Abstract
Evidence from a small number of studies suggests that longer telomere length measured in peripheral leukocytes is associated with an increased risk of non-Hodgkin lymphoma (NHL). However, these studies may be biased by reverse causation, confounded by unmeasured environmental exposures and might miss time points for which prospective telomere measurement would best reveal a relationship between telomere length and NHL risk. We performed an analysis of genetically inferred telomere length and NHL risk in a study of 10 102 NHL cases of the four most common B-cell histologic types and 9562 controls using a genetic risk score (GRS) comprising nine telomere length-associated single-nucleotide polymorphisms. This approach uses existing genotype data and estimates telomere length by weighing the number of telomere length-associated variant alleles an individual carries with the published change in kb of telomere length. The analysis of the telomere length GRS resulted in an association between longer telomere length and increased NHL risk [four B-cell histologic types combined; odds ratio (OR) = 1.49, 95% CI 1.22–1.82, P-value = 8.5 × 10−5]. Subtype-specific analyses indicated that chronic lymphocytic leukemia or small lymphocytic lymphoma (CLL/SLL) was the principal NHL subtype contributing to this association (OR = 2.60, 95% CI 1.93–3.51, P-value = 4.0 × 10−10). Significant interactions were observed across strata of sex for CLL/SLL and marginal zone lymphoma subtypes as well as age for the follicular lymphoma subtype. Our results indicate that a genetic background that favors longer telomere length may increase NHL risk, particularly risk of CLL/SLL, and are consistent with earlier studies relating longer telomere length with increased NHL risk.
Introduction
Telomeres are repetitive AGGGTT nucleotide sequences that protect the ends of chromosomes from degradation and shorten during each round of cell division (1,2). Excessive telomere shortening may lead to cellular senescence, genetic instability and apoptosis (3). Excessively long telomere length and upregulated telomerase activity may result in immortalized cells with unlimited potential for growth and proliferation (4,5). The standard approach to measuring telomere length in large study populations is to measure circulating leukocyte telomere length by multiplex quantitative polymerase chain reactions (6). Family studies suggest that telomere length is highly heritable (7,8). Recently, genome-wide association studies have identified nine common single-nucleotide polymorphisms (SNPs) that are robustly associated with circulating leukocyte telomere length (Table 1) (9–11). When extrapolating association data with qPCR-based telomere length measurements back to Southern blot data, SNPs tagging these loci could explain up to 731 bp of telomere length. This is equivalent to an approximately 20 year or greater difference in age-related telomere attrition (10). While the total variance in telomere length explained by these variants is limited (∼1%), recent studies suggest that genetic risk scores (GRSs) of these variants have utility as surrogate measures of peripheral leukocyte telomere length in deciphering associations with coronary artery disease (9), Alzheimer's disease (12), melanoma (13) and lung cancer (14,15).
Table 1.
Previously published variants associated with circulating leukocyte telomere length
| SNP | Position (GRCh37/hg19) | Nearby gene | Short allele | Long allelea | MAF | Published βb | Published P-value | Reference |
|---|---|---|---|---|---|---|---|---|
| rs10936599 | chr3:169492101 | TERC | T | C | 0.25 | 0.117 | 2.5 × 10−31 | Codd et al. (9) |
| rs2736100 | chr5:1286516 | TERT | A | C | 0.49 | 0.094 | 4.4 × 10−19 | Codd et al. (9) |
| rs7675998 | chr4:164007820 | NAF1 | A | G | 0.22 | 0.090 | 4.3 × 10−16 | Codd et al. (9) |
| rs9420907 | chr10:105676465 | OBFC1 | A | C | 0.14 | 0.083 | 6.9 × 10−11 | Codd et al. (9) |
| rs8105767 | chr19:22215441 | ZNF208 | A | G | 0.30 | 0.058 | 1.1 × 10−9 | Codd et al. (9) |
| rs755017 | chr20:62421622 | RTEL1 | A | G | 0.12 | 0.074 | 6.7 × 10−9 | Codd et al. (9) |
| rs11125529 | chr2:54475866 | ACYP2 | C | A | 0.14 | 0.067 | 4.5 × 10−8 | Codd et al. (9) |
| rs6772228 | chr3:58376019 | PXK | A | T | 0.05 | 0.120 | 3.9 × 10−10 | Pooley et al. (10) |
| rs3027234 | chr17:8136092 | CTC1 | T | C | 0.23 | 0.057 | 2.3 × 10−8 | Mangino et al. (11) |
MAF, minor allele frequency.
aLong allele is allele associated with longer telomere length.
bβ-estimate is reported in telomere kb per long allele.
A few studies have investigated the association of telomere length measured in peripheral circulating leukocytes with non-Hodgkin lymphoma (NHL) risk. A prospective study by Lan et al. (16) of a group of 107 incident male NHL cases and 107 matched controls found evidence that suggested longer relative telomere length, as measured by monochrome multiplex qPCR, might be associated with an increased risk of NHL and this association was similar across common NHL subtypes. A subsequent nested case–control study of 464 lymphoma cases and 464 matched controls from the European Prospective Investigation into Cancer (EPIC) cohort also found evidence that suggests longer telomere length is associated with an increased risk of B-cell lymphoma, particularly for diffuse large B-cell lymphoma (DLBCL) and follicular lymphoma (FL) (17). Further evidence from Epstein–Barr virus-infected B-lymphocytes suggests that these cells express a phenotype of progressively increasing telomere length accompanied by the accumulation of promyelocytic leukemia nuclear bodies (18). It is important to note that these studies investigated the relationship between telomere length and NHL risk, which is distinct from the studies that investigated the relationship between telomere length and NHL prognosis (19,20).
Accumulating evidence indicates that longer telomere length may be associated with increased NHL risk although existing observational studies on telomere length can be confounded by insufficient adjustment for exposures that affect both NHL risk and telomere length; for example, Epstein–Barr infection affects telomere length (18) and NHL risk (21), but was not adjusted for in prospective studies on telomere length and NHL risk (16,17). Existing studies on telomere length and NLH risk also might not capture the biological time points most relevant for determining NHL risk since single measurements of telomere length can be influenced by changes in psychological stress (22), exercise (23) and nutrition (24). In addition, the aforementioned studies (16,17) lacked the statistical power to consider potential subtype-specific associations with relative telomere length. Here, we used telomere length-associated genetic variants to investigate an association between genetically inferred telomere length and B-cell NHL in a large pooled study population that included four major histologic subtypes of B-cell NHL. Specifically, we used associations with individual telomere length-associated variants as well as an aggregate GRS to examine whether telomere length could modify the risk of four major types of B-cell NHL. Improved understanding of how telomere length is linked to NHL risk may provide a better insight into lymphomagenesis and could serve as a biomarker for future NHL risk assessment.
Results
Our analysis consisted of a pooled sample of 10 102 NHL cases and 9562 controls from samples of European descent (Table 2). The NHL cases included 3104 chronic lymphocytic leukemia or small lymphocytic lymphoma (CLL/SLL), 3652 DLBCL, 2521 FL and 825 marginal zone lymphoma (MZL) cases. CLL and SLL are grouped together by REAL and WHO classifications since they are both different manifestations of the same neoplastic immunophenotype in which SLL is the solid phase and CLL is the circulating phase (25,26). There were approximately equal numbers of men and women for NHL cases overall; however, there were more men with CLL/SLL and fewer men with MZL than women. The overall age range of participants was 15–94 years, with an overall median age of 64 years.
Table 2.
Descriptive characteristics of study participants with complete covariate information
| CLL/SLL |
DLBCL |
FL |
MZL |
Combined NHL |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | Cases | Controls | |
| Sex | ||||||||||
| Males | 1796 | 5374 | 1861 | 5452 | 1168 | 5209 | 334 | 4527 | 5159 | 6119 |
| Females | 1308 | 2297 | 1791 | 2617 | 1353 | 2551 | 491 | 1694 | 4943 | 3443 |
| % Males | 58% | 70% | 51% | 68% | 46% | 67% | 40% | 73% | 51% | 64% |
| 3104 | 7671 | 3652 | 8069 | 2521 | 7760 | 825 | 6221 | 10 102 | 9562 | |
| Age | ||||||||||
| 10–20 | 0 | 5 | 25 | 6 | 0 | 20 | 1 | 4 | 26 | 21 |
| 20–30 | 5 | 49 | 118 | 60 | 18 | 98 | 11 | 41 | 152 | 113 |
| 30–40 | 36 | 181 | 268 | 222 | 138 | 273 | 25 | 129 | 467 | 332 |
| 40–50 | 277 | 509 | 420 | 633 | 387 | 652 | 81 | 316 | 1165 | 954 |
| 50–60 | 644 | 1323 | 790 | 1526 | 739 | 1416 | 170 | 968 | 2343 | 1957 |
| 60–70 | 1169 | 2956 | 1031 | 3083 | 753 | 2847 | 274 | 2520 | 3227 | 3452 |
| 70–80 | 849 | 2286 | 799 | 2205 | 415 | 2127 | 209 | 1953 | 2272 | 2364 |
| 80–90 | 123 | 357 | 195 | 330 | 69 | 323 | 54 | 286 | 441 | 364 |
| 90–100 | 1 | 5 | 6 | 4 | 2 | 4 | 0 | 4 | 9 | 5 |
| Median | 66 | 67 | 63 | 66 | 60 | 66 | 66 | 67 | 64 | 65 |
| 3104 | 7671 | 3652 | 8069 | 2521 | 7760 | 825 | 6221 | 10 102 | 9562 | |
An overall excess of SNP associations with P < 0.05 was observed for the four NHL types combined (exact binomial P-value = 0.008) and for CLL/SLL and FL subtypes (exact binomial P-value = 0.001 and 0.008, respectively; Table 3). Associations were found for NHL subtypes combined (rs10936599, rs9420907 and rs11125529), CLL/SLL (rs10936599, rs2736100, rs7675998 and rs9420907), DLBCL (rs3027234) and FL (rs10936599, rs9420907 and rs755017). No evidence of association was observed between any of the nine telomere length-associated variants and MZL risk. Associations generally suggested that longer telomere length increased risk for certain NHL subtypes, with the exception of the associations between NHL subtypes combined and rs11125529, and FL and rs755017 where associations decreased risk.
Table 3.
Associations of telomere length-associated variants and NHL risk
| SNP (ref./alternate) | All NHL subtypes |
CLL/SLL | DLBCL |
FL |
MZL |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | OR | 95% CI | P-value | |
| rs10936599 (T/C) | 1.10 | 1.05–1.15 | 5.67e−5* | 1.17 | 1.09–1.25 | 2.33e−5* | 1.04 | 0.98–1.11 | 0.2158 | 1.09 | 1.01–1.18 | 0.0250* | 1.06 | 0.94–1.20 | 0.3577 |
| rs2736100 (A/C) | 1.03 | 0.99–1.07 | 0.1719 | 1.13 | 1.06–1.20 | 5.86e−5* | 1.01 | 0.95–1.06 | 0.8508 | 0.97 | 0.91–1.03 | 0.3081 | 1.03 | 0.92–1.14 | 0.6314 |
| rs7675998 (A/G) | 1.04 | 0.99–1.10 | 0.0819 | 1.12 | 1.04–1.20 | 0.0023* | 1.04 | 0.98–1.12 | 0.2071 | 1.01 | 0.94–1.09 | 0.7602 | 1.00 | 0.88–1.13 | 0.9515 |
| rs9420907 (A/C) | 1.12 | 1.05–1.18 | 0.0002* | 1.15 | 1.05–1.25 | 0.0014* | 1.02 | 0.94–1.11 | 0.6504 | 1.11 | 1.01–1.22 | 0.0243* | 1.11 | 0.96–1.28 | 0.1675 |
| rs8105767 (A/G) | 0.99 | 0.95–1.04 | 0.7452 | 1.02 | 0.95–1.09 | 0.6254 | 1.00 | 0.94–1.06 | 0.8817 | 0.99 | 0.92–1.06 | 0.7947 | 0.97 | 0.86–1.09 | 0.6392 |
| rs755017 (A/G) | 0.94 | 0.89–1.00 | 0.0561 | 0.97 | 0.88–1.06 | 0.5091 | 0.99 | 0.91–1.08 | 0.7729 | 0.85 | 0.77–0.94 | 0.0022* | 0.91 | 0.77–1.07 | 0.2553 |
| rs11125529 (C/A) | 0.94 | 0.89–1.00 | 0.0352* | 0.96 | 0.88–1.05 | 0.3658 | 0.95 | 0.88–1.03 | 0.2514 | 0.97 | 0.88–1.06 | 0.4997 | 0.90 | 0.77–1.05 | 0.1803 |
| rs6772228 (A/T) | 1.03 | 0.94–1.12 | 0.5516 | 1.05 | 0.92–1.19 | 0.5023 | 1.07 | 0.95–1.22 | 0.2729 | 1.02 | 0.88–1.18 | 0.8157 | 1.06 | 0.83–1.34 | 0.6381 |
| rs3027234 (T/C) | 1.04 | 0.99–1.09 | 0.1076 | 1.05 | 0.97–1.12 | 0.2236 | 1.07 | 1.00–1.15 | 0.0413* | 1.01 | 0.93–1.09 | 0.8195 | 1.02 | 0.90–1.16 | 0.7738 |
| Aggregate testa | – | – | 5.00e−7* | – | – | 4.71e−9* | – | – | 0.3327 | – | – | 0.0095* | – | – | 0.6644 |
| Genetic risk scoreb | 1.49 | 1.22–1.82 | 8.54e−5* | 2.60 | 1.93–3.51 | 3.96e−10* | 1.28 | 0.97–1.70 | 0.0844 | 1.10 | 0.79–1.53 | 0.5640 | 1.16 | 0.69–1.95 | 0.5810 |
| MR (IVW)c | 1.49 | 1.22–1.81 | 1.01e−4* | 2.60 | 1.93–3.51 | 3.29e−10* | 1.28 | 0.97–1.70 | 0.0864 | 1.10 | 0.79–1.53 | 0.5659 | 1.17 | 0.69–1.98 | 0.5673 |
| MR (likelihood)d | 1.50 | 1.22–1.85 | 1.27e−4* | 2.68 | 1.96–3.67 | 8.20e−10* | 1.29 | 0.97–1.71 | 0.0841 | 1.11 | 0.79–1.54 | 0.5572 | 1.17 | 0.69–1.99 | 0.5627 |
| Heterogeneitye | – | – | 1.93e−4* | – | – | 0.0380* | – | – | 0.5180 | – | – | 0.0070* | – | – | 0.6170 |
Bold values designate statistically significant associations with a P-value <0.05
ref., short allele; alternate, long allele.
aAggregate test is a log likelihood ratio test comparing a model having all telomere length-associated SNPs and covariates with a null model with only sex and age.
bGenetic risk score ORs refer to a 1-kb increase in telomere length.
cInverse-variance weighted Mendelian randomization estimate for a 1-kb increase in telomere length.
dPooled estimate for the likelihood-based Mendelian randomization method for a 1-kb increase in telomere length.
eTest for significant heterogeneity across the nine SNP instruments used in the Mendelian randomization analysis.
*Statistical significance at the P < 0.05 level.
Aggregate tests that compared a null model having only sex and age with an expanded model that included all nine telomere length-associated variants were also fit for the combined NHL types and for each NHL subtype (Table 3). Statistically significant aggregate associations of the nine telomere length-associated variants were observed for the combined NHL subtypes and for CLL/SLL and FL subtypes (P-values = 5.0 × 10−7, 4.7 × 10−9 and 0.01, respectively). DLBCL and MZL subtypes were not significantly associated with the nine variants in aggregate. These tests of the association with NHL overall and CLL/SLL and FL subtypes suggest that, in aggregate, one or more of the nine telomere length-associated variants are associated with overall NHL, CLL/SLL and FL risk, but give no information about the individual SNPs driving the association, the direction of the association or the effect size.
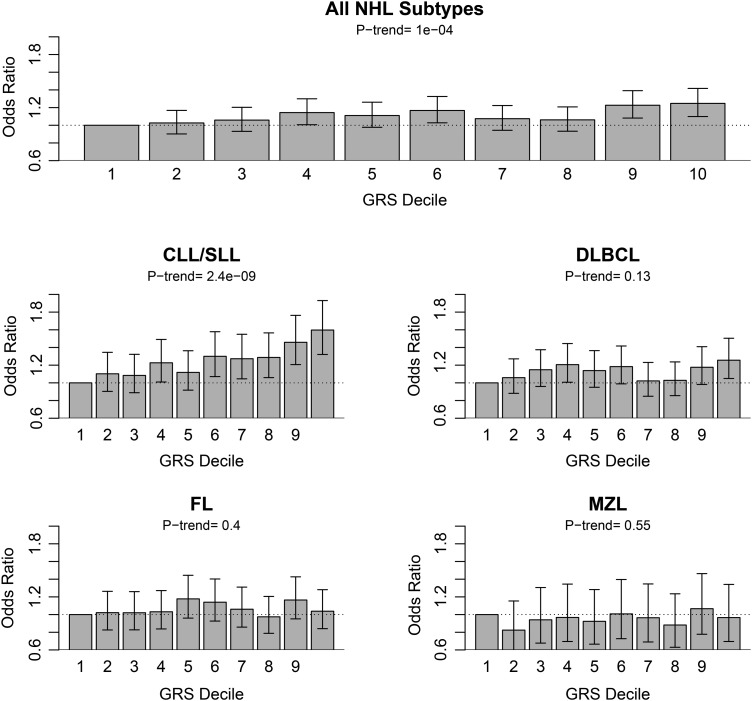
Analyses investigating the associations between a telomere length-associated GRS and NHL risk were conducted in which a higher GRS indicates longer circulating leukocyte telomere length and a lower GRS indicates shorter telomere length (Table 3). Adjusting for sex and age, positive associations of the GRS were observed (i.e. indicating an increased risk with longer telomere length) for the four NHL types combined and for CLL/SLL (P-values = 8.5 × 10−5 and 4.0 × 10−10, respectively). When modeled as a decile of telomere length-associated GRS, a per decile increase in GRS was significantly associated with an increased risk of the four NHL subtypes combined (per decile OR = 1.02, 95% CI 1.01–1.03, P-value = 0.0001) and with CLL/SLL (per decile OR = 1.05, 95% CI 1.03–1.06, P-value = 2.37 × 10−9), but not with the other NHL subtypes (Fig. 1). Comparing the highest with the lowest decile of GRS, the ORs for the combined NHL subtypes and for CLL/SLL were 1.25 (95% CI 1.10–1.42, P-value = 0.0006) and 1.60 (95% CI 1.32–1.93, P-value = 1.32 × 10−6), respectively. When excluding CLL/SLL subtypes and doing a combined DLBCL, FL and MZL analysis, the odds ratio (OR) for the telomere length-associated GRS was 1.16 (95% CI 0.93–1.45, P-value = 0.18).
Figure 1.
ORs for each telomere length-associated GRS decile by NHL overall and subtype. The lowest GRS decile is used as the reference of comparison.
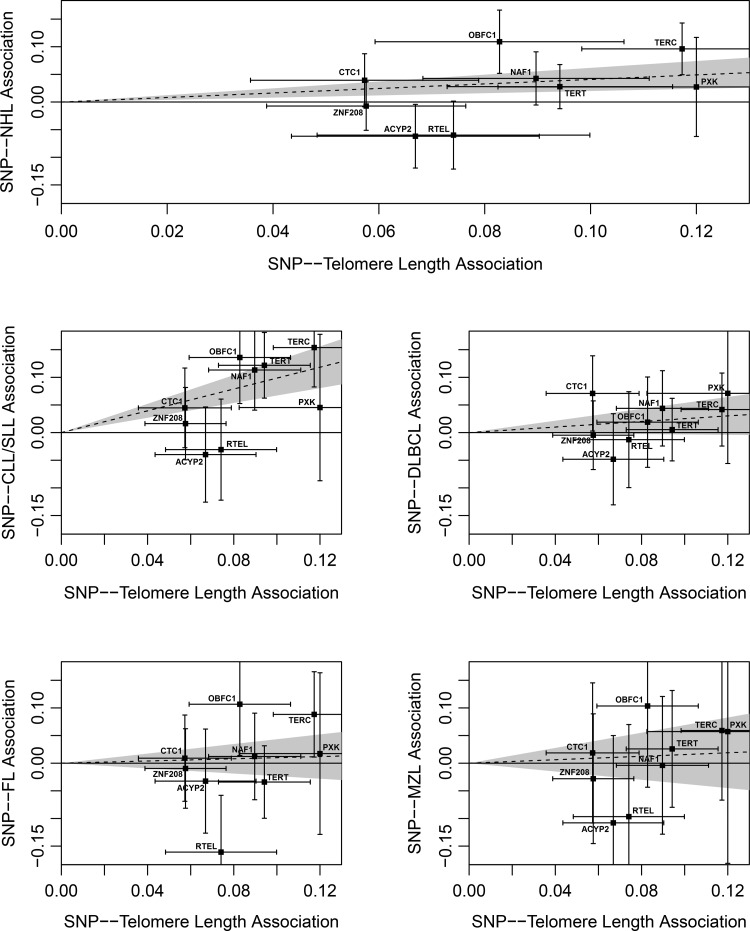
The Mendelian randomization analysis produced similar effect estimates as the GRS associations. In addition, the four NHL subtypes combined (likelihood-based method: OR = 1.50, 95% CI 1.22–1.85, P-value = 1.3 × 10−4) and CLL/SLL subtypes (likelihood-based method: OR = 2.68, 95% CI 1.96–3.67, P-value = 8.2 × 10−10) were significantly associated with increasing telomere length (Table 3 and Fig. 2). No significant Mendelian randomization effect estimates were observed for DLBCL, FL or MZL subtypes of NHL. Heterogeneity tests were conducted to assess if the telomere length-associated variant effects on telomere length were proportional to their effects on NHL risk. Significant evidence for heterogeneity was observed for the four NHL subtypes combined, CLL/SLL and FL subtypes (P-values = 1.9 × 10−4, 0.04 and 0.01, respectively; Table 3), with the rs755017 (RTEL1) and rs11125529 (ACYP2) telomere length-associated variants displaying the largest departures from expectation.
Figure 2.
Plots of the effect of each variant on telomere length and NHL risk overall and by the subtype. The X axis (G–X association) plots the previously published linear regression β-estimates for a 1-kb change in telomere length for each telomere length-associated variant (Table 1). The Y axis (G–Y association) plots the β-estimate from the logistic regression model for the association of each variant with NHL risk overall and by the subtype (Table 3). Error bars around each β-estimate indicate the uncertainty of effect estimates. A best fit regression line and 95% confidence interval are plotted for NHL overall and each subtype using Mendelian randomization likelihood-based estimates. P-values are from the Mendelian randomization likelihood-based method.
In the ASSET analysis, we further explored which of the four examined subtypes of NHL are associated with telomere length-associated variants and identified CLL/SLL as the predominant B-cell NHL subtype included in the majority of the observed associations (Table 4). For example, CLL/SLL appeared in the optimal subset for six of the nine telomere length-associated SNPs and in all four of the significant SNP associations. In addition, the strongest P-value association with the telomere length-associated GRS was observed for the CLL/SLL subtype.
Table 4.
ASSET meta-analysis of all possible NHL subtype subsets
| SNP (ref./alternate) | ||||
|---|---|---|---|---|
| OR | 95% CI | P-value | NHL subsets | |
| rs10936599 (T/C) | 1.13 | 1.06–1.20 | 4.33 × 10−5 | CLL/SLL, FL, MZL |
| rs2736100 (A/C) | 1.12 | 1.05–1.19 | 9.76 × 10−4 | CLL/SLL |
| rs7675998 (A/G) | 1.11 | 1.01–1.22 | 0.04 | CLL/SLL |
| rs9420907 (A/C) | 1.18 | 1.08–1.28 | 1.57 × 10−4 | CLL/SLL, MZL |
| rs8105767 (A/G) | 0.96 | 0.00–Inf. | 1.00 | MZL |
| rs755017 (A/G) | 0.89 | 0.79–1.01 | 0.06 | FL, MZL |
| rs11125529 (C/A) | 0.93 | 0.85–1.02 | 0.11 | CLL/SLL, DLBCL, MZL |
| rs6772228 (A/T) | 1.05 | 0.00–Inf | 1.00 | DLBCL, MZL |
| rs3027234 (T/C) | 1.05 | 0.96–1.16 | 0.30 | CLL/SLL, DLBCL |
| Genetic risk score | 2.47 | 1.82–3.35 | 7.19 × 10−9 | CLL/SLL |
Bold values designate statistically significant associations with a P-value < 0.05
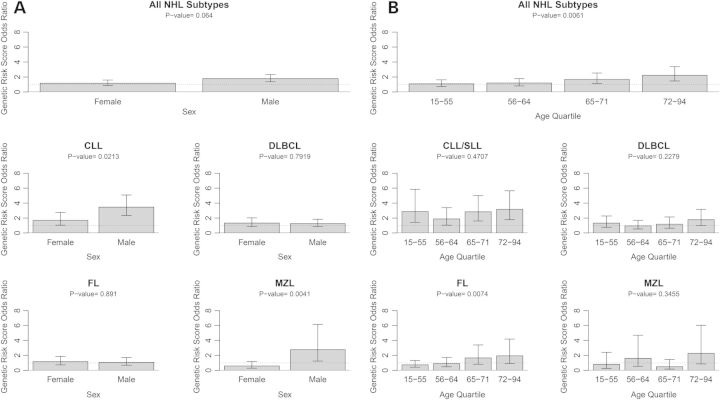
To better characterize the association of telomere length with NHL risk, we investigated the magnitude of the association of the telomere length-associated GRS with NHL risk by sex and age groups. These associations were first investigated for all NHL subtypes combined. Men had a stronger OR for the telomere length-associated GRS (OR = 1.78, 95% CI 1.37–2.32, P-value = 2.0 × 10−5) than women did (OR = 1.17, 95% CI 0.86–1.59, P-value = 0.33); however, the interaction between telomere length-associated GRS and sex was marginally non-significant (P-value = 0.064; Fig. 3A). This sex difference in the GRS–NHL association was more pronounced for the CLL/SLL subtype, for which men had an OR of 3.47 (95% CI 2.36–5.09, P-value = 2.25 × 10−10), whereas women had an OR of 1.70 (95% CI 1.05–2.76, P-value = 0.03), with a statistically significant interaction (P-value = 0.02). An interaction between the telomere length-associated GRS and sex was also detected for the MZL subtype (P-value = 0.004), for which men had a large telomere length-associated GRS effect (OR = 2.76, 95% CI 1.24–6.15, P-value = 0.01), whereas for women the effect was significantly lower (OR = 0.59, 95% CI 0.29–1.19, P-value = 0.14). Additionally, age-related effect modification was suggested across quartiles of age in which older age groups had a higher OR for the telomere length-associated GRS association with a risk of combined NHL subtypes (interaction P-value = 0.006; Fig. 3B). In the youngest age quartile (individuals aged 15–55), the OR for telomere length-associated GRS was 1.09 (95% CI 0.73–1.62, P-value = 0.69), whereas in the oldest age quartile (individuals aged 72–94) the OR was 2.23 (95% CI 1.47–3.38, P-value = 0.0002). This interaction of telomere length-associated GRS and age was also statistically significant for the FL subtype (interaction P-value = 0.007).
Figure 3.
Telomere length GRS associations by strata of sex and age quartile. P-values are for interactions between telomere length-associated GRS and sex or age group.
Discussion
Our study of genetically inferred telomere length provides a proxy measure of circulating leukocyte telomere length that suggests a positive association between telomere length and risk of four major types of B-cell NHL. While evidence suggests that this association may be present in multiple NHL subtypes, our associations with telomere length were most consistent and strongest for CLL/SLL. Subset analyses by sex and age indicate that telomere length may be particularly important for risk of some NHL subtypes for men and for older individuals although further replication is needed.
Recent genetic studies investigating the relationship between telomere length and risk of cancer have suggested that longer telomere length is associated with increased cancer risk. A prior study by our group and another independent group found evidence linking longer genetically inferred telomere length with increased risk of lung cancer (14,15) and another group has linked longer genetically predicted telomere length with increased risk of melanoma (13). We now provide evidence that longer genetically predicated telomere length may also be associated with increased NHL risk, particularly for CLL/SLL. Excessively long telomere length and upregulated telomerase activity may be important for cancer risk since longer telomeres result in greater replicative potential (4,5). Cells with a genetic ability to maintain long telomeres may result in phenotypes with increased potential for growth and proliferation which, when not adequately regulated by cellular growth mechanisms, could lead to greater carcinogenic potential.
Studies of measured telomere length are at risk for reverse causation bias by early undetected disease and can be confounded by shared environmental exposures if the mechanism of the exposure disease relationship is not mediated through alterations in telomere length. To our knowledge, our study is the first to investigate the relationship between telomere length and NHL risk using genetic proxies of telomere length. By using a combination of nine telomere length-associated variants to infer telomere length, our study suggests a genetic background that favors longer telomere length to be associated with increased NHL risk. Our genetic proxy of telomere length estimates exposure to telomere length over an individual's lifetime and may have advantages over traditional measures of telomere length that generally focus on one sample collection time point. Our study has the added advantage of using variants associated with leukocyte telomere length, the same progenitor cell type for NHL. Also, by using a genetic proxy for telomere length, it may be possible to isolate the effect of the genetically determined variance in TL from the effects of the variance in TL due to non-genetic factors, such as aging, oxidative damage and other relevant processes caused by environmental exposures, or unknown genetic factors. Such studies could be instrumental for identifying environmental or genetic risk factors that when appropriately targeted by a focused intervention could reduce NHL risk by its associated effect on telomere length.
The relationship between genetically predicted telomere length and NHL reported in our study for NHL overall and CLL/SLL subtypes is consistent with prior evidence from studies using telomere length measured in white blood cell DNA. For NHL overall, we detected quartile ORs (95% CI) of 1.0, 1.07 (0.98–1.16), 1.07 (0.99–1.16) and 1.17 (1.08–1.26). A prior study of measured telomere length and NHL risk by Lan et al. (16) (107 NHL cases and 107 matched controls) found quartile ORs (95% CI) of 1.0, 1.1 (0.4–2.7), 1.8 (0.7–4.9) and 3.6 (1.4–8.9). Likewise, a study of measured telomere length and NHL risk in the EPIC cohort (464 NHL cases and 464 matched controls) found quartile ORs (95% CI) of 1.00, 1.66 (0.99–2.78), 1.80 (1.05–3.11) and 3.20 (1.71–5.98) (17). While our quartile estimates for the relationship between telomere length and NHL were of lesser magnitude, as one might expect given that a relatively small amount of variance in measured telomere length explained by the GRS, the trend was consistent with that seen in studies of measured telomere length and the 95% confidence intervals generally overlapped. Differences in sample size, sample collection time points, measurement techniques and covariates included in the model may account for the differences observed between the studies that directly measured telomere length and our approach that used genetically predicted telomere length.
Our investigation is one of the first to establish a connection between telomere length-associated genetic variants and risk of NHL. A recent CLL GWAS detected evidence for a signal at the TERC locus (rs10936599) (27). The SNP associated with CLL at this locus is the same telomere length-associated SNP reported by Codd et al. (9) (rs10936599) and was included in our telomere length-associated GRS. Our study provides replication of this association between rs10936599 and CLL/SLL risk (P-value = 2.3 × 10−5). Additionally, the TERT locus (rs10069690) has also been associated with CLL/SLL risk (27,28). Our investigation also observed an association at the TERT locus for CLL/SLL (rs2736100, P-value = 5.9 × 10−5), but the two TERT variants are weakly correlated and may tag different TERT signals (D′ = 0.85, R2 = 0.28). When performing a follow-up analysis that removes the TERC and TERT SNPs from the GRS and separately adjusts for them as covariates in the analysis, the seven SNP telomere length-associated GRS remains statistically significant (OR = 1.93, 95% CI 1.29–2.89, P-value = 0.0014), suggesting that an additional novel signal associated with CLL risk remains in the telomere length-associated GRS. To our knowledge, no prior associations with telomere length-associated variants have been reported for DLBCL, FL and MZL subtypes of NHL. Additional variation in telomere length important for NHL risk may be attributable to other genetic variants that remain to be discovered. An interesting observation from our study is that not all published telomere length-associated variants have associations with NHL subtypes that are proportional to their associations with telomere length. This is particularly true for the rs755017 (RTEL1) and rs11125529 (ACYP2) variants. Such variants may impact telomere length through mechanisms that are not important for risk of the four common NHL subtypes we studied and as such the Mendelian randomization method to estimate the effect of overall telomere length on NHL risk that uses all nine telomere length-associated variants may not be the optimal approach to accurately estimate the magnitude of the true underlying causal effect of telomere length on NHL risk. Future studies are needed to further refine effect estimates for the rs755017 (RTEL1) and rs11125529 (ACYP2) variants and leukocyte telomere length as well as explore the heterogeneity we observed in the association of the nine telomere length-associated variants and risk of the four common NHL subtypes. In addition to telomere length, other aspects of telomeres, such as maintenance of genome stability, chromosomal repair or distinct biological processes tagged by the nine telomere length-associated variants, may also be important for NHL risk.
NHL comprises several subtypes that may have different associations with telomere length. A clear association was observed with longer telomere length and increased CLL/SLL risk in our analysis. DLBCL and FL results were less clear. For DLBCL, a single SNP association was detected for one telomere length-associated variant, and a marginally statistically significant association was observed for the GRS association. The FL subtype showed evidence for three SNP associations and had a significant aggregate association test, but the GRS association was null. No evidence of single SNP associations was observed for telomere length-associated variants with MZL risk, as perhaps we were underpowered to detect associations for this subtype. Due to sample size limitations, we were also not able to examine potential associations of telomere length-associated polymorphisms with other less common B-cell or T-cell NHL subtypes.
Associations were found to differ by sex and age. If replicated, these results may be useful for screening populations where telomere length is more strongly associated with NHL risk. The association of the telomere length-associated GRS with risk of NHL was particularly strong for men, especially for the CLL/SLL and MZL subtypes. Additionally, results from individuals aged 72 years and over suggest that older individuals have elevated estimates for their GRS associations. The biological rationale for these observations is not well understood. Future studies of telomere length and NHL risk that sample men and women across a wide range of ages are needed to confirm these findings.
The abundance of single SNP associations with NHL risk, the dose–response relationship by the decile of GRS and the agreement in directionality of the GRS association with prior evidence from prospective studies suggest that variation in telomere length tagged by the nine telomere length-associated variants is important for four of the most common B-cell lymphoma subtypes. Future functional studies investigating the biological mechanisms in telomere length captured by these genetic variants and their haplotypes will be instrumental in better understanding telomere biology. In addition, an improved understanding of the key molecular pathways responsible for telomere length may also be instrumental in identifying important preventive strategies for NHL as well.
Materials and Methods
Participants and data for this study originate from four previously published subtype-specific NHL genome-wide association studies (GWASs) which included CLL/SLL (28), DLBCL (29), FL (30) and MZL (31). Each NHL subtype-specific GWAS was a collection of cases of European descent from 22 studies of NHL, including 9 prospective cohort studies, 8 population-based case–control studies and 5 hospital or clinic-based case–control studies (Supplementary Material, Table S1). All studies obtained informed consent from the participants and were approved by the respective Institutional Review Boards.
In total, 3104 cases of CLL/SLL, 3906 cases of DLBCL, 2731 cases of FL and 825 cases of MZL were extracted from stage 1 of the respective GWAS analyses. NHL diagnoses were verified by medical and pathology reports to meet InterLymph and World Health Organization (WHO) criteria (32,33). A set of 9562 cancer-free controls was also extracted from stage 1 of the NHL GWASs. Controls included cancer-free representatives from each participating NHL study population as well as additional cancer-free individuals from the α-Tocopherol, β-Carotene Lung Cancer Prevention Study (ATBC), the American Cancer Society Cancer Prevention Study II Cohort (CPS-II) and the Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial (PLCO). Further details on study makeup and participants are described in the original GWAS publications (28–31).
Briefly, DNA was isolated from participants and genotyped on commercially available Illumina SNP genotyping microarrays at participating study centers. Standard quality control and filtering was applied to called genotypes to ensure well-performing samples and high-quality genotypes were reported. Genotype imputation was performed with IMPUTE2 (34) using a hybrid of the 1000 Genomes Project version 2 (February 2012 release) (35) and the Division of Cancer Epidemiology and Genetics (DCEG) European reference panels (36).
Genotypes were extracted for the nine previously identified common SNPs associated with circulating leukocyte telomere length (rs10936599, rs2736100, rs7675998, rs9420907, rs8105767, rs755017, rs11125529, rs6772228 and rs3027234). Depending on the genotyping platform of the contributing study different combinations of telomere length-associated SNPs needed to be imputed for each study. All imputed SNPs had IMPUTE2 info scores >0.75 (Supplementary Material, Table S2), indicating that the imputation for these variants had a high degree of accuracy. There was no evidence of significant departures from Hardy–Weinberg proportions for control participants (P-values >0.01).
GRSs were calculated for telomere length-associated variants. To calculate a GRS for the ith individual from the nine telomere length-associated variants we used the following formula:
where xij is the number of risk alleles for the jth SNP of the ith subject (xij = 0, 1 or 2) and wj is the weight or coefficient for the jth SNP. For the weighted coefficients, wj, of each telomere length-associated allele, we used the previously published telomere length-associated β-estimates scaled to kb of telomere length per length allele (Table 1) (9–11). Weighting typically results in greater specificity of the GRS by assigning more weight to variants with stronger effects.
Only participants with complete genotyping, histology (for cases) and covariate (age and sex) information were included in the analysis (10 102 NHL cases and 9562 controls). Logistic regression models calculating ORs and 95% confidence intervals (95% CI) to investigate combined and subtype-specific NHL risk were adjusted for sex and a continuous variable for age, unless otherwise noted. Likelihood ratio tests were used to assess statistical significance of aggregations of the nine telomere length-associated variants on NHL risk by comparing null models with fitted models of the telomere length-associated variants.
In addition to the GRS approach, we estimated the effect of telomere length on risk of the four subtypes of NHL using two different Mendelian randomization methods that use summary association for each SNP: an inverse-variance weighting method and a likelihood-based method. The methods use average summary association estimates to estimate causal effects and are described in greater detail by Burgess et al. (37). Both the inverse-variance and likelihood-based methods give similar estimates and precision to the least squares method for individual-level data, but have the advantage of using effect estimates from other studies (37). We accessed the online web tool by Burgess et al. at https://sb452.shinyapps.io/summarized/ on 16 December 2015 to calculate both Mendelian randomization estimates. Several conditions are necessary for these Mendelian randomization effect estimates to have a causal interpretation: (1) the telomere length-associated variants need to be associated with telomere length in leukocytes, (2) the telomere length-associated variants are not associated with other factors that are associated with both telomere length and NHL risk and (3) the telomere length-associated SNPs only affect NHL risk through telomere length. While these conditions are not readily testable, tests of heterogeneity can be conducted to assess if a telomere length-associated variant's effect on telomere length is proportional to its effect on NHL risk.
An additional analysis using ASSET (38) meta-analysis software further investigates which of the four subtypes of NHL were associated with each telomere length-associated SNP as well as the overall GRS. ASSET finds the optimal subset of NHL subtypes that are associated with a SNP or GRS by performing meta-analyses that span all possible combinations of NHL subtypes and efficiently adjusts for multiple comparisons. All ASSET analyses were adjusted for age and sex.
All plotting and statistical analyses were performed on a 64-bit Windows build of R version 3.0.1 ‘Good Sport’ (39). Exact binomial tests were carried out using the binom.test function in R. All statistical tests were two-sided with P < 0.05 considered statistically significant.
Supplementary Material
Supplementary Material is available at HMG online.
Acknowledgements
Support for individual studies:
ATBC: This research was supported in part by the Intramural Research Program of the NIH and the National Cancer Institute. Additionally, this research was supported by US Public Health Service contracts N01-CN-45165, N01-RC-45035, N01-RC-37004 and HHSN261201000006C from the National Cancer Institute, Department of Health and Human Services.
BC: Canadian Institutes for Health Research (CIHR); Canadian Cancer Society and Michael Smith Foundation for Health Research.
CPS-II: The Cancer Prevention Study II (CPS-II) Nutrition Cohort is supported by the American Cancer Society. Genotyping for all CPS-II samples were supported by the Intramural Research Program of the National Institutes of Health, NCI, Division of Cancer Epidemiology and Genetics. The authors would also like to acknowledge the contribution to this study from central cancer registries supported through the Centers for Disease Control and Prevention National Program of Cancer Registries, and Cancer Registries supported by the National Cancer Institute Surveillance Epidemiology and End Results program.
ELCCS: Leukaemia & Lymphoma Research.
ENGELA: Association pour la Recherche contre le Cancer (ARC), Institut National du Cancer (INCa), Fondation de France, Fondation contre la Leucémie, Agence nationale de sécurité sanitaire de l'alimentation, de l'environnement et du travail (ANSES).
EPIC: Coordinated Action (contract #006438, SP23-CT-2005-006438); HuGeF (Human Genetics Foundation), Torino, Italy; Cancer Research UK.
EpiLymph: European Commission (grant references QLK4-CT-2000-00422 and FOOD-CT-2006-023103); the Spanish Ministry of Health (grant references CIBERESP, PI11/01810, PI14/01219, RCESP C03/09, RTICESP C03/10 and RTIC RD06/0020/0095), the Marató de TV3 Foundation (grant reference 051210), the Agència de Gestiód'AjutsUniversitarisi de Recerca – Generalitat de Catalunya (grant reference 2014SRG756) who had no role in the data collection, analysis or interpretation of the results; the NIH (contract NO1-CO-12400); the Compagnia di San Paolo—Programma Oncologia; the Federal Office for Radiation Protection grants StSch4261 and StSch4420, the José Carreras Leukemia Foundation grant DJCLS-R12/23, the German Federal Ministry for Education and Research (BMBF-01-EO-1303); the Health Research Board, Ireland and Cancer Research Ireland; Czech Republic supported by MH CZ – DRO (MMCI, 00209805) and RECAMO, CZ.1.05/2.1.00/03.0101; Fondation de France and Association de Recherche Contre le Cancer.
GEC/Mayo GWAS: National Institutes of Health (CA118444, CA148690 and CA92153). Intramural Research Program of the NIH, National Cancer Institute. Veterans Affairs Research Service. Data collection for Duke University was supported by a Leukemia & Lymphoma Society Career Development Award, the Bernstein Family Fund for Leukemia and Lymphoma Research, and the National Institutes of Health (K08CA134919), National Center for Advancing Translational Science (UL1 TR000135).
HPFS: The HPFS was supported in part by the National Institutes of Health grants CA167552, CA149445, CA098122, CA098566 and K07 CA115687 (B.M.B.). We thank the participants and staff of the Health Professionals Follow-up Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA and WY. The authors assume full responsibility for analyses and interpretation of these data.
Iowa-Mayo SPORE: NCI Specialized Programs of Research Excellence (SPORE) in Human Cancer (P50 CA97274); National Cancer Institute (P30 CA086862 and P30 CA086862) and Henry J. Predolin Foundation.
Italian GxE: Italian Association for Cancer Research (AIRC, Investigator Grant 11855) (PC); Fondazione Banco di Sardegna 2010–2012, and Regione Autonoma della Sardegna (LR7 CRP-59812/2012) (MGE).
Mayo Clinic Case-Control: National Institutes of Health (R01 CA92153) and National Cancer Institute (P30 CA015083).
MCCS: The Melbourne Collaborative Cohort Study recruitment was funded by VicHealth and Cancer Council Victoria. The MCCS was further supported by Australian NHMRC grants 209057, 251553 and 504711 and by infrastructure provided by Cancer Council Victoria. Cases and their vital status were ascertained through the Victorian Cancer Registry (VCR) and the Australian Institute of Health and Welfare (AIHW), including the National Death Index and the Australian Cancer Database.
MD Anderson: Institutional support to the Center for Translational and Public Health Genomics.
MSKCC: Geoffrey Beene Cancer Research Grant, Lymphoma Foundation (LF5541); Barbara K. Lipman Lymphoma Research Fund (74419); Robert and Kate Niehaus Clinical Cancer Genetics Research Initiative (57470); U01 HG007033; ENCODE; U01 HG007033.
NCI-SEER: Intramural Research Program of the National Cancer Institute, National Institutes of Health and Public Health Service (N01-PC-65064, N01-PC-67008, N01-PC-67009, N01-PC-67010 and N02-PC-71105).
NHS: The NHS was supported in part by National Institutes of Health grants CA186107, CA87969, CA49449, CA149445, CA098122, CA098566 and K07 CA115687. We thank the participants and staff of the Nurses' Health Study for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA and WY. The authors assume full responsibility for analyses and interpretation of these data.
NSW: NSW was supported by grants from the Australian National Health and Medical Research Council (ID990920), the Cancer Council NSW and the University of Sydney Faculty of Medicine.
NYU-WHS: National Cancer Institute (R01 CA098661 and P30 CA016087); National Institute of Environmental Health Sciences (ES000260).
PLCO: This research was supported by the Intramural Research Program of the National Cancer Institute and by contracts from the Division of Cancer Prevention, National Cancer Institute, NIH, DHHS.
SCALE: Swedish Cancer Society (2009/659). Stockholm County Council (20110209) and the Strategic Research Program in Epidemiology at Karolinska Institute. Swedish Cancer Society grant (02 6661). National Institutes of Health (5R01 CA69669-02); Plan Denmark.
UCSF2: The UCSF studies were supported by the NCI, National Institutes of Health, CA1046282 and CA154643. The collection of cancer incidence data used in this study was supported by the California Department of Health Services as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute's Surveillance, Epidemiology, and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention's National Program of Cancer Registries, under agreement #1U58 DP000807-01 awarded to the Public Health Institute. The ideas and opinions expressed here are those of the authors, and endorsement by the State of California, the California Department of Health Services, the National Cancer Institute or the Centers for Disease Control and Prevention or their contractors and subcontractors is not intended nor should be inferred.
UTAH: National Institutes of Health CA134674. Partial support for data collection at the Utah site was made possible by the Utah Population Database (UPDB) and the Utah Cancer Registry (UCR). Partial support for all datasets within the UPDB is provided by the Huntsman Cancer Institute (HCI) and the HCI Cancer Center Support grant P30 CA42014. The UCR is supported in part by NIH contract HHSN261201000026C from the National Cancer Institute SEER Program with additional support from the Utah State Department of Health and the University of Utah. Partial support for data collection in Sheffield, UK, was made possible by funds from Yorkshire Cancer Research. We thank the NCRI Haemato-oncology Clinical Studies Group, colleagues in the North Trent Cancer Network the North Trent Haemato-oncology Database.
WHI: WHI investigators are: Program Office—(National Heart, Lung, and Blood Institute, Bethesda, Maryland) Jacques Rossouw, Shari Ludlam, Dale Burwen, Joan McGowan, Leslie Ford and Nancy Geller; Clinical Coordinating Center—(Fred Hutchinson Cancer Research Center, Seattle, WA) Garnet Anderson, Ross Prentice, Andrea LaCroix and Charles Kooperberg; Investigators and Academic Centers—(Brigham and Women's Hospital, Harvard Medical School, Boston, MA) JoAnn E. Manson; (MedStar Health Research Institute/Howard University, Washington, DC) Barbara V. Howard; (Stanford Prevention Research Center, Stanford, CA) Marcia L. Stefanick; (The Ohio State University, Columbus, OH) Rebecca Jackson; (University of Arizona, Tucson/Phoenix, AZ) Cynthia A. Thomson; (University at Buffalo, Buffalo, NY) Jean Wactawski-Wende; (University of Florida, Gainesville/Jacksonville, FL) Marian Limacher; (University of Iowa, Iowa City/Davenport, IA) Robert Wallace; (University of Pittsburgh, Pittsburgh, PA) Lewis Kuller; (Wake Forest University School of Medicine, Winston-Salem, NC) Sally Shumaker; Women's Health Initiative Memory Study—(Wake Forest University School of Medicine, Winston-Salem, NC) Sally Shumaker. The WHI program is funded by the National Heart, Lung, and Blood Institute, National Institutes of Health, US Department of Health and Human Services through contracts HHSN268201100046C, HHSN268201100001C, HHSN268201100002C, HHSN268201100003C, HHSN268201100004C and HHSN271201100004C.
YALE: National Cancer Institute (CA62006) and National Cancer Institute (CA165923).
Conflict of Interest statement. The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the National Institute of Health. The authors report no conflicts of interest.
References
- 1.Blackburn E.H. (1991) Structure and function of telomeres. Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E.H. (1990) Telomeres and their synthesis. Science, 249, 489–490. [DOI] [PubMed] [Google Scholar]
- 3.Blasco M.A. (2005) Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet., 6, 611–622. [DOI] [PubMed] [Google Scholar]
- 4.Hackett J.A., Greider C.W. (2002) Balancing instability: dual roles for telomerase and telomere dysfunction in tumorigenesis. Oncogene, 21, 619–626. [DOI] [PubMed] [Google Scholar]
- 5.Kim N.W., Piatyszek M.A., Prowse K.R., Harley C.B., West M.D., Ho P.L., Coviello G.M., Wright W.E., Weinrich S.L., Shay J.W. (1994) Specific association of human telomerase activity with immortal cells and cancer. Science, 266, 2011–2015. [DOI] [PubMed] [Google Scholar]
- 6.Cawthon R.M. (2009) Telomere length measurement by a novel monochrome multiplex quantitative PCR method. Nucleic Acids Res., 37, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slagboom P.E., Droog S., Boomsma D.I. (1994) Genetic determination of telomere size in humans: a twin study of three age groups. Am. J. Hum. Genet., 55, 876–882. [PMC free article] [PubMed] [Google Scholar]
- 8.Broer L., Codd V., Nyholt D.R., Deelen J., Mangino M., Willemsen G., Albrecht E., Amin N., Beekman M., de Geus E.J. et al. (2013) Meta-analysis of telomere length in 19,713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur. J. Hum. Genet., 21, 1163–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Codd V., Nelson C.P., Albrecht E., Mangino M., Deelen J., Buxton J.L., Hottenga J.J., Fischer K., Esko T., Surakka I. et al. (2013) Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet., 45, 422–427, 427e421–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pooley K.A., Bojesen S.E., Weischer M., Nielsen S.F., Thompson D., Amin Al Olama A., Michailidou K., Tyrer J.P., Benlloch S., Brown J. et al. (2013) A genome-wide association scan (GWAS) for mean telomere length within the COGS project: identified loci show little association with hormone-related cancer risk. Hum. Mol. Genet., 22, 5056–5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mangino M., Hwang S.J., Spector T.D., Hunt S.C., Kimura M., Fitzpatrick A.L., Christiansen L., Petersen I., Elbers C.C., Harris T. et al. (2012) Genome-wide meta-analysis points to CTC1 and ZNF676 as genes regulating telomere homeostasis in humans. Hum. Mol. Genet., 21, 5385–5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhan Y., Song C., Karlsson R., Tillander A., Reynolds C.A., Pedersen N.L., Hagg S. (2015) Telomere length shortening and Alzheimer disease—a Mendelian Randomization Study. JAMA Neurol., 72, 1202–1203. [DOI] [PubMed] [Google Scholar]
- 13.Iles M.M., Bishop D.T., Taylor J.C., Hayward N.K., Brossard M., Cust A.E., Dunning A.M., Lee J.E., Moses E.K., Akslen L.A. et al. (2014) The effect on melanoma risk of genes previously associated with telomere length. J. Natl. Cancer Inst., 106, Pii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Machiela M.J., Hsiung C.A., Shu X., Seow W.J., Wang Z., Matsuo K., Hong Y., Seow A., Wu C., Hosgood H.D. III et al. (2015) Genetic variants associated with longer telomere length are associated with increased lung cancer risk among never-smoking women in Asia: a report from the female lung cancer consortium in Asia. Int. J. Cancer, 137, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang C., Doherty J.A., Burgess S., Hung R.J., Lindstrom S., Kraft P., Gong J., Amos C.I., Sellers T.A., Monteiro A.N. et al. (2015) Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum. Mol. Genet., 24, 5356–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan Q., Cawthon R., Shen M., Weinstein S.J., Virtamo J., Lim U., Hosgood H.D. III, Albanes D., Rothman N. (2009) A prospective study of telomere length measured by monochrome multiplex quantitative PCR and risk of non-Hodgkin lymphoma. Clin. Cancer Res., 15, 7429–7433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hosnijeh F.S., Matullo G., Russo A., Guarrera S., Modica F., Nieters A., Overvad K., Guldberg P., Tjonneland A., Canzian F. et al. (2014) Prediagnostic telomere length and risk of B-cell lymphoma—results from the EPIC cohort study. Int. J. Cancer, 135, 2910–2917. [DOI] [PubMed] [Google Scholar]
- 18.Kamranvar S.A., Chen X., Masucci M.G. (2013) Telomere dysfunction and activation of alternative lengthening of telomeres in B-lymphocytes infected by Epstein-Barr virus. Oncogene, 32, 5522–5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roos G., Krober A., Grabowski P., Kienle D., Buhler A., Dohner H., Rosenquist R., Stilgenbauer S. (2008) Short telomeres are associated with genetic complexity, high-risk genomic aberrations, and short survival in chronic lymphocytic leukemia. Blood, 111, 2246–2252. [DOI] [PubMed] [Google Scholar]
- 20.Lin T.T., Letsolo B.T., Jones R.E., Rowson J., Pratt G., Hewamana S., Fegan C., Pepper C., Baird D.M. (2010) Telomere dysfunction and fusion during the progression of chronic lymphocytic leukemia: evidence for a telomere crisis. Blood, 116, 1899–1907. [DOI] [PubMed] [Google Scholar]
- 21.Alexander D.D., Mink P.J., Adami H.O., Chang E.T., Cole P., Mandel J.S., Trichopoulos D. (2007) The non-Hodgkin lymphomas: a review of the epidemiologic literature. Int. J. Cancer, 120(Suppl 12), 1–39. [DOI] [PubMed] [Google Scholar]
- 22.Epel E.S., Blackburn E.H., Lin J., Dhabhar F.S., Adler N.E., Morrow J.D., Cawthon R.M. (2004) Accelerated telomere shortening in response to life stress. Proc. Natl Acad. Sci. USA, 101, 17312–17315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puterman E., Lin J., Blackburn E., O'Donovan A., Adler N., Epel E. (2010) The power of exercise: buffering the effect of chronic stress on telomere length. PLoS One, 5, e10837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ornish D., Lin J., Daubenmier J., Weidner G., Epel E., Kemp C., Magbanua M.J., Marlin R., Yglecias L., Carroll P.R. et al. (2008) Increased telomerase activity and comprehensive lifestyle changes: a pilot study. Lancet Oncol., 9, 1048–1057. [DOI] [PubMed] [Google Scholar]
- 25.Harris N.L., Jaffe E.S., Stein H., Banks P.M., Chan J.K., Cleary M.L., Delsol G., De Wolf-Peeters C., Falini B., Gatter K.C. et al. (1994) A revised European-American classification of lymphoid neoplasms: a proposal from the International Lymphoma Study Group. Blood, 84, 1361–1392. [PubMed] [Google Scholar]
- 26.Campo E., Swerdlow S.H., Harris N.L., Pileri S., Stein H., Jaffe E.S. (2011) The 2008 WHO classification of lymphoid neoplasms and beyond: evolving concepts and practical applications. Blood, 117, 5019–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Speedy H.E., Di Bernardo M.C., Sava G.P., Dyer M.J., Holroyd A., Wang Y., Sunter N.J., Mansouri L., Juliusson G., Smedby K.E. et al. (2014) A genome-wide association study identifies multiple susceptibility loci for chronic lymphocytic leukemia. Nat. Genet., 46, 56–60. [DOI] [PubMed] [Google Scholar]
- 28.Berndt S.I., Skibola C.F., Joseph V., Camp N.J., Nieters A., Wang Z., Cozen W., Monnereau A., Wang S.S., Kelly R.S. et al. (2013) Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat. Genet., 45, 868–876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerhan J.R., Berndt S.I., Vijai J., Ghesquieres H., McKay J., Wang S.S., Wang Z., Yeager M., Conde L., de Bakker P.I. et al. (2014) Genome-wide association study identifies multiple susceptibility loci for diffuse large B cell lymphoma. Nat. Genet., 46, 1233–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skibola C.F., Berndt S.I., Vijai J., Conde L., Wang Z., Yeager M., de Bakker P.I., Birmann B.M., Vajdic C.M., Foo J.N. et al. (2014) Genome-wide association study identifies five susceptibility loci for follicular lymphoma outside the HLA region. Am. J. Hum. Genet., 95, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vijai J., Wang Z., Berndt S.I., Skibola C.F., Slager S.L., de Sanjose S., Melbye M., Glimelius B., Bracci P.M., Conde L. et al. (2015) A genome-wide association study of marginal zone lymphoma shows association to the HLA region. Nat. Commun., 6, 5751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morton L.M., Turner J.J., Cerhan J.R., Linet M.S., Treseler P.A., Clarke C.A., Jack A., Cozen W., Maynadie M., Spinelli J.J. et al. (2007) Proposed classification of lymphoid neoplasms for epidemiologic research from the Pathology Working Group of the International Lymphoma Epidemiology Consortium (InterLymph). Blood, 110, 695–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner J.J., Morton L.M., Linet M.S., Clarke C.A., Kadin M.E., Vajdic C.M., Monnereau A., Maynadie M., Chiu B.C., Marcos-Gragera R. et al. (2010) InterLymph hierarchical classification of lymphoid neoplasms for epidemiologic research based on the WHO classification (2008): update and future directions. Blood, 116, e90–e98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howie B.N., Donnelly P., Marchini J. (2009) A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet., 5, e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genomes Project Consortium Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A. (2010) A map of human genome variation from population-scale sequencing. Nature, 467, 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z., Jacobs K.B., Yeager M., Hutchinson A., Sampson J., Chatterjee N., Albanes D., Berndt S.I., Chung C.C., Diver W.R. et al. (2012) Improved imputation of common and uncommon SNPs with a new reference set. Nat. Genet., 44, 6–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burgess S., Butterworth A., Thompson S.G. (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol., 37, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhattacharjee S., Rajaraman P., Jacobs K.B., Wheeler W.A., Melin B.S., Hartge P., GliomaScan C., Yeager M., Chung C.C., Chanock S.J. et al. (2012) A subset-based approach improves power and interpretation for the combined analysis of genetic association studies of heterogeneous traits. Am. J. Hum. Genet., 90, 821–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.R Development Core Team. (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.