SUMMARY
Glycogen synthase kinase 3 (GSK3) is a key regulator in signaling pathways in both animals and plants. Three Arabidopsis thaliana GSK3s are shown to be related to brassinosteroid (BR) signaling. In a phenotype-based compound screen we identified bikinin, a small molecule that activates BR signaling downstream of the BR receptor. Bikinin directly binds the GSK3 BIN2 and acts as an ATP competitor. Furthermore, bikinin inhibits the activity of six other Arabidopsis GSK3s. Genome-wide transcript analyses demonstrate that simultaneous inhibition of seven GSK3s is sufficient to activate BR responses. Our data suggest that GSK3 inhibition is the sole activation mode of BR signaling and argues against GSK3-independent BR responses in Arabidopsis. The opportunity to generate multiple and conditional knockouts in key regulators in the BR signaling pathway by bikinin represents a useful tool to further unravel regulatory mechanisms.
INTRODUCTION
Mammals possess three closely related isoforms of glycogen synthase kinase 3 (GSK3α, GSK3β, and GSK3β2) whereas Drosophila generates several isoforms from a single Shaggy-locus by differential splicing. GSK3s play a major role in several pathways including Wnt, Hedgehog, and insulin signaling, mitosis, and apoptosis (Meijer et al., 2004). In contrast, plants appear to have a much larger set of divergent GSK3s. Analysis of the Arabidopsis thaliana genome revealed the existence of ten GSK3s, designated as ASKs for Arabidopsis SHAGGY-like kinases, divided in four groups (Jonak and Hirt, 2002). Several lines of evidence indicate that plant GSK3s have numerous roles in development and stress responses (Jonak and Hirt, 2002). However, in plants, the only known signal transduction pathway mediated by GSK3s is brassinosteroid (BR) signaling (Li et al., 2001; Vert and Chory, 2006; Zhao et al., 2002). BRs are a group of polyhydroxylated steroid hormones implicated in multiple developmental processes, including stem elongation, leaf expansion, vascular development, seed germination, and resistance to biotic and abiotic stresses (Bishop and Koncz, 2002; Caño-Delgado et al., 2004). Genetic defects in biosynthesis or perception of BRs result in dwarfism, dark-green and curled leaves, reduced seed germination and fertility, and de-etiolation in the dark (Bishop and Koncz, 2002; Clouse et al., 1996). However, BR-overproducing plants or plants treated with brassinolide (BL; Figure 1A) display an increase in hypocotyl and petiole length and in overall plant growth (Choe et al., 2001). BRs are perceived at the cell surface by direct binding to the plasma membrane-localized BRI1 receptor (Bishop and Koncz, 2002). The next elucidated step in the pathway is the inhibition of BIN2, which belongs to the group II GSK3s. This results in the dephosphorylation of two plant-specific transcription factors, BES1 and BZR1, that accumulate in the nucleus and regulate expression of many known BR-responsive genes (Li and Jin, 2007). Gain-of-function bin2 mutations or overexpression of the wild-type BIN2 gene result in a phenotype resembling that of BR-deficient or BR-signaling mutants (Choe et al., 2002; Li and Nam, 2002; Pérez-Pérez et al., 2002), whereas a triple mutant lacking BIN2 and its two closest homologs is morphologically similar to mutants with a constitutively active BR signaling pathway (Vert and Chory, 2006). Although the triple knockout for group II GSK3s displays constitutive BR responses, BES1 is not completely dephosphorylated, implying that other kinases might be involved (Vert and Chory, 2006). Yet, it still remains unknown whether other GSK3s or unrelated kinases besides group II GSK3s regulate BR signaling.
Figure 1. BR-Constitutive Responses Induced by Bikinin.
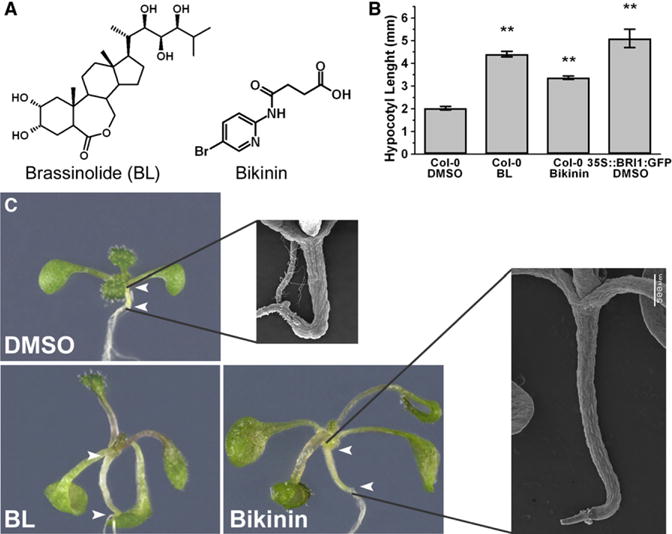
(A) Chemical structure of BL and bikinin.
(B) Hypocotyl lengths of 3-day-old seedlings treated with DMSO, 1 μM BL, or 30 μM bikinin for 3 days (means ± standard error (SE); **p value < 0.001 compared with Col-0 DMSO as determined with standard two-tailed t test). Overexpressing BRI1-GFP plants were included as a positive control.
(C) Phenotypes of 3-day-old light-grown wild-type plants treated with 0.5% DMSO, BL (1 μM), or bikinin (30 μM) for 3 days (insets represent scanning electron microscopy pictures of same treatments). Experiments were repeated three times. Arrowheads indicate the boundaries of the hypocotyl.
By using a chemical genetics approach to identify compounds inducing constitutive BR responses, we identified bikinin as a strong activator of BR signaling. We show that seven Arabidopsis GSK3s are potential targets for bikinin and that bikinin directly binds BIN2 and acts as an ATP-competitor. The specificity and the inhibitory strength of bikinin toward particular Arabidopsis GSK3s are determined by specific residues in the ATP-binding pockets of the GSK3s. Genome-wide transcript analysis demonstrates that simultaneous inhibition of seven GSK3s by bikinin results in a vast transcriptional overlap compared with BL treatment. This confirms that GSK3 inhibition is the sole activation mode of BR signaling and strongly argues against the possibility of GSK3-independent BR responses. In addition our data do not exclude that other GSK3s, apart of group II, could have a potential role in BR signaling.
RESULTS
Bikinin Activates BR Signaling Downstream of BRI1
We screened a commercial 10,000 compound library to identify small molecules that caused constitutive BR responses in Arabidopsis thaliana Col-0 seedlings, similar to those found in plants treated with the most potent brassinosteroid, BL, or plants overexpressing the BR biosynthetic gene DWF4 or the BR signaling gene BRI1 (Wang et al., 2001). One compound (4-[(5-bromo-2-pyridinyl) amino]-4-oxobutanoic acid), designated bikinin (Figure 1A), induced constitutive BR responses, including a significant increase in hypocotyl length (Figures 1B and 1C and insets), long and bending petioles, and blade-shaped, pale-green leaves (Figure 1C). Furthermore, comparable to a treatment with BL at micromolar concentrations, lateral root density was reduced (see Figure S1 available online). Despite the strong resemblance between the respective phenotypes, there was no structural similarity between bikinin and BL (Figure 1A) or its biosynthetic intermediates. To identify the active core structure and assess the requirement of the functional groups of bikinin, we measured the biological activity of five derived molecules (Figure S1) on lateral root density. The structure-activity analysis revealed that removing the halogen, aromatic nitrogen, or carboxyl group either completely abolished or significantly reduced bikinin activity, implying that these functional groups are essential for in planta bikinin activity (Figure S1). Inversely, substitution of bromine with chlorine slightly increased potency.
To determine whether bikinin induces constitutive BR responses by controlling the same subset of BR target genes, we analyzed the effect of bikinin treatment on the RNA levels of the BR feedback-regulated biosynthetic genes DWF4 (Choe et al., 1998), CPD (Szekeres et al., 1996), ROT3 (Tanaka et al., 2005), BR6OX1 (Shimada et al., 2003), and BR6OX2 (Shimada et al., 2003), the genes encoding BR signaling components BRI1 (Clouse et al., 1996), BIN2 (Li and Nam, 2002; Li et al., 2001), BSU1 (Mora-García et al., 2004), BES1 (Li and Deng, 2005), and BZR1 (Li and Deng, 2005), and the BR-inducible genes SAUR-AC1 (Vert et al., 2005) and BAS1 (Neff et al., 1999). For all genes, the expression changes closely resembled those of a BL treatment (Figure S2) indicating that bikinin promotes BR responses through a common transcriptional growth-regulatory module.
To get insight into the compound’s site of action, mutants in BR biosynthesis (loss-of-function, cpd) (Vert et al., 2005), BR signaling (loss-of-function, bri1-116) (Vert et al., 2005), and gain-of-function bin2-1 (Li and Nam, 2002; Li et al., 2001) were treated with BL, bikinin, or an inactive bikinin variant (Var2). In agreement with previous reports, addition of BL rescued the light-grown cpd dwarfed phenotype and the dark-grown de-etiolated phenotype (Figures 2A and 2B) (Szekeres et al., 1996; Vert et al., 2005), but not the bri1-116 loss-of-function and bin2-1 gain-of-function mutants (Figures 2A and 2B). Interestingly, bikinin treatment induced petiole and hypocotyl elongation in all mutants both under light- and dark-grown conditions (Figures 2A and 2B), suggesting that bikinin acts downstream of the BRI1 receptor, more precisely at the level or downstream of BIN2.
Figure 2. Dwarfed Phenotypes of Light- and Dark-Grown BR Mutants Rescued by Bikinin.
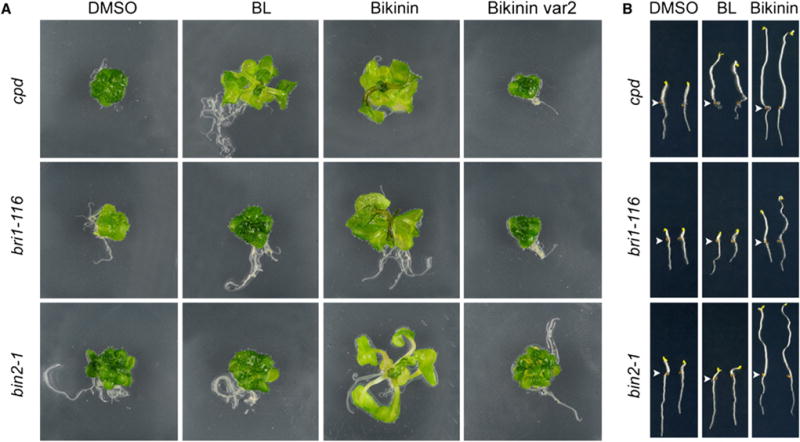
(A) Phenotypes of five day old light-grown cpd, bri1-116 and bin2-1 mutants treated with 0.5 % DMSO, BL (1 μM), or bikinin (30 μM) for 3 more days.
(B) Phenotypes of three-day-old dark-grown cpd, bri1-116 and bin2-1 mutants germinated on 0.5 % DMSO, BL (1 μM), or bikinin (30 μM).
All experiments were repeated three times. Arrowheads indicate root-to-hypocotyl transition.
BIN2 negatively regulates the BR pathway by phosphorylation of the closely homologous BZR1 and BES1/BZR2 nuclear proteins (Vert and Chory, 2006). To show that bikinin induces BR responses by activating these downstream components of the BR signaling, we monitored the phosphorylation status of BES1 in mock-, BL-, or bikinin-treated plants. Consistent with previous reports (Vert and Chory, 2006), in plants grown on control medium, BES1 was preferentially present in its phosphorylated form (Figures 3A and 3B). Addition of BL induced a shift of BES1 to its unphosphorylated state and activated the signaling cascade. As observed with BL, treatment with bikinin resulted in accumulation of unphosphorylated BES1 starting at concentrations of 5–10 μM (Figure 3A). Similar to BL (Yin et al., 2002), BES1 dephosphorylation occurred within 30 min of bikinin application (Figure 3B), suggesting that the effect of bikinin on the phosphorylation state of BES1 is a primary response. These data indicate that bikinin induces BR responses through inhibition of BES1 phosphorylation.
Figure 3. Bikinin Induced BES1 Dephosphorylation.
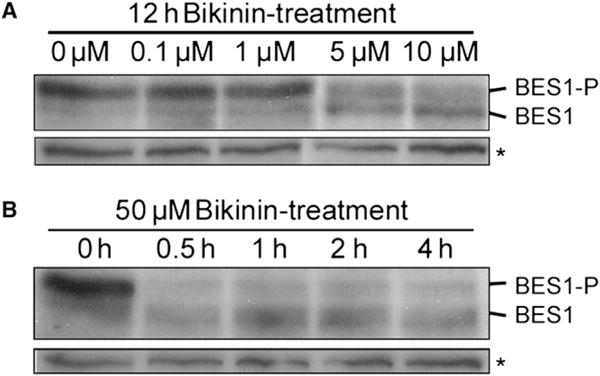
(A) Dose-response analysis of BES1 phosphorylation in response to bikinin treatment. Western blots were performed with extracts from 35S:BES1-GFP plants treated with a concentration range from 0 to 10 μM bikinin for 12 hr and detected with anti-BES1 antibodies.
(B) Time-course analysis of BES1 dephosphorylation following bikinin treatment. Western blots were carried out with extracts from 35S:BES1-GFP plants treated with 50 μM bikinin over a period of 0 to 4 hr and detected with anti-BES1 antibodies. BES1-P, phosphorylated BES1; *, nonspecific antibody-reactive band (loading control).
Bikinin Directly Inhibits BIN2 by Interfering with ATP Binding
As BES1 phosphorylation is regulated by BIN2, we hypothesized that the bikinin-induced shift of BES1 to its dephosphorylated form might be due to direct inhibition of BIN2. Therefore, we examined by in vitro kinase assays whether bikinin interferes directly with BIN2 kinase activity. Bikinin reduced BIN2 kinase activity toward its substrate BES1 in a dose-dependent manner (Figure 4A). The inhibitory effect of bikinin was already apparent at 2.5 μM and became dramatic at 10 μM. The biologically inactive Var2 did not inhibit BIN2 kinase activity, even at high concentrations (Figure S3). To demonstrate direct binding of bikinin with BIN2, we carried out surface plasmon resonance (SPR) experiments. Recombinant GST-BIN2 protein was immobilized on a sensor chip through amine coupling and increasing concentrations of bikinin were injected over the sensor surface. Bikinin interacted with immobilized GST-BIN2 in a dose-dependent manner (Figure 4B), with an increase in response starting from a concentration of 10 μM. Interestingly, the inactive Var2 compound was also able to bind GST-BIN2, although the affinity of this interaction was calculated to be about 2000 times lower (Figure S4). In summary, these observations combined with expression and mutant analyses allowed us to conclude that BIN2 represents a direct target of bikinin.
Figure 4. Binding and Specific Inhibition of Kinase Activity of BIN2 by Bikinin.
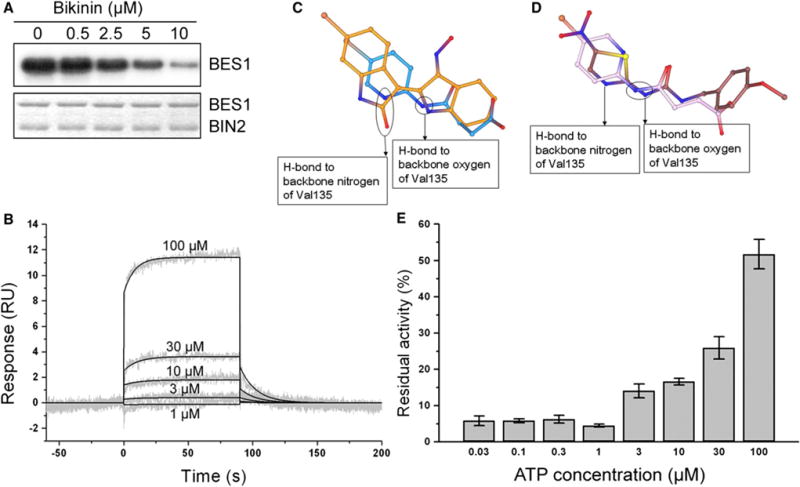
(A) Autoradiography of a kinase assay with GST-BIN2 and MaBP-BES1 on a concentration range of 0 to 10 μM bikinin. Coomassie staining was used as loading control.
(B) SPR sensorgrams for bikinin binding to GST-BIN2 after injection of different bikinin concentrations as indicated. The binding curves are overlaid by calculated curves resulting from the global fits of the data to a 1:1 interaction model (χ2 = 0.126). Reference and blank data are subtracted. The experiment was done in triplicate.
(C) Docking of bikinin in GSK3β. The best-ranked docking position of the bikinin molecule (blue) in the GSK3β crystal structure 1UV5 (Bhat et al., 2003) compared with that of the inhibitor 6-bromoindirubin-3′-oxime (orange). The nitrogens of the bikinin molecule are in a position where they can make similar hydrogen bonds with the backbone of Val135.
(D) The best ranked docking position of the bikinin molecule (pink) in the GSK3β crystal structure 1Q5K (Meijer et al., 2003) compared with the position of the inhibitor AR-A014418 (brown). The amide of the bikinin molecule is in a position where it can make a similar hydrogen bond with the backbone oxygen of Val135. Flipping of the bikinin pyridinyl ring will bring the pyridinyl nitrogen in a position where it can form a hydrogen bond with the backbone nitrogen of Val135.
(E) ATP competition experiment. Residual kinase activity of GST-BIN2 and MBP as a substrate is shown using 10 μM bikinin and a concentration range of 0.03 to 100 μM ATP. Error bars represent mean ± SE.
To get insight into the binding mode and mechanism of inhibition of bikinin, we performed docking simulations of bikinin into the crystal structure of human GSK3β. The docking solutions for bikinin were compared with the position and interactions of two known GSK3β inhibitors into two different crystal structures of human GSK3β (1UV5 for 6-bromoindirubin-3′-oxime [Meijer et al., 2003], Figure 4C; 1Q5K for ARA014418 [Bhat et al., 2003], Figure 4D). The best-ranked, lowest-energy-requiring solution for docking of bikinin into both crystal structures was very similar (Figures 4C and 4D). Comparison with the binding interactions of the inhibitors in these structures shows that, like the known inhibitors, bikinin can make two hydrogen bonds with the backbone amide oxygen and nitrogen of Val135 of GSK3β (Val118 in BIN2), which has a major contribution to the binding energy. Furthermore, again similar to known inhibitors, bikinin has a planar ring at the position where the kinase binds the purine ring of ATP, suggesting that bikinin might interfere with kinase activity by competing with ATP. To validate this finding, we tested the in vitro kinase activity of BIN2 in the presence of 10 μM bikinin and increasing concentrations of ATP. The inhibitory effect of bikinin was reduced with increasing concentrations of ATP (Figure 4E), demonstrating that the inhibition of BIN2 kinase activity by bikinin is mediated by competing with ATP in the ATP-binding pocket.
Bikinin Inhibits a Subset of Arabidopsis GSK3 Kinases
To determine the specificity of bikinin toward different Arabidopsis GSK3 subfamilies, we analyzed the kinase activity of nine ASKs in the presence of bikinin by using MBP as a general substrate. Bikinin strongly inhibited the activity of the closely related groups I and II (Figure 5A; Figure S5) with some residual activity (6%–8%) for group I kinases and total inhibition (1%–2% residual activity) for members of group II. Surprisingly, one member of group III, ASKθ, was moderately inhibited (20% residual activity), whereas the activity of the other member, ASKβ, was not affected by bikinin. The activity of ASKδ, a member of group IV, was not inhibited. Furthermore, the compound had no effect on the activity of three other Arabidopsis Ser/Thr kinases (AtMPK4, AtMPK6, and AtAURORA1; Figure 5B). These data indicate that the activity of bikinin is ASK specific, with additional specificity for certain subgroups.
Figure 5. Specificity of Bikinin for ASK Subgroups.
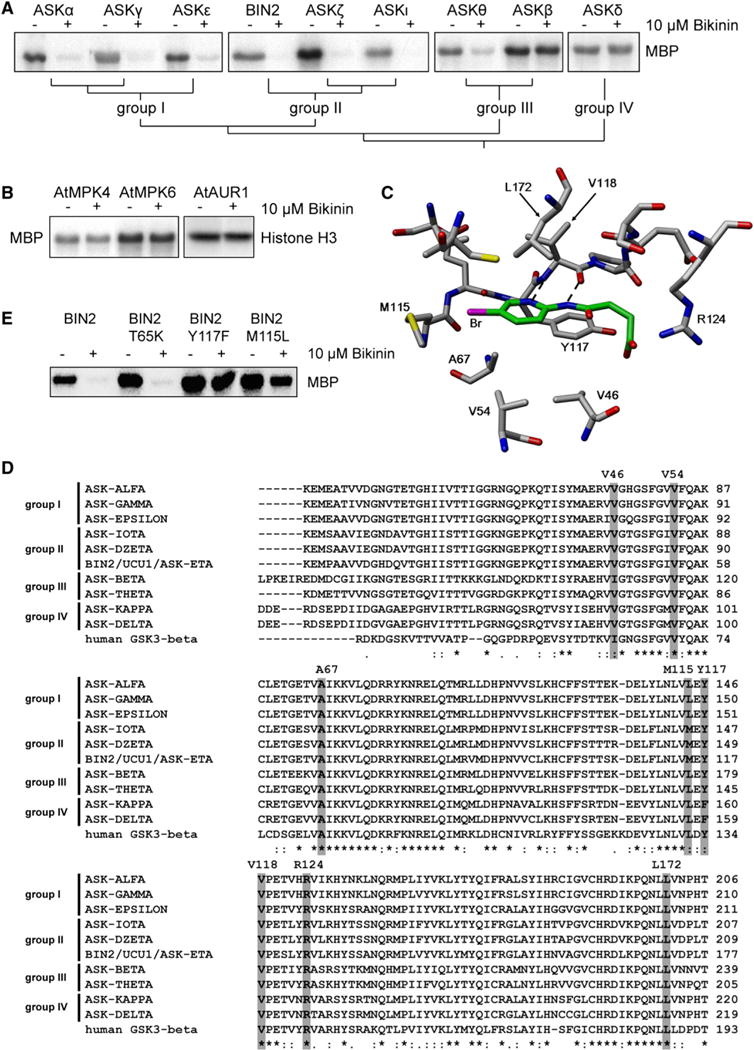
(A) Autoradiography of kinase assays with nine ASKs with MBP as a substrate in the presence or absence of 10 μM bikinin. The second member of group IV, ASKκ, had no kinase activity and was not included in the analysis.
(B) Autoradiography of kinase assays with AtMPK4, AtMPK6 and MBP as a substrate (left) and with AtAUR1 kinase and histone H3 as a substrate (right) in the presence or absence of 10 μM bikinin.
(C) Model of bikinin in a homology model of BIN2. The bromo atom interacts with Met115. The nitrogen atoms hydrogen bond to the backbone of Val118 (dashed lines). The pyridinyl ring is sandwiched between Ala67 and Leu172. In different models, the carboxyl group interacts with Arg 124 and/or hydrogen bonds with Tyr117.
(D) Clustal-W (Larkin et al., 2007) multiple sequence alignment of the conserved region of the Arabidopsis ASKs and human GSK3β. Residues in gray are particularly important to the interaction with bikinin in the ATP-binding pocket.
(E) Autoradiography of kinase assays with BIN2 and in vitro mutated versions of BIN2 using MBP as a substrate in the presence or absence of 10 μM bikinin.
To determine whether the action of bikinin is plant specific, we tested the effect of bikinin on a panel of 77 human kinases (Table S1) at two concentrations. In general, all human kinases were poorly affected by bikinin. The human homolog of BIN2, GSK3β, was inhibited by only ~40% at 10 μM bikinin (Table S1), conditions in which BIN2 activity was completely abolished (Figure S5). Together, these data indicate that the strong in vitro effect of bikinin on kinase activity is limited to specific subgroups of plant GSK3s.
In Xenopus laevis, Wnt signaling mediated by the action of GSK3β was shown to be essential for dorsal differentiation during dorsoventral axis formation (He et al., 1995). To assess the in vivo effect of bikinin on the proper patterning of Xenopus, we applied bikinin to developing embryos, but found no observable effect on dorsoventral axis formation (Figure S6), consistent with the poor inhibition of GSK3β in vitro. These data suggest that bikinin is not sufficiently potent in in vivo vertebrate systems to inhibit GSK3β and to cause subsequent developmental defects.
Given its capacity to act as an ATP-competitive inhibitor and the highly conserved ATP-binding pockets of GSK3s, the specificity of bikinin for certain subgroups is remarkable and provides a unique tool to study the different GSK3 subgroups. To further understand which residues determine the specificity of bikinin, homology models were built for BIN2 and the other ASKs, with bikinin in a position determined by the docking simulations (Figure 5C). We examined the conservation of the bikinin binding site in the different ASK models/sequences and human GSK3β. Strikingly, the residue that interacts with the bikinin bromo atom is a Met115 in group II ASKs, and a Leu in the other ASKs and GSK3β (Figures 5C and 5D). In addition, several other residues localized in the bikinin-binding site are not conserved between the different ASKs and GSK3β, including Tyr117 of BIN2, which is replaced by Phe in ASKδ (Figures 5C and 5D). To confirm these predicted interaction sites, we introduced the mutations M115L or Y117F in the ATP binding pocket of BIN2. As predicted, these mutations reduced the strong inhibition of BIN2 activity by bikinin (Figure 5E), whereas mutating an unrelated T65 residue outside the binding pocket to Lys did not affect inhibition by bikinin. Taken together, the predicted residues in the ATP-binding pocket have an effect on bikinin binding and, consequently, on the inhibitory strength of bikinin for the different ASKs and the reduced potency toward the human GSK3β.
BL and Bikinin Have Largely Overlapping Transcriptional Responses
Because bikinin inhibited seven plant GSK3s from which only three (group II) were previously implicated in BR signaling (Vert and Chory, 2006), we performed a genome-wide transcript analysis after BL and bikinin treatment to evaluate if BR responses are entirely dependent on the group II GSK3s. Wild-type Col-0 seedlings were treated with 1 μM BL, 30 μM bikinin, or dimethyl sulfoxide (DMSO) for 30 min or 2 hr. Shoots were collected for RNA isolation. After normalization and statistical analysis of expression levels, 272 genes were identified as significantly differentially regulated by bikinin and/or BL compared with the DMSO control (p ≤ 0.05 after Benjamini and Hochberg’s multiple-comparison correction and minimal 2-fold change) (Figure 6A; Table S2). A subset of Gene Ontology (GO) terms (BiNGO) (Maere et al., 2005) was used to identify the functional trends in the 272 responsive genes. This analysis showed that genes encoding proteins involved in BR metabolism, BR biosynthesis, hormone-mediated signaling, and transcription were significantly enriched, consistent with the role of BL and bikinin in the BR signaling cascade (Table S3A). Interestingly, genes expressed in response to auxin and abiotic stimuli were also overrepresented.
Figure 6. Scheme of the Genome-wide Transcript Analysis and Simplified Schematic Representation of the BR-Signaling Pathway and Points of Action of BL and Bikinin.
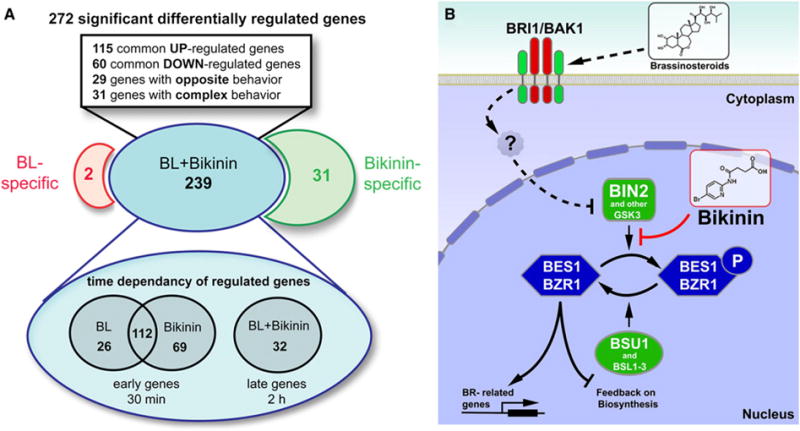
(A) Overview of the distribution of the 272 significant differentially regulated genes and their time dependency.
(B) Activation of the BR signaling cascade (dashed arrows) triggered by binding of BRs to the membrane-localized BRI1 receptor. Through a still unknown mechanism, this signal is transported to the nucleus where it inhibits the BIN2 kinase. Bikinin acts by direct inhibition of the activity of the group II ASK family (red arrow). The inhibition of BIN2 kinase activity, either through an unknown signal after binding of BRs to the BRI1 receptor or after direct inhibition by bikinin, depletes the pool of phosphorylated BES1/BZR1 proteins. By the inhibition of BIN2 and the action of the BSU1 phosphatase, non-phosphorylated BES1/BZR1 proteins accumulate, which in turn activate the transcription of BR-related genes and the feedback inhibition on the BR biosynthesis.
Fold-change values normalized to the control treatment were used to further analyze the significantly modulated genes. Bikinin and BL transcriptional responses substantially overlapped as 88% (239 of 272 genes) of the genes were affected by both treatments. A total of 179 genes (66%) responded similarly to BL and bikinin, indicating that none of these genes were regulated in a BL- or bikinin-independent fashion. However, 60 genes (22%) were regulated differently; 29 genes were regulated in an opposite manner and 31 genes had a complex behavior (Figure 6A; Table S4A). Furthermore, another 31 genes were affected only by bikinin and 2 genes were specific for BL (Figure 6A; Tables S4B and S4C). Bikinin-specific responses were anticipated based on the observation that bikinin was able to inhibit the activity of not only BR-specific group II GSK3s but also group I and one member of group III. BiNGO analysis (Maere et al., 2005) of the 31 bikinin-specific genes showed overrepresentation of genes encoding proteins with transporter activity (Table S3B). In total, 207 of the 239 commonly affected genes responded within 30 min, whereas for the remaining 32 genes transcript changes were observed after 2 hr (Figure 6A; Table S4). Interestingly, bikinin affected the expression of more genes (181) than BL (138) and, in general, the bikinin-induced changes in expression occurred faster. Approximately 25% of the commonly 239 (BL and bikinin) affected genes were previously reported to be regulated by BL in the global microarray analysis performed by Nemhauser et al. (2006). Consistent with the negative feedback regulation model of BR biosynthesis (Mathur et al., 1998), the expression of BR biosynthesis genes (CPD, DWF4, and BR6OX2) was downregulated (Table S4A). Both bikinin and BL treatment resulted in significant downregulation of genes involved in the auxin pathway (e.g., PIN7, IAA29, and IAA2), as previously reported (Goda et al., 2004; Goda et al., 2002; Müssig et al., 2002) (Table S4A). The expression of 26 genes was affected early by BL and only later by bikinin (Figure 6A). Among these are several early auxin-inducible genes from the SAUR family (SAUR-AC1, SAUR14, SAUR10, and SAUR16) and genes involved in auxin transport (PID and WAG2) (Table S4A). This observation is consistent with previous microarray studies showing that BL induces the expression of the auxin-inducible SAUR, GH3, and IAA gene families (Goda et al., 2004; Nemhauser et al., 2004, 2006), in a period of 30 to 60 min.
DISCUSSION
Identification of Bikinin
The GSK3β homolog BIN2 is a key player in the intracellular BR signal transduction cascade (Belkhadir and Chory, 2006). Although a role for proteasome-mediated protein degradation of BIN2 has been suggested as a mechanism for posttranslational BIN2 regulation (Peng et al., 2008), it is still unknown how BIN2 activity is regulated in response to binding of BRs to BRI1. Yet it remains unclear if other GSK3 subgroups besides the group II GSK3s are involved in transducing BR signals, and if GSK3-independent activation pathways of BR signaling exist. Here, we screened a chemical library for compounds that induced constitutively active BR responses. We identified bikinin as the first nonsteroidal molecule to modulate the BR signaling cascade downstream of the BRI1 receptor. Application of bikinin partially restored the phenotype of the bri1 loss-of-function mutant and the bin2 gain-of-function mutant to wild-type. Notably, besides the typical dwarfism seen in other BR mutants like bri1 and cpd, bin2 also shows extremely downward curled leaves, a phenotype reminiscent of some auxin mutants (Choe et al., 2002). Also, BIN2 was recently linked to the auxin signaling pathway (Vert et al., 2008), suggesting that this protein sits at the intersection between several signaling pathways and implying that bin2 harbors more than only the typical BR phenotype. Interestingly, bikinin clearly rescues the dwarfism of the hypocotyls and the petioles of the bin2 gain-of-function mutant (Figure 2A). However, the downward curled leaves remain visible, suggesting that bikinin is fully rescuing the BR-related phenotypes of this mutation while having no effect on the auxin-related phenotypes.
A combination of BES1 phosphorylation analysis, kinase assays, SPR-binding studies, and microarray analysis showed that bikinin directly targets BIN2 (Figure 6B) and activates BR-dependent gene expression.
Binding Mode and Specificity of Bikinin
Up to date, a large number of animal GSK3 inhibitors have been identified as potential drugs against different diseases, and several of them were cocrystallized with GSK3β (Meijer et al., 2004). Despite their wide chemical diversity, most act as an ATP competitor (Bhat et al., 2003; Meijer et al., 2003). Because the ATP-binding pockets of GSK3s or other closely related kinases such as CDKs and MAP kinases are highly similar, selectivity is a key issue. This is especially important when inhibitors are used as pharmacological tools to demonstrate the involvement of the GSK3 in a cellular process. Bikinin showed high selectivity and potency toward Arabidopsis GSK3s. All other Arabidopsis kinases (AtMPK4, AtMPK6, and AtAUR1) tested were not inhibited by bikinin, revealing that the compound has certain specificity toward the GSK3 subfamily of protein kinases in Arabidopsis. It is worth noting that the MAP kinases AtMPK4 and AtMPK6 are structurally similar to GSK3 and belong to the same subclass of protein kinases (Hanks and Hunter, 1995). However, it cannot be excluded that bikinin affects other kinases based on the limited number of Arabidopsis kinases tested in this study. Interestingly, bikinin had a minor inhibitory effect on human GSK3β in vitro and it did not affect the developmental processes in vertebrates in vivo, as opposed to the GSK3β inhibitor LiCl. An ATP-competition assay with BIN2 and modeling of bikinin into the crystal structure of the human BIN2 homolog, GSK3β, revealed that bikinin acts as an ATP-competitive kinase inhibitor. It has been shown that the kinase domains and key residues in both the activation domain and the ATP-binding pocket are highly conserved between GSK3β and ASKs (Jonak and Hirt, 2002). Therefore, specific bikinin-interacting residues in the ATP-binding pocket of BIN2 were predicted and confirmed experimentally. The differences in potential bikinin-binding residues could account for the different levels of inhibition of GSK3β and different ASK family members. Additional understanding of the mechanisms of compound-protein interactions will allow modulating strength and specificity of these interactions. This way, it will be possible to further unravel the role of different GSK3s in plant development and create tools for molecular modeling of new GSK3β specific inhibitors.
Potential Function of Other GSK3s in BR Signaling
Compromising BIN2 activity by a loss-of-function mutation in the BIN2 gene has a minor effect on BR responses. This observation has led to the conclusion that other GSK3s, among the ten found in the Arabidopsis genome, might act redundantly with BIN2 in the BR pathway (Vert and Chory, 2006). Although individual loss-of-function mutants of the two closest BIN2 homologs had no effect on BR signaling, gain-of-function mutations indicated that the two other members of group II GSK3s function in BR signaling as well. This was further confirmed by the constitutive BR response phenotype of the triple knockout for group II GSK3s (Vert and Chory, 2006), which suggests that all members of group II GSK3s are implicated in BR signaling. By contrast, the absence of total dephosphorylation of BES1 in the triple group II GSK3 mutant suggests that BES1 might be targeted by additional kinases (Vert and Chory, 2006). In agreement with the group II GSK3 triple knockout, application of bikinin resulted in a constitutive BR response phenotype. However, in the case of bikinin, members of group I and ASKθ were also strongly inhibited in vitro, a new finding that could not be revealed earlier by mutant analysis. The inhibitory effect of bikinin in vitro for four additional GSK3s, the very specific BR response phenotype induced by bikinin and the large overlap of downstream transcriptional regulation between bikinin and BL are compelling and suggest that the group I GSK3s and ASKθ might be involved in BR signaling as well. This conclusion is further supported by the fact that group I GSK3s and ASKθ, in addition to group II GSK3s, have overlapping expression patterns and are highly expressed in young seedlings (Charrier et al., 2002), a developmental stage at which BL and bikinin exerted strong phenotypic effects in the chemical genetics screen.
Inhibition of Seven GSK3s by Bikinin Argues against GSK3-Independent BR Signaling
Previous studies demonstrated a functional conservation of GSK3s as negative regulators in signal transduction pathways in plants and animals (Jonak and Hirt, 2002). Nevertheless, it remains unknown to what extent BR signaling is entirely dependent on GSK3s, or whether GSK3-independent signaling cascades downstream of the receptor exist. The transcriptome analysis of the early effect of BL and bikinin treatments presented here demonstrates that nearly all genes regulated by BL are regulated by bikinin as well. Thus, activation of the BR pathway by binding of BL to the BRI1 receptor is highly comparable to the direct inhibition of GSK3s by bikinin. Taking into consideration the high specificity of bikinin toward GSK3 subgroups, these data suggest that transducing the BL-induced signal depends on inhibition of specific GSK3s, and argues against the existence of BR signaling pathways that act independently of GSK3s.
EXPERIMENTAL PROCEDURES
Bikinin, Derivatives, and Other Compounds
24-Epibrassinolide (BL) was purchased from Fuji Chemical Industries (Toyama, Japan) and bikinin and all derivative molecules from ChemBridge Corporation (ID for Bikinin: 5122035, Var1: 5122029, Var2: 5133967, Var3: 5843203, Var4: 5121777 and Var5: 5310341).
Chemical Genetics Screening and Growth Conditions
A commercial 10,000 compound library (DIVERSet, ChemBridge Corporation) was screened for a combination of phenotypes indicative of a constitutive BR response at a final concentration of 50 μM in 2% DMSO. Seeds (three or four) of Arabidopsis thaliana (L.) Heynh. were sown in 96-well filter plates (Multi-screen HTS MSBVS1210; Millipore) in liquid medium derived from standard Murashige and Skoog (MS) medium and incubated in a growth chamber under continuous light (110 μE.m−2·s−1 photosynthetically active radiation, supplied by cool-white fluorescent tungsten tubes; Osram) at 22°C. Three days after germination, compounds were added to the 96-well plates. Plants incubated in 2% DMSO and 1 μM BL were used as negative and positive control, respectively. Plants were phenotypically analyzed 6 days after germination. For all subsequent phenotypic analyses, plants were grown on plates (Greiner Labortechnik) with solid medium derived from standard MS medium under the same conditions. For the hypocotyl elongation assay, plants were grown in the dark at 22°C.
Scanning Electron Microscopy
For scanning electron microscopy pictures, plants were treated as indicated in Figure 1 and scanned using a Hitachi TM1000 table top scanning electron microscope after fixation in 1% OsO4 for 2 hr and subsequent dehydration with ethanol.
Western Blotting
For protein extraction, 6-day-old seedlings were grown under standard conditions as described above on solid medium. Subsequently, plants were soaked in liquid MS medium supplemented with the indicated compounds or mock (for concentrations and periods, see Figure 3). Plants were frozen in liquid nitrogen, ground, and homogenized in ice-cold homogenization buffer (25 mM Tris-HCl [pH 8], 5 mM EDTA, 1 mM β-mercapto-ethanol, 15 mM MgCl2, 85 mM NaCl, 0.1% Tween 20, and 1 protease inhibitor tablet/50 ml Complete [Roche Diagnostics]). The homogenate was centrifuged twice (5 min; 20,800 × g) in an Eppendorf centrifuge 5417 at 4°C. After addition of loading buffer, the samples were heated for 10 min at 95°C, centrifuged again, separated on a 12% acrylamide gel or a 4%–20% gradient precast gel (Bio-Rad), and blotted on nitrocellulose membranes (Hybond-C super, GE-Healthcare) in 190 mM glycine and 25 mM Tris-HCl with a mini-blotting system (Bio-Rad) for 2 hr. Membranes were blocked overnight at 4°C in blotting buffer (0.01 M Tris-HCl [pH 8.0], 0.15 M NaCl, 3% skim milk [BD Difco], and 0.1% Triton). For immunodetection, anti-BES1 antibodies at 1:2000 and anti-GFP at 1:1000 dilutions were used as primary antibodies. Secondary antibodies were used at 1:10,000 dilutions. The proteins were detected by chemiluminescence (Perkin-Elmer).
Quantitative Real-Time Polymerase Chain Reaction
RNA was extracted with the RNeasy kit (QIAGEN). Poly(dT) cDNA was prepared from 1 μg total RNA with Superscript III reverse transcriptase (Invitrogen) and analyzed on an LightCycler 480 apparatus (Roche Diagnostics) with the SYBR Green I Master kit (Roche Diagnostics) according to the manufacturer’s instructions. Targets were quantified with specific primer pairs designed with the Beacon Designer 4.0 (Premier Biosoft International) (Table S5). All individual reactions were done in triplicate. Data were analyzed with qBase (Hellemans et al., 2007). Expression levels were normalized to those of EEF1α4 and CDKA1;1.
Kinase Assays
The BIN2 kinase assay with GST-BIN2 and maltose binding protein (MaBP) fused to BES1 (MaBP-BES1) as a substrate was done as reported elsewhere (Yin et al., 2002). Kinase assays of the ASKs, AtMPK4 and AtMPK6 with myelin basic protein (MBP), AURORA1 with histone H3, and of the human kinases as described previously (Bain et al., 2003; Demidov et al., 2005; Kiegerl et al., 2000). Comparable activities of the different ASKs were used. For one particular kinase equal amounts (ranging from 20 to 100 ng depending on the innate kinase activity) were used in the assays with and without bikinin.
In Vitro Mutagenesis
For in vitro mutagenesis of BIN2, polymerase chain reaction (PCR) was performed according to the protocol for Pfu Ultra High Fidelity DNA Polymerase from Stratagene. The primers used for in vitro mutagenesis of BIN2 are listed in Table S6.
SPR Analysis
A Biacore T100 instrument was used to analyze the interaction of bikinin with BIN2. Using amine coupling, purified GST-BIN2 was immobilized in the flow cell of a Series S CM5 Sensor Chip (Research Grade, Biacore AB). HBS-EP (Biacore AB) was used as running buffer, flow rate was set at 5 μl/min. The surface of the chip was activated by injecting a mixture of EDC (0.2 M) and NHS (0.05 M) for 10 min. Subsequently, 20 μg/ml GST-BIN2 in 10 mM sodium acetate buffer (pH 6.0) was injected for 20 min. The immobilization level of GST-BIN2 was ≈20,000 RU. The chip was flushed with 1 M ethanolamine (pH 8.5) for 10 min to deactivate the surface. A flow cell treated with a cycle of activation and deactivation without immobilized protein was used as a reference. Bikinin was bound to GST-BIN2 in HBS-EP running buffer (Biacore AB) supplemented with 10 mM MgCl2. Different concentrations of bikinin dissolved in running buffer were injected at a flow rate of 30 μl/min over the reference and the GST-BIN2 flow cell for 90 s, followed by 180 s of buffer flow (dissociation phase). Zero concentration samples were used as blanks. The absolute response level in the reference cell (background) was ≈40,000 RU, the absolute response level in the flow cell with immobilized GST-BIN2 was z60,000 RU. The flow cell temperature was set to 25°C. Biacore T100 evaluation software (version 1.1.) was used for curve fitting, assuming a 1:1 binding model.
Bikinin Modeling
A bikinin molecule was built in molecular operating environment (chemical computing group) and energy minimized (Amber99 forcefield with Born solvation). eHits (Zsoldos et al., 2007) was used to dock the bikinin molecule into the apo structures of GSK3β with Protein Data Bank codes 1UV5 and 1Q5K. The docking results were visualized and evaluated with CheVi (Sim BioSys). Homology models for all Arabidopsis GSK3s were built with automodel in modeler (Šali and Blundell, 1993) and t_coffee (Notredame et al., 2000) multiple alignment, with the 1UV5 structure of GSK3β as a template.
Microarray Analysis and Statistical Data Processing
Arabidopsis thaliana (L). Heynh., ecotype Columbia (Col-0) seeds were germinated vertically on solid medium derived from standard MS medium for 7 days under 16 hr light/8 hr dark cycles. The seedlings were immersed in liquid medium containing 1 μM BL, 30 μM bikinin or DMSO as a mock control for 0.5 and 2 hr. Shoots were collected for RNA isolation. All sampling points were performed in three independent experiments. RNA was extracted as described for quantitative real-time PCR analysis. Total RNA (200 μg per array) was used to hybridize ATH1 Affymetrix Arabidopsis arrays according to standard procedures. The expression values were RMA normalized (Irizarry et al., 2003) with R (www.r-project.org) and the Bioconductor package affylmGUI (http://bioinf.wehi.edu.au/affylmGUI/) and subsequently a two-factor analysis of variance was performed in TMEV4.0 (Saeed et al., 2003). The overrepresentation of GO groups on sets of differentially expressed genes was analyzed with the BiNGO software (Maere et al., 2005).
Supplementary Material
SIGNIFICANCE.
The availability of a synthetic chemical that activates BR signaling downstream of the BRI1 receptor, more specifically at the level of the GSK3s, will allow to further unravel the role of these GSK3s in BR signaling. Also, further understanding of the mechanisms of the compound-protein interactions will open new possibilities to modulate the strength and/or specificity of these interactions and will support further studies on the role of the different GSK3s in plant development. More generally, the specificity of bikinin for a subset of GSK3s offers the opportunity to study other effects of specifically inhibiting GSK3s in Arabidopsis thaliana. Furthermore, identification of the amino acids that are crucial for the specificity of bikinin will provide a better understanding of specific GSK3 inhibition mechanisms in plants, which could also be extrapolated to the mammalian homolog GSK3β and can be potentially useful for the development of specific GSK3β inhibitors.
Acknowledgments
We thank Jenny Bain, Hilary McLaughlan, and Philip Cohen for analyzing the effect of bikinin on a panel of 77 human kinases, Eveline Van De Slijke for help with western blots, Natalie De Jonge for helpful discussions on the SPR experiments, Karel Spruyt for graphics support, and Martine De Cock for help in preparing the manuscript. This work was supported in part by a grant from the U.S. National Science Foundation (to J.C.), by the Austrian Science foundation (grant to C. J.), the Interuniversity Attraction Poles Programme (IUAP VI/33), initiated by the Belgian State, Science Policy Office, and the Special Research Fund of Ghent University (predoctoral fellowship to B.D.R.). D.A. is a Postdoctoral Fellow of the Research Foundation-Flanders (FWO). D.A. and L.N. are part of the VIB Compound Screening Facility.
Footnotes
SUPPLEMENTAL DATA
Supplemental Data include six figures and six tables and can be found with this article online at http://www.cell.com/chemistry-biology/supplemental/S1074-5521(09)00141-0.
References
- Bain J, McLauchlan H, Elliott M, Cohen P. The specificities of protein kinase inhibitors: an update. Biochem J. 2003;371:199–204. doi: 10.1042/BJ20021535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkhadir Y, Chory J. Brassinosteroid signaling: a paradigm for steroid hormone signaling from the cell surface. Science. 2006;314:1410–1411. doi: 10.1126/science.1134040. [DOI] [PubMed] [Google Scholar]
- Bhat R, Xue Y, Berg S, Hellberg S, Ormö M, Nilsson Y, Radesäter AC, Jerning E, Markgren PO, Borgegård T, et al. Structural insights and biological effects of glycogen synthase kinase 3-specific inhibitor AR-A014418. J Biol Chem. 2003;278:45937–45945. doi: 10.1074/jbc.M306268200. [DOI] [PubMed] [Google Scholar]
- Bishop GJ, Koncz C. Brassinosteroids and plant steroid hormone signaling. Plant Cell. 2002;14:S97–S110. doi: 10.1105/tpc.001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caño-Delgado A, Yin Y, Yu C, Vafeados D, Mora-Garcia S, Cheng JC, Nam KH, Li J, Chory J. BRL1 and BRL3 are novel brassinosteroid receptors that function in vascular differentiation in Arabidopsis. Development. 2004;131:5341–5351. doi: 10.1242/dev.01403. [DOI] [PubMed] [Google Scholar]
- Charrier B, Champion A, Henry Y, Kreis M. Expression profiling of the whole Arabidopsis shaggy-like kinase multigene family by real-time reverse transcriptase-polymerase chain reaction. Plant Physiol. 2002;130:577–590. doi: 10.1104/pp.009175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Dilkes BP, Fujioka S, Takatsuto S, Sakurai A, Feldmann KA. The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell. 1998;10:231–243. doi: 10.1105/tpc.10.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA. Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J. 2001;26:573–582. doi: 10.1046/j.1365-313x.2001.01055.x. [DOI] [PubMed] [Google Scholar]
- Choe S, Schmitz RJ, Fujioka S, Takatsuto S, Lee MO, Yoshida S, Feldmann KA, Tax FE. Arabidopsis brassinosteroid-insensitive dwarf12 mutants are semidominant and defective in a glycogen synthase kinase 3β-like kinase. Plant Physiol. 2002;130:1506–1515. doi: 10.1104/pp.010496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC. A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 1996;111:671–678. doi: 10.1104/pp.111.3.671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov D, Van Damme D, Geelen D, Blattner FR, Houben A. Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell. 2005;17:836–848. doi: 10.1105/tpc.104.029710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Shimada Y, Asami T, Fujioka S, Yoshida S. Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2002;130:1319–1334. doi: 10.1104/pp.011254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks SK, Hunter T. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9:576–596. [PubMed] [Google Scholar]
- He X, Saint-Jeannet JP, Woodgett JR, Varmus HE, Dawid IB. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- Hellemans J, Mortier GR, De Paepe A, Speleman F, Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8:R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jonak C, Hirt H. Glycogen synthase kinase 3/SHAGGY-like kinases in plants: an emerging family with novel functions. Trends Plant Sci. 2002;7:457–461. doi: 10.1016/s1360-1385(02)02331-2. [DOI] [PubMed] [Google Scholar]
- Kiegerl S, Cardinale F, Siligan C, Gross A, Baudouin E, Liwosz A, Eklöf S, Till S, Bögre L, Hirt H, Meskiene I. SIMKK, a mitogen-activated protein kinase (MAPK) kinase, is a specific activator of the salt stress-induced MAPK, SIMK. Plant Cell. 2000;12:2247–2258. doi: 10.1105/tpc.12.11.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li J, Jin H. Regulation of brassinosteroid signaling. Trends Plant Sci. 2007;12:37–41. doi: 10.1016/j.tplants.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH. Regulation of brassinosteroid signaling by a GSK3/SHAGGY-like kinase. Science. 2002;295:1299–1301. doi: 10.1126/science.1065769. [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH, Vafeados D, Chory J. BIN2, a new brassinosteroid-insensitive locus in Arabidopsis. Plant Physiol. 2001;127:14–22. doi: 10.1104/pp.127.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Deng XW. It runs in the family: regulation of brassinosteroid signaling by the BZR1-BES1 class of transcription factors. Trends Plant Sci. 2005;10:266–268. doi: 10.1016/j.tplants.2005.04.002. [DOI] [PubMed] [Google Scholar]
- Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- Mathur J, Molnár G, Fujioka S, Takatsuto S, Sakurai A, Yokota T, Adam G, Voigt B, Nagy F, Maas C, et al. Transcription of the Arabidopsis CPD gene, encoding a steroidogenic cytochrome P450, is negatively controlled by brassinosteroids. Plant J. 1998;14:593–602. doi: 10.1046/j.1365-313x.1998.00158.x. [DOI] [PubMed] [Google Scholar]
- Meijer L, Skaltsounis AL, Magiatis P, Polychronopoulos P, Knockaert M, Leost M, Ryan XP, Vonica CA, Brivanlou A, Dajani R, et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol. 2003;10:1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- Meijer L, Flajolet M, Greengard P. Pharmacological inhibitors of glycogen synthase kinase 3. Trends Pharmacol Sci. 2004;25:471–480. doi: 10.1016/j.tips.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Mora-García S, Vert G, Yin Y, Caño-Delgado A, Cheong H, Chory J. Nuclear protein phosphatases with Kelch-repeat domains modulate the response to brassinosteroids in Arabidopsis. Genes Dev. 2004;18:448–460. doi: 10.1101/gad.1174204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müssig C, Fischer S, Altmann T. Brassinosteroid-regulated gene expression. Plant Physiol. 2002;129:1241–1251. doi: 10.1104/pp.011003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, Chory J. BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA. 1999;96:15316–15323. doi: 10.1073/pnas.96.26.15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Peng P, Yan Z, Zhu Y, Li J. Regulation of the Arabidopsis GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 through proteasome-mediated protein degradation. Mol Plant. 2008;1:338–346. doi: 10.1093/mp/ssn001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Pérez JM, Ponce MR, Micol JL. The UCU1 Arabidopsis gene encodes a SHAGGY/GSK3-like kinase required for cell expansion along the proximodistal axis. Dev Biol. 2002;242:161–173. doi: 10.1006/dbio.2001.0543. [DOI] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Šali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Shimada Y, Goda H, Nakamura A, Takatsuto S, Fujioka S, Yoshida S. Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol. 2003;131:287–297. doi: 10.1104/pp.013029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Németh K, Koncz-Kálmán Z, Mathur J, Kauschmann A, Altmann T, Rédei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Asami T, Yoshida S, Nakamura Y, Matsuo T, Okamoto S. Brassinosteroid homeostasis in Arabidopsis is ensured by feedback expressions of multiple genes involved in its metabolism. Plant Physiol. 2005;138:1117–1125. doi: 10.1104/pp.104.058040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vert G, Chory J. Downstream nuclear events in brassinosteroid signalling. Nature. 2006;441:96–100. doi: 10.1038/nature04681. [DOI] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J. Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol. 2005;21:177–201. doi: 10.1146/annurev.cellbio.21.090704.151241. [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci USA. 2008;105:9829–9834. doi: 10.1073/pnas.0803996105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZY, Seto H, Fujioka S, Yoshida S, Chory J. BRI1 is a critical component of a plasma-membrane receptor for plant steroids. Nature. 2001;410:380–383. doi: 10.1038/35066597. [DOI] [PubMed] [Google Scholar]
- Yin Y, Wang ZY, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- Zhao J, Peng P, Schmitz RJ, Decker AD, Tax FE, Li J. Two putative BIN2 substrates are nuclear components of brassinosteroid signaling. Plant Physiol. 2002;130:1221–1229. doi: 10.1104/pp.102.010918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zsoldos Z, Reid D, Simon A, Sadjad SB, Johnson AP. eHiTS: a new fast, exhaustive flexible ligand docking system. J Mol Graph Model. 2007;26:198–212. doi: 10.1016/j.jmgm.2006.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


