Abstract
In this issue are assembled 10 fascinating, well-researched papers that describe the emerging field centered on the microbiome of vertebrate animals and how these complex microbial populations play a fundamental role in shaping homeostasis of the host. The content of the papers will deal with bacteria and, because of relative paucity of information on these organisms, will not include discussions on viruses, fungus, protozoa, and parasites that colonize various animals. Dissecting the number and interactions of the 500–1000 bacterial species that can inhabit the intestines of animals is made possible by advanced DNA sequencing methods, which do not depend on whether the organism can be cultured or not. Laboratory animals, particularly rodents, have proven to be an indispensable component in not only understanding how the microbiome aids in digestion and protects the host against pathogens, but also in understanding the relationship of various species of bacteria to development of the immune system. Importantly, this research elucidates purported mechanisms for how the microbiome can profoundly affect initiation and progression of diseases such as type 1 diabetes, metabolic syndromes, obesity, autoimmune arthritis, inflammatory bowel disease, and irritable bowel syndrome. The strengths and limitations of the use of germfree mice colonized with single species of bacteria, a restricted flora, or most recently the use of human-derived microbiota are also discussed.
Keywords: animal, human microbiome, mice, microbiota, models, review
Defining the Microbial Landscape
The microbiome of hosts, also known as microflora or microbiota, is routinely defined as all the microorganisms inhabiting a specific environment, and these terms are often used interchangeably. The term microbiota has been used historically, and, most likely, the suffix “-biota” was used to define “living organisms in an ecosystem.” The term microbiome was coined in the “-ome” and “-omics” era by Lederberg and McCray “to signify the ecological community of commensal, symbiotic, and pathogenic microorganisms that literally share our body space and have been all but ignored as determinants of health and disease” (2001). Although “-om” and “-ome” surely have meanings in linguistics and biology, the authors proposed that “the -ome idea is borrowed from the multitude of terms already ensconced into English or the scientific lingua franca,” rather than being derivations from Greek or Sanskrit (Lederberg and McCray 2001). In contrast, others proposed to define microbiota as the microbial taxa and the term microbiome as the catalog of these microbes and their genes (Ursell et al. 2012). Either term can be used to describe microbial communities, and the holistic “-ome” approach also includes their genetic information.
Research in this field has attracted considerable interest in the scientific community over the last decade (Figure 1). Reasons for this are the increasing realization that the microbiota has an enormous impact on the phenotype of various animal models and, secondly, that molecular methods have become available, allowing the analysis of complex microbial communities to a degree that was not obtainable using traditional culture methods. Importantly, guidelines for analyses of microbiota via bacterial culture or sequencing platforms are reviewed in this issue by Hiergeist and colleagues (2015).
Figure 1.
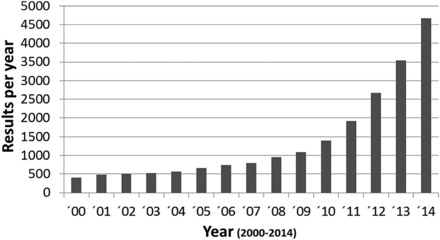
PubMed Search using terms microbiome or microflora, total hits per year.
The Indigenous Microbiota
It is well established that mammals are colonized by microorganisms that outnumber the host in respect to human cells by a factor of 10–100. For example, the human intestine contains 1014 bacteria with approximately 106 microbial genes. Both microbes and the host benefit in a mutualistic way, the first from specific habitats, the last from the microbial activity (e.g., degradation of xenobiotics, epithelial homeostasis, protection against pathogens) (Eberl 2010). At birth, humans are colonized by approximately 100 bacterial species, increasing to 700 at weaning and 1000 in adulthood.
In humans, several factors influence microbiota composition, including age, method of delivery, breast versus bottle feeding, as well as environmental factors like medication, diet, and stress. As an example, the effect of xenobiotics and their effect on intestinal microbiota and resulting metabolomic profile are reviewed in this issue by Lu and colleagues (2015). Compositional changes in the microflora may lead to dysbiosis, possibly contributing to development of various diseases like inflammatory bowel disease, diabetes, asthma, or obesity, to name a few (reviewed by Nicholson et al. 2012).
Composition of the Microbiota
On the phylum level, the gut bacteria are similar in mammals, for example, in humans and mice. However, this does not apply to the species level (Figure 2). Presence and importance of species-specific microbiota, even within rodent species, was demonstrated three decades ago and was also shown more recently (Boot et al. 1985, 1989; Chung et al. 2012; Heidt et al. 1990; Koopman et al. 1984). Of the more than 50 bacterial phyla (29 of which have cultured representatives) (Youssef et al. 2015), humans and mice are colonized mainly by Firmicutes (a phylum containing bacteria like clostridia, lactobacilli, streptococci and staphylococci), Bacteriodetes (like Bacteroides, Porphyromonas), Actinobacteria (like Actinomyces, Streptomyces), and Proteobacteria (which contain Enterobacteriaceae like E. coli or Helicobacter spp.). Composition and complexity vary at different regions of the body, with the highest number of species being found in the colon (mainly Firmicutes and Bacteroidetes in mice and humans) and only a few species in the acid-secreting stomach or the genital tract (Dethlefsen et al. 2007; Sheh and Fox 2013).
Figure 2.
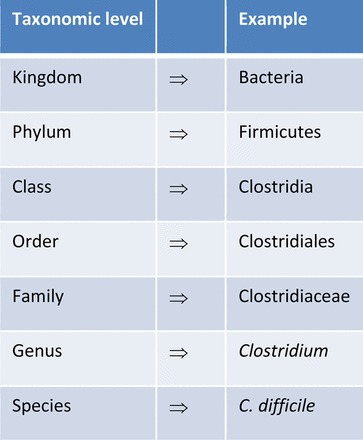
Taxonomic classification of bacteria. Descriptions of the gastrointestinal microbiota focus on the levels of phylum and genus (modified from Sheh and Fox 2013).
In mice, considerable variation has been detected in microbiota composition between mice housed in different facilities, different barriers within a given facility, and even different cages and strains within a barrier in which the mice are housed (Büchler et al. 2012; Hufeldt et al. 2010; Yang et al. 2013). Gut microbiota composition in mice mainly depends on the type of barrier (degree of protection from environmental microbes) and how the barrier was established. For example, the use of germfree mice, or mice colonized with known bacteria like the altered Schaedler's flora (ASF), which is reviewed in this issue by Brand and colleagues (Brand et al. 2015). Further, acquisition and use of mice from commercial vendors and how mice are imported to a facility (quarantine or embryo derivation) is discussed. Importantly, variables such as diet and treatment of water (e.g., chlorination, acidification) is covered (Bleich and Hansen 2012; Wolf et al. 2014). Each of these issues is emphasized and reviewed in this issue by Hansen and colleagues (2015).
Impact of the Microbiota on Animal Models
The impact of the microbiota on host physiology is readily observed when comparing conventional to germfree mice. The enlarged cecum in germfree animals is a characteristic finding evident to even a casual observer. The enlarged cecum occurs from osmosis due to nondegraded mucopolysaccharides that bind sodium and enhance intestinal atonia. A variety of mucosal parameters differ in germfree mice like decreased epithelial renewal, enzyme production, and mucosa thickness. Microbiota play a role in bile salt metabolism, production of short-chain fatty acids, or vitamin K and B-complex vitamins in the large intestine. Immunological changes in germfree animals, such as reduced populations of various innate and adaptive immune cells and reduced or altered cell-specific activities, have been reviewed (Round and Mazmanian 2009). Germfree mice are very susceptible to intestinal pathogens, such as Salmonella spp., and are easily colonized with Escherichia coli; even the probiotic bacterium E. coli Nissle can induce significant disease in mice of certain genetic backgrounds (Bleich et al. 2008).
Given these variables, it becomes clear that absence of the microbiota, as in germfree rodents, reviewed in this issue by Nicklas and colleagues, has a considerable impact on animal models as well (2015). While effects on gut inflammatory phenotypes and intestinal disease models could be expected from absence of gut microbiota (Bleich and Mähler 2005), other conditions such as diabetes, obesity, arthritis, allergy, inflammatory pain, or atopic dermatitis can be affected as well (reviewed in Bleich and Hansen 2012). Even changes in microbiota more subtle than presence or absence lead to phenotypic variation. Limited microbial diversity might be of concern in newly established barrier mouse colonies that were initiated using gnotobiotic mice (usually mice colonized with known bacterial species like the ASF). It has been shown that ASF-colonized mice share more similarities to germfree than to conventionally colonized mice with respect to a number of metabolistic characteristics (Norin and Midtvedt 2010). Therefore, it is possible that a limited diversity could lead to artificial or even a loss of phenotypes in widely used animal models. Further, variation of complex microbiota might lead to phenotypic alterations. A well-known example is the interleukin-10-deficient mouse model of inflammatory bowel disease, which, when maintained at different institutions, develops either spontaneous or Helicobacter-induced disease at differing rates and severity (Keubler et al. 2015; Mähler and Leiter 2002; Yang et al. 2013). Intestinal polyp development was recently described as being reduced in HB-EGFR-transgenic mice after antibiotic treatment (Bongers et al. 2014), and diabetes incidence was reduced in nonobese diabetic (NOD) mice after they received acidified water (Wolf et al. 2014). In principle, all of these models are influenced by variation in the microflora, and the number of published models being affected by microbial variation continues to grow. These models and the importance of the microbiome in the etiopathogenesis of immune-mediated diseases are reviewed in this issue by Hörmannsperger and colleagues and Becker and colleagues (Becker et al. 2015; Hörmannsperger et al. 2015).
Analyzing the Effects of Microbiota
Different strategies are used to identify relevant bacteria as well as mechanisms of microbiota-related phenotype variation. Approaches range from analyzing the effect of single species, simplified communities, or even complex microbiota (e.g., in gnotobiotic animals, reviewed in this issue by Ericsson and Franklin [2015]). Using models of intestinal inflammation, protective effects have been observed using Lactobacillus spp. (Madsen et al. 1999; Schultz et al. 2002; Ukena et al. 2007), Bifidobacterium spp., or E. coli Nissle (Ukena et al. 2007), which are considered to be probiotic bacteria, whereas proinflammatory effects were observed using Enterococcus faecalis, E. coli (Balish and Warner 2002; Kim et al. 2005), Bacteroides vulgatus (Rath et al. 1999), Helicobacter spp. (Fox et al. 1994, 1995, 2011), and murine norovirus (Basic et al. 2014; Cadwell et al. 2010). Segmented filamentous bacteria (SFB) are now known to induce Th17 cells in the intestine, and mice from barriers without SFB may have low Th17 responses compared with SFB-positive, barrier-maintained mice that have strong Th17 responses; and the effect is transferrable with transfer of SFB (Gaboriau-Routhiau et al. 2009; Ivanov et al. 2009). Simplified bacterial communities maintained in gnotobiotic mice, or even complex microbiota administered to germfree animals, revealed pathophysiological effects that vary with the microbial communities (Eun et al. 2014; Faith et al. 2014; Slezak et al. 2014; Wos-Oxley et al. 2012). Molecular analysis of microbiomes using sequencing approaches is increasingly used to elucidate the effect of complex communities on the phenotype expressed in different animal models (Yang et al. 2013).
What is the Relevance of the Microbiome for Scientists Using Animals?
Scientists should be aware of the importance of the intestinal microbiota on the pathophysiology of the host and on the development of various diseases. Alterations in the microbiota can lead to variations even more dramatic than pathogenic organisms. Furthermore, the microbiota will be increasingly important when evaluating the health status of various mouse colonies. Health profiles traditionally and continuously focus on pathogens or potential pathogens (Figure 3a). Screening for specific microorganisms in health-monitoring programs depends on their possible detrimental effect on animal or human health or certain types of research (Mähler et al. 2014; Pritchett-Corning et al. 2014; Shek in press). New concepts in the broader appreciation of microbe–host interactions are reviewed in this issue by Hornef (2015). The future of microbial surveillance and standardization may include some characterization of the complexity and diversity of the microbiota and the presence or absence of phenotypically relevant (but not pathogenic) agents such as the SFB (Figure 3b). Technologies for such analyses are available and are becoming increasingly feasible in rodent health and surveillance programs.
Figure 3.
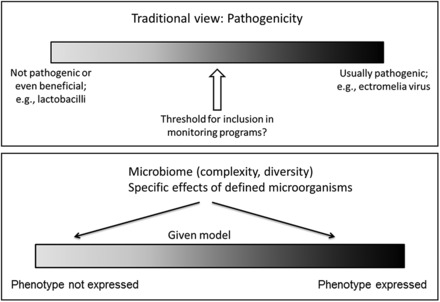
Concepts for microbial surveillance and standardization in laboratory animals. (a) Traditional and current focus of health monitoring is based on virulence of microorganisms (symbiotic, opportunistic, pathogenic) and aims to exclude or report pathogens (and potential pathogens), depending on the barrier level and specified excluded agents. (b) Microbial relevance (other than pathogenicity) to the host is increasingly recognized, as is the complexity and diversity of the microbiota.
Institutional Animal Care and Use Committee (IACUC) Considerations
For most, if not all, IACUCs, there are currently no requirements for detailing the microbiome of animals being used for research or testing, other than perhaps a list of pathogenic organisms excluded from specific pathogen-free colonies. The issue of the microbiome regarding specified research questions is clearly part of an increasingly popular research theme and is considered in the context of individual research protocols, where principal investigators have the expertise to probe a particular question regarding the microbiome. To contemplate IACUC review and approval of microbiome profiles in animal models is premature and fraught with unforeseen problems.
Organization of the Current Issue
This issue of ILAR Journal offers 10 current, concise, and informative reviews on the microbiota's impact in shaping the host's immune response, in the central role of the microbiota in determining susceptibility to a variety of diseases, and on research considerations in animal models. The issue starts with descriptive information and definitions, followed by reviews of the function of microbiota, and it concludes with technical aspects. In the first review, Hornef provides information on the definition of microorganisms according to their virulence potential (2015). This is followed by a description of the microbiota of mammals, summarized in the informative paper by Nelson (2015). Defined floras are a valuable tool to model complex microbiota–host interactions, which is elaborated upon by Brand and colleagues, who review the history, composition, and use of the ASF (2015). Effects of the intestinal microbiota on the host are reviewed by Becker and colleagues and Hörmannsperger and colleagues (Becker et al. 2015; Hörmannsperger et al. 2015). Ericsson and Franklin elaborate on approaches to manipulate the gut microbiota of research animals (2015). The interaction of xenobiotics with the gut microbiota is reviewed by Lu and colleagues (2015). Technical aspects are covered by Hiergeist and coworkers, who provide standards for analyzing the microbiome (2015), and Nicklas and colleagues, who describe the maintenance and monitoring of gnotobiotic rodents (2015). Finally, Hansen and colleagues conclude with applied veterinary aspects with regard to the impact of the gut microbiota on rodent models (2015).
References
- Balish E, Warner T. 2002. Enterococcus faecalis induces inflammatory bowel disease in interleukin-10 knockout mice. Am J Pathol 160:2253–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basic M, Keubler LM, Buettner M, Achard M, Breves G, Schroder B, Smoczek A, Jorns A, Wedekind D, Zschemisch NH, Günther C, Neumann D, Lienenklaus S, Weiss S, Hornef MW, Mähler M, Bleich A. 2014. Norovirus triggered microbiota-driven mucosal inflammation in interleukin 10-deficient mice. Inflamm Bowel Dis 20:431–443. [DOI] [PubMed] [Google Scholar]
- Becker C, Neurath MF, Wirtz S. 2015. The intestinal microbiota in inflammatory bowel disease. ILAR J 56:192–204. [DOI] [PubMed] [Google Scholar]
- Bleich A, Hansen AK. 2012. Time to include the gut microbiota in the hygienic standardisation of laboratory rodents. Comp Immunol Microbiol Infect Dis 35:81–92. [DOI] [PubMed] [Google Scholar]
- Bleich A, Mähler M. 2005. Environment as a critical factor for the pathogenesis and outcome of gastrointestinal disease: experimental and human inflammatory bowel disease and helicobacter-induced gastritis. Pathobiology 72:293–307. [DOI] [PubMed] [Google Scholar]
- Bleich A, Sundberg JP, Smoczek A, von Wasielewski R, de Buhr MF, Janus LM, Julga G, Ukena SN, Hedrich HJ, Gunzer F. 2008. Sensitivity to Escherichia coli Nissle 1917 in mice is dependent on environment and genetic background. Int J Exp Pathol 89:45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bongers G, Pacer ME, Geraldino TH, Chen L, He Z, Hashimoto D, Furtado GC, Ochando J, Kelley KA, Clemente JC, Merad M, van Bakel H, Lira SA. 2014. Interplay of host microbiota, genetic perturbations, and inflammation promotes local development of intestinal neoplasms in mice. J Exp Med 211:457–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boot R, Koopman JP, Kruijt BC, Lammers RM, Kennis HM, Lankhorst A, Mullink JW, Stadhouders AM, De Boer H, Welling GW. 1985. The ‘normalization’ of germfree rabbits with host-specific caecal microflora. Lab Anim 19:344–352. [DOI] [PubMed] [Google Scholar]
- Boot R, Koopman JP, Kruijt BC, Lammers RM, Kennis HM, Lankhorst A, Welling GW, Hectors MP. 1989. The ‘normalization’ of germfree guineapigs with host-specific caecal microflora. Lab Anim 23:48–52. [DOI] [PubMed] [Google Scholar]
- Brand MW, Wannemuehler MJ, Phillips GJ, Proctor A, Overstreet A-M, Jergens AE, Orcutt RP, Fox JG. 2015. The Altered Schaedler Flora: Continued applications of a defined murine microbial community. ILAR J 56:169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Büchler G, Wos-Oxley ML, Smoczek A, Zschemisch NH, Neumann D, Pieper DH, Hedrich HJ, Bleich A. 2012. Strain-specific colitis susceptibility in IL10-deficient mice depends on complex gut microbiota-host interactions. Inflamm Bowel Dis 18:943–954. [DOI] [PubMed] [Google Scholar]
- Cadwell K, Patel KK, Maloney NS, Liu TC, Ng AC, Storer CE, Head RD, Xavier R, Stappenbeck TS, Virgin HW. 2010. Virus-plus-susceptibility gene interaction determines Crohn's disease gene Atg16L1 phenotypes in intestine. Cell 141:1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung H, Pamp SJ, Hill JA, Surana NK, Edelman SM, Troy EB, Reading NC, Villablanca EJ, Wang S, Mora JR, Umesaki Y, Mathis D, Benoist C, Relman DA, Kasper DL. 2012. Gut immune maturation depends on colonization with a host-specific microbiota. Cell 149:1578–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, McFall-Ngai M, Relman DA. 2007. An ecological and evolutionary perspective on human-microbe mutualism and disease. Nature 449:811–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G. 2010. A new vision of immunity: homeostasis of the superorganism. Mucosal Immunol 3:450–460. [DOI] [PubMed] [Google Scholar]
- Ericsson AC, Franklin CL. 2015. Manipulating the gut microbiota: Methods and challenges. ILAR J 56:205–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun CS, Mishima Y, Wohlgemuth S, Liu B, Bower M, Carroll IM, Sartor RB. 2014. Induction of bacterial antigen-specific colitis by a simplified human microbiota consortium in gnotobiotic interleukin-10-/- mice. Infect Immun 82:2239–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. 2014. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med 6:220ra11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Dewhirst FE, Tully JG, Paster BJ, Yan L, Taylor NS, Collins MJ Jr., Gorelick PL, Ward JM. 1994. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J Clin Microbiol 32:1238–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Ge Z, Whary MT, Erdman SE, Horwitz BH. 2011. Helicobacter hepaticus infection in mice: models for understanding lower bowel inflammation and cancer. Mucosal Immunol 4:22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Yan LL, Dewhirst FE, Paster BJ, Shames B, Murphy JC, Hayward A, Belcher JC, Mendes EN. 1995. Helicobacter bilis sp. nov., a novel Helicobacter species isolated from bile, livers, and intestines of aged, inbred mice. J Clin Microbiol 33:445–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaboriau-Routhiau V, Rakotobe S, Lecuyer E, Mulder I, Lan A, Bridonneau C, Rochet V, Pisi A, De Paepe M, Brandi G, Eberl G, Snel J, Kelly D, Cerf-Bensussan N. 2009. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity 31:677–689. [DOI] [PubMed] [Google Scholar]
- Hansen AK, Krych Ł, Nielson DS, Friis Hansen CH. 2015. A review of applied aspects of dealing with gut microbiota impact on rodent models. ILAR J 56:250–264. [DOI] [PubMed] [Google Scholar]
- Heidt PJ, Koopman JP, Kennis HM, van den Logt JT, Hectors MP, Nagengast FM, Timmermans CP, de Groot CW. 1990. The use of a rat-derived microflora for providing colonization resistance in SPF rats. Lab Anim 24:375–379. [DOI] [PubMed] [Google Scholar]
- Hiergeist A, Gläsner J, Reischl U, Gessner A. 2015. Analyses of intestinal microbiota: Culture versus Sequencing. ILAR J 56:228–240. [DOI] [PubMed] [Google Scholar]
- Hörmannsperger G, Schaubeck M, Haller D. 2015. Intestinal microbiota in animal models for inflammatory diseases. ILAR J 56:179–191. [DOI] [PubMed] [Google Scholar]
- Hornef M. 2015. Pathogens, commensal symbionts, and pathobionts: discovery and functional effects on the host. ILAR J 56:159–162. [DOI] [PubMed] [Google Scholar]
- Hufeldt MR, Nielsen DS, Vogensen FK, Midtvedt T, Hansen AK. 2010. Family relationship of female breeders reduce the systematic inter-individual variation in the gut microbiota of inbred laboratory mice. Lab Anim 44:283–289. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, Tanoue T, Imaoka A, Itoh K, Takeda K, Umesaki Y, Honda K, Littman DR. 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139:485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keubler LM, Buettner M, Haeger C, Bleich A. 2015. A multi-hit model: colitis lessons from the interleukin-10-deficient mouse. Inflamm Bowel Dis 21:1967–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Tonkonogy SL, Albright CA, Tsang J, Balish EJ, Braun J, Huycke MM, Sartor RB. 2005. Variable phenotypes of enterocolitis in interleukin 10-deficient mice monoassociated with two different commensal bacteria. Gastroenterology 128:891–906. [DOI] [PubMed] [Google Scholar]
- Koopman JP, Kennis HM, Mullink JW, Prins RA, Stadhouders AM, de Boer H, Hectors MP, van der Logt JT. 1984. Association of germfree rats with different microfloras. Z Versuchstierkd 26:49–56. [PubMed] [Google Scholar]
- Lederberg J, McCray A. 2001. Ome sweet 'omics: -- A genealogical treasury of words. Scientist 15:7. [Google Scholar]
- Lu K, Mahbub R, Fox JG. 2015. Xenobiotics: Interaction with the Intestinal Microflora. ILAR J 56:218–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madsen KL, Doyle JS, Jewell LD, Tavernini MM, Fedorak RN. 1999. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 116:1107–1114. [DOI] [PubMed] [Google Scholar]
- Mähler M, Berard M, Feinstein R, Gallagher A, Illgen-Wilcke B, Pritchett-Corning K, Raspa M. 2014. FELASA recommendations for the health monitoring of mouse, rat, hamster, guinea pig and rabbit colonies in breeding and experimental units. Lab Anim 48:178–192. [DOI] [PubMed] [Google Scholar]
- Mähler M, Leiter EH. 2002. Genetic and environmental context determines the course of colitis developing in IL-10-deficient mice. Inflamm Bowel Dis 8:347–355. [DOI] [PubMed] [Google Scholar]
- Nelson KE. 2015. An update on the status of current research on the mammalian microbiome. ILAR J 56:163–168. [DOI] [PubMed] [Google Scholar]
- Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S. 2012. Host-gut microbiota metabolic interactions. Science 336:1262–1267. [DOI] [PubMed] [Google Scholar]
- Nicklas W, Keubler L, Bleich A. 2015. Maintaining and monitoring the defined microbiota status of gnotobiotic rodents. ILAR J 56:241–249. [DOI] [PubMed] [Google Scholar]
- Norin E, Midtvedt T. 2010. Intestinal microflora functions in laboratory mice claimed to harbor a “normal” intestinal microflora. Is the SPF concept running out of date? Anaerobe 16:311–313. [DOI] [PubMed] [Google Scholar]
- Pritchett-Corning KR, Prins JB, Feinstein R, Goodwin J, Nicklas W, Riley L. 2014. AALAS/FELASA working group on health monitoring of rodents for animal transfer. J Am Assoc Lab Anim Sci 53:633–640. [PMC free article] [PubMed] [Google Scholar]
- Rath HC, Wilson KH, Sartor RB. 1999. Differential induction of colitis and gastritis in HLA-B27 transgenic rats selectively colonized with Bacteroides vulgatus or Escherichia coli. Infect Immun 67:2969–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, Mazmanian SK. 2009. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol 9:313–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz M, Veltkamp C, Dieleman LA, Grenther WB, Wyrick PB, Tonkonogy SL, Sartor RB. 2002. Lactobacillus plantarum 299V in the treatment and prevention of spontaneous colitis in interleukin-10-deficient mice. Inflamm Bowel Dis 8:71–80. [DOI] [PubMed] [Google Scholar]
- Sheh A, Fox JG. 2013. The role of the gastrointestinal microbiome in Helicobacter pylori pathogenesis. Gut Microbes 4:505–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shek W. in press. Microbiological quality control for laboratory rodents and lagomorphs. In: Fox JG, Anderson LC, Otto G, Pritchett-Corning KR, Whary M, eds. Laboratory Animal Medicine. 3rd ed.New York: Academic Press. [Google Scholar]
- Slezak K, Krupova Z, Rabot S, Loh G, Levenez F, Descamps A, Lepage P, Dore J, Bellier S, Blaut M. 2014. Association of germfree mice with a simplified human intestinal microbiota results in a shortened intestine. Gut Microbes 5:176–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, Bleich A, Bruder D, Franzke A, Rogler G, Suerbaum S, Buer J, Gunzer F, Westendorf AM. 2007. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS ONE 2:e1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursell LK, Metcalf JL, Parfrey LW, Knight R. 2012. Defining the human microbiome. Nutr Rev 70(Suppl 1):S38–S44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf KJ, Daft JG, Tanner SM, Hartmann R, Khafipour E, Lorenz RG. 2014. Consumption of acidic water alters the gut microbiome and decreases the risk of diabetes in NOD mice. J Histochem Cytochem 62:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wos-Oxley M, Bleich A, Oxley AP, Kahl S, Janus L, Smoczek A, Nahrstedt H, Pils M, Taudien S, Platzer M, Hedrich HJ, Medina E, Pieper DH. 2012. Comparative evaluation of establishing a human gut microbial community within rodent models. Gut Microbes 3:234–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I, Eibach D, Kops F, Brenneke B, Woltemate S, Schulze J, Bleich A, Gruber AD, Muthupalani S, Fox JG. 2013. Intestinal microbiota composition of interleukin-10 deficient C57BL/6J mice and susceptibility to Helicobacter hepaticus-induced colitis. PLoS One 8:e70783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssef NH, Couger MB, McCully AL, Guerrero Criado AE, Elshahed MS. 2015. Assessing the global phylum level diversity within the bacterial domain: A review. J Adv Res 6:269–282. [DOI] [PMC free article] [PubMed] [Google Scholar]


