Abstract
Isothermal titration calorimetry and surface plasmon resonance were tested for their ability to study substrate binding to the active site (AS) and to the secondary binding site (SBS) of Bacillus subtilis xylanase A separately. To this end, three enzyme variants were compared. The first was a catalytically incompetent enzyme that allows substrate binding to both the AS and SBS. In the second enzyme, binding to the SBS was impaired by site-directed mutagenesis, whereas in the third enzyme, the AS was blocked using a covalent inhibitor. Both techniques were able to show that AS and SBS have a similar binding affinity.
Keywords: Isothermal titration calorimetry, Surface plasmon resonance, Xylanase, Xylooligosaccharides, Mechanism-based inhibitor
In various glycoside hydrolases, substrate molecules can bind not only to the active site (AS)1 but also to remote sites on the surface of the catalytic module. These sites are referred to as secondary binding sites (SBSs) [1]. Several putative roles have been attributed to them, many of which are analogous to functions of carbohydrate-binding modules (CBMs) in modular enzymes [1]. However, characterization of separate binding events at different binding sites (AS and SBSs) in these glycoside hydrolases is challenging. Surface plasmon resonance (SPR) experiments showed that substrate binding affinity changed upon site-directed mutagenesis of the two SBSs in barley α-amylase [2,3]. Nuclear magnetic resonance (NMR)-monitored titration experiments demonstrated that binding of xylooligo-saccharides to the Bacillus circulans xylanase AS and SBS occurs independently, whereas binding of longer chains to both sites occurs cooperatively [4]. The binding affinity constants of short xylooligo-saccharides were similar for the AS and SBS, but the on- and off-rates of substrate binding were estimated to be at least 10-fold higher for the SBS [4]. In the study presented here, the potential of isothermal titration calorimetry (ITC) and SPR, two commonly used techniques quantifying binding, was explored for their ability to quantify the strength of substrate binding to the AS and SBS of the glycoside hydrolase family (GH)-11 Bacillus subtilis xylanase (XBS) separately. XBS differs only one residue from the B. circulans xylanase, and the SBSs of the two enzymes are identical [5]. The SBS plays an important role for XBS in binding and hydrolysis of polymeric substrates and in substrate targeting [6,7].
In contrast to NMR-monitored titration, both ITC and SPR do not allow assessment of binding to the two sites individually in a direct way. Therefore, three enzyme variants were produced, and their purity was verified by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) and silver staining analogous to previous descriptions [6,7]. The first one, XBS_E172A, was a catalytically incompetent mutant that can still bind substrate to the AS and SBS [5]. A two-site binding model was used to fit the data obtained with XBS_E172A. In the second variant, XBS_E172A_AAA, catalytic activity was eliminated and binding to the SBS was also impaired by mutation of three important SBS residues: G56A, T183A, and W185A [6]. Previously, using NMR-monitored titration, Ludwiczek and coworkers [4] could not detect oligosaccharide binding to the SBS of a similar mutant enzyme in which also three important SBS residues were mutated to alanine. Because we obtained similar results for activity and binding affinity when testing both of these mutant enzymes (results not shown), a single-site binding model was used to fit the data obtained for XBS_E172A_AAA. In the third variant, binding in the AS was blocked using the mechanism-based inhibitor 2,3-epoxypropyl β-D-xylopyranoside, which was covalently attached to the catalytic nucleophile of XBS (E78) [8]. The inactivation of the enzyme was performed analogously to what was described by Ntarima and coworkers [8]. After complete inactivation of the enzyme, as assessed by measurement of the residual activity on Xylazyme AX analogous to the previous description [6], the residual unbound inhibitor was washed out using Vivaspin 15R (Sartorius, Aubagne, France). A 1:1 coupling of inhibitor to XBS was verified by electrospray ionization mass spectrometry. A single-site binding model was used to fit the data obtained for XBS with the blocked AS. Both ITC and SPR experiments were performed in McIlvaine buffer (0.10 M citrate and 0.20 M disodium hydrogen phosphate, pH 6.0) at 25 °C. For the experiments, xylohexaose (X6) was chosen as substrate in these experiments because (i) it can fill all six subsites in the AS of XBS, (ii) it is large enough to span all SBS subsites, and (iii) it is too short to bind both the AS and SBS simultaneously [5].
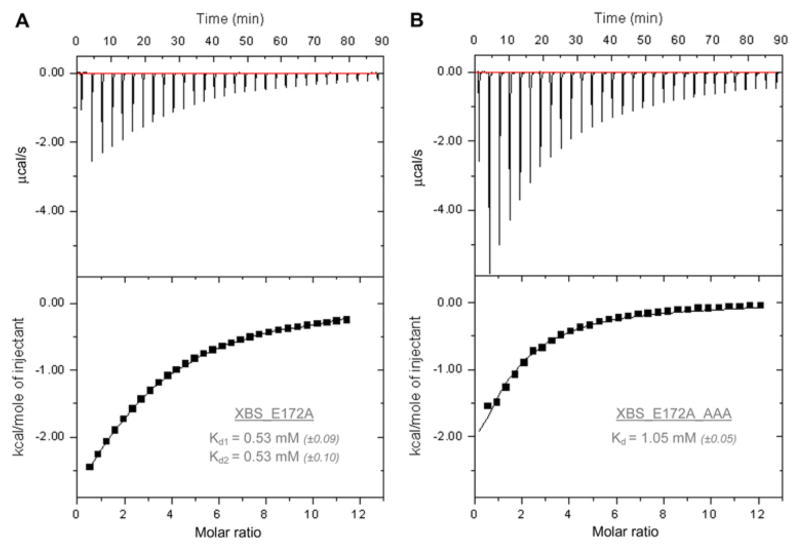
The ITC experiments were performed using an ITC200 calorimeter (GE Healthcare, Uppsala, Sweden). Both XBS_E172A and XBS_E172A_AAA were tested (Fig. 1). The results shown are representative data obtained from three independent measurements on each enzyme. The XBS epoxyalkyl derivative could not be used in these experiments because of the large amount of inactivated protein required for ITC. The presented results are obtained on injection of 1.3 μl aliquots of 10 mM X6 (Megazyme, Bray, Ireland) into 0.17 mM XBS_E172A solution or 27 mM X6 into 0.46 mM XBS_E172A_AAA solution at time intervals of 3 min. The data were corrected for the heat of dilution. Due to the weak binding constant, the stoichiometry needed to be fixed to permit reliable determination of the dissociation constant (Kd) [9]. Binding of X6 to the AS of XBS_E172A, on the one hand, and binding of X6 to the SBS, on the other, were not visually distinguishable from each other in the obtained thermograms. Two identical Kd values of 0.53 mM were obtained using a binding model for two independent sites. For binding of X6 to XBS_E172A_AAA, a Kd value of 1.05 mM was obtained. Because this enzyme is believed to be incapable of binding substrate in the SBS, this value characterizes the binding event in the AS. A Kd value for the SBS alone could not be obtained with ITC because the XBS variant with a blocked AS could not be tested due to enzyme availability limitations. Significantly less heat was released per mole of injectant in ITC experiments with XBS_E172A_AAA than with XBS_E172A, indicating that the titrations of XBS_E172A and XBS_E172A_AAA are indeed characterized by different stoichiometries (Fig. 1A and B). Use of a two-site binding model for XBS_E172A did not allow estimating a realistic enthalpy of binding (ΔH) or deconvolution of ΔH values for the separate binding sites. Fitting a one-site binding model to XBS_E172A_AAA data resulted in ΔH = −6.5 kcal mol−1 and TΔS = −2.5 kcal mol−1 for binding to the AS.
Fig.1.
ITC results of the titration of XBS_E172A (A) and XBS_E172A_AAA (B) with X6. The upper graphs show the raw binding heats on titration, and the lower graphs show the integrated binding heat levels. The inset tables give Kd values (standard errors on the fits are in parentheses).
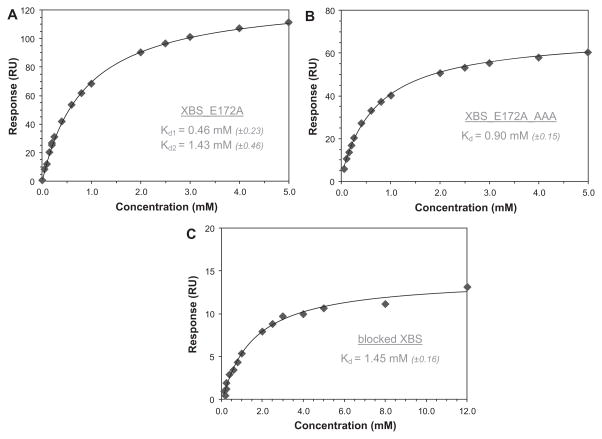
The SPR experiments were carried out on a Biacore T100 (GE Healthcare) and were conducted on all three variants (Fig. 2). Using an amine coupling kit, the xylanases were immobilized on a carboxymethylated dextran chip (GE Healthcare) in 10 mM sodium acetate buffer (pH 5.5). The obtained immobilization levels of XBS_E172A, XBS_E172A_AAA, and the XBS epoxyalkyl derivative on the biosensor chips were 3079, 2431, and 783 response units (RU), respectively. Because 1 RU equals 1 pg/mm2, these values correspond to 0.15 × 10−12, 0.12 × 10−12, and 0.04 × 10−12 mol/mm2. Sensorgrams were collected in the presence of 0.005% (v/v) P20 surfactant at a flow of 30 μl min−1, a contact time of 90 s, and a dissociation time of 100 to 240 s. Both XBS_E172A and XBS_E172A_AAA were immobilized in duplicate on different chips, whereas XBS with the covalent inhibitor attached was immobilized only once due to enzyme availability limitations. On each chip, sensorgrams were made in triplicate. The SPR analysis for the binding of X6 to XBS_E172A gave rise to Kd values of 0.46 and 1.43 mM. The sensorgram was added as a Supplementary figure (see Supplementary material). Based on the experiments with XBS_E172A alone, it was impossible to determine which value belongs to the AS and which one belongs to the SBS. For this, SPR experiments with the other two enzymes needed to be carried out. Experiments with XBS_E172A_AAA gave a Kd value of 0.74 mM, which represents binding to the AS, whereas experiments with the inhibitor-bound XBS resulted in a Kd value of 1.45 mM for binding to the SBS. The estimated maximal responses on binding of X6 to XBS_E172A, XBS_E172A_AAA, and the XBS epoxyalkyl derivative were 135, 66, and 15 RU, respectively. This allowed calculation of the amounts of X6 bound to the chip on saturation. These were 0.16 × 10−12, 0.08 × 10−12, and 0.02 × 10−12 mol/mm2, respectively. The ratio of X6 bound on saturation over the amount of immobilized enzyme for XBS_E172A was 1.10, which is much higher than the ratios of 0.67 for XBS_E172A_AAA and 0.48 for XBS with a blocked AS. This again provides a strong indication that binding to the different proteins is characterized by different stoichiometries. It also indicates that roughly half of the immobilized enzymes remain capable of binding X6. On- and off-rates could not be retrieved from these SPR experiments. For low-affinity oligosaccharide binding, such as the binding of X6 to XBS, both association and dissociation processes are typically too fast and, therefore, only steady-state data are obtained (see Supplementary figure).
Fig.2.
SPR results for the binding of X6 to XBS_E172A (A), XBS_E172A_AAA (B), and XBS with a blocked AS (C). The inset tables give Kd values (standard errors on the fits are in parentheses).
In general, the obtained Kd values from ITC and SPR were in good agreement with each other. They are slightly lower but on the same order of magnitude as those obtained from NMR-monitored titration of B. circulans xylanase with X6, for which Kd values of 1.0 and 4.7 mM have been reported for binding to the AS and SBS, respectively [4]. All three techniques indicated that binding to the AS and binding to the SBS are characterized by similar affinity constants and, thereby, confirm the significance of the SBS for XBS, as demonstrated previously [6]. The binding affinities were also in good agreement with the previously reported binding strength of xylooligosaccharides to a GH-11 Chainia xylanase [10].
In conclusion, both ITC and SPR showed that binding to the AS and binding to the SBS both are characterized by Kd values in the low millimolar (mM) range. Although NMR-monitored titration is probably a more powerful technique, it is also the most time-consuming and technically most demanding technique. Additional experiments are needed for initial NMR peak assignment to distinguish between simultaneous binding events at different sites. This requires the use of 15N or 13C isotope-labeled protein. Moreover, its application is also limited to relatively small proteins (<30–40 kDa) [11]. Therefore, the results presented here show that both ITC and SPR provide valuable alternatives that are, for most researchers, easier in execution and interpretation when attempting to characterize separate binding events on the same glycoside hydrolase.
Supplementary Material
Acknowledgments
Bart Devreese, Kathleen Piens, and Patricia Ntarima (Laboratory for Protein Biochemistry and Biomolecular Engineering, Ghent, Belgium) are thanked for supplying the epoxyalkyl glycoside inhibitor and performing electrospray ionization mass spectrometry analysis. The “Fonds voor Wetenschappelijk Onderzoek-Vlaanderen” (FWO, Brussels, Belgium) is acknowledged for the postdoctoral fellowship of E.D. This study is part of the Methusalem program “Food for the Future” at Katholieke Universiteit Leuven. The Biacore T100 SPR instrument was purchased with a grant from the Danish Research Council for Natural Science.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.ab.2011.09.005.
Footnotes
Abbreviations used: AS, active site; SBS, secondary binding site; CBM, carbohydrate-binding module; SPR, surface plasmon resonance; NMR, nuclear magnetic resonance; ITC, isothermal titration calorimetry; GH, glycoside hydrolase family; XBS, Bacillus subtilis xylanase; SDS–PAGE, sodium dodecyl sulfate–polyacrylamide gel electrophoresis; X6, xylohexaose; RU, response units.
References
- 1.Cuyvers S, Dornez E, Delcour JA, Courtin CM. Occurrence and functional significance of secondary carbohydrate binding sites in glycoside hydrolases. Crit Rev Biotechnol. doi: 10.3109/07388551.2011.561537. in press. [DOI] [PubMed] [Google Scholar]
- 2.Bozonnet S, Jensen MT, Nielsen MM, Aghajari N, Jensen MH, Kramhøft B, Willemoës M, Tranier S, Haser R, Svensson B. The “pair of sugar tongs” site on the non-catalytic domain C of barley α-amylase participates in substrate binding and activity. FEBS J. 2007;274:5055–5067. doi: 10.1111/j.1742-4658.2007.06024.x. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen MM, Bozonnet S, Seo ES, Mótyán JA, Andersen JM, Dilokpimol A, Abou Hachem M, Gyémánt G, Næsted H, Kandra L, Sigurskjold BW, Svensson B. Two secondary carbohydrate binding sites on the surface of barley α-amylase 1 have distinct functions and display synergy in hydrolysis of starch granules. Biochemistry. 2009;48:7686–7697. doi: 10.1021/bi900795a. [DOI] [PubMed] [Google Scholar]
- 4.Ludwiczek ML, Heller M, Kantner T, McIntosh LP. A secondary xylan-binding site enhances the catalytic activity of a single-domain family 11 glycoside hydrolase. J Mol Biol. 2007;373:337–354. doi: 10.1016/j.jmb.2007.07.057. [DOI] [PubMed] [Google Scholar]
- 5.Vandermarliere E, Bourgois TM, Rombouts S, Van Campenhout S, Volckaert G, Strelkov SV, Delcour JA, Rabijns A, Courtin CM. Crystallographic analysis shows substrate binding at the −3 to +1 active-site subsites and at the surface of glycoside hydrolase family 11 endo-1,4-xylanases. Biochem J. 2008;410:71–79. doi: 10.1042/BJ20071128. [DOI] [PubMed] [Google Scholar]
- 6.Cuyvers S, Dornez E, Rezaei MN, Pollet A, Delcour JA, Courtin CM. Secondary substrate binding strongly affects activity and binding affinities of Bacillus subtilis and Aspergillus niger GH11 xylanases. FEBS J. 2011;278:1098–1111. doi: 10.1111/j.1742-4658.2011.08023.x. [DOI] [PubMed] [Google Scholar]
- 7.Cuyvers S, Hendrix J, Dornez E, Engelborghs Y, Delcour JA, Courtin CM. Both substrate hydrolysis and secondary substrate binding determine xylanase mobility as assessed by FRAP. J Phys Chem B. 2011;115:4810–4817. doi: 10.1021/jp110963f. [DOI] [PubMed] [Google Scholar]
- 8.Ntarima P, Nerinckx W, Klarskov K, Devreese B, Bhat MK, Van Beeumen J, Claeyssens M. Epoxyalkyl glycosides of d-xylose and xylo-oligosaccharides are active-site markers of xylanases from glycoside hydrolase family 11, not from family 10. Biochem J. 2000;347:865–873. [PMC free article] [PubMed] [Google Scholar]
- 9.Turnbull WB, Daranas AH. On the value of c: can low affinity systems be studied by isothermal titration calorimetry? J Am Chem Soc. 2003;125:14859–14866. doi: 10.1021/ja036166s. [DOI] [PubMed] [Google Scholar]
- 10.Hegde SS, Kumar AR, Ganesh KN, Swaminathan CP, Khan MI. Thermodynamics of ligand (substrate/end product) binding to endoxylanase from Chainia sp. (NCL-82–5-1): isothermal calorimetry and fluorescence titration studies. Biochim Biophys Acta. 1998;1388:93–100. doi: 10.1016/s0167-4838(98)00163-0. [DOI] [PubMed] [Google Scholar]
- 11.Holdgate GA, Anderson M, Edfeldt F, Geschwindner S. Affinity-based, biophysical methods to detect and analyze ligand binding to recombinant proteins: matching high information content with high throughput. J Struct Biol. 2010;172:142–157. doi: 10.1016/j.jsb.2010.06.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.