Abstract
Internalization of cell surface receptors, followed by either recycling back to the plasma membrane or degradation, is crucial for receptor homeostasis and signaling. The plant brassinosteroid (BR) receptor, BRASSINOSTEROID INSENSITIVE 1 (BRI1), undergoes constitutive cycling between the plasma membrane and the internal membranes. We show that protein phosphatase 2A (PP2A) dephosphorylated BRI1 and that Arabidopsis thaliana rcn1, a mutant for a PP2A subunit, caused an increase in BRI1 abundance and BR signaling. We report the identification, in A. thaliana, of a suppressor of bri1, sbi1, which caused selective accumulation of BR-activated BRI1, but not the BR co-receptor BAK1 (BRI1-ASSOCIATED KINASE 1), in the membranous compartment. SBI1 mRNA was induced by BRs, and SBI1 encodes a leucine carboxylmethyltransferase (LCMT) that methylated PP2A and controlled its membrane-associated subcellular localization. We propose that BRs increase production of SBI1, which methylates PP2A, thus facilitating its association with activated BRI1. This leads to receptor dephosphorylation and degradation, and thus to the termination of BR signaling.
INTRODUCTION
Brassinosteroids (BRs), the polyhydroxylated steroid hormones of plants, regulate almost every phase of plant growth and development (1, 2). In Arabidopsis thaliana, most BR signaling components are known, and a pathway linking steroid recognition at the cell surface to changes in nuclear gene expression has been proposed (3). The BR receptor BRI1 (BRASSINOSTEROID INSENSITIVE 1), a cell surface dual-specificity receptor kinase, is a Toll-like receptor kinase composed of a large extra-cellular ligand-binding domain containing 25 leucine-rich repeats (LRRs), a transmembrane segment, and a cytoplasmic kinase domain (2, 4–9). Previous studies indicate that a small region embedded within BRI1’s LRRs binds the steroid hormone, which then induces a conformational change in the receptor, leading to its activation and autophosphorylation (2, 6–9). This increases the affinity of BRI1 for its co-receptor, BAK1 (BRI1-ASSOCIATED KINASE 1), a receptor kinase with five LRRs (10, 11). Extensive cross-phosphorylation events between the kinase domains of the receptor and the co-receptor then lead to a fully activated signaling complex (10–12). Although the precise order of events is not yet clear, upon BR binding, besides BAK1, BRI1 also phosphorylates at least two other proteins: the positive regulator BSK1 (BR-SIGNALING KINASE 1), which may be a serine-threonine kinase, and the negative regulator BKI1 (BRI1 KINASE INHIBITOR 1) (13–15). Downstream from the activated receptor, BIN2 (BRASSINOSTEROID INSENSITIVE 2), a glycogen synthase kinase 3 (GSK3)–related protein, negatively regulates the pathway (16), whereas BSU1 (BRI1 SUPPRESSOR 1), a serine-threonine-tyrosine phosphatase counteracts the effects of BIN2 (3, 17). Dephosphorylation of the related transcription factors BES1 (BRI1-EMS suppressor 1) and BZR1 (BRASSINAZOLE-RESISTANT 1) allows them to multimerize either on their own or together with other transcription factors to bind target promoters (18–24). Despite these advances, however, little information is available regarding how activated BR signal is attenuated.
The receptor internalization from and the subsequent either recycling back to the cell surface, or trafficking to hydrolytic compartments for degradation is a major mechanism that controls receptor abundance and the intensity and duration of receptor signaling. Receptors can be internalized by either ligand-dependent or ligand-independent pathways (25–27). Like transforming growth factor–β (TGF-β) receptors in metazoans, BRI1 in plants is internalized constitutively in a ligand-independent manner (26, 28, 29). Internalization of receptors from the cell surface is often associated with posttranslational modifications that affect the binding of the receptors to their partners and allow the receptors to be recognized by sorting components, which determine whether the receptors are recycled back to the cell surface or are degraded (27, 30). Proteomic studies reveal that more than one-third of the almost 600 human kinases are linked to endocytosis, suggesting that kinase activities are important for receptor internalization and sorting (31). For receptors with intrinsic kinase activity, such as the epidermal growth factor receptor (EGFR), TGF-β receptor, and BRI1, ligand binding leads to phosphorylation and activation of the receptors. Although the involvement of receptor phosphorylation in receptor internalization and sorting is supported by many independent studies, whether ligand-activated, phosphorylated receptors or ligand-unbound, inactive unphosphorylated receptors are sorted for degradation is less certain (24, 27, 30, 32–37). In addition, an alternative model proposes that dimerization rather than phosphorylation controls the endocytosis of receptors (37). Therefore, it is important to further analyze the role of phosphorylation in the control of receptor signaling and degradation.
In A. thaliana, a large spectrum of viable BR receptor mutants and well-established molecular readouts of the BR signaling pathway are available, making it a good model system for studying receptor signaling and degradation in plants (1, 2, 23, 24, 29). We investigated the mechanisms that control the abundance of BRI1 in membranous compartments to identify previously unknown mechanisms controlling receptor homeostasis that may also apply to other receptor systems. We identified a suppressor of an A. thaliana bri1 mutant (sbi1, suppressor of bri1) as a mutant that had increased abundance of BR-activated, but not ligand-unoccupied and inactive, BRI1. We determined that SBI1 encoded a leucine carboxylmethyltransferase (LCMT), whose only known substrate is the catalytic subunit of protein phosphatase 2A (PP2A). BR stimulated the expression of SBI1. We show that SBI1 methylated PP2A, leading to an increase in the ratio of methylated to unmethylated PP2A that was associated with the microsomal fraction. We propose a model in which the SBI1 activity induced by BRs initiates a series of events to specify the dephosphorylation and subsequent degradation of activated BRI1, thereby reducing receptor abundance and BR signaling strength.
RESULTS
A bri1-5 suppressor screen identifies SBI1 as a positive regulator of BRI1 degradation
BR signaling controls plant size. Null mutations in BRI1 result in extremely dwarf plants, whereas overexpression of BRI1 increases plant stature, suggesting that the abundance of active receptor can affect BR signaling. BRI1 is a long-lived protein found in the plasma membrane and endosomes. The distribution of BRI1 does not change in response to ligands, and its total protein abundance remains relatively constant throughout rapid growth phases of the seedlings (8, 29). These results are consistent with the fact that the mRNA abundance of BRI1 is constant in the absence of BR signaling, although exogenous application of BRs represses BRI1 transcription (38, 39). Together, these results suggest that BRI1 is probably regulated by a complex mechanism that may include transcriptional and translational, as well as posttranslational events. A mechanism that regulates BRI1 internalization and degradation to offset the biosynthesis and delivery of BRI1 to the plasma membrane has been proposed (29). To find the mechanism that regulates BRI1 degradation, we searched for viable bri1 mutants that altered BRI1 protein abundance and found that a previously reported weak allele of BRI1, bri1-5, which encodes a functionally competent receptor with a mutation in one of the cysteine pairs (C69Y) in the extracellular domain (40), had reduced abundance of bri1-5 protein but not transcripts (fig. S1, A and C). Expression of bri1-5::GFP under the control of the BRI1 promoter in bri1-5 (bri1-5;ProBRI1::bri1-5:GFP) partially rescued the dwarf phenotype of bri1-5 (fig. S1B) (40), indicating that bri1-5 phenotypic alterations were largely caused by a reduction in the amount of bri1-5 protein.
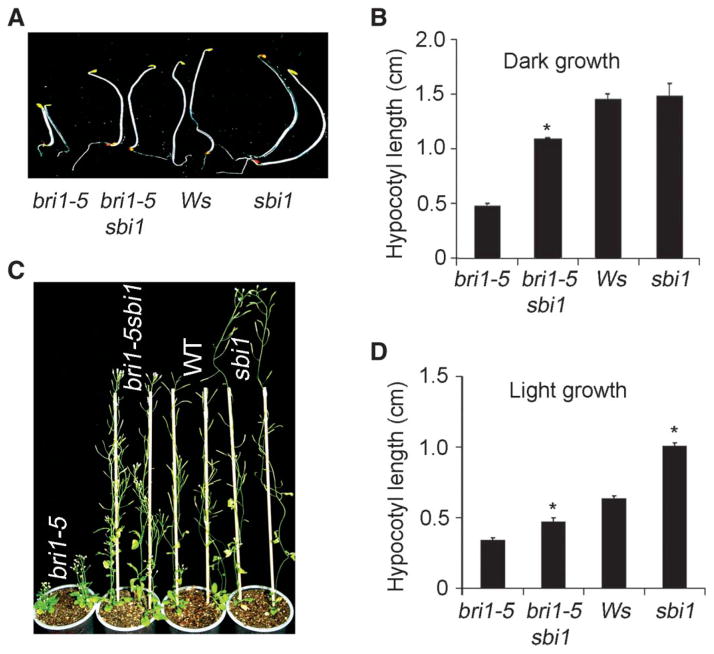
To identify the mechanism that controls BRI1 protein abundance, we performed a sensitized suppressor screen using the bri1-5 mutant. We screened 200,000 ethylmethane sulfonate (EMS)–mutagenized bri1-5 M2 seedlings germinated from M1 seeds, which were propagated directly from EMS-treated bri1-5 seeds (M0) (fig. S2), and identified a putative suppressor, sbi1, that had longer hypocotyls than those of bri1-5 seedlings (Fig. 1, A, B, and D). When the bri1-5 sbi1 double mutant was back-crossed to bri1-5, the resulting F1 plants showed bri1-5 phenotypes, and the F2 plants showed a 3:1 dwarf-to-normal phenotypic segregation (380:126), indicating that sbi1 is a recessive single gene mutation. The sbi1 single mutant had longer hypocotyls and inflorescence stems than did wild-type plants (Fig. 1, C and D) and resembled BRI1-overexpressing plants (7, 41), suggesting that SBI1 may inhibit BR signaling.
Fig. 1.
sbi1 suppressed bri1-5 phenotypes and promoted plant growth in Arabidopsis. (A) Hypocotyls of 5-day-old seedlings grown in the dark. Ws is the wild-type (WT) plant. (B) Quantitation of hypocotyl length of 5-day-old seedlings grown in the dark. Error bars represent SE. *P = 0.01; bri1-5 sbi1 hypocotyls were significantly longer than bri1-5 hypocotyls (n > 20 seedlings). (C) Six-week-old plants. WT plants are Ws. (D) Quantitative measurements of hypocotyls from 8-day-old seedlings grown in continuous light. Error bars represent SE. *P = 0.01; sbi1 and bri1-5 sbi1 hypocotyls were significantly longer than WT (Ws) and bri1-5, respectively (n > 10 seedlings).
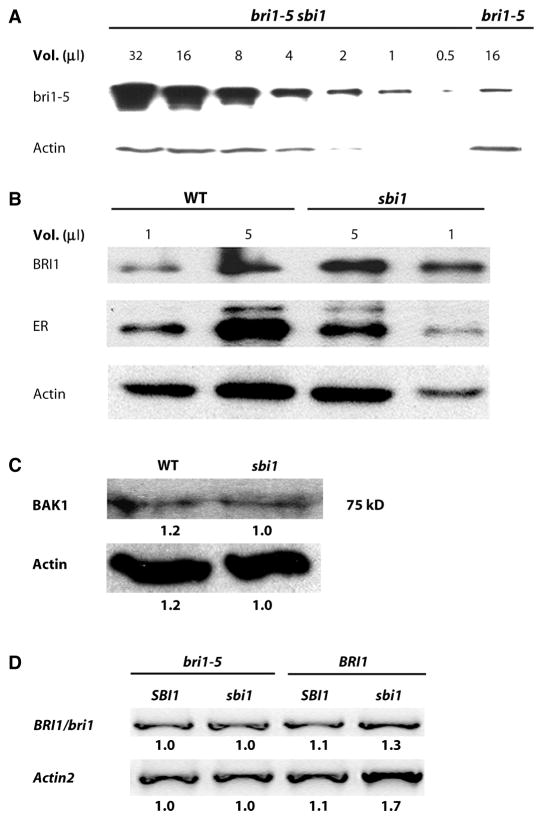
Semiquantitative analysis indicated that the amount of bri1-5 protein in the whole seedlings was increased in bri1-5 sbi1 compared to that in seedlings of bri1-5 (Fig. 2A). The amount of BRI1 was also increased in sbi1 seedlings compared to that in wild-type seedlings (Fig. 2B), indicating that wild-type SBI1 decreased the abundance of wild-type BRI1. In contrast, the abundance of BAK1 was not increased in the sbi1 mutant (Fig. 2C), and the abundance of ER (ERECTA), an unrelated LRR kinase, was slightly decreased in the sbi1 mutant (Fig. 2B). These results suggest that wild-type SBI1 specifically decreases the abundance of BRI1. Reverse transcription–polymerase chain reaction (RT-PCR) using total mRNAs from whole seedlings showed that the amounts of BRI1 and bri1-5 transcripts in either SBI1 or sbi1 backgrounds were similar (Fig. 2D). The Western blotting and RT-PCR data suggest that SBI1 mediated the increase in BRI1 and bri1-5 abundance through a posttranscriptional mechanism.
Fig. 2.
The abundance of BRI1 is increased in sbi1 plants. (A) Semi-quantitative Western blot analysis of a series of twofold dilutions from the extract of bri1-5 sbi1 plants reveals that the abundance of bri1-5 was increased compared to that in extracts from bri1-5 plants. Actin served as the loading control. Extracts were prepared from 14-day-old seedlings. Blot shown is representative of three experiments. (B) Protein abundance was detected by Western blotting with specific antibodies against BRI1, the receptor kinase ERECTA (ER), and actin in WT and sbi1 protein extracts. Actin and ER served as controls. WT plants are Ws. Extracts were prepared from 14-day-old seedlings. Blot shown is representative of three experiments. (C) BAK1 was detected by Western blotting with antibodies against the kinase domain of BAK1. Actin served as the loading control. Extracts were prepared from 14-day-old seedlings. The numbers below each band represent the relative band intensities, where the lowest density sample in each row is set to 1. Blot shown is representative of four experiments. (D) WT (Ws), sbi1, bri1-5, and bri1-5 sbi1 plants have similar amounts of bri1-5 mRNAs as shown by RT-PCR. Actin2 was used as an internal control. BRI1/bri1 indicates either the WT or the mutant protein. Relative integrated density is shown below each band, where the lowest-density sample in each row is set to 1. Blot shown is representative of three experiments.
SBI1 inhibits the BR signaling pathway
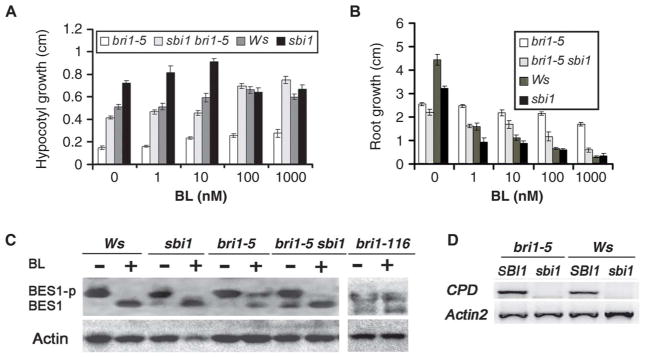
To determine whether BR responses were altered in the sbi1 plants, we used a dose-response assay for the effect of brassinolide (BL, the most active BR) on hypocotyl elongation and root growth. In response to a concentration range of 0 to 1000 nM BL, bri1-5 sbi1 and sbi1 showed increased hypocotyl length (Fig. 3A) and greater root growth inhibition (Fig. 3B) relative to bri1-5 and wild-type seedlings, respectively. BR treatment leads to the dephosphorylation of the BR-specific transcription factor BES1 in wild-type plants (18), but this dephosphorylation is compromised in bri1-116, a null allele of bri1 (Fig. 3C) (2). Thus, BES1 dephosphorylation is a marker for BR signaling (18, 40). BL-stimulated dephosphorylation of BES1 was compromised in bri1-5, and dephosphorylation of BES1 was similar to that in wild type for both the double mutant bri1-5 sbi1 and sbi1 (Fig. 3C). A negative feedback mechanism (22) represses genes involved in BR biosynthesis in response to BR treatments or situations of enhanced BR signaling (13, 14, 42). RT-PCR analysis showed that the expression of CPD (CONSTITUTIVE PHOTOMORPHOGENESIS DWARF), a BR biosynthetic gene, was reduced in sbi1 compared to the expression of CPD in wild-type plants (Fig. 3D), indicating that SBI1 normally plays a negative role in the BR signaling pathway.
Fig. 3.
SBI1 inhibits the BR signaling pathway. (A) Hypocotyl lengths of 8-day-old bri1-5, bri1-5 sbi1, WT (Ws), and sbi1 seedlings grown in continuous light with specified concentrations of BL. Error bars represent SE (n > 12 seedlings). (B) Root length of 8-day-old bri1-5, bri1-5 sbi1, Ws, and sbi1 seedlings grown in continuous light with specified amount of BL. Error bars represent SE (n > 12 seedlings). (C) Phosphorylated BES1 (BES1-p) and nonphosphorylated BES1 were detected with BES1 antibodies in extracts of 14-day-old seedlings of the indicated genotypes. Where indicated, the plants were treated with 1 μM BL for 1 hour before preparation of the extracts. Actin served as the loading control. Blot shown is representative of three experiments. bri-116 is a null allele. (D) RT-PCR for CPD was performed with mRNAs from bri1-5, bri1-5 sbi1, WT (Ws), and sbi1. Actin2 served as an internal control. mRNA was obtained from 14-day-old seedlings. Blot shown is representative of three experiments.
SBI1 regulates BR-activated but not inactive BRI1
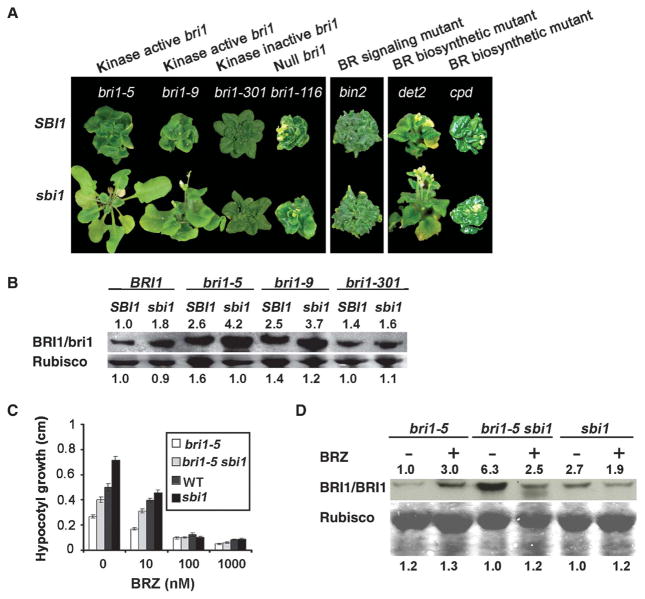
To place SBI1 in the BR signaling cascade, we crossed sbi1 to bin2-1, which is a gain-of-function allele of the gene BIN2 encoding a negative regulator of BRI1 signaling (16). The bin2-1 plants have a retarded growth phenotype like that of bri1 plants. We found that sbi1 did not rescue the growth retardation phenotype of bin2-1, indicating that sbi1 does not activate BR signaling downstream of BIN2 and that SBI1 may act upstream of BIN2 (Fig. 4A, middle panel).
Fig. 4.
sbi1-dependent regulation requires ligand-activated BRI1. (A) Five-week-old plants of the indicated genotypes of BR-responsive mutants and BR biosynthetic mutants in the sbi1 mutant and WT SBI1 background are shown. (B) BRI1 and bri1 were detected by Western blotting. Extracts of WT (Ws), sbi1, bri1-5, bri1-5 sbi1, bri1-9, bri1-9 sbi1, bri1-301, and bri1-301 sbi1 were from 14-day-old seedlings. Rubisco served as the loading control. Relative integrated density is shown above or below each band where the density of WT sample in each row is set to 1. Blot shown is representative of three experiments. (C) Hypocotyl length of 8-day-old bri1-5, bri1-5 sbi1, WT (Ws), and sbi1 seedlings grown in continuous light with the specified amount of BRZ. Error bars represent SE (n > 12 seedlings). (D) BRI1 and bri1 were detected by Western blotting. Extracts of sbi1, bri1-5, and bri1-5 sbi1 were from seedlings grown in the ½ MS medium with or without 10 μM BRZ for 2 weeks. Rubisco served as the loading control. Relative integrated density was shown above or below each band where the lowest-density sample in each row is set to 1. Blot shown is representative of three experiments.
To investigate whether functional BRI1 proteins were required for sbi1 to enhance BR signaling, we crossed sbi1 with mutants carrying the null bri1-116 allele, the bri1-9 allele that harbors a mutation in the extracellular domain of BRI1 but otherwise encodes a functionally competent receptor that can be activated by BR, or the bri1-301 allele that encodes a protein with reduced kinase activity due to a mutation in the kinase domain (43). We found that sbi1 suppressed the retarded growth phenotype of bri1-9 but did not suppress the retarded growth phenotype of either bri1-116 or bri1-301 (Fig. 4A, left panel) (2, 43). Consistent with these phenotypic assays, in whole seedlings, the amount of wild-type BRI1, bri1-5, and bri1-9, but not the amount of bri1-301, was increased in the sbi1 background compared to that in the SBI1 background (Fig. 4B). Our results suggest that SBI1 regulates the abundance of kinase-active BRI1.
To test whether endogenous BRs were required for sbi1-dependent enhancement of BR signaling, we crossed sbi1 to det2 (de-etiolated 2), a weak BR biosynthetic mutant (44), and to cpd, a strong BR biosynthetic mutant (45). We found that sbi1 partially rescued the growth retardation phenotype of det2, but it had no effect on cpd (Fig. 4A, right panel). The BR biosynthesis inhibitor BRZ inhibited the growth of bri1-5 sbi1 and sbi1 hypocotyls in a concentration-dependent manner (Fig. 4C) and caused a small increase in the amount of wild-type BRI1 or bri1-5 protein in SBI1 plants (Fig. 4D and fig. S3), which could be due to a transcriptional or translational regulation of BRI1 (38, 39). However, BRZ treatment completely abolished the increased abundance of wild-type BRI1 or mutant bri1 proteins in the sbi1 mutant background compared to that in the untreated sbi1 plants (Fig. 4D and figs. S3 and S4). As indicated by several small-size bands that were still specifically recognized by the BRI1 antibodies, this effect is likely due to the increase in BRI1 degradation rather than a decrease in BRI1 transcription or translation (figs. S3 and S4). Our results indicate that the ligand must be present for the sbi1-dependent accumulation of BRI1 and bri1-5.
SBI1 encodes an LCMT
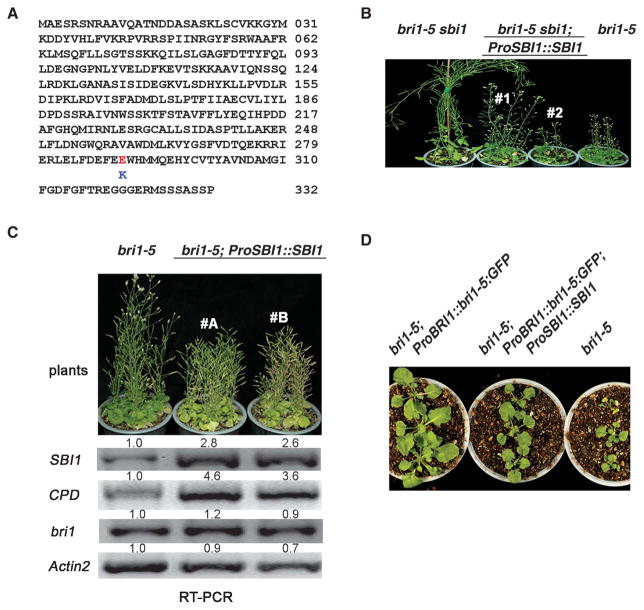
We cloned the sbi1 gene by positional cloning and identified a single G-to-A change in At1g02100, which encodes a putative LCMT (fig. S5). This mutation changed a glutamic acid to a lysine at amino acid residue 290 (E290K) (Fig. 5A and fig. S6). To confirm that we had identified the correct gene, we cloned the At1g02100 genomic fragment and transformed it into the bri1-5 sbi1 plants. Two independent transgenic lines showed phenotypes similar to bri1-5, confirming that At1g02100 is indeed SBI1 (Fig. 5B).
Fig. 5.
SBI1 encodes an LCMT that methylates and regulates PP2A. (A) Amino acid sequence of SBI1; the E290K mutation is indicated. Abbreviations for the amino acids are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr. (B) Complementation of bri1-5 sbi1 with a genomic SBI1 fragment (ProSBI1::SBI1). Two independent transgenic lines (#1 and #2) are shown. (C) Two independent transgenic lines (#A and #B) overexpressing ProSBI1::SBI1 in bri1-5 are shown. The abundance of the indicated transcripts from 14-day-old seedlings was detected by RT-PCR. Relative integrated density is shown above each band where the density of bri1-5 sample in each row is set to 1. Blot shown is representative of three experiments. (D) Three-week-old seedlings of the indicated genotypes. ProSBI1::SBI1 was overexpressed in bri1-5 transgenics with ProBRI1::bri1-5:GFP fusion gene (bri1-5 and GFP fusion gene under BRI1 promoter).
SBI1 is coexpressed with BRI1 and CPD in most tissues; however, in mature organs, SBI1 transcripts accumulate at higher levels than those of either BRI1 or CPD (fig. S7). We expressed SBI1 under its own promoter (ProSBI1::SBI1) in bri1-5 to test the in vivo function of SBI1. Two independent transgenic lines exhibited enhanced bri1-5 phenotypes with reduced apical dominance, shorter inflorescence stems and branches, and early senescence (Fig. 5C), suggesting that SBI1 negatively regulates plant growth. Consistent with these phenotypes, transcripts for CPD were increased in both transgenic lines (Fig. 5C), which is consistent with reduced BR signaling. Expression of ProBRI1::bri1-5:GFP in bri1-5 (bri1-5;ProBRI1::bri1-5:GFP) enhanced BR signaling and promoted plant growth (fig. S1B). We reasoned that if SBI1 negatively regulated BRI1 signaling, the expression of ProSBI1::SBI1 in bri1-5;ProBRI1::bri1-5:GFP plants would counteract this enhanced BR signaling caused by overexpressing bri1-5::GFP. In fact, we observed that the phenotype of the transgenic plants (bri1-5;ProBRI1::bri1-5:GFP; ProSBI1::SBI1) was similar to that of bri1-5 plants (Fig. 5D). These results suggest that SBI1 genetically interacts with bri1-5 (and BRI1) and inhibits BR signaling in vivo.
Methylation of PP2A by SBI1 controls PP2A distribution between the membrane and the cytosol
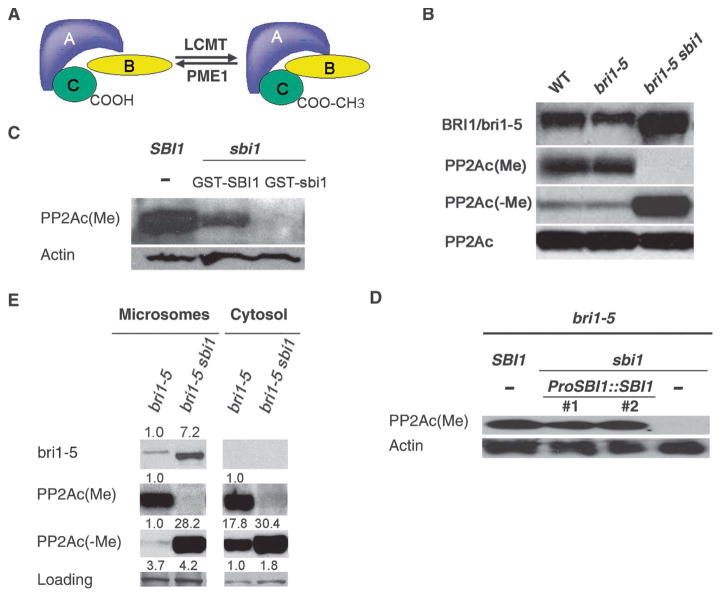
The phosphatase PP2A is composed of three subunits: a catalytic subunit (PP2Ac), a core scaffolding subunit (PP2Aa), and a regulatory subunit PP2Ab (Fig. 6A). PP2Ac subunits are the only known substrates of LCMTs (Fig. 6A and fig. S8) (46, 47), and reversible methylation of the PP2Ac is a conserved regulatory mechanism for PP2A function (46, 48). Methylation of the C-terminal leucine of PP2Ac enhances the affinity of the PP2A core enzyme consisting of PP2Aa and PP2Ac for some, but not all regulatory subunits, suggesting that PP2A methylation could modulate the specificity and activity of PP2A in vivo (46, 48). Using commercially available antibodies against methylated PP2Ac [PP2Ac(Me)], we detected PP2Ac(Me) in wild type, bri1-5, bri1-9, and bri1-301, but not in any line carrying the sbi1 mutation (Fig. 6B and fig. S9). SBI1 methylated PP2Ac in vitro (Fig. 6C), and expression of genomic SBI1 (ProSBI1::SBI1) in bri1-5 sbi1 rescued the methylation activity toward PP2Ac (Fig. 6D). Thus, PP2Acs appear to be bona fide substrates of SBI1, and SBI1 may regulate BRI1 through its activity toward PP2A.
Fig. 6.
SBI1 methylates PP2A to control its distribution between the membrane and the cytosol. (A) Diagram of the structure and regulation of PP2A. PP2A consists of scaffold subunit A (PP2Aa), regulatory subunit B (PP2Ab), and catalytic subunit C (PP2Ac) and is regulated by reversible methylation. LCMT methylates PP2Ac, whereas PME1 (PP2A methylesterase) demethylates PP2Ac. Methylation of PP2Ac affects PP2A substrate selectivity by PP2Ab (46, 48). (B) Western blot analysis with antibodies specific for methylated PP2Ac [PP2Ac(Me)] and unmethylated PP2Ac [PP2Ac(−Me)]. Total PP2Ac was detected with antibodies for unmethylated PP2Ac after removal of the methyl group from the PP2Ac that was bound to the nitrocellular membrane by a 30-min treatment with 0.2 N sodium hydroxide. Samples were prepared from 14-day-old seedlings. Blot shown is representative of five experiments. (C) Extracts of 14-day-old seedlings of the indicated genotypes were incubated with or without recombinant WT SBI1 or mutant sbi1 proteins and methylated PP2Ac was detected by Western blotting. Actin served as the loading control. Blot shown is representative of three experiments. (D) Western blot analysis for PP2Ac(Me) and PP2Ac(−Me) of extracts of 14-day-old seedlings of the indicated genotypes. The two transgenic lines of ProSBI1::SBI1 in sbi1 bri1-5 are the same as those shown in Fig. 5B. (E) The abundance of the indicated proteins in microsomal and cytosolic fractions of extracts of 14-day-old seedlings of the indicated genotypes was detected by Western blotting. Nonspecific bands were used as loading controls. Relative integrated density was shown above each band where the sample with the lowest detectable density in each row is set to 1. Blot shown is representative of three experiments.
Methylation of PP2A regulates its subcellular targeting and substrate specificity (46, 48). To determine whether SBI1 affected the subcellular localization of PP2A, we isolated the microsomal and cytosolic fractions from young seedlings of bri1-5 and bri1-5 sbi1 mutants. Most of the bri1-5 protein was found in the microsomal fraction (Fig. 6E and fig. S10). We found that in the absence of functional SBI1, unmethylated PP2Ac [PP2Ac(−Me)] localized equally to the microsomal and cytosolic fractions (Fig. 6E). In the presence of active SBI1, there was less PP2Ac(−Me) present in the microsomal fraction, and similar amounts of PP2Ac(Me) were detected in both fractions (Fig. 6E), suggesting that SBI1 reduces the amount of PP2Ac(−Me) in the microsomal fraction, probably by converting unmethylated, membrane-associated PP2Ac to PP2Ac(Me). Consistent with an activity at cellular membranes, SBI1::GFP partially associated with membranous compartments (fig. S11). It has been reported that protein carboxylmethylation is required for its efficient plasma membrane targeting (49), suggesting that methylation enhances protein affinity for the membrane. Because SBI1 was also detected in the cytoplasm (fig. S11), it is possible that SBI1 methylates PP2Ac in the cytosol followed by the replacement of PP2Ac(−Me) with PP2Ac(Me) in the membranous compartments due to the higher affinity of PP2Ac(Me) than that of PP2Ac(−Me) for the membrane. These localization studies suggest that SBI1 controls PP2Ac localization.
PP2A dephosphorylates BRI1, which decreases BRI1 abundance, reducing the activity of the BR signaling pathway
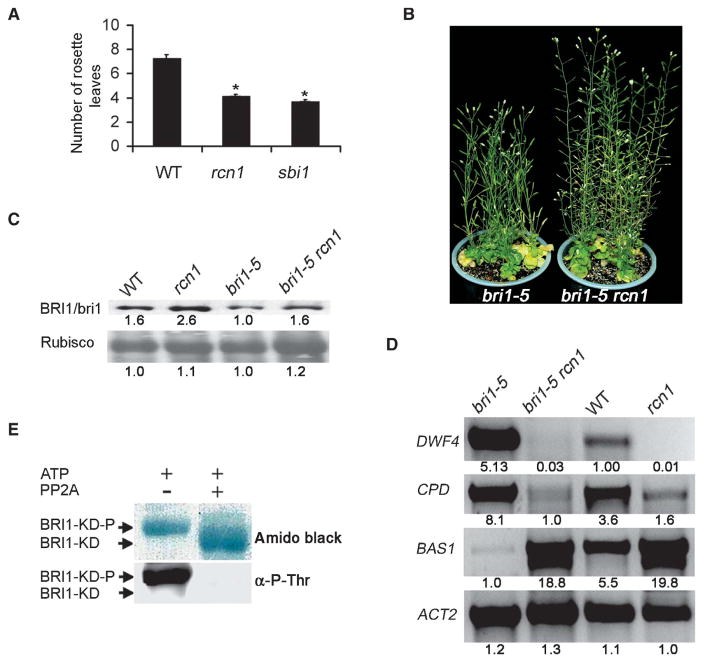
To investigate the relationship between SBI1 and PP2A through genetic experiments, we compared the phenotypes of sbi1 and a previously characterized pp2a mutant. We observed that both sbi1 and a transferred DNA (T-DNA) knockout mutant of pp2aa1 named rcn1 (root curling with NPA, an auxin transporter inhibitor) (50) exhibited similar phenotypes: increased apical dominance, larger cauline leaves, fewer rosette leaves, and earlier flowering than wild-type plants (Fig. 7A and fig. S12), suggesting that SBI1 and PP2A are involved in similar biological processes.
Fig. 7.
PP2A reduces BR signaling and dephosphorylates BRI1 in vitro. (A) Both PP2Aa mutant rcn1 (pp2aa1) and sbi1 plants (5-week-old) have early flowering phenotypes. Number of rosette leaves (n) produced before bolting in WT (Ws), rcn1 (pp2aa1), and sbi1 plants is shown. Error bars represent SE. *P = 0.01. The number of rosette leaves in both rcn1 and sbi1 was significantly different from that of WT plants (n > 14). (B) Partial suppression of bri1-5 phenotypes by rcn1 shown by 2-month-old plants. (C) BRI1 or bri1-5 was detected by Western blotting of extracts of 14-day-old plant seedlings of the indicated genotypes. Wild-type (WT) is Ws. Relative integrated density was shown below each band where the lowest density sample in each row is set to 1. Blot shown is representative of three experiments. (D) The abundance of transcripts or proteins for the BR biosynthesis genes (DWF4 and CPD) and a BR inactivation gene (BAS1) was detected by RT-PCR of extracts of 14-day-old plant seedlings of the indicated genotypes. Relative integrated density was shown below each band where the visible band with lowest density in each row is set to 1. Blot shown is representative of three experiments. (E) In vitro dephosphorylation of the kinase domain of BRI1 (BRI1-KD) with human PP2A. Protein blot stained with amido black (upper panel); Western blot with antibody against phosphothreonines (α-P-Thr) (lower panel). BRI1-KD-P, phosphorylated BRI1-KD.
To address PP2A function in BR signaling, we crossed rcn1 to bri1-5 and found that rcn1 partially suppressed the bri1-5 phenotype (Fig. 7B), suggesting that a PP2A regulates BR signaling. However, the abundance of BRI1 and bri1-5 proteins was only slightly increased in rcn1 and bri1-5 rcn1 compared to that in wild type and bri1-5, respectively (Fig. 7C). Because reduced expression of BR biosynthesis genes, such as DWF4 and CPD, and increased expression of BR inactivation genes, such as BAS1, are sensitive markers for enhanced BR signaling (13, 14, 42), we tested these markers in the mutants. The transcripts of DWF4 and CPD were decreased, whereas the transcripts of BAS1 were increased in rcn1 and bri1-5 rcn1 plants compared to wild-type and bri1-5 plants, respectively (Fig. 7D). We noticed that sbi1 and rcn1 exhibited similar, but not identical, phenotypes (Figs. 1C and 7B and fig. S12) (50). For example, double PP2Aa mutants rcn1 pp2aa2 and rcn1 pp2aa3 had severe dwarf phenotypes (50), whereas sbi1 had no apparent growth defect (Fig. 1C). One explanation for these differences may be that SBI1 affects only a subset of PP2A functions, such as BRI1 degradation, or that PP2A regulates multiple components in the BR signaling pathway. Notably, the transcripts of SBI1, but not RCN1, were increased in A. thaliana seedlings treated with BRs, but not in seedlings treated with other hormones (fig. S13), suggesting that of these two genes only SBI1 is a BR-responsive gene. Therefore, it is possible that only the PP2A activity that is specified by BR-dependent activity of SBI1 but not the overall PP2A activity regulates BR signaling pathway.
Purified Arabidopsis PP2A is not available, but PP2A is conserved in eukaryotes. Therefore, to determine whether BRI1 could be a substrate of PP2A, we used a purified mammalian PP2A consisting of the catalytic subunit PP2Ac and the scaffold subunit PP2Aa to dephosphorylate the BRI1 kinase domain (BRI1-KD) that had been phosphorylated in vitro. Half of the phosphorylated sample was treated with PP2A, and the other half was used as the control. In the PP2A-treated half, we detected a faster-moving band in SDS–polyacrylamide gel electrophoresis (SDS-PAGE) corresponding to BRI1-KD that was not detected with antibodies that recognize proteins that have phosphorylated threonines (Fig. 7E), indicating that PP2A can dephosphorylate BRI1 in vitro. However, we failed to detect an interaction between PP2Ac and BRI1 in vivo by coimmunoprecipitation with antibodies that recognize BRI1 in wild-type plants or with antibodies that recognize green fluorescent protein (GFP) in plants harboring the fusion proteins BRI1::GFP or bri1-5::GFP. Substrates of PP2A interact with PP2Ac through the regulatory subunit PP2Ab, and the lack of a detectable interaction by immunoprecipitation may be because the regulatory subunit PP2Ab that confers substrate specificity is loosely bound to the PP2Aa and PP2Ac core (Fig. 6A) (46, 48), as has been reported in mammalian and yeast systems (46, 48).
DISCUSSION
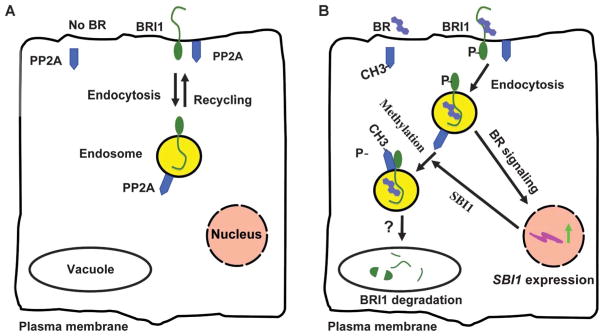
We report the identification of SBI1, an LCMT, and show that it plays a role in the BR signaling pathway through its action on PP2Ac. Methylation of PP2Ac by SBI1 leads to an enrichment of methylated PP2A at membranes. We propose that this relocation of methylated PP2A brings the active PP2A into proximity to BR-activated BRI1 and leads to BRI1 dephosphorylation. This dephosphorylation targets the BR-activated BRI1 pool for degradation, thereby inhibiting BR signaling and controlling BRI1 receptor abundance through a negative feedback loop (Fig. 8).
Fig. 8.
A working model for a negative feedback mechanism that controls BRI1 abundance and BR signaling. (A) In the absence of BRs, inactive BRI1 molecules are constitutively internalized and recycled between the endosomes and the plasma membrane. (B) Binding of BRs by BRI1 at the cell surface leads to BRI1 activation and phosphorylation. Activated BRI1 initiates the BR signaling cascade, including the induction of SBI1 transcription. SBI1 methylates and targets PP2A to the endosomes, which in turn dephosphorylates BRI1. Dephosphorylated BRI1 molecules are then degraded (possibly in the vacuole), thus terminating BR signaling. A question mark represents a process that is postulated to occur, but has not been shown. Our model indicates BR signaling is initiated from the cell surface, but this model would not exclude the possibility that BR signaling can be initiated from intra-cellular membranous compartments. For simplicity, other components of the pathway, such as BAK1, BSK1, BKI1, BIN2, BES1, BZR1, and BSU1, are not shown.
Although we could not definitely show that BRI1 was a bona fide PP2A substrate in vivo, several lines of evidence suggest that BRI1 is a PP2A substrate. First, SBI1 acted upstream of BIN2 (Fig. 4A, middle panel) and required BRI1 for its function in the BR pathway (Fig. 4A, left panel), suggesting that SBI1 acts at the level of the receptor in the BR pathway. Second, kinase activity of BRI1 (or mutants thereof) was required for sbi1-dependent BRI1 or bri1 accumulation, suggesting that SBI1 targets active BRI1 (Fig. 4B). Third, ligands that directly bind and activate BRI1 were required for the increased accumulation of active BRI1 or bri1 in sbi1 (Fig. 4D), supporting the idea that the effect of SBI1 on BR signaling depends on BRI1 activation (Fig. 4, B and D). Fourth, the amount of the bri1-301 protein that lacks kinase activity was not increased in the sbi1 background (Fig. 4B), indicating that unphosphorylated BRI1 is not a target for SBI1 regulation. Furthermore, we did not detect a change in the basal amounts of BRI1 or bri1 in plants with the sbi1 mutation in the absence of BRs (Fig. 4D and fig. S3). In addition, SBI1 did not alter the abundance of the BR co-receptor BAK1 (Fig. 2C). Together, our results suggest that SBI1 promotes the dephosphorylation of BRI1 or bri1 (deactivation of BR-activated and autophosphorylated BRI1 or bri1) rather than affects unphosphorylated BRI1 or bri1. Consistent with this dephosphorylation model, PP2A, the only known substrate of LCMTs such as SBI1, completely dephosphorylated phosphothreonine residues on BRI1 in vitro, and the reduction of PP2A activity (in rcn1 mutant plants) promoted BRI1 accumulation and BR signaling, suggesting that PP2A inhibits the BR signaling pathway by degrading BRI1 (Fig. 7).
In our model, unbound BRI1 constitutively cycles between the plasma membrane and the internal membranes (Fig. 8A), but dephosphorylation of ligand-activated BRI1 at internal membranes results in receptor degradation (Fig. 8B). This suggests that the cell can recognize the difference between unphosphorylated BRI1 that has not been activated and BRI1 that has been dephosphorylated subsequent to activation. Our model bears some commonalities with the proposed mechanisms for the down-regulation of EGFRs and TGF-β receptors. The relationship of EGFR phosphorylation to its cellular trafficking remains an area of study, with some reports showing that activation of tyrosine kinase activity of the EGFR leads to receptor signaling and recycling and that inactive unphosphorylated receptors are destined for degradation (27, 30–32). Others show the opposite or no effect of phosphorylation on EGFR trafficking and stability (32, 35–37). In some of the studies that report a role for phosphorylation in EGFR trafficking, ligand-activated EGFRs with phosphorylated serines or threonines are selected for endocytosis and degradation (33, 34). Similar to BRI1, PP2A can dephosphorylate an activated EGFR (51); however, whether and how PP2A activity affects EGFR degradation is not known (27, 30, 32, 46, 48, 51).
Like BRI1, TGF-β receptor molecules are constitutively internalized and recycled in a ligand-independent manner, and TGF-β receptor degradation is enhanced by ligand binding (26, 52). In addition, phosphatase inhibitors can decrease the binding of TGF-β in the plasma membrane. Furthermore, TGF-β receptors directly interact with the regulatory PP2Ab subunits (52), suggesting that PP2A may play a role in the regulation of TGF-β receptor degradation (26). Our results are consistent with the involvement of receptor serine-threonine kinase activity in the control of receptor degradation, although the detailed mechanisms may be different. Our model indicates that neither the phosphorylated receptors nor the unphosphorylated ones are destined for degradation; rather, the receptors phosphorylated first and dephosphorylated subsequently are marked for degradation. Therefore, BRI1 down-regulation seems to share some, but not all, aspects of mechanisms that control EGFR and TGF-β receptor down-regulation. Our work thus provides a conceptual framework for further study of cell surface receptor kinases in eukaryotes, but it remains to be tested how widespread this mechanism is among plants and metazoans.
In animals, PP2A activity accounts for most serine-threonine phosphatase activity in cells, and its substrates are diverse (46, 48). Plant genomes are predicted to have a greater number of PP2Aa and PP2Ac subunits than are predicted for animals; thus, the potential for a role for PP2A in many plant signaling pathways exists (46, 47). KAPP, a protein phosphatase 2C (PP2C), interacts with many Arabidopsis receptor kinases; however, KAPP mutant phenotypes are weak, and convincing in vivo data for KAPP function in the regulation of BRI1 or other kinases have not been obtained (53–55). Thus, KAPP does not appear to be a major regulator of the other more than 600 predicted receptor kinases in Arabidopsis (5).
Little is known about the function of PP2As in plant signaling. A PP2A plays a role in localizing the PIN auxin transporters in the plasma membrane, and the plasma membrane–associated receptor kinase phototropin 2 (phot2) has been identified as a PP2A substrate (56, 57). It is now key to test whether other plant receptor kinases are also the substrates of PP2A, as well as the role of methylation in regulating substrate specificity in plants.
MATERIALS AND METHODS
Plant material handling and growth conditions
A. thaliana ecotype Wassilewskija-2 (Ws) was used as wild-type controls. rcn1 (Ws) and cpd (45) were obtained from the Arabidopsis Biological Research Center at Ohio State University. Mutant bri1-5 is in the Ws background and was provided by F. Tax (58). bri1-301 is in the Col background and was a gift from J. Li (43). Seeds were germinated on either ½ Murashige & Skoog (MS) medium plus 1% sucrose then transferred to the soil or directly in soil. Plants were grown under long daylight conditions (16-hour light/8-hour dark cycles) or continuous light. Images of seedlings or plants were taken at specified developmental stages. Seedlings for measuring BR-dependent responses were germinated on ½ MS medium with or without BL (CIDtech Research Inc.) or BRZ in the dark or in the light.
Isolation of suppressor and cloning of the mutated gene
Mutagenesis on bri1-5 seeds was performed as previously described (18, 58). EMS mutagenized seeds of bri1-5 (M0) were grown in the greenhouse in 20 flats with about 1000 plants in each flat to propagate M1 seeds, which germinated to produce M2 seedlings. The screen is outlined in fig. S2. Suppressor bri1-5 sbi1 was crossed to Col, and bri1-5–like seedlings with shorter hypocotyls were selected in the segregating F2 population and grown in the greenhouse to obtain F3 seeds. These F2 plants were further genotyped to identify bri1-5 homozygous individuals. To map the mutated locus, we screened for seedlings with long hypocotyls in the F3 generation. The Monsanto Arabidopsis Polymorphism database (http://www.arabidopsis.org/browse/Cereon/index.jsp) was used for designing simple sequence length polymorphisms (SSLPs), cleaved amplified polymorphic sequences (CAPSs), and derived cleaved amplified polymorphic sequence (dCAPS) markers for fine-mapping of 4000 chromosomes in 2000 plants. The final 54-kb fragment was sequenced to identify the mutated gene (Fig. 5).
Gene expression, plasmid constructs, and transgenic plants
The construct ProBRI1::bri1-5:GFP was generated from pPZP212-ProBRI1::BRI1:GFP (41) by site-directed mutagenesis with a Stratagene QuikChange II XL kit. SBI1 or sbi1 complementary DNA (cDNA) was amplified by RT-PCR using Pfu DNA polymerase (Stratagene) with the first-strand cDNA generated from wild-type or sbi1 RNAs. SBI1 and sbi1 cDNA was cloned into the binary vector CHF3:GFP or pGEX-5X (18). A SBI1 genomic fragment containing 1.5 kb of promoter and 300 base pairs (bp) downstream of the stop codon was amplified from genomic DNA purified from wild-type plants by PCR and cloned into a PCambia vector. All clones were confirmed by DNA sequencing. The resulting constructs were first transformed into Agrobacterium tumefaciens, strain GV3101, followed by plant transformation with the floral dip method (59). Transformed seeds were germinated and selected on ½ MS medium containing kanamycin (50 mg/liter).
In vitro methylation of PP2Ac by recombinant SBI1
Following the manufacturer’s instruction (GE Healthcare), we purified glutathione S-transferase (GST)–SBI1 and GST-sbi1 proteins from BL21 strain transformed with pGEX::SBI1 and pGEX::sbi1, respectively. Plant seedlings (200 mg) of sbi1 mutant were homogenized in 0.5 ml of lysis buffer [2.5% glycerol, 50 mM tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, and protease inhibitor] on ice. The mixture was then transferred to a microfuge tube and centrifuged at 12,000g at 4°C for 10 min to remove the debris. The supernatant was transferred to a new tube and 4 mg of AdoMet (A7007, Sigma) was added to the supernatant. The supernatant was partitioned into two aliquots; each was mixed with 5 μg of either GST-SBI1 or GST-sbi1 proteins. After 30 min of incubation in RT, the reactions were terminated by adding 10 volumes of ethanol and stored in −80°C for 2 hours to precipitate the proteins. Protein pellets were obtained by centrifugation at 12,000g at 4°C for 10 min to remove the supernatant and were resuspended in 0.1 ml of SDS running buffer. The proteins were then resolved by SDS-PAGE, followed by Western blot detection with antibodies that recognized methylated PP2Ac.
Western blot analysis of plant proteins
Microsomal and cytosolic fractions were purified as previously described (6). Crude protein extracts were obtained directly from seedlings with or without BL treatment for 1 hour. The protein extracts were then resolved in SDS-PAGE. Western blot detection was performed with the specified antibodies. Antibodies for BRI1 and BES1 were provided by Y. Yin (18) and antibodies for ER were a gift from K. Torii (60). Y. Jaillais provided the antibody that recognizes BAK1. Antibodies that recognize actin were purchased from Santa Cruz Biotechnology. Other antibodies were purchased from Millipore.
Transcript analysis by RT-PCR
Total mRNAs were isolated from whole plant seedlings with RNeasy Plant Mini kit (Qiagen, #74904). The mRNA concentration was estimated by spectrophotometer. First-strand cDNA was synthesized from 2 μg of total RNA with SuperScript II RT (Invitrogen) following the manufacturer’s instruction. The cDNA was then amplified by PCR with gene-specific primers for bri1-5, BRI1, SBI1, CPD, DWF4, BAS1, and ACT2 at an annealing temperature of 55°C for 30 cycles (42). Primers used for RT-PCR are listed in Table 1.
Table 1.
Primers used for RT-PCR.
| Gene name | Locus | Primer sequences |
|---|---|---|
| DWF4 | At3g50660 | 5′-TACCTCTTCTTCTTCTCCCATCGC-3′ |
| 5′-CGAGAAACCCTAATAGGCAAACCG-3′ | ||
| CPD | At5g05690 | 5′-TCCTCCTCCTCTCTTCCATCGCCG-3′ |
| 5′-ACGGCGCTTCACGAAGATCGGGTA-3′ | ||
| BAS1 | At2g26710 | 5′-GCTCTCCTTTTTGTGTTTTCTCTCT-3′ |
| 5′-AGTCCGGAACAAATTTTTGACCGTT-3′ | ||
| SBI1 | At1g02100 | 5′-ATGGCGGAATCTCGCAGCAA-3′ |
| 5′-CAGGTGATGACGCTGATGA-3′ | ||
| BRI1 or bri1 | At4g39400 | 5′-GGTAGAGAGATGAGGAAGAGA-3′ |
| 5′-CTCATAATTTTCCTTCAGGAACTTC-3′ | ||
| ACT2 | At5g09810 | 5′-TTCCGCTCTTTCTTTCCAAGCTCA-3′ |
| 5′-AAGAGGCATCAATTCGATCACTCA-3′ |
Dephosphorylation of BRI1 by PP2A
The construct of GST:BRI1 kinase domain (GST–BRI1-KD) was previously described (18) and the resulting fusion protein was purified with glutathione agarose beads. The purified GST–BRI1-KD attached to the agarose beads was phosphorylated in a kinase buffer [20 mM tris (pH 7.5), 100 mM NaCl, and 12 mM MgCl2] with 1 mM of adenosine 5′-triphosphate (ATP) for 1 hour at 37°C. The beads were washed with TBS (tris-buffered saline) buffer (pH 7.5), and then divided into two parts; one with 0.2 U of purified PP2A (Millipore) and the other without PP2A. After incubation at 37°C for 1 hour, the reactions were stopped by adding 2 × SDS sample buffer and the samples were boiled for 5 min. Proteins were resolved by SDS-PAGE and transferred to nitrocellulose membrane, which was first stained with amido black, and then blotted with antibodies that recognize phosphorylated threonine residues to allow detection of phosphorylation differences in BRI1.
To coimmunoprecipitate PP2A with BRI1 or bri1-5, we used a MAC Epitope Tag Protein Isolation kit (Miltenyi Biotec) for GFP or BRI1 following the manufacturer’s instructions with the addition of protease inhibitors (Qiagen) and phosphatase inhibitor cocktail (Sigma). BRI1 or bri1-5 was immunoprecipitated with antibodies that recognize BRI1 from extracts of 10- or 14-day-old seedlings of WT and bri1-5, respectively, whereas BRI1-GFP or bri1-5–GFP was immunoprecipitated with antibodies that recognize GFP from the extracts of 10- or 14-day-old seedlings of ProBRI1::BRI1:GFP or ProBRI1::bri1-5:GFP, respectively, followed by Western blot analysis with antibody that recognizes BRI1 (18) and antibodies that recognize PP2Ac (Millipore).
BFA treatment and confocal microscopy
Five-day-old Arabidopsis seedlings were treated with 10 μM FM4-64 (Invitrogen) with or without 50 μM BFA (Sigma) for 30 min. Confocal microscopy was performed on detached roots from treated seedlings with a Zeiss LSM 5 Pascal or a Zeiss LSM 510 META. GFP and FM4-64 were excited with 488- and 543-nm laser lines, respectively, and detected with BP505-530 and LP560 emission filters, respectively. The resulting images were processed with Zeiss LSM software and Adobe Photoshop.
Data processing
Bands in Western blots and gels from RT-PCR experiments, as well as the length of hypocotyls and roots, were measured on images with Scion software followed by a validation with ImageJ. For Western blot and transcripts, the integrated density was calculated for each band. The relative integrated density was calculated as a ratio of each measurement to a common data point of each protein or gene, and was shown near the band in the blot or gel. Each experiment was repeated at least three times and the one that best represents all different trials was shown. Hypocotyl and root length was measured on the scanned images of A. thaliana seedlings with Scion software. Where appropriate, t test was performed to determine the level of significance, and SE was included in each experiment as an error bar.
Supplementary Material
Acknowledgments
G.W. thanks S. McCormick (Plant Gene Expression Center) for her support during the revision of this manuscript. We thank H. Kasahara, Y. Zhao, N. Geldner, Y. Jaillais, and S. Yamaguchi for helpful discussions. We are grateful to Y. Jaillais, Y. Yin, and K. Torii for antibodies; T. Asami for BRZ; G. Monshausen and S. Swanson for technical help with the confocal microscope; and Y. Zhao, N. Geldner, Y. Jaillais, Y. Belkhadir, and M. Roe for reading earlier versions of the manuscript. We acknowledge the usage of the microscopy facility at Botany Imaging Center, University of Wisconsin. We also thank members of the Chory, Kamiya, and Otegui laboratories for their comments. Funding: These studies were supported by the Howard Hughes Medical Institute and grants from the U.S. Department of Agriculture (National Research Initiative Competitive grant 2006-35304-16586) and National Science Foundation (NSF) (IOS-06-49389) to J.C., NSF grants MCB-0619736 and MCB-0843151 to M.S.O., and Grants-in-Aid for Scientific Research 18-06765 to Y.K. G.W. was supported by Ruth L. Kirschstein NIH and Japan Society for the Promotion of Science Postdoctoral Fellowships.
Footnotes
www.sciencesignaling.org/cgi/content/full/4/172/ra29/DC1
Fig. S1. The abundance of bri1 was reduced in bri1-5, but bri1-5 phenotypes can be rescued by expression of additional bri1-5.
Fig. S2. Scheme for screening for suppressor of bri1-5.
Fig. S3. sbi1-dependent BRI1 or bri1-5 protein accumulation requires BRs.
Fig. S4. BRI1 or bri1-5 proteins in sbi1 are degraded in the absence of BRs.
Fig. S5. Scheme for map-cloning of SBI1.
Fig. S6. LCMTs are conserved in eukaryotes.
Fig. S7. SBI1(At1g02100) is expressed at similar levels as BRI1(At4g39400) and CPD(At5g05690) in young tissues and expression is higher than these two genes in mature organs.
Fig. S8. PP2Acs are conserved in eukaryotes.
Fig. S9. Methylated PP2Acs were undetectable in sbi1 mutant background.
Fig. S10. Distribution of BRI1 or bri1 in subcellular fractions.
Fig. S11. SBI1 partially localizes to membranous compartments.
Fig. S12. rcn1 and sbi1 mutants have similar phenotypes.
Fig. S13. SBI1 was specifically increased by BRs.
Reference
Author contributions: G.W., X.W., and X.L. performed the experiments, and G.W., Y.K., M.S.O., and J.C. designed the experiments, analyzed the data, and wrote the paper.
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 3.Kim TW, Guan S, Sun Y, Deng Z, Tang W, Shang JX, Sun Y, Burlingame AL, Wang ZY. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol. 2009;11:1254–1260. doi: 10.1038/ncb1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shiu SH, Bleecker AB. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci STKE. 2001;2001:re22. doi: 10.1126/stke.2001.113.re22. [DOI] [PubMed] [Google Scholar]
- 5.Becraft PW. Receptor kinase signaling in plant development. Annu Rev Cell Dev Biol. 2002;18:163–192. doi: 10.1146/annurev.cellbio.18.012502.083431. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167–171. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 7.Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Goshe MB, Soderblom EJ, Phinney BS, Kuchar JA, Li J, Asami T, Yoshida S, Huber SC, Clouse SD. Identification and functional analysis of in vivo phosphorylation sites of the Arabidopsis BRASSINOSTEROID-INSENSITIVE1 receptor kinase. Plant Cell. 2005;17:1685–1703. doi: 10.1105/tpc.105.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oh MH, Wang X, Kota U, Goshe MB, Clouse SD, Huber SC. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 11.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110:203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Kota U, He K, Blackburn K, Li J, Goshe MB, Huber SC, Clouse SD. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 13.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 14.Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, Wang R, Burlingame AL, Wang ZY. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jaillais Y, Hothorn M, Belkhadir Y, Dabi T, Nimchuk ZL, Meyerowitz EM, Chory J. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25:232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- 17.Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 19.Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 20.He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- 22.Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 23.Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- 24.Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
- 25.Katzmann DJ, Odorizzi G, Emr SD. Receptor downregulation and multivesicular-body sorting. Nat Rev Mol Cell Biol. 2002;3:893–905. doi: 10.1038/nrm973. [DOI] [PubMed] [Google Scholar]
- 26.Chen YG. Endocytic regulation of TGF-β signaling. Cell Res. 2009;19:58–70. doi: 10.1038/cr.2008.315. [DOI] [PubMed] [Google Scholar]
- 27.Gonzalez-Gaitan M. The garden of forking paths: Recycling, signaling, and degradation. Dev Cell. 2008;15:172–174. doi: 10.1016/j.devcel.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 28.Russinova E, Borst JW, Kwaaitaal M, Caño-Delgado A, Yin Y, Chory J, de Vries SC. Heterodimerization and endocytosis of Arabidopsis brassinosteroid receptors BRI1 and AtSERK3 (BAK1) Plant Cell. 2004;16:3216–3229. doi: 10.1105/tpc.104.025387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Geldner N, Hyman DL, Wang X, Schumacher K, Chory J. Endosomal signaling of plant steroid receptor kinase BRI1. Genes Dev. 2007;21:1598–1602. doi: 10.1101/gad.1561307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sorkin A, Goh LK. Endocytosis and intracellular trafficking of ErbBs. Exp Cell Res. 2009;315:683–696. doi: 10.1016/j.yexcr.2008.07.029. [DOI] [PubMed] [Google Scholar]
- 31.Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436:78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- 32.Eden ER, White IJ, Futter CE. Down-regulation of epidermal growth factor receptor signalling within multivesicular bodies. Biochem Soc Trans. 2009;37:173–177. doi: 10.1042/BST0370173. [DOI] [PubMed] [Google Scholar]
- 33.Heisermann GJ, Wiley HS, Walsh BJ, Ingraham HA, Fiol CJ, Gill GN. Mutational removal of the Thr669 and Ser671 phosphorylation sites alters substrate specificity and ligand-induced internalization of the epidermal growth factor receptor. J Biol Chem. 1990;265:12820–12827. [PubMed] [Google Scholar]
- 34.Countaway JL, Nairn AC, Davis RJ. Mechanism of desensitization of the epidermal growth factor receptor protein-tyrosine kinase. J Biol Chem. 1992;267:1129–1140. [PubMed] [Google Scholar]
- 35.Goh LK, Huang F, Kim W, Gygi S, Sorkin A. Multiple mechanisms collectively regulate clathrin-mediated endocytosis of the epidermal growth factor receptor. J Cell Biol. 2010;189:871–883. doi: 10.1083/jcb.201001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Avraham R, Yarden Y. Feedback regulation of EGFR signalling: Decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Villeneuve G, Wang Z. Control of epidermal growth factor receptor endocytosis by receptor dimerization, rather than receptor kinase activation. EMBO Rep. 2005;6:942–948. doi: 10.1038/sj.embor.7400491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nomura T, Bishop GJ, Kaneta T, Reid JB, Chory J, Yokota T. The LKA gene is a BRASSINOSTEROID INSENSITIVE 1 homolog of pea. Plant J. 2003;36:291–300. doi: 10.1046/j.1365-313x.2003.01863.x. [DOI] [PubMed] [Google Scholar]
- 39.Bouquin T, Meier C, Foster R, Nielsen ME, Mundy J. Control of specific gene expression by gibberellin and brassinosteroid. Plant Physiol. 2001;127:450–458. [PMC free article] [PubMed] [Google Scholar]
- 40.Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell. 2008;20:3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Friedrichsen DM, Joazeiro CA, Li J, Hunter T, Chory J. Brassinosteroid-insensitive-1 is a ubiquitously expressed leucine-rich repeat receptor serine/threonine kinase. Plant Physiol. 2000;123:1247–1256. doi: 10.1104/pp.123.4.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 2005;138:1117–1125. doi: 10.1104/pp.104.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu W, Huang J, Li B, Li J, Wang Y. Is kinase activity essential for biological functions of BRI1? Cell Res. 2008;18:472–478. doi: 10.1038/cr.2008.36. [DOI] [PubMed] [Google Scholar]
- 44.Li J, Nagpal P, Vitart V, McMorris TC, Chory J. A role for brassinosteroids in light-dependent development of Arabidopsis. Science. 1996;272:398–401. doi: 10.1126/science.272.5260.398. [DOI] [PubMed] [Google Scholar]
- 45.Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 46.Janssens V, Longin S, Goris J. PP2A holoenzyme assembly: In cauda venenum (the sting is in the tail) Trends Biochem Sci. 2008;33:113–121. doi: 10.1016/j.tibs.2007.12.004. [DOI] [PubMed] [Google Scholar]
- 47.Farkas I, Dombrádi V, Miskei M, Szabados L, Koncz C. Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 2007;12:169–176. doi: 10.1016/j.tplants.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 48.Shi Y. Serine/threonine phosphatases: Mechanism through structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Rodriguez-Concepción M, Toledo-Ortiz G, Yalovsky S, Caldelari D, Gruissem W. Carboxyl-methylation of prenylated calmodulin CaM53 is required for efficient plasma membrane targeting of the protein. Plant J. 2000;24:775–784. doi: 10.1046/j.1365-313x.2000.00924.x. [DOI] [PubMed] [Google Scholar]
- 50.Zhou HW, Nussbaumer C, Chao Y, DeLong A. Disparate roles for the regulatory A subunit isoforms in Arabidopsis protein phosphatase 2A. Plant Cell. 2004;16:709–722. doi: 10.1105/tpc.018994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hernández-Sotomayor SM, Mumby M, Carpenter G. Okadaic acid-induced hyperphosphorylation of the epidermal growth factor receptor. Comparison with receptor phosphorylation and functions affected by another tumor promoter, 12-O-tetradecanoylphorbol-13-acetate. J Biol Chem. 1991;266:21281–21286. [PubMed] [Google Scholar]
- 52.Kang JS, Liu C, Derynck R. New regulatory mechanisms of TGF-β receptor function. Trends Cell Biol. 2009;19:385–394. doi: 10.1016/j.tcb.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 53.Shah K, Russinova E, Gadella TW, Jr, Willemse J, De Vries SC. The Arabidopsis kinase-associated protein phosphatase controls internalization of the somatic embryogenesis receptor kinase 1. Genes Dev. 2002;16:1707–1720. doi: 10.1101/gad.220402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams RW, Wilson JM, Meyerowitz EM. A possible role for kinase-associated protein phosphatase in the Arabidopsis CLAVATA1 signaling pathway. Proc Natl Acad Sci USA. 1997;94:10467–10472. doi: 10.1073/pnas.94.19.10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ding Z, Wang H, Liang X, Morris ER, Gallazzi F, Pandit S, Skolnick J, Walker JC, Van Doren SR. Phosphoprotein and phosphopeptide interactions with the FHA domain from Arabidopsis kinase-associated protein phosphatase. Biochemistry. 2007;46:2684–2696. doi: 10.1021/bi061763n. [DOI] [PubMed] [Google Scholar]
- 56.Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, Schwab R, Weigel D, Meyerowitz EM, Luschnig C, Offringa R, Friml J. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- 57.Tseng TS, Briggs WR. The Arabidopsis rcn1-1 mutation impairs dephosphorylation of Phot2, resulting in enhanced blue light responses. Plant Cell. 2010;22:392–402. doi: 10.1105/tpc.109.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Noguchi T, Fujioka S, Choe S, Takatsuto S, Yoshida S, Yuan H, Feldmann KA, Tax FE. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 60.Torii KU, Mitsukawa N, Oosumi T, Matsuura Y, Yokoyama R, Whittier RF, Komeda Y. The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell. 1996;8:735–746. doi: 10.1105/tpc.8.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.