Summary
Adapted filamentous pathogens such as the oomycetes Hyaloperonospora arabidopsidis (Hpa) and Phytophthora infestans (Pi) project specialized hyphae, the haustoria, inside living host cells for the suppression of host defence and acquisition of nutrients. Accommodation of haustoria requires reorganization of the host cell and the biogenesis of a novel host cell membrane, the extrahaustorial membrane (EHM), which envelops the haustorium separating the host cell from the pathogen. Here, we applied live-cell imaging of fluorescent-tagged proteins labelling a variety of membrane compartments and investigated the subcellular changes associated with accommodating oomycete haustoria in Arabidopsis and N. benthamiana. Plasma membrane-resident proteins differentially localized to the EHM. Likewise, secretory vesicles and endosomal compartments surrounded Hpa and Pi haustoria revealing differences between these two oomycetes, and suggesting a role for vesicle trafficking pathways for the pathogen-controlled biogenesis of the EHM. The latter is supported by enhanced susceptibility of mutants in endosome-mediated trafficking regulators. These observations point at host subcellular defences and specialization of the EHM in a pathogen-specific manner. Defence-associated haustorial encasements, a double-layered membrane that grows around mature haustoria, were frequently observed in Hpa interactions. Intriguingly, all tested plant proteins accumulated at Hpa haustorial encasements suggesting the general recruitment of default vesicle trafficking pathways to defend pathogen access. Altogether, our results show common requirements of subcellular changes associated with oomycete biotrophy, and highlight differences between two oomycete pathogens in reprogramming host cell vesicle trafficking for haustoria accommodation. This provides a framework for further dissection of the pathogen-triggered reprogramming of host subcellular changes.
Introduction
Most plant pathogens colonize tissues in the intercellular space but they depend for their development and proliferation on gaining access to the host cell (O’Connell and Panstruga, 2006). Filamentous biotrophic and hemibiotrophic pathogens penetrate host cells by breaching through the plant cell wall but their hyphae remain surrounded by a host-derived plasma membrane. It has been well documented that plant cells respond to such pathogen penetration with substantial subcellular rearrangements and that these cellular defences are important mechanisms for resisting the pathogen ingress at the level of host cell entry (Lipka et al., 2005). Remodelling of the cytoskeleton architecture, aggregation of the cell cytoplasm, the endoplasmatic reticulum (ER), and the vacuole, focal accumulation of mitochondria, Golgi bodies, peroxisomes and secretory vesicles, as well as polarized plasma membrane microdomains beneath the penetration site are subcellular changes associated with anti-fungal defence, e.g. against the powdery mildews Blumeria graminis f. sp. hordei and Erysiphe cichoracearum in barley and Arabidopsis, respectively, and Colletotrichum species in Arabidopsis (Collins et al., 2003; Bhat et al., 2005; Koh et al., 2005; Lipka et al., 2005; Shimada et al., 2006; Kwon et al., 2008). The role of vesicle secretion in defence at the level of host cell entry is underpinned by the papillary deposition of exocytic compartments and accumulation of callose beneath the fungal penetration hyphae in the extracellular space, and demonstrated by compromised penetration resistance in mutants impaired in the formation of secretory vesicles (An et al., 2006a; Kwon et al., 2008; Bednarek et al., 2009; Meyer et al., 2009). Host cell polarization has also been observed in plant–oomycete interactions. This includes the reorganization of the cytoskeleton, ER aggregation, focal accumulation of Golgi bodies and migration of the nucleus towards the penetration sites of Hyaloperonospora arabidopsidis in Arabidopsis and Phytophthora infestans in potato respectively (Schmelzer, 2002; Takemoto et al., 2003; Schütz et al., 2006). Current evidence suggests that local mechanical pressure and perception of conserved pathogen-associated molecular patterns (PAMPs) are triggers of these host subcellular changes in penetration resistance (Gus-Mayer et al., 1998; Xu et al., 1998).
Successful pathogens overcome the first layer of cellular defence and develop specialized hyphae, referred to as haustoria, which are projected inside host cells (O’Connell and Panstruga, 2006). As the haustorium grows inside the host cell, it remains enveloped by a plant-derived membrane, the extrahaustorial membrane (EHM), which separates the pathogen from the host cell and engulfs the extrahaustorial matrix (EHMx) in between. The EHM constitutes an intimate interface between the host and the pathogen, across which pathogens take up nutrients from the host and deliver effector proteins for the suppression of host defences. The EHM appears as an invagination from the plasma membrane, but its high electron density in electron micrographs indicates a distinct composition and functional differentiation (Koh et al., 2005; Micali et al., 2011). In addition, a number of plasma membrane-resident proteins are excluded from the EHM produced around fungal haustoria further supporting the view that this membrane differs distinctly from the plasma membrane (Koh et al., 2005; Micali et al., 2011). For example, Arabidopsis PEN1 encoding a plasma membrane syntaxin that focally accumulates during penetration resistance against non-adapted powdery mildew fungi was restricted to the neckband at the collar region of haustoria, and absent from the EHM of Colletotrichum hyphae (O’Connell and Panstruga, 2006). This absence of plasma membrane-resident proteins from the EHM has been suggested to result from selective mechanisms during the biogenesis of the EHM (Koh et al., 2005; O’Connell and Panstruga, 2006). One model is that exocytic compartments provide the material for membrane expansion by fusion to the plasma membrane at the collar region followed by sorting at the neckband (An et al., 2006a; Meyer et al., 2009). Alternatively, the EHM could form directly from fusing secretory vesicles since ER/Golgi-type vesicles accumulate at the penetration site of the growing haustorium (Takemoto et al., 2003; Micali et al., 2011). This is further substantiated by the localization pattern of the Arabidopsis powdery mildew resistance protein RPW8.2, which localizes to ER/Golgi-type vesicles and is present at the EHM of Golovinomyces cichoracearum (Wang et al., 2009). However, the subcellular changes underlying accommodation of filamentous pathogen haustoria are yet poorly understood. In addition, most of the modern research on haustorial cell biology has focused so far on fungi, and plant pathogenic oomycetes have been neglected despite their diversity and economic importance (Thines and Kamoun, 2010).
During later stages of the infection, fungal and oomycete haustoria are often encased by a double-layered cup-shaped callose-containing membrane structure, that appears to originate from secretory vesicles and exocytic compartments (Donofrio and Delaney, 2001; Meyer et al., 2009; Micali et al., 2011). Notably, plasma membrane-resident proteins like PEN1, which are absent from the EHM, localize to haustorial encasements (O’Connell and Panstruga, 2006; Meyer et al., 2009). Haustorial encasements constitute another layer of host cellular defence, which is delayed in compatible interactions lagging behind the pathogen proliferation (Van Damme et al., 2009).
Accommodation of haustoria most likely also affects host membrane trafficking. However, studies addressing this connection are limited despite its potential importance in understanding disease and in particular in effector delivery via endocytic processes (Rafiqi et al., 2010; Stassen and Van den Ackerveken, 2011). Ultrastructural analysis revealed a proliferation of multivesicular compartments in response to adapted powdery mildew fungi (An et al., 2006a). No evidence of subcellular changes using electron microscopy, however, was found in haustoria-containing cells infected with the oomycete pathogen Albugo (Soylu, 2004). This is unexpected as oomycetes are some of the most devastating filamentous plant pathogens known to proliferate in leaf tissues between cells and access host cells via haustoria along the growing hyphae. We therefore investigated the subcellular reorganization of host cells in response to the adapted oomycetes H. arabidopsidis (Hpa), an obligate biotrophic pathogen, and the hemibiotroph P. infestans (Pi) in Arabidopsis thaliana and Nicotiana benthamiana, respectively, using live-cell imaging of a large number of fluorescence-tagged proteins. Because the EHM constitutes the interface between the host cell and the pathogen, we focussed on markers labelling the plasma membrane, secretory and endocytic vesicles connecting the host endomembrane system with the plasma membrane.
Our study showed that there are both similarities and differences in the localization of plasma membrane-resident proteins to the EHM that surround Hpa and Pi haustoria. Differences between the two oomycetes were also revealed with markers labelling secretory and endocytic compartments. About half of the tested markers accumulated around Pi haustoria, whereas all markers surrounded Hpa haustoria, suggesting important differences in reprogramming of host cells by oomycete pathogens. Endosomes localizing around Hpa haustoria maintain bidirectional trafficking and recycling, and together with the enhanced susceptibility of mutants altered in endosome-mediated trafficking, this supports a role for endocytic trafficking at the interface between the EHM and the host endomembrane system. In contrast to Pi, Hpa haustoria are frequently encased during later stages of infections, and intriguingly, all tested markers accumulate at Hpa haustorial encasements. Encasements may therefore develop from the recruitment of default trafficking pathways.
Results
Plasma membrane markers are differentially localized to oomycete haustoria
To obtain spatial and temporal information about the host subcellular changes caused by invagination of pathogen haustoria, we performed live-cell imaging using confocal microscopy. Surprisingly little is known about the subcellular rearrangements in plant–oomycete infections and therefore we focussed on two compatible interactions: (i) Hpa isolate Waco 9 infecting A. thaliana (Tör et al., 2002), which allowed us to inspect the large collection of Arabidopsis transgenic lines stably expressing fluorescent-tagged marker proteins with known localizations; and (ii) Pi 88069 infecting N. benthamiana (Chaparro-Garcia et al., 2011) taking advantage of the transient expression of fluorescent subcellular marker proteins (Table S1). The observed differences, however, may depend on the pathogen isolates and the plant genotypes used in this study. Pi infects a several solanaceous plants including the wild tobacco species N. benthamiana (Becktell et al., 2006). Although Pi is not reported to infect N. benthamiana in nature, it responds like a compatible host with typical features of the infection process, and it is established as a useful model to dissect Pi-plant interactions (Chaparro-Garcia et al., 2011). Transgenic Arabidopsis marker lines and constructs transiently transformed in N. benthamiana exhibiting good expression levels in leaves were selected for analysing subcellular changes upon infection with Hpa or Pi respectively. Heterologous overexpression of the subcellular markers and the presence of Agrobacterium, which confers transient transformation of N. benthamiana, may influence plant defence responses to Pi infection. Thus, the use of this transient system and the study of two different host plants infected with two different oomycete pathogens may limit the comparison of subcellular changes triggered by Hpa and Pi. Importantly, despite Agrobacterium-mediated heterologous expression of mostly Arabidopsis-derived subcellular markers in N. benthamiana, the fluorescent-tagged proteins labelled the previously described subcellular compartments and were comparable to those detected in stable Arabidopsis transgenic lines (Tables S1 and S2). This suggests a significant level of conservation of the used subcellular markers between Arabidopsis and N. benthamiana making it suitable for a comparative study.
We first used fluorescent probes to follow the infection of Hpa in Arabidopsis two-week-old leaves 3 days post inoculation. Staining with the lipophilic dye FM4-64 revealed Hpa haustoria projected inside the host cells (Fig. S1A). The cytoplasm of infected cells was aggregated at the haustorial site as detected in 35S::GFP expressing transgenic lines, and the nucleus labelled by Hoechst staining was repeatedly found in close proximity to the Hpa haustorium, as recently described (Caillaud et al., 2011). Evidently, at this stage of the infection, when no sporulation of Hpa has yet occurred, the host cells are already responding with substantial subcellular changes and therefore we used this time point for further study. Along the invading hyphae, we also detected encased Hpa haustoria, which could be identified based on their double layered structure visualized by bright field imaging and confirmed by FM4-64 membrane staining as well as 35S::GFP-labelled cytoplasm surrounding both, the EHM and the encasement (Fig. S1B). For Pi infection, we used a transgenic Pi isolate 88069td expressing the red fluorescent marker tandem dimer RFP (known as tdTomato), which allowed identification of digit-like haustoria in epidermal and mesophyll cells as previously reported (Bozkurt et al., 2011; Chaparro-Garcia et al., 2011). N. benthamiana leaves transiently expressing fluorescent probes were examined 2–4 days post inoculation. At this stage of the Pi–N. benthamiana interaction, the majority of haustoria display either a callosic neckband or no callose deposits, whereas they rarely show callosic encasements (Bozkurt et al., 2011).
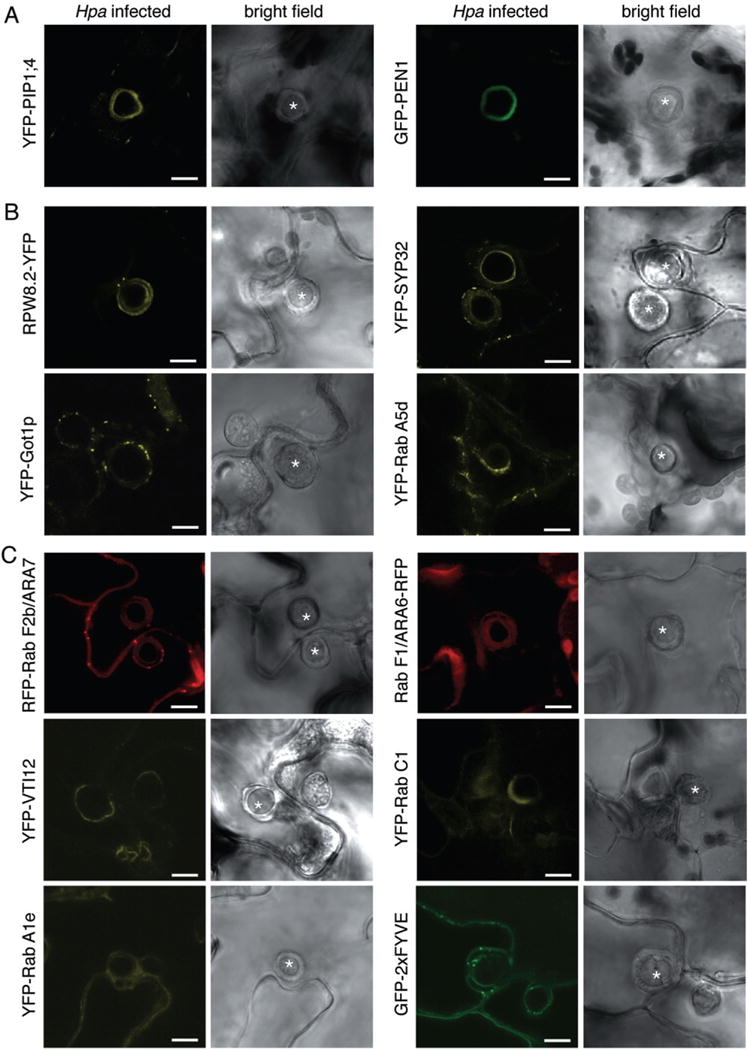
Because previous reports described that most plasma membrane-resident proteins are excluded from the EHM of fungal haustoria (Koh et al., 2005; O’Connell and Panstruga, 2006), we investigated the localization of a number of plasma membrane-resident proteins. The YFP-tagged aquaporin PIP1;4 labelled the host cell plasma membrane during infection but was not present at the EHM of Hpa and Pi haustoria (Figs 1 and 2, Table S2). Similar observations were made with the calcium ATPase ACA8 fused to GFP/YFP. These data show that plasma membrane-resident proteins are excluded from the EHM of oomycete haustoria as reported from fungal haustoria. The sharp exclusion of these plasma membrane-resident proteins from the EHM was observed at a host plasma membrane domain surrounding the haustorial neck. However, not all plasma membrane-localized proteins were excluded from the EHM of oomycete haustoria, as the plant syntaxin PEN1 fused to GFP was detected at the EHM surrounding Hpa haustoria, and GFP/YFP-tagged synaptotagmin SYT1 and the remorin StREM1.3, respectively, were present around Pi haustoria (Fig. 2, Table S2). This implicates an active selection mechanism for the absence or presence of plasma membrane proteins at the haustorial interface. Close inspection of the GFP–PEN1 around Hpa haustoria signal revealed some spot-like appearance. This may occur from exosome-derived secretion to the haustorium as known for accumulation of GFP–PEN1 in the paramural space beneath the attempted site of fungal penetration (Meyer et al., 2009).
Fig. 1.
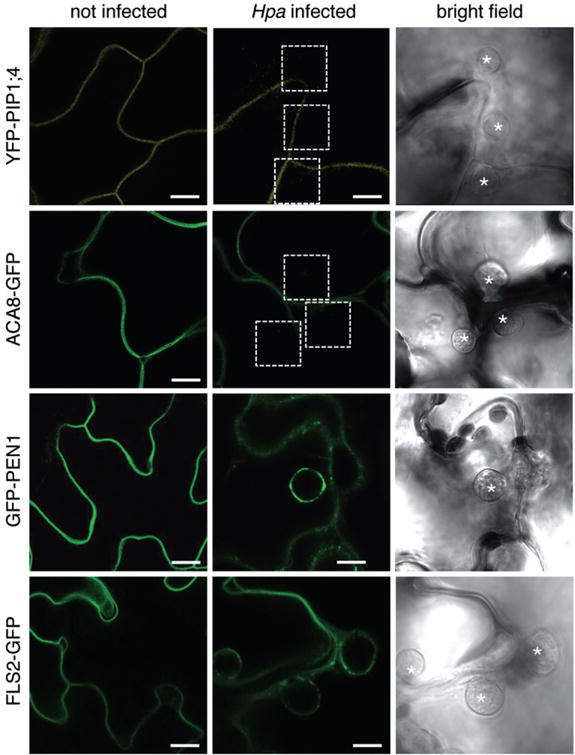
Plasma membrane-resident proteins differentially localize to the Hpa EHM. Arabidopsis Col-0 transgenic lines expressing the indicated fluorescent-tagged proteins were infected with Hpa isolate Waco 9 and haustoria were imaged at 3 dpi. Representative confocal micrographs show cross-sections of non-infected and Hpa infected leaves. Hpa haustoria are shown in bright field images indicated by asterisks. The YFP–PIP1;4 and ACA8–GFP signals are visible at the plant cell plasma membrane but not around haustoria (dashed boxes), while the GFP–PEN1 and FLS2–GFP signals are visible at the EHM of young haustoria. Bar = 10 μm.
Fig. 2.
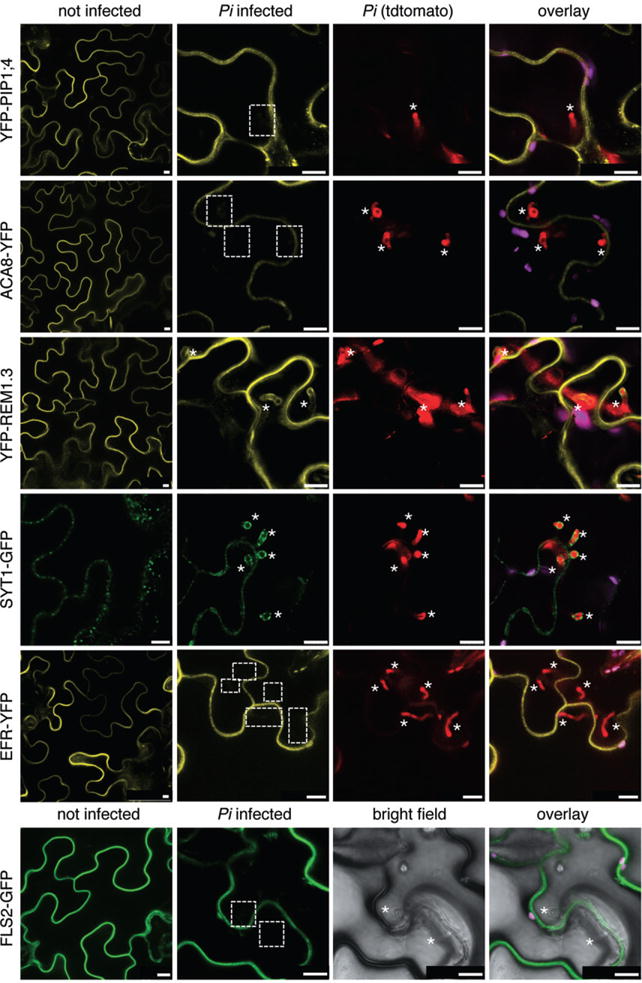
Plasma membrane-resident proteins differentially localize to Pi haustoria. N. benthamiana leaves transiently expressing the indicated fluorescently tagged proteins were infected with Pi 88069td or 88069 (lower panel) and imaged 3 dpi. Representative confocal micrographs show cross-sections of non-infected and Pi infected leaves. Pi haustoria are indicated by asterisks. The YFP–PIP1;4, ACA8–YFP EFR–YFP and FLS2–GFP signals are visible at the plant cell plasma membrane but not around haustoria (dashed boxes), while the YFP–StRem1.3 and SYT1–GFP signals are visible at haustoria. Bar = 10 μm.
We also assayed the plasma membrane receptor kinases FLS2 and EFR that sense PAMPs and have roles in plant immunity (Zipfel et al., 2004; 2006). Remarkably, we observed a differential pattern of accumulation between Hpa and Pi interactions. FLS2–GFP was detected at the EHM surrounding Hpa haustoria, but neither FLS2–GFP nor EFR–YFP signals were present around Pi haustoria (Figs 1 and 2, Table S2). This highlights specific differences in the composition of the host cell membrane enveloping Hpa and Pi haustoria. The depletion of FLS2–GFP and EFR–YFP from the EHM was not associated with a loss-of fluorescence signals at the host cell plasma membrane, which indicates that the general accumulation and delivery of these proteins to the plasma membrane remained intact.
Secretory vesicles are localized to oomycete haustoria
Focal accumulation of proteins localizing to the host cell plasma membrane and secretory vesicles is a first layer of cellular defence that has been associated with resistance to fungi and oomycetes (Schmelzer, 2002; Lipka et al., 2005; Kwon et al., 2008). However, the extent to which recruitment of secretory vesicles to penetration sites contributes to the biogenesis of the haustorial EHM during susceptible interactions remains unknown. We therefore monitored the localization of a set of YFP-tagged proteins that label vesicles of the secretory pathway ranging from Golgi compartments to the trans-Golgi network (TGN) and post-Golgi vesicles (Table S1). In infections with Hpa, we noticed that the pattern of the peripheral or vesicular labelling of the fluorescent markers was mostly unchanged between uninfected and infected cells, and in all cases was surrounding the haustorium (Figs 3A and S2A, Table S2). One marker, the syntaxin SYP32 fused to YFP, showed an enhanced accumulation of vesicle compartments at the Hpa haustorium. We observed a similar accumulation of YFP-tagged Rab GTPases D2a, E1d and A5d at vesicles surrounding Pi haustoria (Figs 3B and S2B). By contrast, GFP–VAMP722 did not accumulate at most haustorial sites but was noted to accumulate around encased Pi haustoria (Fig. 3B). However, not all vesicle markers accumulated at haustoria. We could not observe differences in the subcellular localization of vesicles labelled with the Golgi markers YFP–SYP32 and YFP–Got1p in Pi infected versus non-infected cells, and we did not notice any particular localization around the haustorium. In summary, these data provide further evidence for specific differences between the two oomycetes in the recruitment of vesicle trafficking pathways during infection. In addition, recruitment of secretory vesicles to infection sites can be dependent on the developmental stage of the haustoria, namely whether or not they are encased.
Fig. 3.
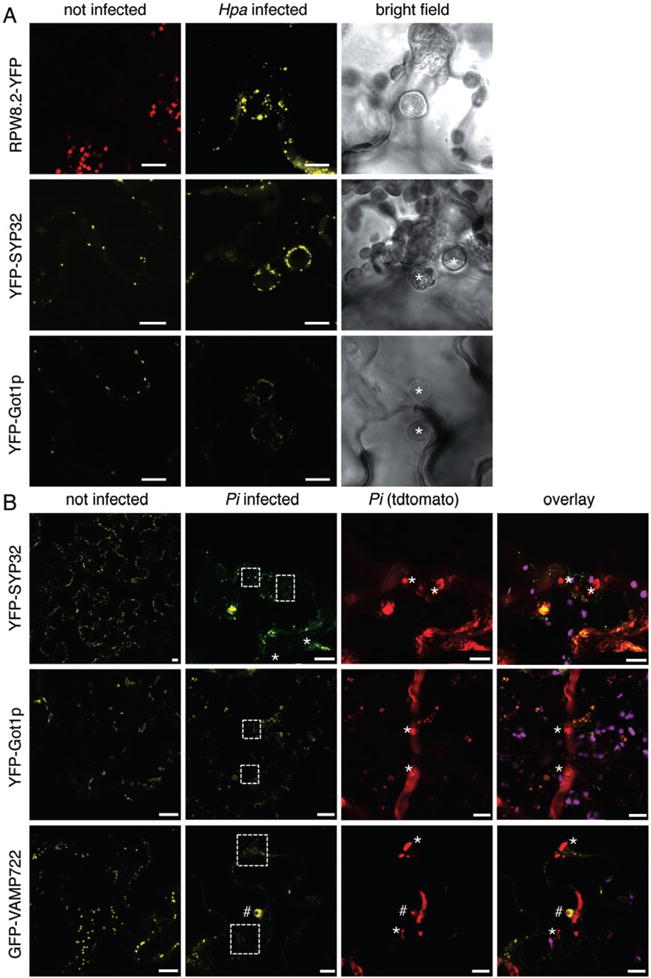
Secretory vesicles differentially localize around Hpa and Pi haustoria. A. Confocal micrographs of Arabidopsis transgenic lines expressing the indicated fluorophore fusions show cross-sections of non-infected and Hpa-infected leaves at 3 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. RPW8.2–YFP, YFP–SYP32 and YFP–Got1p are detected in vesicles around Hpa haustoria. Red signals seen in uninfected RPW8.2–YFP leaves are chlorophyll autofluorescence. Bar = 10 μm. B. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated fluorophore fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. No accumulation of YFP–SYP32, YFP–Got1p or GFP–VAMP722 was observed at haustoria with exception of encased haustoria, where GFP–VAMP722 fluorescence accumulated (#). Bar = 10 μm.
Recent reports showed that the resistance protein RPW8.2, which confers immunity against powdery mildew in Arabidopsis, is inducibly expressed and recruited to the EHM surrounding G. cichoracearum and G. orontii haustoria (Wang et al., 2009; Wang et al., 2010; Micali et al., 2011). Because RPW8.2 is also involved in immunity against oomycetes (Wang et al., 2007), we tested whether RPW8.2 localizes to Hpa haustoria. RPW8.2–YFP expression was induced during infection (Wang et al., 2010), and the protein accumulated to vesicular compartments surrounding the Hpa haustorium (Fig. 3A, Table S2). This is in agreement with the previously reported ER/Golgi-type localization of constitutively expressed RPW8.2 (Wang et al., 2007). However, unlike in powdery mildew interactions, RPW8.2 did not uniformly label the EHM of Hpa haustoria. This difference in subcellular localization indicates that RPW8.2 may have distinct functions in the interaction of fungi and oomycetes with their hosts.
Vesicles of the endocytic pathway localize around oomycete haustoria
To date much attention has been paid to EHM biogenesis from plasma membrane expansion and secretory vesicles (Koh et al., 2005; Micali et al., 2011). The presence of coated vesicles at the haustorial interface and in the surrounding cytoplasm has been noted indicating that endocytosis occurs at the EHM (O’Connell and Panstruga, 2006). Endocytic trafficking can play several roles during plant–pathogen interactions. It provides an additional way of controlling plasma membrane expansion and composition (Dhonukshe et al., 2008; Sousa et al., 2008). Moreover, endocytosis may contribute to pathogen effector trafficking, enabling effectors to cross the EHM and enter into the host cell (Gan et al., 2010; Rafiqi et al., 2010). To address the role of endocytic trafficking in plant–oomycete interactions, we studied the subcellular localization of a range of GFP/RFP/YFP-tagged markers labelling TGN/early endosomal compartments throughout multivesicular body/late endosomal compartments (Table S1).
Similar to the observations we made with markers of the secretory pathway, the overall pattern of subcellular localization between uninfected and Hpa infected cells remained mostly unchanged (Figs 4A and S3A, Table S2). All endosomal markers localized around Hpa haustoria (Figs 4A and S3A, Table S2), indicating that endocytic trafficking also occurs in Arabidopsis at the Hpa haustorial interface. We also noted that endosomal compartments surrounded Pi haustoria similar to Hpa interactions (Figs 4B and S3B, Table S2). Remarkably, two proteins, YFP–Rab F2a and YFP–Rab A1e, showed marked accumulation around Pi haustoria (Figs 4B and S3B). By contrast, the subcellular distribution of the endosomal marker VTI12 fused to YFP was unaltered in haustoria-containing plant cells demonstrating differential recruitment of endosomal compartments to the haustorial site. This highlights specific differences in the employment of endosomal compartments to oomycete haustoria, a similar finding we obtained from monitoring vesicles of the secretory pathway (Table S2). Endocytic trafficking interfaces with the vacuole to direct cargo for degradation. We tested markers labelling late endosomal compartments and/or the tonoplast localized and observed that in all cases these markers are present around the haustoria of Hpa and Pi (Fig. S4A and B), as recently reported (Caillaud et al., 2011).
Fig. 4.
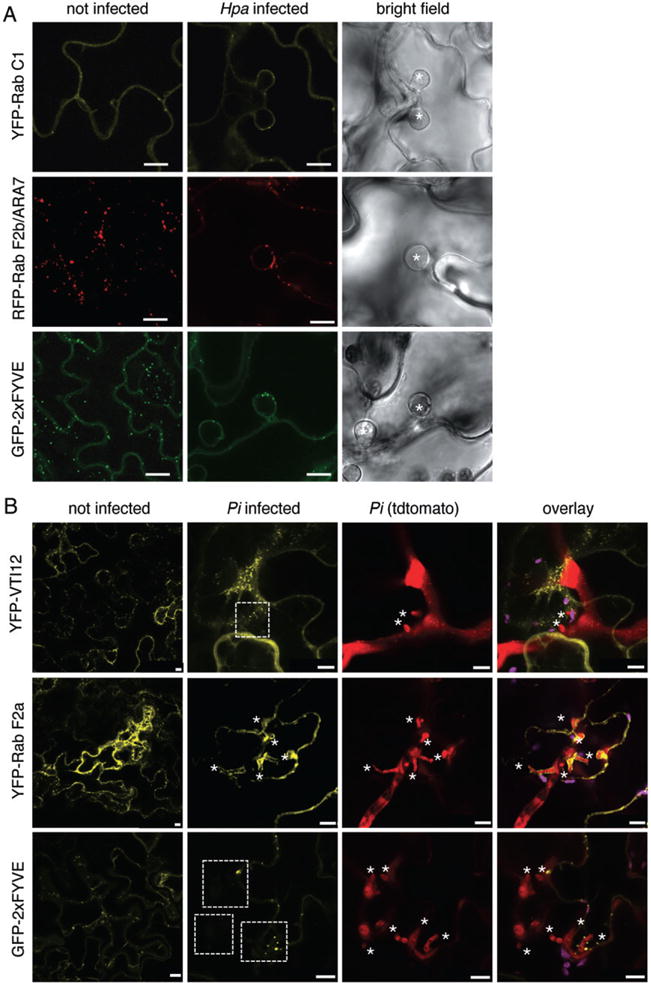
Endosomal compartments differentially localize around Hpa and Pi haustoria. A. Confocal micrographs of Arabidopsis transgenic lines expressing the indicated fluorophore fusions show cross-sections of non-infected and Hpa-infected leaves at 3 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. YFP–Rab C1, RFP–ARA7 and GFP–2xFYVE signals are surrounding Hpa haustoria. Bar = 10 μm. B. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated fluorophore fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. While YFP–Rab F2a accumulates around haustoria, YFP–RabC1 and GFP–2xFYVE signals are surrounding Pi haustoria but do not show a significant accumulation. Bar = 10 μm.
It is well possible that the observed changes in vesicle localization are a result of altered trafficking/turnover. Pathogen effectors could affect the mobility of the vesicles, and/or host cell trafficking machinery may be constrained by accommodation of haustoria (Bozkurt et al., 2011). To examine the trafficking of endosomal compartments in Hpa infected cells, we applied time-lapse imaging and monitored movements of GFP–2xFYVE-labelled vesicles. We found that GFP–2xFYVE-labelled vesicles were highly mobile and trafficked along cytoplasmic strands throughout the cell and at the periphery (Fig. S5A). Notably, vesicle movements occurred towards and away from the haustorial site. This demonstrates active endosomal trafficking in Hpa infected cells with no obvious directed movements, thereby enabling bidirectional interactions between the EHM and the host endosomal pathway. The biosensor GFP–2xFYVE labels late endosomal compartments and also to some extent the plasma membrane through the association with phospho-inositol-3-phosphates (Vermeer et al., 2006). As a weak uniform 2xFYVE–GFP signal was also present around the haustorium, the EHM may contain phosphoinositol-3-phosphates, which are currently discussed as binding sites for exerting effector activities (Gan et al., 2010; Rafiqi et al., 2010; Yaeno et al., 2011).
One possible interface between the EHM and trafficking endosomes could involve endocytic recycling. We therefore addressed whether Hpa infected cells maintain sensitivity to Brefeldin A (BFA). BFA is an inhibitor of endocytic recycling leading to the accumulation of early endosomes in the cytoplasm referred as BFA-bodies (Geldner et al., 2003). Staining with FM4-64 revealed the presence of BFA-bodies in Hpa infected cells (Fig. S5B). The formation of BFA-bodies in Hpa infected cells shows that endocytosis, early and recycling endosome trafficking is functional in haustoria-containing cells and may occur at both sites, the PM and the EHM.
We further focussed on the ENDOSOMAL COMPLEX REQUIRED FOR TRANSPORT (ESCRT), because ESCRT proteins are involved in endosome biogenesis and cargo recognition (Hurley and Hanson, 2010). The ESCRT machinery consists of three subcomplexes I–III with ESCRT-I composed of VACUOLAR PROTEIN SORTING (VPS) 23, VPS28 and VPS37, each of which encoded by at least two gene homologues in Arabidopsis (Spitzer et al., 2006). Recently, VPS28 was shown to localize to the TGN/early endosomal compartment (Scheuring et al., 2011), and to gain insights into the role of TGN-derived vesicle trafficking in oomycete infections, we investigated VPS28 subcellular localization and mutant phenotypes. RFP- or YFP-tagged VPS28-2 labelled vesicles, which were detected around Hpa and Pi haustoria (Fig. 5A and B), in line with the localization pattern of other TGN/early endosomal markers. T-DNA insertion knock-out mutant plants of VPS28-2 exhibited enhanced susceptibility when infected with Hpa isolate Waco 9 (Fig. 5C). Likewise, T-DNA insertion knock-out plants in VPS37-1, another ESCRT-I component, showed enhanced susceptibility to Hpa infection. Both vps28-2 and vps37-1 mutants supported similar growth of Hpa, but less than ENHANCED DISEASE SUSCEPTIBLE 1 (eds1) mutants pointing at genetic redundancy. Nevertheless, the enhanced susceptibility of mutants in ESCRT-I components suggests that endosomal trafficking contributes to plant immunity against invasive pathogens possibly at multiple levels.
Fig. 5.
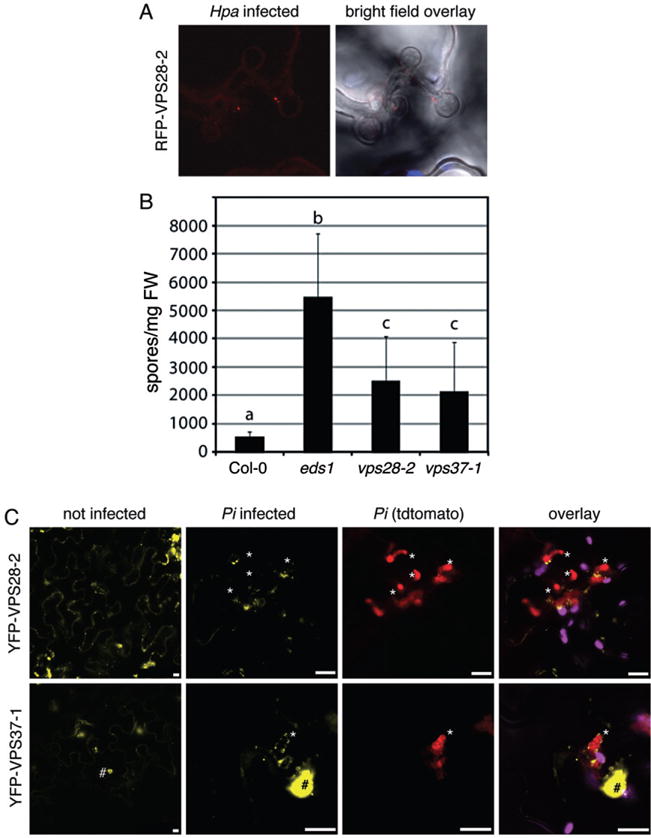
ESCRT-I components contribute to immunity against Hpa infection. A. Representative confocal images of RFP–VPS28-2 expressing plants show cross-sections of leaves infected with Hpa at 4 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. RFP–VPS28-2 vesicles are present at Hpa haustoria. B. Spores of Hpa Waco 9 were quantified at 7 dpi on eight 2-week-old seedlings. Different letters indicate statistically significant of P <0.05 based on multiple pairwise comparisons based on standard post hoc ANOVA analysis. Error bars represent SD. C. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated YFP fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. YFP–VPS28-2 and YFP–VPS37-1 positive compartments localize around Pi haustoria. YFP–VPS37-1 also localizes to subnuclear foci (#). Bar = 10 μm.
Haustorial encasements accumulate membrane compartments by default
To counteract successful pathogens and restrict their access to nutrients, plant cells employ a second layer of cellular defence to fully encase haustoria over time. Fungal haustoria are encased by continuous focal accumulation of secretory vesicles to the penetration site (Meyer et al., 2009). We investigated oomycete encasements by focusing on Hpa haustoria according to their distinct appearance that can be visualized by bright field imaging. All plasma membrane markers including YFP–PIP1;4 and ACA8–GFP, which were absent from the EHM, were detected at encasements (Fig. 6A). This difference in exclusion from the EHM but accumulation at encasements suggests a profound difference in the pathways leading to the formation of the EHM versus encasements. We also found that all markers of secretory compartments label Hpa haustorial encasements (Fig. 6B). Interestingly, RPW8.2 localized more uniformly to encasements compared with its vesicular localization around haustoria, which may be the result of fusion of the secretory vesicles. Likewise, all markers of endosomal compartments labelled encasements (Fig. 6C). Multivesicular compartments have been previously revealed at papillae beneath penetration sites and at haustorial encasements during fungal infections and it is suggested that their deposition employs exocytic pathways (Meyer et al., 2009). Exocytic trafficking may also apply to deliver late endosomal compartments to Hpa haustorial encasements. However, it is less clear by which route early endosomal compartments are targeted to encasements. Taken together, the collective accumulation of all tested markers at encasements points to an overall recruitment of default trafficking pathways for non-selectively sending membrane compartments to Hpa haustoria. Furthermore, the distinct accumulation of VAMP722 to encased haustoria but not to non-encased haustoria suggests a reprogramming of subcellular trafficking dependent on the maturation status of the haustorium (Fig. 3B).
Fig. 6.
Proteins labelling host cell membrane compartments localize to Hpa encasements. Arabidopsis Col-0 transgenic lines expressing the indicated fluorescent-tagged proteins were infected with Hpa isolate Waco 9 and imaged by confocal microscopy. Encased Hpa haustoria were imaged at 3 dpi and shown in bright field images indicated by asterisks. The fluorescent signals of (A) plasma membrane-localized, (B) Golgi-localized and (C) endosomal-localized proteins are visible at encasements of old Hpa haustoria. Bar = 10 μm.
Discussion
The subcellular changes at the haustorial interface of oomycetes have not been comprehensively studied even though the EHM enveloping haustoria is a critical component of disease caused by filamentous biotrophic pathogens, providing the most intimate interface of molecular exchange between the pathogen and the plant. Our findings point to a significant difference between Hpa and Pi infections that may reflect the different lifestyles of these pathogens. In contrast to Hpa, which represents an obligate biotroph, Pi is a hemibiotrophic pathogen and thus likely triggers different and/or additional suppression of plant defences, which may also reflect species-specific differences in the biogenesis and specialization of the EHM between both oomycetes.
Specialization of the oomycete EHM
Previous studies have shown that the EHM that is formed following infection by fungal pathogens such as powdery mildews is a highly specialized membrane structure with most plasma membrane proteins being excluded (Koh et al., 2005; O’Connell and Panstruga, 2006; Micali et al., 2011). In particular, the absence of a number of aquaporins was noted from the EHM of E. cichoracearum (Koh et al., 2005), which is also absent from the EHM of both, Hpa and Pi, highlighting commonalities in the composition of the EHM across microbial kingdoms and between species. The absence of aquaporins might be a functional requirement for the accommodation of haustoria in general; a possibility that deserves to be explored in the future. Similarly, ATPases may be generally excluded from the EHM (O’Connell and Panstruga, 2006), in agreement with the absence of the Ca2+ ATPase ACA8 from the Hpa and Pi EHM.
The selective mechanisms that function to exclude plasma membrane-localized proteins from the EHM but allow normal delivery of proteins such as PIP1;4 and ACA8 to the plasma membrane must prevent the diffusion of proteins from the plasma membrane into the EHM and the delivery of plasma membrane-localized proteins from newly fusing vesicles (Fig. S6). The underlying molecular mechanism, however, remains unknown. Inhibition of lateral diffusion of plasma membrane-localized proteins was recently described for the casparian strip membrane domain (CSD) in the root endodermis, and interestingly, like the EHM, the CSD is highly dense in electron micrographs (Roppolo et al., 2011). Given that both the EHM and CSD are membrane domains in continuum with the plasma membrane, it is possible to speculate that some of the components that prevent lateral diffusion of plasma membrane-localized proteins might be shared between these two specialized membranes.
PEN1 is an important component of the first layer of cellular resistance preventing pathogens from accessing host cells, and focally accumulates at hyphal penetration sites showing some species-specific differences (Collins et al., 2003; Kwon et al., 2008; Meyer et al., 2009). While PEN1 is excluded from the EHM of mature hyphae of adapted Colletotrichum, it localizes to the invaginated plasma membrane around young haustoria of these fungi (Shimada et al., 2006), and is present at the EHM of Hpa, in line with PEN1 being dispensable for immunity against virulent Hpa isolates (Kwon et al., 2008). This differential localization of PEN1 at the haustorial interface of fungi and oomycetes demonstrates EHM specialization in different pathosystems, likely resulting from differences in effector-triggered host subcellular changes and representing an adaptation to the individual requirements of each pathogen.
Focusing on receptor kinases, BRI1 is absent from powdery mildew EHM (Koh et al., 2005), but FLS2 localized to the EHM of Hpa, further demonstrating that plasma membrane-resident proteins are not excluded from the EHM by default, but rather through a selective mechanism. Nevertheless, FLS2 and EFR were not detected around Pi haustoria. So far these two receptors are not known to play a role in immunity against Hpa and Phytophthora, but this needs to be carefully evaluated in the future. Adapted pathogens may interfere with the integration into the EHM of a subset of receptors to prevent the detection of PAMPs, such as chitin and β-1-3-glucans, which are exposed at the surface of powdery mildew haustoria (Micali et al., 2011). The exclusion of pattern recognition receptors from the EHM could be one mechanism employed by pathogens to avoid detection by the plant immune system. Current work in our laboratory aims at determining the extent to which host-translocated effectors of Pi perturb the cellular localization of plant immune receptors and other extracellular defences (Bozkurt et al., 2011). In addition, it is possible that Pi effectors trigger the degradation of PRRs as it was reported for bacterial effectors (Göhre et al., 2008; Gimenez-Ibanez et al., 2009). In such case Pi effectors could specifically act at the haustorial site without much impacting host protein levels at the plasma membrane.
Vesicle trafficking in accommodation of oomycete haustoria
Secretory pathways have been implicated in the biogenesis of the EHM in fungal infections (Takemoto et al., 2003; Koh et al., 2005; Micali et al., 2011), and underlined by our findings that all tested proteins labelling a range of secretory vesicles localize around Hpa haustoria, potentially delivering membrane material to the EHM as the haustorium develops. This is supported by ultrastructural analysis showing small vesicles in near proximity and connected with the EHM of Hpa (Mims et al., 2004). As a result, proteins normally delivered to the plasma membrane such as PEN1, SYT1, StREM1.3 and FLS2 can be present at the EHM surrounding the haustoria of Hpa and Pi. The differences in localization of Golgi markers in Hpa versus Pi haustorium-containing cells demonstrate differential employment of secretory vesicles during EHM biogenesis, which may further account for the specific exclusion of plasma membrane-resident proteins from the EHM (Fig. S6). It also shows that the described reorganization of the host cell cytoskeleton and polarization upon oomycete infection does not simply have a non-selective, global effect on vesicle trafficking (Schmelzer, 2002; Takemoto et al., 2003).
Although previously there was only little evidence for endocytosis employed at the haustorial interface (O’Connell and Panstruga, 2006), the large number of endosomal compartments localizing around haustoria suggest a role in the biogenesis/specialization of the EHM (Fig. S6). Endocytosis is important in counter balancing vesicle fusion in the tip growth of pollen tubes and may play a similar role in the growing EHM (Sousa et al., 2008). Recently, identification of a barley ROP GTPase regulating root tip growth and haustoria accommodation was reported, pointing at shared mechanisms between these two pathways (Hoefle et al., 2011). Endocytosis also provides a mechanism, by which the composition of plasma membrane proteins is regulated, e.g. the polar localization of the PIN proteins (Dhonukshe et al., 2008). Thus, it is possible that the exclusion of plasma membrane-resident proteins from the EHM is regulated by endocytosis as a result of pathogen-mediated reprogramming of host membrane trafficking. However, endosomal trafficking also contributes to plant immunity, as demonstrated by the enhanced susceptibility of mutants in the ESCRT-I components VPS28-2 and VPS37-1 that regulate the sorting of endocytosed cargoes (Spitzer et al., 2006; Hurley and Hanson, 2010; Scheuring et al., 2011). Possible cargoes could be pattern-recognition receptors or pathogen effectors, and inefficient sorting/trafficking of these cargoes could affect plant defences.
Haustorial encasements
Encasements have been described for both fungal and oomycete haustoria and differ from the EHM not only by structure but also in composition, as they are, e.g. rich in callose. Also, plasma membrane-resident PEN1, which is excluded from the EHM of Colletotrichum fungi, localizes to haustorial encasements of non-adapted B. graminis and adapted G. orontii powdery mildews as well as to Hpa, in line with our findings (Meyer et al., 2009). Remarkably, all tested fluorescent-tagged membrane markers accumulate at haustorial encasements of Hpa. This includes proteins depleted from the EHM, such as the aquaporin PIP1;4 and the ATPase ACA8, as well as ER/Golgi-localized RPW8.2. While RPW8.2 localizes to vesicular compartments around the haustorium, it shows a uniform distribution at encasements, possibly a result from vesicle fusion and suggests the presence of membranes, which differ in composition to the EHM and rather resemble plasma membrane features.
Previous ultrastructural studies revealed the presence and proliferation of multivesicular bodies in close proximity to powdery mildew haustoria (An et al., 2006a), which also can accumulate at papillae beneath fungal penetration sites (An et al., 2006b; Böhlenius et al., 2010). We found proteins labelling TGN/early endosomal compartments as well as late endosomes/multivesicular bodies localizing to Hpa encasements, which suggests similar mechanisms in papillae formation and development of encasements. Thus, overlapping but different pathways may act in the biogenesis of the highly specialized EHM and haustorial encasements. Callose-containing encasements of Pi haustoria have also been described in incompatible interactions (Enkerli et al., 1997; Lipka et al., 2005), but were rare in the Pi–N. benthamiana interaction we examined. Interestingly, at non-encased haustoria, we noted a marked accumulation of trafficking vesicles around haustoria that was not restricted to the haustorial neck region (Fig. S6). Such altered vesicle localization could be a consequence of the perturbation of plant secretory pathways by the pathogen consistent with the recent findings that the Pi RXLR-type effector AVRblb2 interferes with focal immunity by preventing the secretion of a host immune protease (Bozkurt et al., 2011). Alternatively, the recruitment of these vesicles to haustorial sites may reflect early stages of encasement development.
Altogether, live-cell imaging using fluorescent-tagged markers of membrane compartments has revealed the dynamic subcellular changes that occur during host cell accommodation of pathogen haustoria. These subcellular changes have been poorly described so far and our study provides a framework for further dissection of pathogen reprogramming of host cells. Our data highlight possible sorting mechanisms acting at a host plasma membrane domain surrounding the haustorial neck and suggest a role for vesicle trafficking, particularly endocytosis, in compatible plant–oomycete interactions (Fig. S6). Moreover, we provide evidence for commonalities and differences between fungal, Hpa and Pi oomycete EHMs. While the commonalities point at underlying mechanisms of biotrophy, the differences may account for the specific requirements of each pathogen and its effector repertoire to cause disease. Future work will reveal the extent to which pathogen effectors perturb the accumulation of plant proteins at the EHM and around haustoria.
Experimental procedures
Plant lines and growth
Arabidopsis Col-0 transgenic lines used in this study were described before: WAVE lines (Geldner et al., 2009), 35S::GFP–PEN1 (Meyer et al., 2009), pRPW8.2::RPW8.2–YFP (Wang et al., 2009), 35S::ACA8–GFP (Lee et al., 2007), pFLS2::FLS2–GFP (Göhre et al., 2008). The FLS2–GFP line is in Nd-0 background. The UBQ10::ARA6-mRFP and UBQ10::mRFP–ARA7 lines were available from K. Schumacher, University Heidelberg, Germany. The GFP–2xFYVE line is in the Arabidopsis Ler background (Voigt et al., 2005). All plants were grown on soil under long day conditions in controlled environments. The 35S::RFP–VPS28-2 lines were generated by Agrobacterium-mediated floral transformation of Col-0 (Clough and Bent, 1998) using the VPS28-2 coding sequence cloned into the pGWB555 binary vector (Nakagawa et al., 2007). Primers used for cloning RFP–VPS28-2 were: 5′-GGG GAC AAG TTT GTA CAA AAA AGC AGG CTT AAT GAT GGA GGT CAA ATT ATG GAA CGA C-3′ and 5′-GGG GAC CAC TTT GTA CAA GAA AGC TGG GTA TTA ATT ACC AGC TTT AGG CAA AGC TGC C-3′. The construct was confirmed by sequencing. Homozygous lines in T2 generation were used for this study. T-DNA insertion lines of VPS28-2 (At4g05000) and VPS37-1 (At3g53120) were obtained from SALK. Primers used for genotyping to select homozygous vps28-2 and vps37-1 knock-out alleles were: 5′-ATG ATG GAG GTC AAA TTA TGG AAC GAC-3′ and 5′-TTA ATT ACC AGC TTT AGG CAA AGC TGC C-3′, and 5′-ATG TTC AAT TTC TGG GGA TC-3′ and 5′-TCA AAT GTT TGA CGT TTT AGC-3′ respectively.
N. benthamiana transient transformation
Additional binary vector T-DNA constructs were described before: 35S::SYT1–GFP (Schapire et al., 2008), 35S::GFP–VAMP722 (Schornack et al., 2009), 35S::YFP–StREM1 (Raffaele et al., 2009). A. tumefaciens suspensions expressing the binary constructs were diluted in infiltration buffer medium (10 mM MgCl2, 5 mM 2-N-morpholino ethanesulfonic acid pH 5.3, and 150 μM acetosyringone) to a final OD600 of 0.4. Five week old N. benthamiana leaves were infiltrated with the agrobacteria and leaves were detached 24 hours post infiltration, washed under de-ionized water, surface dried and placed in closed trays on wetted paper towels to maintain high humidity for subsequent Pi infections.
Hpa infections
For microscopic inspection, two weeks-old Arabidopsis Col-0 seedlings expressing the whole range of fluorescent-labelled subcellular markers were inoculated with 5 × 104 spores ml−1 spore suspensions of H. arabidopsidis isolate Waco 9 (Tör et al., 2002). Likewise, Arabidopsis Ler seedlings expressing the GFP–2xFYVE marker were infected with H. arabidopsidis isolate Cala 2. Infected plants were kept under high humidity at 18°C and long day conditions, and at 3 dpi detached leaves were subjected to confocal microscopy. For Hpa infections, Hpa spore suspensions of 5 × 104 spores ml−1 were spray-inoculated onto two weeks-old seedling and incubated at high humidity at 18°C. At seven dpi, spores of twelve seedlings per genotype (in pools of three) were washed from infected leaves by vortexing in 1 ml ddH2O and quantified in relation to seedling fresh weight. Spores were counted with an improved Neubauer haemocytometer (Brand, Wertheim, Germany).
Pi infections
The following isolates were used in Pi infection assays: P. infestans 88069 (van West et al., 1998) and a transformant expressing a cytosolic tandem DsRed protein (88069td) (Chaparro-Garcia et al., 2011). For Phytophthora infection assays were carried out as described using zoospore droplet inoculation of detached N. benthamiana leaves (Chaparro-Garcia et al., 2011).
Staining and chemical treatments
Detached Arabidopsis leaves were incubated with 30 μM Brefeldin A for 90–120 min, 5 μg ml−1 FM4-64 for 30 min, or 5 μg ml−1 Hoechst dye (all obtained from Sigma-Aldrich) for 30 min after vacuum infiltration.
Confocal microscopy
For imaging Hpa haustoria, Arabidopsis cotyledon leaves were detached three days post infection. Imaging was done using a Leica TCS SP5 confocal microscope (Leica Microsystems, Germany) and a 63 × water immersion objective. For imaging Pi haustoria, discs of infected N. benthamiana leaves were mounted in water and imaged using 20 ×, 40 × air and 63 × water immersion objectives. Samples were excited at the following wavelengths: 488 nm for GFP, 514 nm for eYFP, and 561 nm for mRFP. The following filters for emission spectra were used: GFP at 495–530 nm, for eYFP at 540–580 nm, mRFP at 600–650 nm. Hoechst stained samples were excited at 405 nm and the emission was measured at 450–480 nm. FM4-64 stained samples were excited at 561 nm and the emission was measured at 600–650 nm. Image analysis was done with Leica LAS AF software, ImageJ (1.43u) and Adobe PHOTOSHOP CS4 (11.0).
Supplementary Material
Fig. S1. Imaging Hpa haustoria. Arabidopsis Col-0 wild-type and 35S::GFP transgenic lines were infected with Hpa isolate Waco and haustoria were imaged by confocal microscopy. At 3 dpi wild-type leaves were incubated with the membrane dye FM4-64, or nuclear stain Hoechst respectively. Confocal micrographs show cross-sections of non-infected and Hpa infected leaves. Hpa haustoria are shown in bright field images indicated by asterisks.
A. Membrane FM4-64 and cytosolic 35S::GFP signals are visible at the host cell periphery and around young haustoria. The Hoechst signal is visible in the nucleus in close proximity to the haustorium.
B. FM4-64 membrane and 35S::GFP cytosol labelling shows the double-layered structure of encasements of old Hpa haustoria. Bar = 10 μm.
Fig. S2. Secretory vesicles differentially localize around Hpa and Pi haustoria.
A. Confocal micrographs of Arabidopsis transgenic lines expressing the indicated fluorophore fusions show cross-sections of non-infected and Hpa-infected leaves at 3 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. YFP–Rab E1d and YFP–Rab A5d signals are detected around Hpa haustoria. Bar = 10 μm.
B. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated fluorophore fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. YFP–Rab D2a, YFP–Rab E1d and YFP–Rab A5d are surrounding Pi haustoria. Bar = 10 μm.
Fig. S3. Endosomal compartments differentially localize around Hpa and Pi haustoria.
A. Confocal micrographs of Arabidopsis transgenic lines expressing the indicated fluorophore fusions show cross-sections of non-infected and Hpa-infected leaves at 3 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. YFP–VTI12, YFP–Rab A1e and ARA6–RFP signals are present at Hpa haustoria. Bar = 10 μm.
B. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated fluorophore fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. While YFP–VTI12 does not surround haustoria, YFP–Rab A1e accumulates around Pi haustoria. Bar = 10 μm.
Fig. S4. Proteins labelling the vacuolar tonoplast localize around Hpa and Pi haustoria.
A. Confocal micrographs show cross-sections of Arabidopsis transgenic leaves expressing the indicated fluorescent-tagged proteins non-infected and Hpa-infected at 3 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. YFP–Rab G3f and YFP–VAMP711 signals are surrounding Hpa haustoria. Bar = 10 μm.
B. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated fluorophore fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. YFP–Rab G3c accumulates around Pi haustoria. Bar = 10 μm.
Fig. S5. Hpa haustoria-containing cells maintain endosomal trafficking.
A. Arabidopsis Ler transgenic lines expressing GFP–2xFYVE as a marker for late endosomal compartments were infected with Hpa isolate Cala 2 and haustoria were imaged at 3 dpi. Time-lapse imaging by confocal microscopy shows cross-sections of Hpa infected leaves over 815 ms. Hpa haustoria are indicated by asterisks. The GFP–2xFYVE signal is visible at the host cell periphery, some emerging plasma membrane strands and at the EHM of Hpa haustoria. The arrows indicate vesicle movement towards the haustorial site (upper panel) and away from the haustorium (lower panel). Bar = 10 μM.
B. Arabidopsis Col-0 plants were infected with Hpa isolate Waco 9, and at 3 dpi leaves were treated with BFA and stained with FM4-64. Confocal micrographs show cross-sections of non-infected and Hpa-infected leaves. Hpa haustoria are shown in bright field images indicated by asterisks. The FM4-64 signal is visible at the plant cell plasma membrane, at BFA bodies indicated by arrows, and at the Hpa EHM. The BFA body is detected in a different focal plane of the haustoria-containing cell shown by single plane and maximal projection. Bar = 10 μm.
Fig. S6. Schematic representation of membrane trafficking in oomycete infected plant cells. Secretory vesicles deliver proteins such as PIP1;4 to the plasma membrane, but their lateral diffusion and incorporation into the extrahaustorial membrane must be prevented by the pathogen (EHM; red lines). Secretory vesicles may also deliver proteins like PEN1 to the EHM. Early endosomes (EE) may interact with the EHM and regulate presence of proteins at the EHM via recycling trafficking. Late endosomes (LE)/multivesicular bodies may contribute to enveloping the haustorium but proteins are sorted at the haustorial neck. As a result of recruitment of default membrane trafficking pathways, secretory vesicles, early and late endosomes contribute in the biogenesis of the encasement. TGN: trans-Golgi network; EHMx: extrahaustorial matrix; black arrows: proposed membrane trafficking pathways; molecular markers are highlighted in grey.
Table S1. Markers and lines used in this study. Arabidopsis plants stably expressing fluorescent-tagged markers (At line) were used for Hyaloperonospora arabidopsidis (Hpa) infections; transient expression of the respective markers (construct) in N. benthamiana (Nb) was used for Phytophthora infestans (Pi) infections; plasma membrane (PM), endoplasmatic reticulum (ER), trans-Golgi network (TGN), extrahaustorial membrane (EHM).
Table S2. Summary of marker localization in the absence and presence of Hyaloperonospora arabidopsidis (Hpa) and Phytophthora infestans (Pi) infection in A. thaliana (At) and N. benthamiana (Nb). Detection of fluorescent signals: clear discrete signal (discrete); weak widespread signal (diffuse); at cell periphery (periphery); at spots (vesicles); not at haustoria (excluded); at haustoria (surrounding); at extrahaustorial membrane (EHM); at haustorial encasements (encased); accumulates at haustoria (+); reduced (−); no difference in localization between uninfected and infected (no difference).
Acknowledgments
We thank R. Panstruga (Aachen) for reading the manuscript and Rachael Wilkinson for technical support. We are grateful to B. Voigt, V. Lipka, C. Zipfel for providing constructs and S. Whisson (Dundee) for the Pi RFP strain.
Footnotes
Author contributions
Y.-J.L., S.S., T.S., S.K. and S.R. designed research; N.G., J.C., Sw.S. and K.S. provided materials; Y.-J.L., S.S. and T.S. performed research; Y.-J.L., S.S., T.S. and S.R. analysed the data; S.S., N.G. and S.K. edited the paper; and S.R. wrote the paper.
Supporting information
Additional Supporting Information may be found in the online version of this article:
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- An Q, Ehlers K, Kogel KH, van Bel AJ, Hückelhoven R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 2006a;172:563–576. doi: 10.1111/j.1469-8137.2006.01844.x. [DOI] [PubMed] [Google Scholar]
- An Q, Hückelhoven R, Kogel KH, van Bel AJ. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol. 2006b;8:1009–1019. doi: 10.1111/j.1462-5822.2006.00683.x. [DOI] [PubMed] [Google Scholar]
- Becktell MC, Smart CD, Haney CH, Fry WE. Host-pathogen interactions between Phytophthora infestans and the solanaceous hosts Calibrachoa × hybridus, Petunia × hybrida and Nicotiana benthamiana. Plant Dis. 2006;90:24–32. doi: 10.1094/PD-90-0024. [DOI] [PubMed] [Google Scholar]
- Bednarek P, Pislewska-Bednarek M, Svatos A, Schneider B, Doubsky J, Mansurova M, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA. 2005;102:3135–3140. doi: 10.1073/pnas.0500012102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhlenius H, Mørch SM, Godfrey D, Nielsen ME, Thordal-Christensen H. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell. 2010;22:3831–3844. doi: 10.1105/tpc.110.078063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt TO, Schornack S, Win J, Shindo T, Oliva R, Cano LM, et al. Phytophthora infestans effector AVRblb2 prevents focal secretion of a plant immune protease. Proc Natl Acad Sci USA. 2011;108:20832–20837. doi: 10.1073/pnas.1112708109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud MC, Piquerez SJ, Fabro G, Steinbrenner J, Ishaque N, Beynon J, Jones JD. Subcellular localization of the Hpa RxLR effector repertoire identifies the extrahaustorial membrane-localized HaRxL17 that confers enhanced plant susceptibility. Plant J. 2011;69:252–265. doi: 10.1111/j.1365-313X.2011.04787.x. [DOI] [PubMed] [Google Scholar]
- Chaparro-Garcia A, Wilkinson RC, Gimenez-Ibanez S, Findlay K, Coffey MD, Zipfel C, et al. The receptor-like kinase SERK3/BAK1 is required for basal resistance against the late blight pathogen Phytophthora infestans in Nicotiana benthamiana. PLoS ONE. 2011;6:e16608. doi: 10.1371/journal.pone.0016608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Collins NC, Thordal-Christensen H, Lipka V, Bau S, Kombrink E, Qiu JL, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- van Damme M, Zeilmaker T, Elberse J, Andel A, de Sain-van der Velden M, van den Ackerveken G. Downy mildew resistance in Arabidopsis by mutation of HOMOSERINE KINASE. Plant Cell. 2009;21:2179–2189. doi: 10.1105/tpc.109.066811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Tanaka H, Goh T, Ebine K, Mähönen AP, Prasad K, et al. Generation of cell polarity in plants links endocytosis, auxin distribution and cell fate decisions. Nature. 2008;456:962–966. doi: 10.1038/nature07409. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Donofrio NM, Delaney TP. Abnormal callose response phenotype and hypersusceptibility to Peronospoara parasitica in defence-compromised arabidopsis nim1-1 and salicylate hydroxylase-expressing plants. Mol Plant Microbe Interact. 2001;14:439–450. doi: 10.1094/MPMI.2001.14.4.439. [DOI] [PubMed] [Google Scholar]
- Enkerli K, Hahn MG, Mims CW. Immunogold localization of callose and other plant cell wall components in soybean roots infected with the oomycete Phytophthora sojae. Can J Bot. 1997;75:1509–1517. [Google Scholar]
- Gan PH, Rafiqi M, Ellis JG, Jones DA, Hardham AR, Dodds PN. Lipid binding activities of flax rust AvrM and AvrL567 effectors. Plant Signal Behav. 2010;5:1272–1275. doi: 10.4161/psb.5.10.13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, et al. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Geldner N, Dénervaud-Tendon V, Hyman DL, Mayer U, Stierhof YD, Chory J. Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 2009;59:169–178. doi: 10.1111/j.1365-313X.2009.03851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez-Ibanez S, Hann DR, Ntoukakis V, Petutschnig E, Lipka V, Rathjen JP. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- Göhre V, Spallek T, Häweker H, Mersmann S, Mentzel T, Boller T, et al. Plant pattern-recognition receptor FLS2 is directed for degradation by the bacterial ubiquitin ligase AvrPtoB. Curr Biol. 2008;18:1824–1832. doi: 10.1016/j.cub.2008.10.063. [DOI] [PubMed] [Google Scholar]
- Gus-Mayer S, Naton B, Hahlbrock K, Schmelzer E. Local mechanical stimulation induces components of the pathogen defense response in parsley. Proc Natl Acad Sci USA. 1998;95:8398–8403. doi: 10.1073/pnas.95.14.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefle C, Huesmann C, Schultheiss H, Börnke F, Hensel G, Kumlehn J, Hückelhoven R. A barley ROP GTPase ACTIVATING PROTEIN associates with microtubules and regulates entry of the barley powdery mildew fungus into leaf epidermal cells. Plant Cell. 2011;23:2422–2439. doi: 10.1105/tpc.110.082131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh S, André A, Edwards H, Ehrhardt D, Somerville S. Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 2005;44:516–529. doi: 10.1111/j.1365-313X.2005.02545.x. [DOI] [PubMed] [Google Scholar]
- Kwon C, Neu C, Pajonk S, Yun HS, Lipka U, Humphry M, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- Lee SM, Kim HS, Han HJ, Moon BC, Kim CY, Harper JF, Chung WS. Identification of a calmodulin-regulated autoinhibited Ca2+-ATPase (ACA11) that is localized to vacuole membranes in Arabidopsis. FEBS Lett. 2007;581:3943–3949. doi: 10.1016/j.febslet.2007.07.023. [DOI] [PubMed] [Google Scholar]
- Lipka V, Dittgen J, Bednarek P, Bhat R, Wiermer M, Stein M, et al. Pre-and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- Meyer D, Pajonk S, Micali C, O’Connell R, Schulze-Lefert P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 2009;57:986–999. doi: 10.1111/j.1365-313X.2008.03743.x. [DOI] [PubMed] [Google Scholar]
- Micali CO, Neumann U, Grunewald D, Panstruga R, O’Connell R. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol. 2011;13:210–226. doi: 10.1111/j.1462-5822.2010.01530.x. [DOI] [PubMed] [Google Scholar]
- Mims CW, Richardson EA, Holt BF, Dangl JL. Ultrastructure of the host-pathogen interface in Arabidopsis leaves infected with the downy mildew Hyaloperonospora parasitica. Can J Bot. 2004;82:1001–1008. [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- O’Connell RJ, Panstruga R. Tête à tête inside a plant cell: establishing compatibility between plants and biotrophic fungi and oomycetes. New Phytol. 2006;171:699–718. doi: 10.1111/j.1469-8137.2006.01829.x. [DOI] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, et al. Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell. 2009;21:1541–1555. doi: 10.1105/tpc.108.064279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafiqi M, Gan PH, Ravensdale M, Lawrence GJ, Ellis JG, Jones DA, et al. Internalization of flax rust avirulence proteins into flax and tobacco cells can occur in the absence of the pathogen. Plant Cell. 2010;22:2017–2032. doi: 10.1105/tpc.109.072983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roppolo D, De Rybel B, Denervaude Tendon V, Pfister A, Alassimone J, Vermeer JEM, et al. A novel protein family mediates Casparian strip formation in the endodermis. Nature. 2011;473:380–384. doi: 10.1038/nature10070. [DOI] [PubMed] [Google Scholar]
- Schapire AL, Voigt B, Jasik J, Rosado A, Lopez-Cobollo R, Menzel D, et al. Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell. 2008;20:3374–3388. doi: 10.1105/tpc.108.063859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuring D, Viotti C, Krüger F, Künzl F, Sturm S, Bubeck J, et al. Multivesicular bodies mature from the trans-Golgi network/early endosome in Arabidopsis. Plant Cell. 2011;23:3463–3481. doi: 10.1105/tpc.111.086918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E. Cell polarization, a crucial process in fungal defence. Trends Plant Sci. 2002;7:411–415. doi: 10.1016/s1360-1385(02)02307-5. [DOI] [PubMed] [Google Scholar]
- Schornack S, Fuchs R, Huitema E, Rothbauer U, Lipka V, Kamoun S. Protein mislocalization in plant cells using a GFP-binding chromobody. Plant J. 2009;60:744–754. doi: 10.1111/j.1365-313X.2009.03982.x. [DOI] [PubMed] [Google Scholar]
- Schütz I, Gus-Mayer S, Schmelzer E. Profilin and Rop GTPases are localized at infection sites of plant cells. Protoplasma. 2006;227:229–235. doi: 10.1007/s00709-005-0151-1. [DOI] [PubMed] [Google Scholar]
- Shimada C, Lipka V, O’Connell R, Okuno T, Schulze-Lefert P, Takano Y. Nonhost resistance in Arabidopsis-Colletotrichum interactions acts at the cell periphery and requires actin filament function. Mol Plant Microbe Interact. 2006;19:270–279. doi: 10.1094/MPMI-19-0270. [DOI] [PubMed] [Google Scholar]
- Sousa E, Kost B, Malhó R. Arabidopsis phosphatidylinositol-4-monophosphate 5-kinase 4 regulates pollen tube growth and polarity by modulating membrane recycling. Plant Cell. 2008;20:3050–3064. doi: 10.1105/tpc.108.058826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soylu S. Ultrastructural characterisation of the host-pathogen interface in white blister-infected Arabidopsis leaves. Mycopathologia. 2004;158:457–464. doi: 10.1007/s11046-004-2453-9. [DOI] [PubMed] [Google Scholar]
- Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, et al. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development. 2006;133:4679–4689. doi: 10.1242/dev.02654. [DOI] [PubMed] [Google Scholar]
- Stassen JH, Van den Ackerveken G. How do oomycete effectors interfere with plant life? Curr Opin Plant Biol. 2011;14:407–414. doi: 10.1016/j.pbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Takemoto D, Jones DA, Hardham AR. GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J. 2003;33:775–792. doi: 10.1046/j.1365-313x.2003.01673.x. [DOI] [PubMed] [Google Scholar]
- Thines M, Kamoun S. Oomycete-plant coevolution: recent advances and future prospects. Curr Opin Plant Biol. 2010;13:427–433. doi: 10.1016/j.pbi.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Tör M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Tuerk F, et al. Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell. 2002;14:993–1003. doi: 10.1105/tpc.001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, et al. Visualization of PtdIns3P dynamics in living plant cells. Plant J. 2006;47:687–700. doi: 10.1111/j.1365-313X.2006.02830.x. [DOI] [PubMed] [Google Scholar]
- Voigt B, Timmers AC, Samaj J, Hlavacka A, Ueda T, Preuss M, et al. Actin-based motility of endosomes is linked to the polar tip growth of root hairs. Eur J Cell Biol. 2005;84:609–621. doi: 10.1016/j.ejcb.2004.12.029. [DOI] [PubMed] [Google Scholar]
- Wang W, Devoto A, Turner JG, Xiao S. Expression of the membrane-associated resistance protein RPW8 enhances basal defense against biotrophic pathogens. Mol Plant Microbe Interact. 2007;20:966–976. doi: 10.1094/MPMI-20-8-0966. [DOI] [PubMed] [Google Scholar]
- Wang W, Wen Y, Berkey R, Xiao S. Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell. 2009;21:2898–2913. doi: 10.1105/tpc.109.067587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Berkey R, Wen Y, Xiao S. Accurate and adequate spatiotemporal expression and localization of RPW8.2 is key to activation of resistance at the host-pathogen interface. Plant Signal Behav. 2010;5:1002–1005. doi: 10.4161/psb.5.8.12230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van West P, de Jong AJ, Judelson HS, Emons AM, Govers F. The ipiO gene of Phytophthora infestans is highly expressed in invading hyphae during infection. Fungal Genet Biol. 1998;23:126–138. doi: 10.1006/fgbi.1998.1036. [DOI] [PubMed] [Google Scholar]
- Xu JR, Staiger CJ, Hamer JE. Inactivation of the mitogen-activated protein kinase Mps1 from the rice blast fungus prevents penetration of host cells but allows activation of plant defense responses. Proc Natl Acad Sci USA. 1998;95:12713–12718. doi: 10.1073/pnas.95.21.12713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaeno T, Li H, Chaparro-Garcia A, Schornack S, Koshiba S, Watanabe S, et al. Phosphatidylinositol monophosphate-binding interface in the oomycete RXLR effector AVR3a is required for its stability in host cells to modulate plant immunity. Proc Natl Acad Sci USA. 2011;108:14682–14687. doi: 10.1073/pnas.1106002108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T. Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 2004;428:764–767. doi: 10.1038/nature02485. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Imaging Hpa haustoria. Arabidopsis Col-0 wild-type and 35S::GFP transgenic lines were infected with Hpa isolate Waco and haustoria were imaged by confocal microscopy. At 3 dpi wild-type leaves were incubated with the membrane dye FM4-64, or nuclear stain Hoechst respectively. Confocal micrographs show cross-sections of non-infected and Hpa infected leaves. Hpa haustoria are shown in bright field images indicated by asterisks.
A. Membrane FM4-64 and cytosolic 35S::GFP signals are visible at the host cell periphery and around young haustoria. The Hoechst signal is visible in the nucleus in close proximity to the haustorium.
B. FM4-64 membrane and 35S::GFP cytosol labelling shows the double-layered structure of encasements of old Hpa haustoria. Bar = 10 μm.
Fig. S2. Secretory vesicles differentially localize around Hpa and Pi haustoria.
A. Confocal micrographs of Arabidopsis transgenic lines expressing the indicated fluorophore fusions show cross-sections of non-infected and Hpa-infected leaves at 3 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. YFP–Rab E1d and YFP–Rab A5d signals are detected around Hpa haustoria. Bar = 10 μm.
B. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated fluorophore fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. YFP–Rab D2a, YFP–Rab E1d and YFP–Rab A5d are surrounding Pi haustoria. Bar = 10 μm.
Fig. S3. Endosomal compartments differentially localize around Hpa and Pi haustoria.
A. Confocal micrographs of Arabidopsis transgenic lines expressing the indicated fluorophore fusions show cross-sections of non-infected and Hpa-infected leaves at 3 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. YFP–VTI12, YFP–Rab A1e and ARA6–RFP signals are present at Hpa haustoria. Bar = 10 μm.
B. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated fluorophore fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. While YFP–VTI12 does not surround haustoria, YFP–Rab A1e accumulates around Pi haustoria. Bar = 10 μm.
Fig. S4. Proteins labelling the vacuolar tonoplast localize around Hpa and Pi haustoria.
A. Confocal micrographs show cross-sections of Arabidopsis transgenic leaves expressing the indicated fluorescent-tagged proteins non-infected and Hpa-infected at 3 dpi. Hpa haustoria are shown in bright field images indicated by asterisks. YFP–Rab G3f and YFP–VAMP711 signals are surrounding Hpa haustoria. Bar = 10 μm.
B. Confocal micrographs of N. benthamiana leaves transiently expressing the indicated fluorophore fusions show cross-sections of non-infected and Pi-infected leaves at 3 dpi. Pi haustoria are indicated by asterisks. YFP–Rab G3c accumulates around Pi haustoria. Bar = 10 μm.
Fig. S5. Hpa haustoria-containing cells maintain endosomal trafficking.
A. Arabidopsis Ler transgenic lines expressing GFP–2xFYVE as a marker for late endosomal compartments were infected with Hpa isolate Cala 2 and haustoria were imaged at 3 dpi. Time-lapse imaging by confocal microscopy shows cross-sections of Hpa infected leaves over 815 ms. Hpa haustoria are indicated by asterisks. The GFP–2xFYVE signal is visible at the host cell periphery, some emerging plasma membrane strands and at the EHM of Hpa haustoria. The arrows indicate vesicle movement towards the haustorial site (upper panel) and away from the haustorium (lower panel). Bar = 10 μM.
B. Arabidopsis Col-0 plants were infected with Hpa isolate Waco 9, and at 3 dpi leaves were treated with BFA and stained with FM4-64. Confocal micrographs show cross-sections of non-infected and Hpa-infected leaves. Hpa haustoria are shown in bright field images indicated by asterisks. The FM4-64 signal is visible at the plant cell plasma membrane, at BFA bodies indicated by arrows, and at the Hpa EHM. The BFA body is detected in a different focal plane of the haustoria-containing cell shown by single plane and maximal projection. Bar = 10 μm.
Fig. S6. Schematic representation of membrane trafficking in oomycete infected plant cells. Secretory vesicles deliver proteins such as PIP1;4 to the plasma membrane, but their lateral diffusion and incorporation into the extrahaustorial membrane must be prevented by the pathogen (EHM; red lines). Secretory vesicles may also deliver proteins like PEN1 to the EHM. Early endosomes (EE) may interact with the EHM and regulate presence of proteins at the EHM via recycling trafficking. Late endosomes (LE)/multivesicular bodies may contribute to enveloping the haustorium but proteins are sorted at the haustorial neck. As a result of recruitment of default membrane trafficking pathways, secretory vesicles, early and late endosomes contribute in the biogenesis of the encasement. TGN: trans-Golgi network; EHMx: extrahaustorial matrix; black arrows: proposed membrane trafficking pathways; molecular markers are highlighted in grey.
Table S1. Markers and lines used in this study. Arabidopsis plants stably expressing fluorescent-tagged markers (At line) were used for Hyaloperonospora arabidopsidis (Hpa) infections; transient expression of the respective markers (construct) in N. benthamiana (Nb) was used for Phytophthora infestans (Pi) infections; plasma membrane (PM), endoplasmatic reticulum (ER), trans-Golgi network (TGN), extrahaustorial membrane (EHM).
Table S2. Summary of marker localization in the absence and presence of Hyaloperonospora arabidopsidis (Hpa) and Phytophthora infestans (Pi) infection in A. thaliana (At) and N. benthamiana (Nb). Detection of fluorescent signals: clear discrete signal (discrete); weak widespread signal (diffuse); at cell periphery (periphery); at spots (vesicles); not at haustoria (excluded); at haustoria (surrounding); at extrahaustorial membrane (EHM); at haustorial encasements (encased); accumulates at haustoria (+); reduced (−); no difference in localization between uninfected and infected (no difference).


