.fr/download/tp53_directsequencing_iarc.pdf). Mutations were identified using Mutation Surveyor (Softgenetics). Benign polymorphisms were identified using the International Agency for Research on Cancer's Portal (http://p53.iarc.fr/TP53GeneVariations.aspx).
Expressing different CRBN proteins in Ba/F3 cells
Variants of human and mouse CRBN were cloned into a modified pRSF91 backbone to generate pRSF91-CRBN-IRES-GFP-T2A-Puro retroviral constructs. 200,000 Ba/F3 cells were infected with ecotropic retrovirus in the presence of 2 μg/ml polybrene. After 24 hours, 1 μg/ml puromycin (Gibco) was added and cells were selected for 3-4 days. Cells were confirmed to be >90% GFP+ by flow cytometry and 1,000,000 cells were plated per 6-well and treated with DMSO or lenalidomide for 24 hours. Protein lysates were harvested and immunoblotted for CK1α as described above.
IKZF1 and IKZF3 luciferase reporter assay
10,000 293T cells were transfected with 50 ng of pCMV-IRES-RenillaLUC-IRES-IKZF1/IKZF3-FireflyLUC reporter plasmid together with 100 ng of a vector expressing hCRBN, mCrbn, different chimeric or mutant CRBN forms or empty control. After 48 hours, cells were treated with DMSO and lenalidomide for 4 hours. Firefly and Renilla luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega) according to the manufacturer's protocol.
Mouse experiments
Mouse experiments were performed according to an IACUC approved protocol at Children's Hospital Boston. Generation and characterization of the conditional Csnk1a1 knockout mouse has been described previously15. Csnk1a1flox/+ mice were crossed with Mx1Cre+ mice on a C57BL/6NTac background to obtain Csnk1a1flox/+ Mx1Cre+ mice. Csnk1a1flox/+ Mx1Cre+ or control Csnk1a1+/+ Mx1Cre+ mice were treated with 3 doses of 200 μg poly(I:C) (Invivogen HMW) at 8-10 weeks of age and gene excision was confirmed where applicable. At least 2 weeks following poly(I:C) treatment, the long bones and spines were harvested and crushed and red blood cells were lysed. Bone marrow from age-matched mice of the same genotype was pooled to create sex-balanced groups. C-Kit+ cells were isolated with a CD117 MicroBead Kit (Miltenyi) and an AutoMacs Pro and grown in SFEM (StemSpan) supplemented with antibiotics and 50 ng/ml mTPO (Peprotech) and 50 ng/ml mSCF (Peprotech) for 24 hours. Ecotropic pseudotyped retrovirus was spun onto Retronectin (Clontech) coated 6-well plates and cells were added in 1 ml of media with 2 μg/ml polybrene. An additional 1 ml of media was added after 24 hours. After 48 hours, GFP+ cells were isolated by FACS sorting (BD FACS Aria II) and CD45.1 and CD45.2 cells were mixed. Cells were treated with various doses of lenalidomide and the percent CD45.1 and CD45.2 cells expressing the fluorescent marker was followed by flow cytometry over time following cell surface staining. Antibodies for flow cytometry were as follows: CD45.1 APC/Cy7 (A20, BioLegend), CD45.2 PE (104, eBioscience), and CD45.2 FITC (104, eBioscience). The number of mice needed for each experiment was calculated by assuming a yield of 5 million c-Kit+ cells per mouse (long bones and spine), a transduction rate of 40%, and a target of 50,000 GFP+ cells per experimental well, which was found to reduce variability in pilot experiments. Due to the experimental design, the genotypes of the mice could not be blinded or randomized.
For in vivo treatment, female C57BL/6 mice were treated with once daily intraperitoneal injection of 10 mg/kg lenalidomide dissolved in DMSO and diluted in 100 μl PBS or with DMSO in 100 μl PBS. Bone marrow was harvested after 14 days and lysed with IP lysis buffer (Pierce). Due to experimental design, the treatment groups could not be blinded.
Quantitative Mass Spectrometry in MDS-L cells
MDS-L multiplexed quantitative mass spectrometry samples were processed and analyzed by the Thermo Fisher Scientific Center for Multiplexed Proteomics at Harvard Medical School. Samples were prepared as previously described38 with the following modification. All solutions are reported as final concentrations. Lysis buffer (8 M Urea, 1% SDS, 50 mM Tris pH 8.5, Protease and Phosphatase inhibitors from Roche) was added to the cell pellets to achieve a cell lysate with a protein concentration between 2-8 mg/ml. A micro-BCA assay (Pierce) was used to determine the final protein concentration in the cell lysate. Proteins were reduced and alkylated as previously described. Proteins were precipitated using methanol/chloroform. In brief, four volumes of methanol was added to the cell lysate, followed by one volume of chloroform, and finally three volumes of water. The mixture was vortexed and centrifuged to separate the chloroform phase from the aqueous phase. The precipitated protein was washed with one volume of ice cold methanol. The washed precipitated protein was allowed to air dry. Precipitated protein was resuspended in 4 M Urea, 50 mM Tris pH 8.5. Proteins were first digested with LysC (1:50; enzyme:protein) for 12 hours at 25 °C. The LysC digestion is diluted down to 1 M Urea, 50 mM Tris pH8.5 and then digested with trypsin (1:100; enzyme:protein) for another 8 hours at 25 °C. Peptides were desalted using a C18 solid phase extraction cartridges as previously described. Dried peptides were resuspended in 200 mM EPPS, pH 8.0. Peptide quantification was performed using the micro-BCA assay (Pierce). The same amount of peptide from each condition was labeled with tandem mass tag (TMT) reagent (1:3; peptide:TMT label) (Pierce). The 6-plex and 10-plex labeling reactions were performed for 2 hours at 25 °C. Modification of tyrosine residue with TMT was reversed by the addition of 5% hydroxyl amine for 15 minutes at 25 °C. The reaction was quenched with 0.5% TFA and samples were combined at a 1:1:1:1:1:1 ratio for 6-plex experiments or 1:1:1:1:1:1:1:1:1:1 for 10-plex experiments. Combined samples were desalted and offline fractionated into 24 fractions as previously described.
Liquid chromatography-MS3 spectrometry (LC-MS/MS) in MDS-L cells
12 of the 24 peptide fraction from the basic reverse phase step (every other fraction) were analyzed with an LC-MS3 data collection strategy on an Orbitrap Fusion mass spectrometer (Thermo Fisher Scientific) equipped with a Proxeon Easy nLC 1000 for online sample handling and peptide separations. Approximately 5 μg of peptide resuspended in 5% formic acid + 5% acetonitrile was loaded onto a 100 μm inner diameter fused-silica micro capillary with a needle tip pulled to an internal diameter less than 5 μm. The column was packed in-house to a length of 35 cm with a C18 reverse phase resin (GP118 resin 1.8 μm, 120 Å, Sepax Technologies). The peptides were separated using a 120 min linear gradient from 3% to 25% buffer B (100% ACN + 0.125% formic acid) equilibrated with buffer A (3% ACN + 0.125% formic acid) at a flow rate of 600 nL/min across the column. The scan sequence for the Fusion Orbitrap began with an MS1 spectrum (Orbitrap analysis, resolution 120,000, 400–1400 m/z scan range, AGC target 2 × 105, maximum injection time 100 ms, dynamic exclusion of 75 seconds). ‘Top speed’ (1 second) was selected for MS2 analysis, which consisted of CID (quadrupole isolation set at 0.5 Da and ion trap analysis, AGC 4 × 103, NCE 35, maximum injection time 150 ms). The top ten precursors from each MS2 scan were selected for MS3 analysis (synchronous precursor selection), in which precursors were fragmented by HCD prior to Orbitrap analysis (NCE 55, max AGC 5 × 104, maximum injection time 150 ms, isolation window 2.5 Da, resolution 60,000 (10-plex experiments) or 15,000 (6-plex experiments))
LC-MS3 data analysis for MDS-L cells
A suite of in-house software tools were used to for .RAW file processing and controlling peptide and protein level false discovery rates, assembling proteins from peptides, and protein quantification from peptides as previously described38. MS/MS spectra were searched against a Uniprot human database (February 2014) with both the forward and reverse sequences. Database search criteria are as follows: tryptic with two missed cleavages, a precursor mass tolerance of 50 ppm, fragment ion mass tolerance of 1.0 Da, static alkylation of cysteine (57.02146 Da), static TMT labeling of lysine residues and N-termini of peptides (229.162932 Da), and variable oxidation of methionine (15.99491 Da). TMT reporter ion intensities were measured using a 0.03 Da window (6-plex) or 0.003 Da window (10-plex) around the theoretical m/z for each reporter ion in the MS3 scan. Peptide spectral matches with poor quality MS3 spectra were excluded from quantitation (<100 summed signal-to-noise across 6 channels and <0.5 precursor isolation specificity for 6-plexes or (<200 summed signal-to-noise across 10 channels and <0.5 precursor isolation specificity for 10-plexes).
A moderated t-test was applied across all proteins to assess statistical significance35. This statistic is similar to the ordinary t-statistic, with the exception that the standard errors are calculated using an empirical Bayes method utilizing information across all proteins, thereby making inference about each individual protein more robust. This test assumes normality and uses an estimated standard deviation intended to handle relatively few replicates per condition. Posterior residual standard deviations are used in place of ordinary standard deviation in the moderated t-test applied. This shrinkage of protein-wise sample variances to a pooled estimate provides more stable inference when sample numbers are reduced. The nominal p-values arising from the moderated t-statistic are corrected for multiple testing by controlling the false discovery rate (FDR), as proposed by Benjamini and Hochberg36. Proteins with an FDR adjusted p-value of less than 0.05 were deemed to be reproducibly regulated.
IMiD compound substrate selectivity in KG-1 and MDS-L cells
Cells (2-4 × 106) were plated in 10cm dishes and incubated overnight (18-24 h). Cells were treated with DMSO, lenalidomide (1-10 μM), CC-122 (1-10 μM), pomalidomide (1-10 μM), or thalidomide (10-100 μM) for 6 hours. Drug- treated cells were collected, washed with PBS and cell pellets were lysed in RIPA buffer containing protease and phosphatase inhibitors for 30-45 minutes followed by sonication and centrifugation. Protein lysates were quantified using BCA protein assay kit and 10-15 μg of protein was used for western analysis. CC-122 and lenalidomide competition experiment was conducted as above except cells were pre-treated with DMSO or 10 μM CC-122 for 90 minutes followed by treatment with lenalidomide (0.3-10 μM) or DMSO for 6 hours.
The synthesis and characterization of CC-122 is described in the Supplementary Methods section.
Extended Data
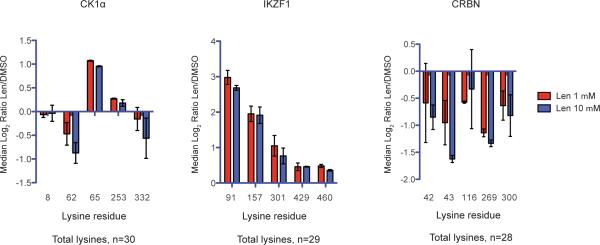
Extended Data Figure 1. Effect of lenalidomide on specific ubiquitination sites.
Median log2 ratios for different lysine residues in CK1α isoform 2, IKZF1 isoform 1, and CRBN isoform 2 for 1 or 10 μM lenalidomide treated KG-1 cells versus DMSO treated cells. SILAC experiments were performed in two biological replicates with flipped SILAC labeling. Only lysine residues detected in both replicates are shown. Error bars are range.
Extended Data Figure 2. Effect of lenalidomide in human cells.
a, Time course of effect of lenalidomide treatment on CK1α protein levels in KG-1 cells. b, Immunoblot of CK1α protein levels in the bone marrow (1,2) and peripheral blood (3,4) mononuclear cells of AML patients treated with lenalidomide as part of a clinical trial. Pre-treatment samples are taken at the screen or before the first treatment (C1D1). Subsequent time points are cycle 1 day 15 (C1D15), cycle 2 day 1 (C2D1) or cycle 1 day 8 (C1D8) of lenalidomide treatment. Further details about these patients (n=4) can be found in Extended Data Table 2. c, MM1S, K562, and Jurkat cells were treated with different concentrations of lenalidomide for 24 hours. CK1α protein levels were detected by western blot and CSNK1A1 mRNA expression levels were measured by RQ-PCR. Data are mean ± s.d., n=3 each with three technical replicates. d, Immunoblot confirming loss of CRBN expression in 293T cells with the CRBN gene disrupted by CRIPSR-Cas9 genome editing. e, Immunoprecipitation with a CRBN-specific antibody in 293T cells treated with DMSO or 10 μM lenalidomide for 5 hours in the presence of 10μM MG132. Results in a, c, d, and e are each representative of two independent experiments. Uncropped blots are shown in Supplementary Figure 1.
Extended Data Figure 3. Sequence determinants of CK1α degradation.
a, 293T cells were transfected with plasmids expressing FLAG-CK1α isoform 1 or isoform 2 together with an hCRBN-expressing plasmid. Cells were treated with DMSO or 10 μM lenalidomide for 16 hours. Cells expressing FLAG-CK1α isoform 1, which contains a nuclear localization domain, were incubated in the absence or presence of the nuclear export inhibitor leptomycin B. b, 293T cells expressing FLAG-CK1α isoform 2 wild-type or two different point mutations identified in patient samples were treated with DMSO or 10 μM lenalidomide for 16 hours. c, Immunofluorescence for CK1α after treatment with DMSO or 10 μM lenalidomide. Enlarged area is indicated by a box in Merge. FITC channel represents staining for CK1α. No changes in CK1α localization are seen upon lenalidomide treatment. Experiment was performed twice in biological duplicate. In each condition, at least 25 cells were assessed. d, Chimeric proteins of casein kinase 1A1 (CK1α) and casein kinase 1E (CK1ε), which shares significant homology with CK1α but is not responsive to lenalidomide, that were used in e to determine the lenalidomide-responsive region in CK1α. e, FLAG-tagged (chimeric) proteins from d were transfected in 293T cells together with a CRBN-expressing plasmid. Cells were treated with 1 μM lenalidomide for 24 hours and protein was detected with a FLAG-specific antibody. Data are representative of two (a, c), three (b) or four (e) independent experiments. Uncropped blots are shown in Supplementary Figure 1.
Extended Data Figure 4. CSNK1A1 knockdown increases lenalidomide sensitivity in hematopoietic cells.
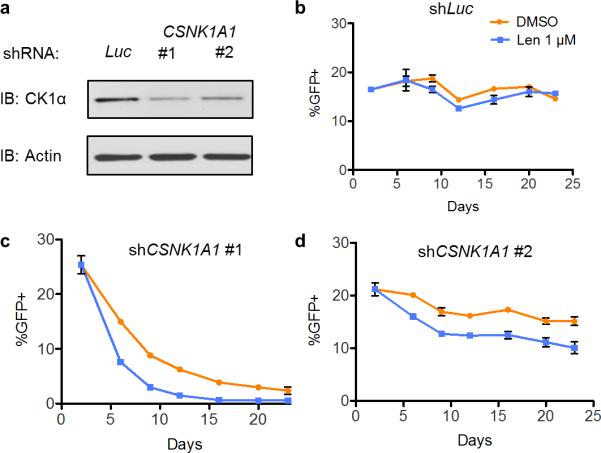
a, Knockdown validation by western blot. b-d, CD34+ cells were transduced with GFP-labeled lentivirus expressing either control shRNA targeting luciferase (b) or shRNA targeting CSNK1A1 (c, d) and treated with DMSO or 1 μM lenalidomide. The percentage of GFP+ cells was assessed by flow cytometry over time. Results are representative of 3 independent experiments each with n=3 biological replicates.
Extended Data Figure 5. Expression of CSNK1A1 and IKZF1 in patient samples.
a, mRNA expression of CSNK1A1 in cord blood CD34+ cells infected with lentivirus expressing hCSNK1A1 or empty vector. CD34+ cells were infected with GFP-tagged lentivirus and GFP+ cells were sorted three days later. Values are mean ± s.d., n=4 biological replicates, each with 3 technical replicates. b, mRNA expression of IKZF1 in cord blood CD34+ cells infected with lentivirus expressing hIKZF1 or empty vector as in a. Values are mean ± s.d., n=3 biological replicates, each with 3 technical replicates. c, CD34+ cells derived from patient or control bone marrow were transduced with a lentivirus expressing IKZF1 and GFP or an empty control vector and treated with DMSO or 1 μM lenalidomide. The percentage of GFP+ cells was assessed by flow cytometry after five days for each vector-drug combination. Results are reported as a ratio of the percentage of GFP+ cells in the lenalidomide condition to the percentage of GFP+ cells in the DMSO condition. Results are combined from three experiments. d, Characteristics of patient samples used for CSNK1A1 and IKZF1 expression experiments. Results of TP53 sequencing, including exons with adequate coverage, is given in the rightmost column. All samples sequenced had wild-type TP53. ND, not done due to limited patient material. WT, TP53 exon sequence has only known benign polymorphisms.
Extended Data Figure 6. Effect of lenalidomide on mouse cells.
a, CK1α protein levels are unaffected in mouse Ba/F3 cells and primary mouse AML cells (MA9) treated with a range of lenalidomide doses. Data are representative of two independent experiments (n=2). b, CK1α expression in bone marrow cells of mice treated with DMSO (n=5) or lenalidomide (n=5). c, CK1α protein levels in Ba/F3 cells transduced with empty vector, mouse Crbn, human CRBN or mCrbnI391V and treated with lenalidomide d, Quantification of CK1α protein levels in Ba/F3 cells using ImageJ. Graphs show the fraction of normalized CK1α protein levels as compared to control (DMSO) treated cells of the respective line. Bars represent mean ± s.e.m. from three independent experiments as in c. e+f, Effect of lenalidomide on an IKZF3- (e) and IKZF1-luciferase fusion protein (f) in 293T cells expressing human, mouse or different chimeras or mutations of CRBN. Data are shown as mean ± s.e.m. (n=3, biological replicates) and are representative of three (f) or five (e) independent experiments. Uncropped blots are shown in Supplementary Figure 1.
Extended Data Figure 7.
Difference electron density map of mouse residue I391 calculated in the absence of a sidechain showing the favored orientation of the residue. The density is contoured at 3.8 σ following a single round of Refmac5 refinement.
Extended Data Figure 8. Comparison of the effects of thalidomide derivatives.
a, Comparison of Log2 ratios for CK1α and IKZF1 in MDS-L cells after treatment with lenalidomide or CC-122 for 24 or 72 hours assessed by tandem mass tag (TMT) quantitative proteomics. Analysis was performed with n=4 for DMSO control and n=3 for each drug treatment time point. b, Adjusted P- values for CK1α and IKZF1 proteomic data in MDS-L cells. c, Western blot validation of IKZF1 and CK1α levels in DMSO (n=4), lenalidomide (n=3) and CC-122 (n=3) treated samples used for MDS-L proteomic analysis. d, Western blot validation of the effects of the different agents on CK1α and IKZF1 protein levels in KG-1 cells. e, Effect of lenalidomide, pomalidomide (Pom), and thalidomide (Thal) on protein levels of CK1α, β-catenin, and IKZF1 in KG-1 cells treated for 24 hours with the indicated drug concentrations. f, Effect of CC-122 and lenalidomide on β-catenin protein levels in KG-1 cells after 72 hours. g, Effect of lenalidomide on CK1α and β-catenin protein levels in HEL cells. Data are representative of two (e, g) or three (c, d) independent experiments. Uncropped blots are shown in Supplementary Figure 1.
Extended Data Table 1. Statistically significant SILAC results with 1 μM lenalidomide.
a, List of significantly regulated K-ε-GG sites with 1 μM lenalidomide vs. DMSO. P-value is adjusted as described in the methods section. b, List of significantly regulated proteins with 1 μM lenalidomide vs. DMSO. P-value is adjusted as described in the methods section. Average log2 fold change of two biological replicates.
| a | |||||
|---|---|---|---|---|---|
| K-ε-GG site | Replicate 1 | Replicate 2 | Average Log2 fold change | adj.P.Val | Direction of change |
| IKZF1 | 2.78 | 3.18 | 2.98 | 7.23E-06 | up |
| IKZF1 | 1.73 | 2.17 | 1.95 | 0.000497 | up |
| MARCH8 | 1.58 | 0.92 | 1.25 | 0.036986 | up |
| CK1α | 1.06 | 1.08 | 1.07 | 0.035843 | up |
| CRBN | −1.07 | −1.21 | −1.14 | 0.026006 | down |
| RNF166 | −1.49 | −1.37 | −1.43 | 0.003102 | down |
| RNF166 | −1.58 | −1.39 | −1.48 | 0.003102 | down |
| b | |||||
|---|---|---|---|---|---|
| Protein | Replicate 1 | Replicate 2 | Average Log2 fold change | adj.P.Val | Direction of change |
| ZNF692 | −1.89 | −2.20 | −2.05 | 0.013806 | down |
| IKZF1 | −1.62 | −1.54 | −1.58 | 0.005638 | down |
| CK1α | −1.59 | −1.53 | −1.56 | 0.005638 | down |
| RNF166 | −1.41 | −1.64 | −1.52 | 0.015257 | down |
| ZFP91 | −0.69 | −0.69 | −0.69 | 0.047677 | down |
| LEMD3 | −0.66 | −0.68 | −0.67 | 0.047677 | down |
| NRM | −0.64 | −0.68 | −0.66 | 0.047677 | down |
| LBR | −0.67 | −0.65 | −0.66 | 0.047677 | down |
| UNC84A.SUN1 | −0.68 | −0.64 | −0.66 | 0.047677 | down |
| C12orf57 | 0.66 | 0.70 | 0.68 | 0.047677 | up |
Extended Data Table 2.
Characteristics of the patient samples from the AML-001 trial used in Extended Data Figure 2b.
| Coded Patient Number (source of cells) | Sex | Primary Race | AML Classification | Peripheral Blood Blast Count | Age at Randomization | Prior MDS History? | MDS Primary Or Secondary | Study Arm Randomized To | Cycle 1 Dosing |
|---|---|---|---|---|---|---|---|---|---|
| 1 (BMMC) | Male | White | AML not otherwise specified | >=1X10^9/L | 71 | Yes | Primary | Lenalidomide | 50 mg daily, except drug withheld days 4-12 |
| 2 (BMMC) | Male | White | AML with myelodysplasia-related changes | >=1X10^9/L | 80 | Yes | Primary | Lenalidomide | 50 mg daily, except drug withheld days 3-6 and 24-28 |
| 3 (PBMC) | Male | Asian | AML with myelodysplasia-related changes | <1X10^9/L | 75 | No | Lenalidomide | 50 mg daily | |
| 4 (PBMC) | Male | White | AML with myelodysplasia-related changes | >=1X10^9/L | 81 | Yes | Primary | Lenalidomide | 50 mg daily |
BMMC: Bone marrow mononuclear cells
PBMC: Peripheral blood mononuclear cells
Supplementary Material
Acknowledgments
We thank Dirk Heckl for cloning of the EGI vector and technical advice, Ronald Mathieu and Mahnaz Paktinat for help with FACS sorting, Michelle Chen for help with colony management and animal care, Simon Köpff for technical support, and Celia Fontanillo for proteomic analysis computational support. Patient samples were provided by the Stem Cell and Xenograft Core of the University of Pennsylvania. This work was supported by the NIH (R01HL082945 and P01CA108631), the Edward P. Evans Foundation, the Gabrielle's Angel Foundation, and a Leukemia and Lymphoma Society Scholar Award to B.L.E. J.K. was supported by the German Research Foundation (DFG, Emmy Noether Fellowship Kr3886/2-1, Kr3886/1-1, and SFB1074) and the Else-Kröner Fresenius Foundation. E.C.F. was supported by award T32GM007753 from the National Institute of General Medical Sciences.
Footnotes
Author contributions:
J.K., E.C.F., and B.L.E. initiated the project. J.K., E.C.F., P.W.H., K.J.M., S.N.H, M.M.C., A.K.G., M.J., and R.K.S. designed and performed cell experiments and protein analysis. E.C.F. designed mouse competition and patient sample experiments. N.D.U., D.R.M., and T.S. performed KG-1 proteomics and analysis. E.G. and M.C. provided patient samples. P.P.C. performed the structural analysis. H.W.M synthesized CC-122. K.J.M. provided MDS-L proteomics. B.E.C., R.C., L.B., S.A.C., and B.L.E. supervised the work. J.K., E.C.F., and B.L.E. wrote the manuscript. All authors assisted in editing the manuscript.
Supplementary Information is linked to the online version of the paper at www.nature.org/nature.
The original mass spectra may be downloaded from MassIVE (http://massive.ucsd.edu) using the identifier: MSV000079014. The data are accessible at ftp://MSV000079014:a@massive.ucsd.edu.
Competing Financial Interests:
P.W.H, K.M, P.P.C, H.W.M, A.K.G, B.E.C, and R.C. are employed by Celgene Corporation. B.L.E. has consulted for Celgene. J.K. received honoraria from Celgene. All other authors have no competing interests to declare.
References
- 1.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010;327:1345–1350. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 2.Krönke J, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science. 2014;343:301–305. doi: 10.1126/science.1244851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu G, et al. The myeloma drug lenalidomide promotes the cereblondependent destruction of Ikaros proteins. Science. 2014;343:305–309. doi: 10.1126/science.1244917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gandhi AK, et al. Immunomodulatory agents lenalidomide and pomalidomide co-stimulate T cells by inducing degradation of T cell repressors Ikaros and Aiolos via modulation of the E3 ubiquitin ligase complex CRL4(CRBN.). Br J Haematol. 2014;164:811–821. doi: 10.1111/bjh.12708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.List A, et al. Lenalidomide in the myelodysplastic syndrome with chromosome 5q deletion. The New England journal of medicine. 2006;355:1456–1465. doi: 10.1056/NEJMoa061292. [DOI] [PubMed] [Google Scholar]
- 6.List A, et al. Efficacy of lenalidomide in myelodysplastic syndromes. N Engl J Med. 2005;352:549–557. doi: 10.1056/NEJMoa041668. [DOI] [PubMed] [Google Scholar]
- 7.Fenaux P, et al. A randomized phase 3 study of lenalidomide versus placebo in RBC transfusion-dependent patients with Low-/Intermediate-1-risk myelodysplastic syndromes with del5q. Blood. 2011;118:3765–3776. doi: 10.1182/blood-2011-01-330126. [DOI] [PubMed] [Google Scholar]
- 8.Pellagatti A, et al. Lenalidomide inhibits the malignant clone and upregulates the SPARC gene mapping to the commonly deleted region in 5qsyndrome patients. Proc Natl Acad Sci U S A. 2007;104:11406–11411. doi: 10.1073/pnas.0610477104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei S, et al. A critical role for phosphatase haplodeficiency in the selective suppression of deletion 5q MDS by lenalidomide. Proc Natl Acad Sci U S A. 2009;106:12974–12979. doi: 10.1073/pnas.0811267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boultwood J, et al. Narrowing and genomic annotation of the commonly deleted region of the 5q- syndrome. Blood. 2002;99:4638–4641. doi: 10.1182/blood.v99.12.4638. [DOI] [PubMed] [Google Scholar]
- 11.Graubert TA, et al. Integrated genomic analysis implicates haploinsufficiency of multiple chromosome 5q31.2 genes in de novo myelodysplastic syndromes pathogenesis. PloS one. 2008;4 doi: 10.1371/journal.pone.0004583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Molecular & cellular proteomics : MCP. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 13.Udeshi ND, Mertins P, Svinkina T, Carr SA. Large-scale identification of ubiquitination sites by mass spectrometry. Nature protocols. 2013;8:1950–1960. doi: 10.1038/nprot.2013.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boultwood J, et al. Gene expression profiling of CD34+ cells in patients with the 5q- syndrome. British journal of haematology. 2007;139:578–589. doi: 10.1111/j.1365-2141.2007.06833.x. [DOI] [PubMed] [Google Scholar]
- 15.Schneider RK, et al. Role of Casein Kinase 1A1 in the Biology and Targeted Therapy of del(5q) MDS. Cancer cell. 2014 doi: 10.1016/j.ccr.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaras M, et al. Csnk1a1 inhibition has p53-dependent therapeutic efficacy in acute myeloid leukemia. J Exp Med. 2014;211:605–612. doi: 10.1084/jem.20131033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huart AS, MacLaine NJ, Meek DW, Hupp TR. CK1alpha plays a central role in mediating MDM2 control of p53 and E2F-1 protein stability. J Biol Chem. 2009;284:32384–32394. doi: 10.1074/jbc.M109.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu S, Chen L, Becker A, Schonbrunn E, Chen J. Casein kinase 1alpha regulates an MDMX intramolecular interaction to stimulate p53 binding. Mol Cell Biol. 2012;32:4821–4832. doi: 10.1128/MCB.00851-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elyada E, et al. CKIalpha ablation highlights a critical role for p53 in invasiveness control. Nature. 2011;470:409–413. doi: 10.1038/nature09673. [DOI] [PubMed] [Google Scholar]
- 20.Knippschild U, et al. The CK1 Family: Contribution to Cellular Stress Response and Its Role in Carcinogenesis. Frontiers in oncology. 2014;4:96. doi: 10.3389/fonc.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Venerando A, Ruzzene M, Pinna LA. Casein kinase: the triple meaning of a misnomer. The Biochemical journal. 2014;460:141–156. doi: 10.1042/BJ20140178. [DOI] [PubMed] [Google Scholar]
- 22.Fratta ID, Sigg EB, Maiorana K. Teratogenic Effects of Thalidomide in Rabbits, Rats, Hamsters, and Mice. Toxicology and applied pharmacology. 1965;7:268–286. doi: 10.1016/0041-008x(65)90095-5. [DOI] [PubMed] [Google Scholar]
- 23.Chesi M, et al. Drug response in a genetically engineered mouse model of multiple myeloma is predictive of clinical efficacy. Blood. 2012;120:376–385. doi: 10.1182/blood-2012-02-412783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer ES, et al. Structure of the DDB1-CRBN E3 ubiquitin ligase in complex with thalidomide. Nature. 2014;512:49–53. doi: 10.1038/nature13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chamberlain PP, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nature structural & molecular biology. 2014 doi: 10.1038/nsmb.2874. [DOI] [PubMed] [Google Scholar]
- 26.Jädersten M, et al. TP53 Mutations in Low-Risk Myelodysplastic Syndromes With del(5q) Predict Disease Progression. Journal of Clinical Oncology. 2011;29:1971–1979. doi: 10.1200/JCO.2010.31.8576. [DOI] [PubMed] [Google Scholar]
- 27.Rauniyar N, Yates JR., 3rd Isobaric labeling-based relative quantification in shotgun proteomics. Journal of proteome research. 2014;13:5293–5309. doi: 10.1021/pr500880b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frei E., 3rd Gene deletion: a new target for cancer chemotherapy. Lancet. 1993;342:662–664. doi: 10.1016/0140-6736(93)91764-d. [DOI] [PubMed] [Google Scholar]
- 29.Nijhawan D, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebert BL, et al. Identification of RPS14 as a 5q-syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narla A, Ebert BL. Ribosomopathies: human disorders of ribosome dysfunction. Blood. 2010;115:3196–3205. doi: 10.1182/blood-2009-10-178129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012;21:723–737. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mertins P, et al. Integrated proteomic analysis of post-translational modifications by serial enrichment. Nature methods. 2013;10:634–637. doi: 10.1038/nmeth.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bland JM, Altman DG. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet. 1986;1:307–310. [PubMed] [Google Scholar]
- 35.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 1995 [Google Scholar]
- 37.Heckl D, et al. Generation of mouse models of myeloid malignancy with combinatorial genetic lesions using CRISPR-Cas9 genome editing. Nature biotechnology. 2014 doi: 10.1038/nbt.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weekes MP, et al. Quantitative temporal viromics: an approach to investigate host-pathogen interaction. Cell. 2014;157:1460–1472. doi: 10.1016/j.cell.2014.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.