Abstract
Aims
Breast cancer (BC) is the most frequent tumour in women, representing 20–30% of all malignancies, and continues to be the leading cause of cancer deaths among European women. Triple-negative (TN) BC biological aggressiveness is associated with a higher dissemination rate, with central nervous system (CNS) metastases common. This study aims to elucidate the association between gene expression profiles of PTGS2, HBEGF and ST6GALNAC5 and the development of CNS metastases in TNBC.
Methods
This is a case-controlled retrospective study comparing patients (pts) with CNS metastases versus patients without them after adjuvant treatment. The selection of the samples was performed including 30 samples in both case and control groups. Formalin-fixed, paraffin-embedded samples were retrieved from the Hospital 12 de Octubre Biobank. Five 10 µm sections from each FFPE sample were deparaffinised with xylene and washed with ethanol, and the RNA was then extracted with the RecoverAll Kit (Ambion). Gene expression was assessed using TaqMan assays.
Results
A total of 53 patients were included in the study. The average age was 55 years (range 25–85). About 47 patients (88.67%) had ductal histology and presented high grade (III) tumours (40 patients; 75.47%). Eight women in the case group presented first distant recurrence in the CNS (34.80%), local recurrence (three patients, 13.04%), lungs (two patients; 8.7%), bone (one patient; 4.34%) and other locations (seven patients; 30.38%). In the control group, first distant recurrence occurred locally (six patients; 46.1%), in bone (two patients; 15.4%), lungs (one patient; 7.7%) and other sites (four patients; 23.1%). RNA was successfully obtained from 53 out of 60 samples. PTGS2, HBEGF, and ST6GALNAC5 expression values were not related to metastasis location.
Conclusion
TN tumours frequently metastasise to the visceral organs, particularly lungs and brain, and are less common in bone. The literature suggests that expression of the three genes of interest (PTGS2, HBEGF, and ST6GALNAC5) could be different in TNBC patients with CNS metastasis when compared to patients without it. We did not find a differential expression pattern in PTGS2, HBEGF, and ST6GALNAC5 genes in primary TNBC showing CNS metastases. Further studies are needed to clarify the role of these genes in CNS metastases in TNBC patients.
Keywords: breast cancer, central nervous system, metastases, triple negative
Introduction
The incidence of breast cancer (BC) is increasing [1] and has now displaced cardiovascular pathology as the leading cause of mortality among women in the Western world. In Spain, its incidence, mortality and five-year prevalence are 29%, 15%, and 41%, respectively (Globocan 2012), and it is the third most deadly neoplasm in Spain [2, 3] with an annual incidence of 61 cases for every 100,000 women. The triple negative (TN) BC subtype represents approximately 10–20% of all cases of BC in Caucasian women. It characteristically affects young women, is associated with poor prognosis pathological characteristics, high rates of early tumoural relapse, high rates of visceral metastasis (20–30%, particularly lung and brain), short survival and the absence of targeted biological therapy. Additionally, it is related to the basal-like group obtained via genetic analysis and to tumours associated with changes in the BRCA1 gene [12–13, 36]. Tumour genetic changes define the conduct of this tumour and could be responsible for the poor prognosis in this type of patient [7]. Recent studies indicate that shorter survival in the group of patients with BC is due to two characteristics: triple-negative (TN) phenotype and central nervous system (CNS) metastasis [6, 19]. A recent study suggests that the differential expression of some genes is related to the appearance of cerebral metastasis [7].
Genetic expression studies based on studies of levels of mRNA such as the PAM50 (RT-PCR) assay [11] have identified at least four molecular subtypes of BC with different clinical behaviour: Luminal A, luminal B, HER2 and basal-like [4]. Correlating this mRNA study-based classification with an immunohistochemical-based classification shows that most tumours that belong to the basal-like subtype lack expression of oestrogen receivers (ER), progesterone receivers and HER-2 and hence are called triple-negative (TN) tumours [5]. Even though tumours that share an aggressive course are grouped together in the TN group, it is not entirely homogenous; there are at least six subgroups of tumours with different gene expression patterns [12].
The only study (BOS and Col.) that has evaluated additional markers related to the development of metastasis at the cerebral level finds that RE-negative BC cells express three genes (PTGS2, HBEGF, and ST6GALNAC5) in an altered form: these cells have a predisposition to spread at the CNS level. In normal physiological conditions, expression of ST6GALNAC5 is only at the cerebral level [38, 39].
In the present work, we will analyse the expression of three genes of interest (PTGS2, HBEGF, and ST6GALNAC5) in the FFPE tissue of primary TN phenotype breast tumours with CNS metastasis compared to patients who do not present with cerebral metastasis.
Materials and methods
Sample design and selection
The study is an analytical observational case (30 samples with CNS metastasis) – control (30 samples without CNS metastasis) study, chosen for similar characteristics in each case. The present study was evaluated and approved by the Hospital 12 de Octubre of Madrid’s institutional ethics committee.
Patient selection
Patients diagnosed with BC between January 1, 1994 and December 31, 2012 with clinical information and updated clinical follow-up available in the Hospital 12 of October’s archive and documentation service.
Sample collection
The selection of the biological samples was taken from the Hospital 12 of October Pathological Anatomy Department’s paraffinised tissue bank.
Cases with the TN phenotype with adequate follow-up and with a formalin-fixed and paraffin-embedded (FFPE) tumour sample with at least 50% tumour cells were selected from the pathological anatomy archive. Subsequently, 30 cases that subsequently metastasised to the CNS and 30 cases that did not were taken.
Sample analysis
Sample processing
About 10 micron sections of the FFPE tissue blocks were made using a microtome. The RNA was isolated using the Life Technologies RecoverAll Kit following the manufacturer’s protocol. Therefore, the nucleic acids isolated were quantified using UV spectrophotometry.
Gene expression analysis using RT-qPCR
Quantitative PCR was used to analyse the expression of three genes of interest (PTGS2, HBEGF, and ST6GALNAC5) and two reference genes (IPO8 and POLR2A) validated for use as reference genes for breast cancer using paraffinised tissue [45] using TaqMan assays (ThermoFisher Scientific).
Drawing from the 50–100 ng of RNA extracted from the FFPE samples, a total reverse transcription of 1 ug of RNA was carried out with the Life Technologies High Capacity cDNA Reverse Transcription Kit following the aforementioned company’s TaqManGene Expression Assays Protocol.
| Gene | Assay ID |
|---|---|
| IPO8 | Hs00183533_m1 |
| POLR2A | Hs00172187_m1 |
| PTGS2 | Hs00153133_m1 |
| HBEGF | Hs00181813_m1 |
| ST6GALNAC5 | Hs00229612_m1 |
Statistical analysis
These expression data (Ct values) were obtained in triplicate for each sample, and the mean Ct was calculated. The missing values were replaced with a maximum Ct value set at 40. Subsequently, the expression values of the genes of interest were standardised using the ΔCt method, which consists of calculating relative expression values as differences between a normalisation factor, which in this case is the geometric mean of the expression of both reference genes, and the mean CT value of each gene. Later, a constant was added and a relative expression value for each gene in each sample was obtained, in which an increase in one supposes double the expression. In order to evaluate whether there is a differential expression of the genes of interest in TN breast cancer patients with CNS metastasis vis-à-vis the group without CNS metastasis, the Mann–Whitney U-test was applied.
Results
Clinical characteristics of the patients
The median age of the patients in the study was 55 years (range 25–85). In this group of patients, 47 (88.67%) were of the ductal histologic type and 40 patients (75.47%) were histologic Grade III. Tumour size and nodal involvement are summarised in Table 1.
Table 1. Clinical characteristics of the tumour.
| Patients (pts) | % | |
|---|---|---|
| T1 | 9 | 16.98 |
| T2 | 34 | 64.15 |
| T3 | 6 | 11.32 |
| T4 | 4 | 7.54 |
| NO | 20 | 37.73 |
| N1 | 13 | 24.52 |
| N2 | 6 | 11.32 |
| N3 | 14 | 26.41 |
In the case group, eight patients (34.80%) had cerebral metastasis as the first site of recurrence, three patients (13.04%) had local recurrence, two patients (8.7%) lung, one patient (4.34%) bone, and seven patients (30.08%) other locations. In the control group, six patients (46.1%) had local relapse as the first site of remote recurrence, two patients (15.4%) bone, one patient (7.7%) lung, and four patients (23.1%) other sites.
Evaluation of gene expression (PTGS2, HBEGF, ST6GALNAC5)
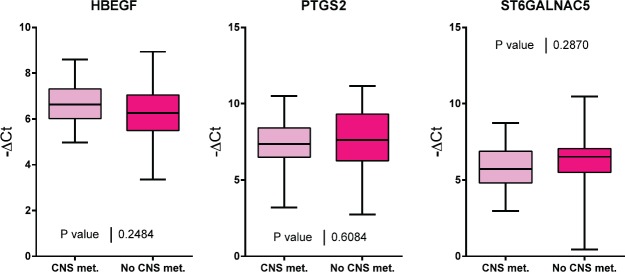
Of the 60 selected samples, a sufficient amount of RNA was extracted to carry out experiments on 53 samples. PTGS2, HBEGF, and ST6GALNAC5 gene expression was evaluated for the two groups of interest (Figure 1). No significant differences were found in the expression of these three genes between both groups of patients.
Figure 1. A comparison of HBEGF, PTGS2 and ST6GALNAC5 gene expression in tumour samples from patients with and without CNS metastasis.
Discussion
BC, as previously mentioned, is a genetically heterogeneous disease and its clinical incidence, characteristics, and prognosis differ significantly by ethnicity and race [21]. Studies carried out in North America find that Latina patients have a lower incidence but also greater mortality from BC than Caucasian patients. Also, the rate of TN tumours is significantly greater in this group of patients [22–34].
The metastasis process is complex and includes cellular intravasation, survival in the circulation, extravasation to a distant organ, angiogenesis and uninhibited growth in the host tissue [8]. Tumour gene expression studies find that the expression of some genes enables tumour cells and predisposes them to dissemination in a specific form to organs like the lungs [9–10].
Exploration of this pathology has begun to produce results and a variety of studies have found responses, albeit modest ones, to drugs like anti-EGFR (cetuximab and erlotinib), SRC inhibitors (dasatinib) and anti-angiogenics (bevacizumab) [14]. Additionally, drugs that target the PI3K/PTEN/AKT pathway and Notch survivin are presently being studied [14, 16, 17].
The epidemiological evaluation made by our group of the patients with BC seen at the 12 de Octubre University Hospital found that the TN phenotype, in concurrence with the American reports, represents around 20% of breast tumours [22].
The development of cerebral metastasis differs from that of other locations due to particularities like the special composition and density of the cerebral parenchyma and the high impermeability of the blood–brain barrier (BBB) produced by the complexity of the structures that form it, including tight junctions, the absence of fenestrations and very low pinocytic activity, as well as an extracellular matrix, pericytes, and astrocytic foot processes. The cerebral capillaries also have a high electrical resistance that increases the impermeability of this membrane to the polar and ionic substrata. Added to this is a set of extraction transporters that includes p-glycoprotein, MRP-1 to 6, breast cancer-resistant protein (BCRP), and organic anion and cation transporters [20]. All this causes systemic therapies to pass through the BBB, and therefore, their access to metastasised parts of the brain is low and insufficient to guarantee the effectiveness of most of the treatments available.
Bos et al selected RE-negative BC cells with a high predisposition to infiltrate the brain and evaluated the expression of more than 240 genes in them. The process included the inoculation of these cells in murine models, and the selection of those cells with high capacity to develop cerebral metastasis. After identifying genes through an in vivo model, their role was evaluated on the basis of breast tumour data and a group of genes related to the development of cerebral metastasis in RE-negative breast tumours was thus selected. They found that the altered expression of 17 genes was associated with the development of cerebral metastasis. Three of these genes had high expression: cyclooxygenase COX2, an EGFR ligand and the α2, 6-sialyltransferase ST6GALNAC5 gene. The identification of the COX2 gene in this group demonstrates the importance of the inflammation process in the development of brain metastasis, whereas genes related to the epithelial growth factor (EGF) are related to the replication capacity of the tumour cell. Finally, sialyltransferases are related to cell–cell interactions and change in this capacity could be related to the capacity to produce remote metastasis [7].
TN tumours frequently metastasise to the visceral organs, particularly the lungs and brain, and less frequently at the bone level. The literature suggests that the expression of the three genes of interest (PTGS2, HBEGF, and ST6GALNAC5) must be different in patients with TNBC that developed metastasis at the cerebral level when compared to patients who do not develop CNS metastasis.
We did not find a differential expression pattern in these genes (PTGS2, HBEGF, ST6GALNAC5) in the primary breast tumour that developed cerebral metastasis. This could be due to the fact that cerebral metastases host genetic alterations different from the ones observed in the primary tumours; this is still unknown. Priscilla et al sequenced the exome of 86 cerebral metastases paired with the primary tumour tissue and normal tissue, finding clinically informative changes in the cerebral metastasis that were not found in the primary tumour tissue sample in 53% of the cases. The genes involved in the development of cerebral metastasis with greater frequency were TP53, PIK3CA, GATA3 and other mutated genes in lesser frequency including AKT1, CDH1, MAP3K1, PTEN, CDH1, RB1, and CDKN1B. They concluded that activation of the PI3K/AKT/mTOR pathway and CDK may be involved in the development of CNS metastasis [40].
The study of patients with TNBC who carry the BRCA-1 and BRCA-2 mutation is one analysis pathway, as these tumours are sensitive to PARP (polyadenosine diphosphate ribose polymerase 1) inhibitors like Olaparib, which penetrates the BBB [15, 44].
Other authors have suggested that androgens, directly activating astrocytes in the cerebral microenvironment, can facilitate the establishment of neoplastic cells originating in TN breast tumours. This would explain why younger TN patients, with greater oestrogen levels, have a greater risk of cerebral metastasis [41]. These findings could justify the absence of differences in gene expression between cases with the presence of cerebral metastasis and those that do not develop it, since the development of these metastases will depend more on the hormonal profile of the patient than on intrinsic factors of the tumour cell.
Other researchers also suggest elements external to the tumour itself as they show that the methylation level is unchanged in TN breast tumour cells in comparison with other BC phenotypes when they metastasise to the brain [42]. Stirzaker et al indicate that characterising methylation patterns could help us identify predictive biomarkers in the future [43].
Conclusion
In conclusion, our findings highlight that not only are the differences in gene expression important when it comes to predicting the risk of cerebral metastasis but also other aspects like oestrogen level could play an important role. Future studies are needed to clarify the role of these genes in primary breast tumours in patients with TNBC who develop cerebral metastases.
References
- 1.Smigal C, Jemal A, Ward E, et al. Trends in breast cancer by race and ethnicity: update 2006. CA Cancer J Clin. 2006;56:168–183. doi: 10.3322/canjclin.56.3.168. [DOI] [PubMed] [Google Scholar]
- 2.Ferlay J, Soerjomataram I, Ervik M, et al. Cancer Incidence and Mortality Worldwide: IARC; 2012. Incidencia y mortalidad para todas las edades Prevalencia a 5 años sólo en población adulta. GLOBOCAN 2012 v1.0. [Google Scholar]
- 3.Bray F, Ren JS, Masuyer E, et al. Estimates of global cancer prevalence for 27 sites in the adult population in 2008. Int J Cancer. 2013;132(5):1133–1145. doi: 10.1002/ijc.27711. Epub 2012 Jul 26. [DOI] [PubMed] [Google Scholar]
- 4.Sorlie T, Perou CM, Tibshirani R, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carey LA, Perou CM, Livasy CA, et al. Race, breast cancer subtypes, and survival in the Carolina breast cancer study. JAMA. 2006;295:2492–2502. doi: 10.1001/jama.295.21.2492. [DOI] [PubMed] [Google Scholar]
- 6.Niwinska A, Murawska M, Pogoda K. Breast cancer brain metastases: differences in survival depending on biological subtype, RPA RTOG prognostic class and systemic treatment after whole-brain radiotherapy (WBRT) Ann Oncol. 2010;21:942–948. doi: 10.1093/annonc/mdp407. [DOI] [PubMed] [Google Scholar]
- 7.Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chambers AF, Groom AC, MacDonald IC. Dissemination and growth of cancer cells in metastatic sites. Nat Rev Cancer. 2002;2:563–572. doi: 10.1038/nrc865. [DOI] [PubMed] [Google Scholar]
- 9.Castaneda CA, Agullo-Ortuno MT, Fresno Vara JA, et al. Implication of miRNA in the diagnosis and treatment of breast cancer. Expert Rev Anticancer Ther. 2011;11:1265–1275. doi: 10.1586/era.11.40. [DOI] [PubMed] [Google Scholar]
- 10.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27:1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehmann BD, Bauer JA, Chen X, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121:2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lachapelle J, Foulkes WD. Triple-negative and basal-like breast cancer: implications for oncologists. Curr Oncol. 2011;18:161–164. doi: 10.3747/co.v18i4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reddy KB. Triple-negative breast cancers: an updated review on treatment options. Curr Oncol. 2011;18:e173–e179. doi: 10.3747/co.v18i4.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Shaughnessy J, Osborne C, Pippen JE, et al. Iniparib plus chemotherapy in metastatic triple-negative breast cancer. N Engl J Med. 2011;364:205–214. doi: 10.1056/NEJMoa1011418. [DOI] [PubMed] [Google Scholar]
- 16.Castaneda CA, Cortes-Funes H, Gomez HL, et al. The phosphatidyl inositol 3-kinase/AKT signaling pathway in breast cancer. Cancer Metastasis Rev. 2010;29:751–759. doi: 10.1007/s10555-010-9261-0. [DOI] [PubMed] [Google Scholar]
- 17.National Cancer Institute. (Access to Investigational Drugs) www.cancer.gov/about-cancer/treatment/drugs/investigational-drugaccess-fact-sheet.
- 18.Lin NU, Bellon JR, Winer EP. CNS metastases in breast cancer. J Clin Oncol. 2004;22:3608–3617. doi: 10.1200/JCO.2004.01.175. [DOI] [PubMed] [Google Scholar]
- 19.Nam BH, Kim SY, Han HS, et al. Breast cancer subtypes and survival in patients with brain metastases. Breast Cancer Res. 2008;10:R20. doi: 10.1186/bcr1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deeken JF, Loscher W. The blood-brain barrier and cancer: transporters, treatment, and Trojan horses. Clin Cancer Res. 2007;13:1663–1674. doi: 10.1158/1078-0432.CCR-06-2854. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien KM, Cole SR, Tse CK, et al. Intrinsic breast tumor subtypes, race, and long-term survival in the Carolina breast cancer study. Clin Cancer Res. 2010;16:6100–6110. doi: 10.1158/1078-0432.CCR-10-1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vallejos CS, Gomez HL, Cruz WR, et al. Breast cancer classification according to immunohistochemistry markers: subtypes and association with clinicopathologic variables in a peruvian hospital database. Clin Breast Cancer. 2010;10:294–300. doi: 10.3816/CBC.2010.n.038. [DOI] [PubMed] [Google Scholar]
- 23.Chlebowski RT, Chen Z, Anderson GL, et al. Ethnicity and breast cancer: factors influencing differences in incidence and outcome. J Natl Cancer Inst. 2005;97:439–448. doi: 10.1093/jnci/dji064. [DOI] [PubMed] [Google Scholar]
- 24.Gapstur SM, Dupuis J, Gann P, et al. Hormone receptor status of breast tumors in black, Hispanic, and non-Hispanic white women An analysis of 13,239 cases. Cancer. 1996;77:1465–1471. doi: 10.1002/(SICI)1097-0142(19960415)77:8<1465::AID-CNCR7>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 25.Li CI, Malone KE, Daling JR. Differences in breast cancer stage, treatment, and survival by race and ethnicity. Arch Intern Med. 2003;163:49–56. doi: 10.1001/archinte.163.1.49. [DOI] [PubMed] [Google Scholar]
- 26.Hausauer AK, Keegan TH, Chang ET, et al. Recent breast cancer trends among Asian/Pacific Islander, Hispanic, and African-American women in the US: changes by tumor subtype. Breast Cancer Res. 2007;9:R90. doi: 10.1186/bcr1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parise CA, Bauer KR, Caggiano V. Variation in breast cancer subtypes with age and race/ethnicity. Crit Rev Oncol Hematol. 2010;76:44–52. doi: 10.1016/j.critrevonc.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Shavers VL, Harlan LC, Stevens JL. Racial/ethnic variation in clinical presentation, treatment, and survival among breast cancer patients under age 35. Cancer. 2003;97:134–147. doi: 10.1002/cncr.11051. [DOI] [PubMed] [Google Scholar]
- 29.Gonzalez Burchard E, Borrell LN, Choudhry S, et al. Latino populations: a unique opportunity for the study of race, genetics, and social environment in epidemiological research. Am J Public Health. 2005;95:2161–2168. doi: 10.2105/AJPH.2005.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Millikan RC, Newman B, Tse CK, et al. Epidemiology of basal-like breast cancer. Breast Cancer Res Treat. 2008;109:123–139. doi: 10.1007/s10549-007-9632-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pike MC, Kolonel LN, Henderson BE, et al. Breast cancer in a multiethnic cohort in Hawaii and Los Angeles: risk factor-adjusted incidence in Japanese equals and in Hawaiians exceeds that in Whites. Cancer Epidemiol Biomarkers Prev. 2002;11:795–800. [PubMed] [Google Scholar]
- 32.Probst-Hensch NM, Pike MC, McKean-Cowdin R, et al. Ethnic differences in post-menopausal plasma oestrogen levels: high oestrone levels in Japanese-American women despite low weight. Br J Cancer. 2000;82:1867–1870. doi: 10.1054/bjoc.1999.1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Looker AC, Dawson-Hughes B, Calvo MS, et al. Serum 25-hydroxyvitamin D status of adolescents and adults in two seasonal subpopulations from NHANES III. Bone. 2002;30:771–777. doi: 10.1016/S8756-3282(02)00692-0. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh MM, Everhart JE, Byrd-Holt DD, et al. Prevalence of neutropenia in the U.S. population: age, sex, smoking status, and ethnic differences. Ann Intern Med. 2007;146:486–492. doi: 10.7326/0003-4819-146-7-200704030-00004. [DOI] [PubMed] [Google Scholar]
- 35.Castaneda CA, G H, Vallejos C, Cortes-Funes, et al. Comparison between Spanish and Peruvian Patients with early breast cancer. In: San Antonio Breast Cancer Meeting.
- 36.Heitz F, Harter P, Lueck HJ, et al. Triple-negative and HER2-overexpressing breast cancer exhibit an elevated risk and an earlier occurrence of cerebral metastases. Eur J Cancer. 2009;45:2792–2798. doi: 10.1016/j.ejca.2009.06.027. [DOI] [PubMed] [Google Scholar]
- 37.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N England. 363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 38.Nguyen Don X, Bos Paula D, Massagué Joan. Metastasis: from dissemination to organ-specific colonization. Nature Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 39.Bollig-Fischer A, Michelhaugh SK, Ali-Fehmi R, et al. The molecular genomics of metastatic brain tumours. OA Mol Oncol. 2013;1(1):6. doi: 10.13172/2052-9635-1-1-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brastianos PK, Carter SL, Santagata S, et al. Genomic characterization of brain metastases reveals branched evolution and potencial therapeutic targets. Cancer Discov. 2015;5(11):1–14. doi: 10.1158/2159-8290.CD-15-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sartorius CA, Hanna CT, Grill B, et al. Estrogen promotes the brain metastatic colonization of triple negative breast cancer cells via an astrocyte-mediated paracrine mechanism. Oncogene. 2015;28:1–12. doi: 10.1038/onc.2015.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salhia B, Kiefer J, Ross JT, et al. Integrated genomic and epigenomic analysis of breast cancer brain metastasis. PLoS One. 2014;9:e85448. doi: 10.1371/journal.pone.0085448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stirzaker C, Zotenko E, Song JZ, et al. Methylome sequencing in triple-negative breast cancer reveals distinct methylation clusters with prognostic value. Nat Commun. 2015;6:5899. doi: 10.1038/ncomms6899. [DOI] [PubMed] [Google Scholar]
- 44.Chalmers AJ. Overcoming resistance of Glioblastoma to conventional cytotoxic therapies by the addition of PARP inhibitors. Anticancer Agents Med Chem. 2010;10:520–533. doi: 10.2174/187152010793498627. [DOI] [PubMed] [Google Scholar]
- 45.Sánchez-Navarro I, Gámez-Pozo A, González-Barón M, et al. Comparison of gene expression profiling by reverse transcriptionquantitative PCR between fresh frozen and formalin-fixed, paraffin-embedded breast cancer tissues. Biotechniques. 2010;48(5):389–97. doi: 10.2144/000113388. [DOI] [PubMed] [Google Scholar]