Abstract
Network science provides theoretical, computational, and empirical tools that can be used to understand the structure and function of the human brain in novel ways using simple concepts and mathematical representations. Network neuroscience is a rapidly growing field that is providing considerable insight into human structural connectivity, functional connectivity while at rest, changes in functional networks over time (dynamics), and how these properties differ in clinical populations. In addition, a number of studies have begun to quantify network characteristics in a variety of cognitive processes and provide a context for understanding cognition from a network perspective. In this review, we outline the contributions of network science to cognitive neuroscience. We describe the methodology of network science as applied to the particular case of neuroimaging data and review its uses in investigating a range of cognitive functions including sensory processing, language, emotion, attention, cognitive control, learning, and memory. In conclusion, we discuss current frontiers and the specific challenges that must be overcome to integrate these complementary disciplines of network science and cognitive neuroscience. Increased communication between cognitive neuroscientists and network scientists could lead to significant discoveries under an emerging scientific intersection known as cognitive network neuroscience.
INTRODUCTION
The conceptual frameworks that we use to understand the brain and guide empirical and theoretical investigations have evolved slowly over several centuries. Phrenology gave way to a focus on the interactions between brain areas or smaller computational units (connectionism) and the symbolic language of thought itself (computationalism). During this evolution, cognitive psychologists reached out to mathematical frameworks developed in other disciplines—physics, mathematics, and engineering—to capture the brain’s function in formal models. Artificial neural networks, for example, provided an early means of simulating information processing paradigms inspired by biological neural systems.
The landscape of potential frameworks and mathematical tools to examine complex dynamical systems like the human brain changed dramatically in the last few decades with the popularization and further development of network science (Newman, 2010). The use of networks in neuroimaging has provided new means to investigate key questions in cognitive neuroscience. In this scheme, brain regions are treated as network nodes and the anatomical connections or putative functional interactions between these regions are treated as network edges (Figure 1). The network representation provides a parsimonious description of heterogeneous interaction patterns thought to underlie the information processing mechanisms of the human brain. Moreover, the mathematical formalism is both generalizable (not being limited to applications to a single type of data or at a single spatial or temporal resolution) and flexible (enabling group comparisons, statistical inference, and model development).
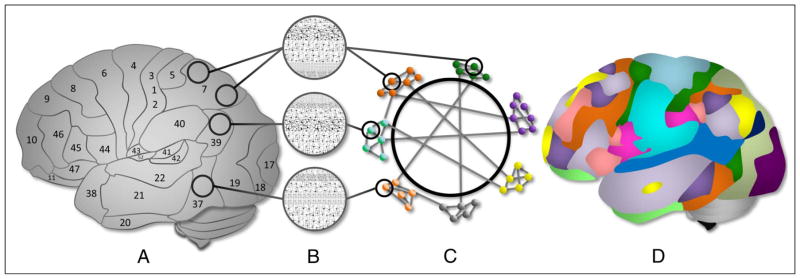
Figure 1.
From nodes to networks. (A) Brain regions are organized into cytoarchitectonically distinct areas. (B) Each cytoarchitectural configuration has structural properties with different implications for computational functions. (C) Cytoarchitectural regions can be represented as nodes in a network. The nodes have functional associations, represented as edges, that extend beyond spatial boundaries evident in cytoarchitectural organization. Subsystems can be described as network modules. Modules have varying intraconnectivity and intermodule connectivity in the human brain. (D) An example topology of the modular organization of functional brain networks demonstrating the communication between computational resources of different types. Panel D adapted with permission of Yeo et al. (2011).
As with any new conceptual or mathematical framework, it is critical to determine whether the novel approach is actually enlightening. Scientific enlightenment can take one of three forms: (i) the discovery of fundamental principles that govern observed phenomena; (ii) validated relationships with other known variables; and (iii) utility in uncovering novel processes, structures, or phenomena that assist us in interpreting (but cannot simply be explained by) prior empirical or principled knowledge (Woodward, 2014). In the first case (fundamental principles), it may be that there are governing attributes of dynamical networks in general that apply to the special case of brains and the minds that depend upon them, a notion to which we will return in Current Frontiers, below. In the second case (validation), confidence can be afforded by demonstrated network correlates of behavior (Reijmer, Leemans, Brundel, & Biessels, 2013), network alterations in psychiatric conditions or neurological disorders (Basset, Yang, Wymbs, & Grafton, in press; Fornito, Zalesky, Pantelis, & Bullmore, 2012; Bassett & Bullmore, 2009; He, Chen, Gong, & Evans, 2009), and network predictors of future brain function or behavioral performance (Ekman, Derrfuss, Tittgemeyer, & Fiebach, 2012; Heinzle, Wenzel, & Haynes, 2012; Bassett, Wymbs, et al., 2011). In the third case (novel utility), network-based approaches provide new information about brain function that cannot be derived from what we already know about a person and their psychological, clinical, or other status. In this case, the application of network science allows us to observe new phenomena, rather than explaining an already-observed phenomenon.
A strong criterion for achieving enlightenment is whether fundamental mechanisms have been identified. Mechanisms are “entities and activities organized in such a way that they are responsible for the phenomenon” (Illari & Williamson, 2012). Mechanism discovery proceeds gradually, and we propose that network science has the potential to uncover fundamental mechanisms in cognitive neuroscience. In principle, it is uniquely able to represent the brain in its complexity. A major advantage of network techniques is the explicit representation and assessment of both neural components (neurons or brain regions) and their interactions with one another (synapses or functional connections).
The application of network techniques to neuroimaging data entails a coarse-grained perspective of lower-level dynamical processes. Such techniques have begun to characterize brain network features relevant to cognition that cannot be observed from the sole perspective of functional localization. Numerous studies have applied network methods to brain structural data and functional neuroimaging data during rest. Applications of network methods to understand cognition have been relatively few. We suggest that cognitive network neuroscience is in an early phase of enlightenment and that increased communication between cognitive neuroscientists and network scientists can lead to substantial discoveries.
We review here some early successes in this new field and outline the potential for cognitive network neuroscience to enrich our understanding of human cognition. This complements several other recent reviews on the intrinsic functional connectivity (see Seeley, Menon, Schatzberg, Keller, & Glover, 2007 regarding the use of this term) of the brain (see Raichle, 2011); relationship between the brain’s intrinsic functional networks, organization during cognition, and underlying structure (Smith et al., 2009); methodological approaches to examining brain networks (Craddock et al., 2013); and relevance of network approaches to cognitive neuroscience (Sporns, 2014). In this review, we demonstrate that neuroimaging studies to date have uncovered many new network phenomena in the human brain and their associations with cognitive processes. We first provide a comprehensive description of the conceptual and mathematical framework of networks as applied within cognitive neuroscience in the study of what has become known as the human connectome (Sporns, 2011, 2012; Sporns, Tononi, & Kötter, 2005; see also Kopell, Gritton, Whittington, & Kramer, 2014). Then, we address key questions in cognitive neuroscience using noninvasive neuroimaging measurements in humans. We conclude with a discussion of exciting new frontiers and important theoretical and methodological considerations.
CONCEPTUAL AND MATHEMATICAL FRAMEWORK
Conceptual Framework
Many network systems comprise complex and diverse interactions (Newman, 2010). Network representations have the unique advantages of (i) enabling the quantitative analysis of these heterogeneous interactions within a unified mathematical framework and (ii) enabling the examination of higher order multivariate patterns rather than simply pairwise interactions. These advantages are particularly useful in the study of the human brain, where different brain areas have different structural properties (cytoarchitectural configuration, volume, shape, white matter tracts) and dynamics and are known to play distinct roles in cognitive function.
Conceptually, brain networks are simplified representations of region–region relationships. Functional brain networks capture temporal relationships between activity in different brain regions (e.g., based on estimates of functional or effective connectivity; Friston, 1994), anatomical brain networks capture white matter links between brain regions (e.g., using diffusion tractography; Hagmann et al., 2008), and morphometric brain networks capture structural relationships between brain regions based on covariation between regional volume (Bassett et al., 2008), cortical thickness (He, Chen, & Evans, 2007), surface area (Sanabria-Diaz et al., 2010), and curvature (Ronan et al., 2012) over subjects. Although structural and functional networks can be produced on a subject-by-subject basis, morphometric networks rely on data from multiple subjects. For example, to ascertain a connection (or correlation) between gray matter thickness in brain region x and in brain region y requires multiple measurements of gray matter thickness in both regions and hence multiple subjects. An analogous approach could be taken within subjects over time during brain development to maturity. These three prongs of investigation stem from three types of neuroimaging measurements: functional imaging (fMRI, EEG; and MEG), diffusion imaging (diffusion spectrum imaging, diffusion tensor imaging), and structural imaging (structural MRI, sMRI).
Network representations in neuroimaging data have a different meaning than traditional representations in the cognitive sciences, computational neuroscience, and cognitive neuroscience. Cognitive science tends to reduce cognitive systems to models of representations paired with processes. Measured variables in empirical cognitive studies are often behavioral indices. It typically describes the symbol level architectures of cognitive processes irrespective of their physical instantiation. Measured behavioral variables are in turn used as evidence for or against the predictive capabilities of a cognitive model, which is in turn modified to better predict behavioral data. In computational neuroscience, operations performed over various levels of neural tissue organization are modeled, including operations proposed to support cognition. In cognitive neuroscience, brain structures composed of complex organizations of neurons are assumed to support cognitive functions. It describes the neural localization of cognitive processes in the brain. Neuroimaging variables are used to predict behavioral indices to make inferences about the operations of the underlying neural substrate. Relationships between behavior and imaging variables are used to modify the understanding of how functions are represented in particular brain structures. In contrast to these traditional approaches, cognitive network neuroscience focuses on complex interactions between spatially discrete brain regions, represented by graphs, and seeks to link these patterns of interaction to measured behavioral variables. One key epistemological consequence of using network representations is that they can describe and uncover higher-level complexity that depends on the interacting elements of the networked system, which traditional approaches cannot provide. As neuroscience progresses, these disciplines will converge on a common scientific understanding of how cognition is represented in the human brain.
Mathematical Framework
Mathematically, a brain network can be defined as a graph G composed of N nodes (brain regions) and E edges (region–region relationships). In network science, the term graph refers to the join-the-dots pattern of connections (edges) between nodes, rather than to a visual representation of data on axes. We examine the pattern of edges linking nodes by quantifying the graph’s structure using a variety of diagnostics, which each provide complementary but not necessarily independent information (Valente, Coronges, Lakon, & Costenbader, 2008). In this review, we will describe a few of these diagnostics to illustrate the types of structures that one can probe but we point the reader to Newman’s recent textbook Networks: An Introduction (Newman, 2010) for a more comprehensive list and associated descriptions and mathematical formulae and to Sporns’ book Networks of the Brain (Sporns, 2010) for intuitive descriptions of several diagnostics in the context of neuroscience.
Network diagnostics can be used to probe the organization of functional or structural connections in the brain across a spectrum of spatial scales from the neighborhood surrounding a single brain area (local) to the summary statistics of the connectivity in the whole brain (global). Diagnostics that describe the organization of connections in between these two scales are referred to as mesoscale diagnostics.
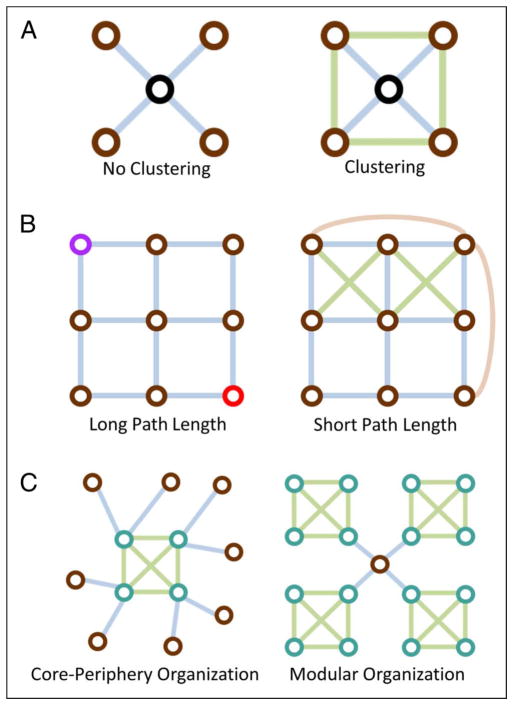
A common example of a local diagnostic is the clustering coefficient, which can be defined as triangles in which a node participates, divided by the number of connected triples in which a node participates (Figure 2A). In brain networks, it is thought that the clustering coefficient might indirectly measure the degree of local information integration (Sporns, 2010; Bullmore & Sporns, 2009).
Figure 2.
Network diagnostics. (A) The clustering coefficient is a diagnostic of local network structure. The left panel contains a network with zero connected triangles and therefore no clustering, whereas the right panel contains a network in which additional edges (green) have been added to close the connected triples (i.e., 3 nodes connected by 2 edges) to form triangles (i.e., 3 nodes connected by 3 edges), thereby leading to higher clustering. (B) The average shortest path length is a diagnostic of global network structure. The left panel contains a network with a relatively long average path length. For example, to move from the purple node (top left) to the red node (bottom right) requires one to traverse at least 4 edges. The right panel contains a network in which addition edges have been added to form triangles (green) or to link distant nodes (peach), thereby leading to a shorter average path length in comparison. (C) Mesoscale network structure can take many forms. The left panel contains a network with a core of densely connected nodes (green circles; green edges) and a periphery of sparsely connected nodes (brown circles; gray edges). The right panel contains a network with four densely connected modules (green circles; green edges) and a connector hub (brown circle; gray edges) that links these modules to one another.
A common example of a global diagnostic is the average shortest path length L. The shortest path between node i and node j is the fewest number of connections that must be traversed to get from node i to node j, and the average shortest path is the mean of this value over all possible pairs of nodes in the graph (Figure 2B). In brain networks, it is thought that the average shortest path length might indirectly measure the degree of global information segregation (Sporns, 2010; Bullmore & Sporns, 2009). A related concept—network efficiency (Latora & Marchiori, 2001, 2003)—is also calculated based on shortest paths through a network, but in this case, a network will have high efficiency if it has a short path length and the network will have low efficiency if it has a long path length. In the brain, this network efficiency is often interpreted to underlie efficiency of information processing (Sporns, 2010; Achard & Bullmore, 2007).
Centrality measures, which quantify the relative influence or rank of a node in a network, include both local and global types. The degree centrality of a node, for example, is given by the number of connections with that node and therefore quantifies a local property of the network. The betweenness centrality of a node, v, is a more global diagnostic, given by the number of shortest paths between any two nodes in the network (e.g., node i and j) that must pass through node v.
Mesoscale organization can take various forms (Rombach, Porter, Fowler, & Mucha, 2012; Fortunato, 2010; Porter, Onnela, & Mucha, 2009). Two interesting types of mesoscale organization are (i) core–periphery organization (Borgatti & Everett, 1999), in which a set of nodes forms a densely connected core and a second set of nodes forms a sparsely connected periphery (Figure 2C, left), and (ii) modular organization (Newman & Girvan, 2004), in which sets of nodes form densely connected modules (Figure 2C, right). In brain networks, both types of organization appear to exist at both the structural (Bassett et al., 2013; van den Heuvel & Sporns, 2011) and functional levels (Bassett, Brown, Deshpande, Carlson, & Grafton, 2011; Meunier, Lambiotte, Fornito, Ersche, & Bullmore, 2009). Core–periphery organization could play a role in conferring robustness to the brain’s structural core (van den Heuvel & Sporns, 2011) and in enabling a balance between stability and adaptivity in brain dynamics (Bassett et al., 2013). Modular organization provides a natural substrate for the combined integration and segregation of information processing arguably required during healthy brain function (Bullmore & Sporns, 2009).
Although many network properties—such as high clustering, short path length, core–periphery structure, and modularity—have consistently been found to characterize connectivity patterns extracted from many types of noninvasive neuroimaging data, these properties all vary among people. There is mounting evidence that one can reliably identify individual differences in structural (Owen et al., 2013; Dennis et al., 2012; Bassett, Brown, et al., 2011) and functional (Braun et al., 2012; Wang et al., 2011; Telesford et al., 2010; Deuker et al., 2009) brain network organization, suggesting that individual variation in such architectures can be linked to individual variation in cognitive performance.
APPLICATIONS IN NEUROIMAGING
In this section, we describe recent applications of network-based methods to questions in cognitive neuroscience. For each area of enquiry, we summarize the studies that have applied network methods to questions in cognitive neuroscience. We then highlight evidence that we feel particularly illustrates the epistemological gains that network neuroscience has brought to that area.
A Taxonomy for Nodes and Edges
The definition of nodes in a network is a necessary step with consequences for network modeling and interpretation. Since 1909, Brodmann’s areas (BAs) have served as a useful reference point in the neurosciences (Brodmann, 1909). These histologically defined cytoarchitectonic brain areas support functionally distinct operations. In principle, differences between BAs may imply different dynamical properties (at the level of cortical columns or smaller) that support critical operations during cognition. In functional neuroimaging, many hundreds or thousands of cortical columns may be contained in sampled regions used to define graphs. Despite this, maintaining distinctions between the cytoarchitecture sampled can provide a guiding taxonomy for qualitatively different nodes (see Figure 1). It is fundamentally important to cognition that the neural microstructures underlying regions sampled in neuroimaging studies vary in a highly organized manner. Unfortunately, there is no modern atlas that validly represents all BAs in probabalistic space, although one is under development (Eickhoff et al., 2007). If a valid probabalistic atlas becomes available, the considerable variance in the size and morphology of BAs across individuals will remain problematic. In light of these considerations, we refer to estimated BAs where noted in primary references and mention other spatial reference systems (e.g., gross morphological areas) when they are used in primary sources. The study and modeling of variation in cytoarchitectonics will ultimately prove to be critical in understanding how brain networks support cognition.
Subcortical structures do not have an analogous system to Brodmann’s mapping but have observably different microstructures and associated functions. Subcortical structures are often more obviously distinguishable from one another and the cortex on standard anatomical scans. Each subcortical structure tends to contain homogeneous circuits with fewer layers relative to cortical systems. These circuits tend to be composed of parallel, closed-loop projections (McHaffie, Stanford, Stein, Coizet, & Redgrave, 2005), and link anatomically with multiple other subcortical and cortical regions. It is important to note that atlases used in network analyses have over-sampled the cortex relative to subcortical structures despite the fact that subcortical structures contain well over half of the neurons in the brain. Most subcortical neurons are in the cerebellum, which has 3.6 times the number of neurons as the neocortex (Herculano-Houzel, 2010).
Finally, the definition of edges in this context typically refers to measures of structural or functional connections between nodes. Structural edges may include measures of fractional anisotropy or streamline counts along white matter pathways between regions. Functional edges may include any of a number of measures of relatedness between signals from standard signal processing approaches. Important for cognitive network neuroscience is that the type of information communicated along edges differs as a result of computations performed in local regions, which are transmitted along white matter pathways. The weight, number, and configuration of edges represent patterns of information flow within the brain.
Anatomical Network Correlates of Cognitive Processes
Discussion of associations between brain structure and cognition provide an important context for understanding brain function. Anatomical brain networks represent the mediating architecture over which functional dynamics operate. Diffusion weighted imaging data provides quantitative measurements of white matter microstructure, whose integrity is crucial for healthy cognitive function (Roberts, Anderson, & Husain, 2013). Age-related individual differences in cognitive performance—particularly perceptual speed and executive functioning—are accompanied by variations in white matter integrity across neural systems that display an anterior–posterior gradient across the lifespan (see Madden, Bennett, & Song, 2009, for a recent review). White matter fiber bundles in temporal lobe projections (uncinate fasciculus, fornix, cingulum, inferior longitudinal fasciculus, and superior longitudinal cortex) were associated with better executive function. Global network compromise was related to deficits in processing speed.
Injury to white matter can immediately alter cognitive function in multiple domains (Silver, McAllister, & Arciniegas, 2009) and can interact with normal aging to drive later cognitive decline (Moretti et al., 2012). Changes in white matter microstructure correlate with cognitive impairments in processing speed (Niogi et al., 2008), working memory (Palacios et al., 2012; Kinnunen et al., 2011; Wu et al., 2010), and motor skills (Leunissen et al., 2013; Farbota et al., 2012). The efficiency of larger-scale network structure correlates with switching scores on an executive function task (Caeyenberghs et al., 2012, 2014). These studies have demonstrated that the structure of single brain areas or tracts are not the only, and perhaps not even the best, predictors of cognitive function. Instead, cognition is supported by a pattern of connections between distributed sets of brain areas.
Functional Network Correlates of Cognitive Processes
Cognitive processes are associated with altered brain activity and—by extension—functional connectivity (Siebenhuhner, Weiss, Coppola, Weinberger, & Bassett, 2013; Yu et al., 2013; Bassett, Nelson, Mueller, Camchong, & Lim, 2012; Zalesky, Fornito, & Bullmore, 2012). By identifying changes in functional connectivity patterns induced by experimental tasks, we can begin to uncover the distributed network processes underlying mental function and behavioral performance. See Figure 3 for an overview of the representation of brain networks during cognitive states.
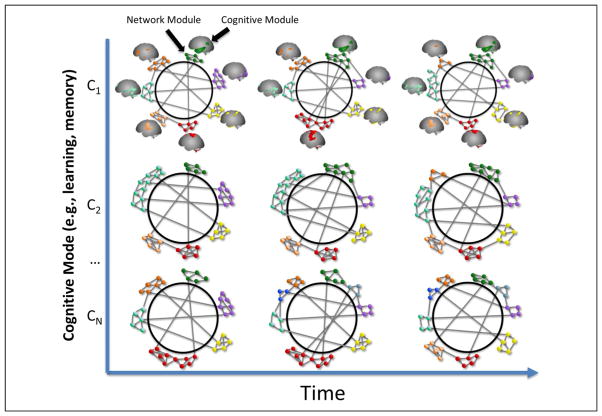
Figure 3.
Cognitive network neuroscience (C = Cognitive state). A schematic representation of functional brain networks during cognition. Cognitive modules are indicated by collections of identically colored nodes organized into network modules. The organization of brain networks varies across cognitive states and time. Some features of functional network organization may remain relatively stable as a system “core,” and others may vary substantially. Modules may merge and separate. Connections within and between modules may change in strength, configuration, and number. Network organization may change over time as a function of learning processes.
In the following paragraphs, we will review recent studies that have focused on characterizing whole-brain functional connectivity patterns in four main areas: (i) vision, audition and motion; (ii) memory; (iii) learning; (iv) emotion; (v) language; and (vi) attention and cognitive control. These domains are by no means exhaustive of cognition but represent the constructs studied in the majority of network analyses to date. Although methods based on both seed-based correlation (Biswal, Yetkin, Haughton, & Hyde, 1995) and independent component analyses (McKeown et al., 1998) have also been applied to task-based data (Michael, Calhoun, Andreasen, & Baum, 2008; Hampson et al., 2006; Calhoun et al., 2002), here we constrain ourselves to network- or graph-based approaches.
From Intrinsic Functional Networks to Cognition
Understanding the intrinsic organization of brain networks provides a context for the reconfigurations observed during tasks. Intrinsic functional connectivity has been found to be more stable than the synchrony of elicited activation in a number of tasks (Cao et al., 2014), suggesting, surprisingly, that there may be greater constraints on how the brain must globally organize to be at rest than on how it must allocate resources to achieve a specific task. Between-atlas comparisons have demonstrated reliable global and local network properties in intrinsic connectivity and task-related conditions across network parcellation methodologies (Cao et al., 2014).
An interesting question is how intrinsic functional networks subtly reconfigure to support cognitive functions. In comparisons between task and nontask states, functional network organization can appear strikingly similar at a global level (Cole, Bassett, Power, Braver, & Petersen, 2014), suggesting that a stable core network is necessary for healthy general cognitive function (Buckner et al., 2009; Bassett, Meyer-Lindenberg, Achard, Duke, & Bullmore, 2006). However, the spatial layout of functional wiring and other local properties of functional connectivity can simultaneously be drastically altered, representing local reconfigurations to meet task demands (Bassett et al., 2006). For example, the intrinsic functional connectivity involves predominantly short and by extension potentially efficient (Bullmore & Sporns, 2012) functional wiring, which is lengthened to perform simple motor tasks such as finger tapping (Bassett et al., 2006). In addition, the local connectivity of individual nodes has been shown to vary across cognitive states, though nodes known to be recruited to manage general tasks demonstrate reliable node degree across task conditions (Cao et al., 2014). Thus, functional systems in the brain are perhaps best understood as a stable organization that supports a number of relatively minor state reconfigurations that enable cognitive functions. The role of some brain regions in the network changes flexibly to address specific task demands, whereas others are reliably interactive with the rest of the network to manage global demands.
Primary Sensorimotor Regions and Their Relationship to Cognitive Hubs
What roles do specific brain regions play within this stable organization, and how might those roles change during cognitive functions? Perhaps the simplest set of areas in which to answer this question are the primary sensory and motor regions, whose functions have been well delineated for over a century. Using network-based techniques, these early-evolving regions have been shown to display intrinsic functional connectivity patterns that converge on cognitive “hubs.”
Functional connections from sensorimotor regions are partially integrated in a multimodal network between sensorimotor systems and cognitive hubs (Sepulcre, 2012, 2014). The multimodal network consists of portions of the superior parietal cortex, parietal operculum, anterior insula in the frontal operculum, dorsal ACC/SMA (BA 6/24/32), dorsolateral pFC (BA 10/46), and the confluence of BA 19/22/37/39 in the TPJ (Sepulcre, 2012, 2014). Following partial integration in the multimodal network, functional connections all converge in cognitive hubs. Cognitive hubs are those with high centrality in the brain overall (Buckner et al., 2009) or between many brain subsystems (Warren et al., 2014). Hubs have been identified within the frontoparietal and default mode systems and include the posterior cingulate, lateral temporal, lateral parietal, and medial/lateral prefrontal cortices (van den Heuvel & Sporns, 2013; Buckner et al., 2009). Damage to cognitive hubs is implicated in many diseases (Crossley et al., 2014) and can result in generalized and catastrophic failures in cognitive function (Warren et al., 2014).
Overall, this initial work has formed an important basis for understanding the relationships between networked subsystems in the brain. These discoveries suggest an overarching organization of multiple sensory and motor processes integrated within a set of mediating multimodal systems. In turn, multimodal systems are supervised by a complex frontoparietal network and the default mode network. This is consistent with a view of cognition in which the gross organization is a parallel distribution of extrinsic inputs to the brain, intermediate parallel associating mechanisms that potentially subserve the “binding” of sensory input, and an overarching supervisory network. Within each layer of this hierarchy, distinct computational operations presumably occur across varying cytoarchitectural mechanisms. Thus, the major organization of human functional systems forms a robust heterarchy with a balance of information segregating and integrating functions (see Bressler & Richter, 2015; Buckner & Krienen, 2013; Passingham, Rowe, & Sakai, 2013). To better elucidate the network basis of cognition within this organization, we now turn to network analyses in specific cognitive behavioral domains that have identified associations with functional network features.
Motion, Vision, and Audition
Whereas traditional approaches examine the topological organization and activity of the sensorimotor cortices in association with behavior, network analyses have discovered underlying functional interactions within and between these regions related to variation in motor function. For example, the consistency of community structures in the sensorimotor cortex and density of connections with the insula and superior temporal gyrus predicted mobility across the lifespan (Mishkin & Ungerleider, 2014).
Furthermore, a study of motor execution and motor imagination revealed that the SMA was a key node with high betweenness centrality during motor execution and the right premotor area had high betweenness centrality during imagination (Xu et al., 2014). In a study of self-initiated finger movements, network connectivity strengths predicted striatal activity. The connectivity strength between the dorsolateral pFC and striatum was negatively correlated with RTs, whereas the connectivity between the ventrolateral pFC and striatum was positively correlated to RTs (Nagano-Saito, Martinu, & Monchi, 2014).
Thus, network studies may extend our classical understanding of the organization of the motor system with a focus on the interactive processes between the motor system and the rest of the brain. It is possible that motor regions serve as conditionally recruited processing hubs in conjunction with frontal control regions. Network studies have supported this notion by dissociating between intrinsic and extrinsic distributed processes mediated by motor regions (Xu et al., 2014) and identifying opponent interactions involving frontal control regions (Nagano-Saito et al., 2014).
With respect to vision and audition, network studies have shown that the strengthening of key functional connections underlies tasks that demand sensory integration. The spatial distribution of network modules in auditory and visual cortex and of hub-like areas became more constrained to traditional anatomical boundaries in a multisensory task and displayed less variability across subjects (Moussa et al., 2011). These results were qualitatively corroborated by Ma, Calhoun, Eichele, Du, and Adali (2012) who showed task-induced increases in the clustering coefficient during an auditory oddball task in comparison to rest. In preparatory intervals just before the performance of a trial in a visual discrimination task, functional networks extracted from fMRI data showed a dynamic adjustment in core–periphery interactions, in which task-relevant visual areas move toward the core of densely and mutually interconnected regions (Ekman et al., 2012). Notably, this reconfiguration predicted successful task performance.
Network approaches can therefore contextualize the local functions of primary sensory areas within systems that support dynamic sensory integration and consolidation. Such approaches suggest that tasks with a heavy emphasis on sensory processing and integration appear to depend on tightly communicating cognitive hubs and sensorimotor regions (Ma et al., 2012; Moussa et al., 2011). Validation for a network view is suggested by the finding that increasing functional network integration results in better performance (Ekman et al., 2012). Sensory tracking and discrimination may require robust sustained communication between primary sensory and nearby regions.
Memory
Network approaches have begun to enlighten our understanding of the distributed processes supporting effective memory performance. One fMRI study involving a yes–no odor recognition task demonstrated that the hippocampus, caudate nucleus, and anterior cingulate gyrus more frequently belonged to the same functional module during hits than all other conditions. Network-wide modularity values were negatively correlated with memory in the hit condition and positively related to bias scores in hit and false alarm conditions (Meunier et al., 2014), meaning that increasing segregation of the network resulted in inaccuracy. Thus, integration in this memory subsystem may be critical to some discriminative decision processes. In particular, achievement of true positive memories may require a cooperation of coordinating, error monitoring, and memory indexing processes that is optimized within sufficiently organized brain networks.
In addition, network analyses have brought insight into the brain dynamics underlying working memory. They have demonstrated that the efficient organization of functional networks in specific frequency domains supports working memory function. During visual memory maintenance, a combined EEG/MEG study revealed that alpha and beta band networks were more clustered and small-world like with smaller global efficiency than delta-and theta band networks. The alpha and beta band networks had truncated power law degree distributions (Palva, Monto, & Palva, 2010). Sustained phase synchrony during retention was found among frontoparietal and visual areas in alpha, beta, and gamma frequency bands (Palva, Monto, Kulashekhar, & Palva, 2010). Overall, these findings suggest that working memory processes are predominantly supported by an efficiently organized network predominantly in the alpha and beta frequency regimes.
Specific aspects of functional network configurations are associated with variance in working memory capacity and performance. Network modularity and small-worldness in intrinsic functional connectivity following a working memory test predicted capacity (Stevens, Tappon, Garg, & Fair, 2012). Individuals with higher memory accuracy in a 2-back working memory task tended to have more cost-efficient functional network architecture, as measured by MEG in the beta frequency band (12–20 Hz; Bassett, Meyer-Lindenberg, Weinberger, Coppola, & Bullmore, 2009). Synchrony in frontoparietal regions increased with load, and synchrony in a hub within the intraparietal sulcus predicted individual visual working memory capacity (Palva, Monto, Kulashekhar, et al., 2010; Palva, Monto, & Palva, 2010). With increasing cognitive load (0-back to 2-back), networks became more globally efficient, less clustered, and less modular, with more long-distance synchronization between brain regions. This configuration was greater in faster performing individuals than in slower performing individuals for the beta frequency (Kitzbichler, Henson, Smith, Nathan, & Bullmore, 2011). Functional networks extracted from BOLD data demonstrated that increased connection pruning (Ginestet & Simmons, 2011) and decreased clustering (He et al., 2012) are associated with swift and accurate performance. Decreased memory encoding recognition and encoding in aging individuals was associated with increased path length and decreased efficiency (Wang, Li, Metzak, He, & Woodward, 2010).
Taken together, network findings suggest that working memory is arbitrated in the frontoparietal system. Network studies have led to the discovery that working memory may fundamentally rely on sustained and efficiently organized frontoparietal and visual interactions especially in the alpha and beta frequency regimes (Palva, Monto, Kulashekhar, et al., 2010; Palva, Monto, & Palva, 2010). Validation for a network perspective was found within this organization: increases in brain-wide small-world organization support increased working memory capacity (Stevens et al., 2012), and increased network efficiency results in better memory performance (Kitzbichler et al., 2011; Bassett et al., 2009).
Learning
Learning takes place over multiple timescales, from hours to days to months. Network findings have begun to clarify brain-wide functional changes that support learning across these timescales. For example, in a comparison between pretraining (Day 0) and posttraining (Day 5) sessions for a bimanual motor learning task, improved performance was related to increased clustering coefficients, network degree, connection strength, and shortened path lengths across the brain in auditory and feedback conditions (Heitger et al., 2012).
Several recent studies have begun to capture dynamic patterns of functional connectivity at finer temporal scales, from temporal networks (Holme & Saramäki, 2012) extracted from contiguous 2–3 min windows of fMRI data (Bassett et al., in press; Bassett et al., 2013; Mantzaris et al., 2013; Bassett, Wymbs, et al., 2011). Dynamic community detection techniques (Mucha, Richardson, Macon, Porter, & Onnela, 2010) can uncover changes in clusters of brain regions linked by strong functional connectivity (i.e., putative functional modules; Bassett, Porter, et al., 2012). Individuals with greater network flexibility (changes in module allegiance over time) tended to learn a motor sequence more effectively (Bassett, Wymbs, et al., 2011; Figure 4). Using dynamic centrality techniques for the same task, Mantzaris and colleagues (2013) demonstrated that broadcast and receive centralities, which together measure the flow of functional connectivity changes over time, decrease strikingly over the course of the experiment. In an extended 6-week learning experiment, the presence of a stable temporal core of primary task-related areas and a flexible temporal periphery of multimodal association areas was identified (Bassett et al., 2013). Individuals with a greater separation between their core and periphery learned better during the following 10 days of practice. As learning progresses, motor and visual modules become increasingly autonomous, and the disengagement of a fronto-cingulate cognitive control network predicts individual differences in learning rate (Bassett et al., in press).
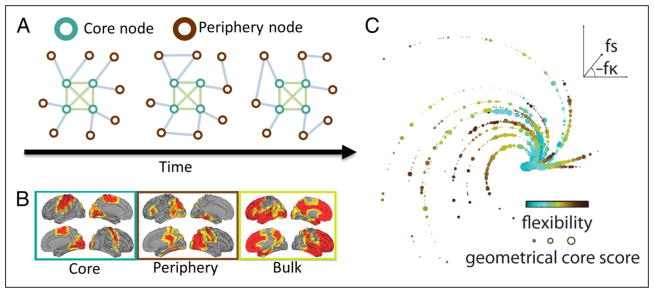
Figure 4.
Brain network dynamics during learning. (A) The flexibility of brain network dynamics—defined as the frequency of a brain region when it changes its allegiances to network modules over time—predicts individual differences in learning: More flexible individuals learn better than less flexible individuals (Bassett, Wymbs, et al., 2011). Moreover, brain regions differ in their flexibility. Regions with greater flexibility form a temporal network core, whereas regions with less flexibility form a temporal network periphery. (B) The distribution of the temporal core and periphery nodes in the brain during learning. “Bulk” nodes are those that do not significantly differ from a temporal network null model. (C) The relationship between region flexibility (f ), core–periphery separation (s), and learning (parameterized by κ). Brain regions are represented using data points located at the polar coordinates (fs,fκ). Color indicates flexibility: Blue nodes have lower flexibility, and brown nodes have higher flexibility. Poor learners (straighter spirals) tend to have a small separation between core and periphery (short spirals), whereas good learners (curvier spirals) tend to have large separation between core and periphery (longer spirals). The separation between core and periphery is a good predictor of individual differences in learning success. Adapted with permission from Bassett et al. (2013).
Thus, network neuroscience has discovered that learning processes require sufficient network flexibility around a stable core system during learning. In terms of validation, learning was strongly related to brain flexibility (Bassett et al., 2013; Bassett & Gazzaniga, 2011), the growing autonomy of motor and visual systems, and the decreasing engagement of cognitive control regions (Bassett et al., in press). Whether these findings generalize to tasks that more heavily emphasize declarative memory remains to be seen.
Emotion
The involvement of distributed patterns of functional connectivity is perhaps unsurprising in higher order cognitive functions that require cooperative activity from multiple brain systems. Perhaps as a result, “higher” cognitive domains have received considerable attention in network studies. Network studies of emotion are to date rare but informative. In response to emotion cues, Kinnison, Padmala, Choi, and Pessoa (2012) demonstrated increased global efficiency as well as decreased modularity. In a network analysis of vulnerability to reduced mood, resilient individuals demonstrated decreased global connectivity and hub-like properties in the right ventrolateral pFC and decreased local connectivity in the dorsal ACC. Susceptible individuals demonstrated decreased local connectivity of the amygdala and ventrolateral pFC and decreased hub properties of the amygdala and the dorsal ACC. Self-reported severity of early life stressors correlated negatively with global network connectivity and hub properties of the left dorsolateral pFC (Cisler et al., 2012). In depressed individuals, the centrality of the right dorsolateral pFC was negatively correlated with the duration of disease. Centrality in the right inferior frontal gyrus (pars triangularis) and efficiency in the right superior frontal gyrus were positively related to depression severity (Qin et al., 2014).
Network analyses in emotion tasks have begun to extend our understanding of the representation of distributed emotion regulatory processes in the brain. The dorsolateral pFCs (Koenigs & Grafman, 2009) and inferior frontal gyrus (Shamay-Tsoory, Aharon-Peretz, & Perry 2008) have been implicated as nodes in a distributed emotion processing system with differentiable functional roles. Thus far, network approaches have discovered network-wide processes in support of an emerging view that a competing balance of function between the right and left dorsolateral pFC has important implications for mood regulation (Qin et al., 2014; Cisler et al., 2012; Grimm et al., 2008). Validity was suggested by the finding that as components of the right frontal cortex increasingly dominate function across the network, negative mood results. In addition, environmentally induced reductions in broad network communication may result in vulnerability to emotion dysregulation over the lifespan (Cisler et al., 2012).
Language
Whereas traditional views of language have placed an emphasis on the left hemisphere, network neuroscience has led to discovery in this domain as well. They have implicated important interactions within supporting mechanisms of both the left hemisphere and the right hemisphere. A graph theoretical analysis of intrinsic functional connectivity data demonstrated significant connectivity between Broca’s area and right hemispheric regions (Muller & Meyer, 2014). This suggests that language processing may be asymmetric but involves systems in both hemispheres. Examining functional network configurations during language tasks and their relationships to language performance has led to additional discoveries. A characteristic of network function underlying language may be the extent to which specific nodes mediate communication across the network and the extent to which hemispheres communicate. In the former case, a study of sentence comprehension revealed that engaging in an explicit task resulted in greater global efficiency and increased betweenness centrality of the left inferior frontal gyrus (Qin et al., 2013). In the latter case, in a split visual field experiment, Doron and colleagues demonstrated that network-based measurements of interhemispheric coordination are generally weakest when lexical stimuli are introduced to the language-dominant (left) hemisphere and strongest when they are introduced to the contralateral hemisphere (Doron, Bassett, & Gazzaniga, 2012). This novel observation of coordination highlights the underlying dynamic nature of brain communication during language processing and specifically that interhemispheric mechanisms can transiently coordinate to subserve processing under challenging conditions.
An ongoing debate concerns the nature of language with respect to other cognitive systems. In particular, one problem is whether language relies on generalized as opposed to exclusive and dedicated mechanisms. It has been proposed that language systems may be organized in functionally specialized cores with conditionally recruited regions on the periphery (see Figure 2C, left; Fedorenko & Thompson-Schill, 2014). In particular, cognitive control and other regions that respond to multiple cognitive demands may be peripheral mechanisms recruited to support language functions depending on task demands (Blank et al., 2014; Fedorenko, 2014).
The application of network approaches has discovered that language processing involves interactions between classically identified language systems, supporting right-hemispheric systems, and cognitive control systems. In particular, linguistic processing may involve dynamic recruitment of homologous right hemispheric resources (Doron et al., 2012). The left inferior frontal gyrus may serve as a hub that mediates between conflicting representations (Thompson-Schill et al., 1998), and this feature may be represented in its fluctuating network betweenness centrality (Qin et al., 2013). These techniques have begun to identify the conditions under which general control mechanisms are recruited to support relatively specialized language functions (Fedorenko & Thompson-Schill, 2014). Examining associations between functional network properties and language performance may further validate network neuroscience approaches in this domain.
Attention and Cognitive Control
Cognitive control and attention presumably subserve numerous brain functions (Braver, 2012; Borgers & Kopell, 2008; Corbetta & Shulman, 2002; Miller & Cohen, 2001; Miyake, Friedman, Emerson, Witzki, & Howerter, 2000). Network approaches have begun to clarify the interactions between attention, cognitive control regions, and other brain systems in the service of cognitive functions.
Attention-demanding tasks activate a frontoparietal network (see Parks & Madden, 2013, for a review), largely subtended by structural wiring (Hermundstad et al., 2013). Using nicotine replacement to modulate attention over a prolonged task duration, Giessing, Thiel, Alexander-Bloch, Patel, and Bullmore (2013) demonstrated that modulations in this network correlated with intraindividual differences in cognitive function. Nicotine replacement induced an increase in task performance accompanied by functional network reconfiguration toward greater efficiency, less clustering, and longer connection distance (Giessing et al., 2013). On the other hand, extended intervals on-task induced a decrease in task performance accompanied by a functional network reconfiguration toward less efficiency, increased clustering, and shorter connection distance.
Some distinctions between the network properties underlying attention and cognitive control have been identified. The relatively global effects on network configuration during external modulation of attention contrast against the region-centered signatures identified in cognitive control studies (Cole, Laurent, & Stocco, 2013; Cole, Yarkoni, Repovs, Anticevic, & Braver, 2012; Dosenbach et al., 2007). Dosenbach and colleagues (2007) demonstrated dissociable network structure of two distinct sets of brain areas that appear to coordinate adaptive (cingu-lo-opercular network) and stable (frontoparietal network) task control on different timescales and using different mechanisms. Power and Petersen (2013) extended this work and found that control-related regions tend to display start-cue, sustained, and error-related activity during cognitive tasks. Employing a network analysis of a working memory task with high cognitive control demands, Cole and colleagues (2012) identified the lateral pFC as the single region whose functional connectivity within and outside of the frontoparietal system displayed a selective relationship with individual differences in fluid intelligence. EEG recordings acquired during performance of a mathematical task with high cognitive control demands found that overall connectivity in the frontoparietal system was differentially engaged. During transitions from subitizing (rapid counting of small numbers of objects) to retrieval, increased local and global network efficiency were observed in the delta band. Difficult mathematics resulted in increased cliquishness in delta and theta bands (Klados et al., 2013).
Overall, these early network discoveries in attention and cognitive control support and extend the view that these processes are frontally mediated (Szczepanski & Knight, 2014). Externally cued attention recruits efficient global network activity, whereas dissociable cognitive control demands rely upon dual networks that manage shifting and sustained control processes (Dosenbach et al., 2007). Validation with performance was suggested by the finding that the left pFC engages in dynamic activity with the rest of the brain to support general cognitive flexibility and fluid reasoning (Cole et al., 2012). Moreover, network approaches have led to the discovery of a spatiotemporal nesting of processes: cognitive control may be mediated through distributed network processes via increased modularization in dissociable frequency bands. In distinction to working memory, cognitive control may be executed across brain networks in the delta and theta bands (Klados et al., 2013). Limits to sustaining cognitive control may result from network failure to maintain difficult-to-reach states (Giessing et al., 2013). Notably, tasks used in network studies to date have not systematically decomposed certain cognitive control contributions (see Miyake et al., 2000). How specific components of attention and cognitive control represented in particular nodes influence the rest of the brain remains an area open for rigorous investigation.
CURRENT FRONTIERS
Intersections with Cognitive Theory
So far, we have reviewed progress in an early phase of cognitive network neuroscience. A prevailing challenge for network neuroscience is to provide the basis for formal integration with established cognitive theories (Sporns, 2014). That is: How do neural networks produce cognitive states? Although we have described the techniques and potential advantages of examining cognition with network neuroscience, it is clear that studies to date have only tangentially addressed classical issues in cognitive psychological theories. Additionally, only a small number of cognitive domains have received attention in cognitive network neuroscience. Importantly, as this area develops, several key issues will require exploration. In particular, whether network neuroscience can account for semantics, conscious experience, and complex coordination and competition between cognitive systems at multiple levels of organization will be a stringent test of its explanatory power. Explicit focus on these issues may encourage theoretical innovations and empirical verification. Progressive refinement of, integration with, and testing in light of the explanatory power of existing traditional cognitive architectures (Sporns, 2014; Langley, Laird, & Rogers, 2009; Anderson, 1995) will be necessary. Such investigations will address whether abstract cognitive architectures can accurately represent the mechanisms of mind and whether we can determine the physical basis of such architectures. To motivate this process, we now highlight major frontiers that will define the emergence of cognitive network neuroscience.
Network Dynamics and Cognitive Processes
The human brain is a dynamical system (Deco, Jirsa, & McIntosh, 2011), which has far-reaching implications for both basic neuroscience and clinical translation in, for example, prosthetic design (Shenoy, Kaufman, Sahani, & Churchland, 2011), epileptic control (Ching, Brown, & Kramer, 2012), and the application of control theory to subcortical neural systems (Schiff, 2012). A critical frontier in the application of network-based methods to cognitive neuroscience lies in the development and application of dynamic network methods that characterize the evolution of putative communication patterns over the multiple temporal scales in which cognitive function changes (Kopell et al., 2014). Examples include dynamic centrality (Mantzaris et al., 2013) and dynamic community detection (Bassett, et al., 2011 and in press) in healthy and diseased cohorts, which can be used to predict fundamental human capacities. However, the work in this area is limited, and the field is rich with opportunities for both method development and applications to further questions in cognitive neuroscience.
Network-based Prediction of Cognitive Processes
Although network-based approaches show significant promise to further basic understanding in systems and cognitive neuroscience, they are perhaps more pragmatically accompanied by novel possibilities for classification and prediction. The strength of individual putative functional connections between region pairs (Mokhtari & Hossein-Zadeh, 2013; Shirer, Ryali, Rykhlevskaia, Menon, & Greicius, 2012; Chen et al., 2011; Richiardi, Eryilmaz, Schwartz, Vuilleumier, & Van De Ville, 2011; Shen, Wang, Liu, & Hu, 2010) and more global graph properties themselves (Fekete et al., 2013; Bassett, Nelson, et al., 2012; Heinzle et al., 2012; Supekar, Menon, Rubin, Musen, & Greicius, 2008) form a new set of features that can be used to decode healthy brain states (Mokhtari & Hossein-Zadeh, 2013; Heinzle et al., 2012; Shirer et al., 2012; Richiardi et al., 2011) or to classify diseased and nondiseased cohorts in an effort to develop neurodiagnostics (Fekete et al., 2013; Bassett, Nelson, et al., 2012; Richiardi et al., 2012; Chen et al., 2011; Shen et al., 2010; Supekar et al., 2008) by exploiting robust techniques from machine learning (Figure 5; see Richiardi, Achard, Bunke, & Van De Ville, 2013, for a recent review). Complementary approaches such as mathematical modeling (Raj, Kuceyeski, & Weiner, 2012) and statistical analyses (Bassett et al., 2013; Zhou, Gennatas, Kramer, Miller, & Seeley, 2012; Bassett, Wymbs, et al., 2011) of functional and anatomical network structure have shown promise in predicting individual brain maturity (Dosenbach et al., 2010), disease progression (Raj et al., 2012; Zhou et al., 2012), and potential receptivity for neurorehabilitation efforts (Bassett et al., 2013; Bassett, Wymbs, et al., 2011). The nontrivial heterogeneities in region–region relationships require finely tuned models that are true to underlying microscopic variables. Although most studies have utilized data from only one imaging modality, multimodal studies could arguably provide a more comprehensive understanding of cognitive function and its alteration in disease (Pandit et al., 2013; Sugranyes et al., 2012; Camchong, MacDonald, Bell, Mueller, & Lim, 2011; Steffens, Taylor, Denny, Bergman, & Wang, 2011; Pomarol-Clotet et al., 2010; Jeong, Wible, Hashimoto, & Kubicki, 2009).
Figure 5.
Network-based prediction (M = graph theory metric, S = subject, V = vector). Two complementary approaches to network-based prediction. (A) Trial level prediction. Node time series are sampled from brain regions, and their functional connectivity is estimated. (B) Functional connectivity (e.g., correlation) matrices are created for different trials (e.g., accurate vs. inaccurate trials). The matrices can be reorganized into vectors ( V) representing connectivity values from different trials. (C) Pattern classifiers can be used to associate observed connectivity patterns across subjects to specific performance values. Finally, the connectivity classification can be used to predict whether vectors from a new subject are associated with performance of different types. (D) Graph level prediction of cognitive states. An elaborated approach can be taken at the level of network metrics during cognitive processes. A graph of nodes and edges can be constructed from brain data. Then, graph theoretical metrics for each node (e.g., clustering coefficients, betweenness centrality, node degree) can be calculated. (E) A pattern classifier can be trained on the graph theory metric patterns observed during different cognitive tasks. (F) Summaries of the absolute and relative importance of nodes, measures, and nodes within measures can be used within the pattern classification scheme. (G) Finally, graph theoretical pattern features can be used to predict cognitive states. Note that, in principle, pairwise functional connectivity such as that used to predict trial types (A–C) can be used for cognitive state prediction (D–G) and vice versa. Panels A–C adapted from Heinzle et al. (2012) and Panels D–G adapted from Ekman et al. (2012), with permission.
Networks across Spatial and Temporal Scales
Brain mechanisms are nonlinear and cross-multiple scales of spatiotemporal and qualitative organization (Kopell et al., 2014). As a result, a default theoretical view that explicitly acknowledges the important aspects of this complexity is warranted for a comprehensive account of cognitive mechanisms (Turk-Browne, 2013; Bassett & Gazzaniga, 2011). As in traditional approaches to the brain sciences, the analysis of dynamics on brain network architectures can occur on scales ranging from intracellular mechanics to the macrolevel connectome. Understanding how cognition emerges within such a complex system may necessarily involve the complementary use of simulation and empirical techniques outside of neuroimaging (e.g., intracranial recording, brain stimulation). As network neuroscience develops within levels, a focus should be placed on dynamics that emerge across different levels of organization. For example, intersections between macroscale brain models such as the Virtual Brain (Ritter, Schirner, McIntosh, & Jirsa, 2013) and mesoscale models such as for cortical columns (Chemla & Chavane, 2010) and microscale models examining self-organized criticality (Plenz, Niebur, & Schuster, 2014) could form an important frontier. Without this strategy, some interlevel dynamical properties of the brain relevant to cognitive processes may remain obscured. One additional challenge is to consider the influences of rare events (Taleb, 2007) within and across levels of organization in nonlinear brain systems. Thus, cognitive network neuroscience researchers should seek and benefit from combined innovative technologies and mathematical approaches in network science. Across these efforts, the core challenge is to draw associations between multilevel brain dynamics and cognitive states. It is possible that fundamental rules governing dynamical features of brain function can be identified. These could be manipulated in experimental designs to determine robust mappings from multilevel dynamics and cognition.
Networks Incorporating Subcortical Systems and the Cerebellum
As mentioned previously, most approaches in cognitive network neuroscience have undersampled or entirely omitted subcortical structures, especially the cerebellum. This is a crucial limitation of studies to date in light of increasingly well-described contributions of subcortical regions to cognitive processes (Koziol & Budding, 2009; Stoodley & Schmahmann, 2009) and signaling in brain systems (Buzsaki, 2011). Specifically investigators should consider including data for nodes sampled with atlases that validly represent subcortical systems. This can occur at a level of resolution such as substructures of the thalamus (Behrens et al., 2003) or lobules of the cerebellum (Diedrichsen, Balsters, Flavell, Cussans, & Ramnani, 2009). The software package FSL has useful documentation on additional atlases that include subcortical regions.
METHODOLOGICAL AND INTERPRETATIONAL CONSIDERATIONS
It is necessary to identify and utilize optimal mathematical and computational techniques in each data modality and cognitive domain of interest. In this section, we highlight three sets of key considerations of particular import in current research efforts: those pertaining to network construction, statistical inference, and interpretation.
Network Construction
In the construction of any network, it is necessary to identify system elements (network nodes) and interactions (network edges; Butts, 2009). Such questions remain open in the extraction of anatomical, morphometric, and functional networks from neuroimaging data (Bullmore & Bassett, 2011; Bassett & Bullmore, 2009; Bullmore & Sporns, 2009; Bullmore et al., 2009).
A division of the brain into regions is often termed a parcellation, which can either be derived from known anatomical boundaries or from data-driven clustering of anatomical or state-induced functional connectivity. The former a priori anatomical boundary parcellations have been defined across a range of spatial resolutions (Bassett, Brown, et al., 2011), enable straightforward neurobiological interpretation, and can be utilized similarly across each individual in a study, thereby simplifying group-based interpretations and group comparisons. The latter connectivity-based parcellations have been derived for the whole cortex, individual lobes, and ROIs (see Zhang et al., 2014; Power et al., 2011; Nelson et al., 2010, for examples). Connectivity-based parcellations provide a unique window into individual differences in brain structure and function while somewhat complicating group comparisons. It is important to note that all parcellation schemes can be defined across a range of spatial resolutions, enabling the exploration of multiresolution structure in the human brain (Bassett, Brown, et al., 2011; Bassett et al., 2010; Zalesky et al., 2010; Meunier et al., 2009). The choice of parcellation at any particular resolution affects the value of graph diagnostics but has not been found to impact on qualitative features of network organization (de Reus & van den Heuvel, 2013; Bassett, Brown, et al., 2011; Wang et al., 2009).
The definition of interactions between brain areas depends upon the type of network being constructed. Edges in anatomical networks can represent white matter streamline counts, mean fractional anisotropy, or indirect measurements of myelination (van den Heuvel, Mandl, Stam, Kahn, & Hulshoff Pol, 2010; Hagmann et al., 2008). Edges in functional networks can be defined based on statistical relationships between regional time series (Dawson, Cha, Lewis, Mendola, & Shmuel, 2013; Watanabe et al., 2013; Gates & Molenaar, 2012; Smith et al., 2011; David, Cosmelli, & Friston, 2004), a common choice in fMRI networks is the Pearson correlation (Zalesky et al., 2012), although other measures of signal relatedness can be used, such as covariance, mutual information, and coherence.
The estimated strength of edges in both anatomical and functional networks is often complicated by noise in the underlying measurements. Statistically less significant edges are often removed from the subsequent analysis to maximize power to detect true signals in connectivity patterns. Binary networks (where edges are treated as either present or absent) neglect potentially important biological signatures present in their weighted network counterparts (where edges maintain estimated strengths). The complex interplay between mean edge weight, the density of network connections, and the topology of the network itself (Langer, Pedroni, & Jäncke, 2013; Bassett, Nelson, et al., 2012; Ginestet, Nichols, Bullmore, & Simmons, 2011; van Wijk, Stam, & Daffertshofer, 2010) has inspired a range of methodological developments to accurately capture group differences in network organizations without resorting to arbitrary thresholds on the edge weights themselves. These approaches include functional data analysis (Bassett, Nelson, et al., 2012), iterative cumulative and windowed thresholding (Bassett, Nelson, et al., 2012; Schwarz & McGonigle, 2011), and cost integration (Ginestet et al., 2011).
Statistics
Once networks have been constructed, a researcher must determine whether its observed organization is simply consistent with a null model, that is, a control graph. The choice of a control graph depends on the goal. Often, we are not simply interested in whether a brain network is different from a random network (one would hope so), but whether it is different from a network that shares some of the constraints of a brain but is not a brain network.
The development of null models for brain networks (Bassett, Porter, et al., 2012; Bassett & Gazzaniga, 2011) and networks more generally (Rybarsch & Bornholdt, 2012; Milenkovic, Filippis, Lappe, & Przulj, 2009; Higham, Rasajski, & Przulj, 2008; Kose, Budczies, Holschneider, & Fiehn, 2007; Thorne & Stumpf, 2007) is an area of energetic research. Null models fall into two general categories: models for static networks and models for temporal networks. Common static null models include Erdos-Rényi networks with random connection probability (Bollobás, 2001), Watts-Strogatz networks with small-world properties (Watts & Strogatz, 1998), and fractal networks with hierarchically modular structure (Sporns, 2006). Dynamic null models include those that permute time, node identity, and connection patterns (Bassett et al., 2013; Bassett, Porter, et al., 2012; Doron et al., 2012; Bassett, Wymbs, et al., 2011), for example, using a rewiring rule that maintains the underlying degree distribution (Maslov & Sneppen, 2002). Together, these null models are helpful for use in statistical inference; however, the development of null models with more accurate neurobiological underpinnings will be critical.
Given the complexity of networks, a researcher is also likely to search for interpretative simplicity. One simplifying approach is to create a single network structure or organizational decomposition from a larger set of data, whether that be, for example, a group of subjects in a static network data set or a set of time points in a temporal network data set. Average network structures created by taking the mean of individual networks within the group (Zuo et al., 2012; Song et al., 2009; Achard, Salvador, Whitcher, Suckling, & Bullmore, 2006) often fail to adequately reflect representative topological statistics of the networks used to construct them (Simpson, Moussa, & Laurienti, 2012). A promising alternative is the use of exponential random graph models (Newman, 2010), which appear both accurate and flexible in representing the topological characteristics of a brain network ensemble (Simpson et al., 2012; Simpson, Hayasaka, & Laurienti, 2011). Representative organization decompositions, such as the partition of network nodes into communities or modules, can be identified using the so-called consensus methods that identify reliable community structures. Consensus clustering methods can be utilized to uncover a modular decomposition that represents a group of subjects or time points (Lancichinetti & Fortunato, 2012), and such methods can be made more statistically rigorous with the use of appropriate null models for comparison (Bassett, Porter, et al., 2012). Together, these methods enable the identification of representative and potentially more interpretable structure in sets of networks.
Generalized Utility in Understanding Cognitive Processes
Network-based approaches outside of traditional neuroimaging data analysis could also have implications for our understanding of cognitive function. For example, community structure is a network-based concept that is intuitively comparable to the modular structures observed in behavior, perception, and evolution and development (Gerhart & Kirschner, 2007; Kirschner & Gerhart, 1998). One example is the chunking of motor movements in humans, in which small sets of swift finger movements are separated by pauses in a process similar to how we remember three to five digits of a phone number separately from the next several digits. This temporal chunking can be robustly identified (Bassett, Porter, et al., 2012) in networks of behavior by applying dynamic community detection techniques to time-dependent similarities in intermovement durations and can be differentially linked to the recruitment of sensorimotor putamen and frontoparietal cortex (Wymbs, Bassett, Mucha, Porter, & Grafton, 2012). In a complementary application, the temporal community structure of task events was shown to give rise to discrete neural representations in a manner similar to that thought to underpin the learning of semantic categories (Schapiro, Rogers, Cordova, Turk-Browne, & Botvinick, 2013). Finally, the evolution of modular structure in synaptic genes across a representative sampling of the animal kingdom can provide insight into the formation of complex cellular machines impacting on neural function (Conaco et al., 2012). Together, these and related studies begin to demonstrate the utility of network-based methods to uncover meaningful structure in a wide range of data types with implications for cognitive neuroscience at large.
Interpretational Caveats
We close with a final word of caution in the interpretation of network diagnostics in neuroimaging data. Network diagnostics can be intuitively interpreted in terms of information processing: High clustering can suggest that information is processed in local domains, whereas short path length can suggest that information is being transmitted over longer distances within the network. However, the biological meaning of these interpretations requires a conceptual leap from topological to biological terms, which are semantically equivalent but not necessarily conceptually interchangeable. Biological efficiency, for example, has evolutionary implications, which may not apply to topological or cognitive efficiency (Poldrack, 2014). More generally, such interpretations of network diagnostics require empirical validation demonstrating the relationship between quantitative estimates of information processing or biological efficiency and network characteristics. Until then, the cautious interpretation of network diagnostics need not hamper the utility of these approaches in prediction and classification in the study of system level dynamics underlying cognitive function.
In addition, the dynamic nature of brain network organization begs for knowledge of underlying neurophysiological mechanisms. In a few initial forays into this area, links between the strength of functional connectivity and properties of regional activity time series such as power and entropy have been identified across brain areas (Bassett, Nelson, et al., 2012), within brain areas (Zalesky et al., 2012) and between subjects (Siebenhuhner et al., 2013; Yu et al., 2013). In a similar vein, the magnitude of CBF has also been linked to the strength and topology of functional connectivity in individual regions within a wider network (Liang, Zou, He, & Yang, 2013). Perhaps even more interestingly, recent work has linked regional patterns of aerobic glycolysis to intrinsic functional connectivity (Vaishnavi et al., 2010). Neuroplasticity (Arnsten, Paspalas, Gamo, Yang, & Wang, 2010) and genetic contributors to neuromorphometry, synapse formation, and neuromodulatory systems likely all impact on observed network organization. Indeed, evidence supports the claim that network organization is both heritable (Fornito et al., 2011) and modulated by individual alleles (Brown et al., 2011; Esslinger et al., 2009). Together, these studies circumscribe an open space of fascinating questions yet to be answered about the neurophysiological and genetic drivers of network organization.
CONCLUSION
In this review, we have described how network-based approaches provide a powerful means to reveal neurophysiological correlates of behavior and new organizational principles governing cognitive function. Cognitive network neuroscience is in an early stage of development and is only beginning to demonstrate its utility. The field has started to provide important explanations and validations, and in the future may clarify the rules governing the links between cognition and brain network dynamics. Under the banner of this emerging field, the integration of dynamic systems analysis, mathematical network theory, and classical cognitive neuroscience could lead to a fundamental shift in our understanding of the mind. We encourage increased communication between researchers in each of these fields to capitalize on early progress and promote discovery at this new scientific intersection.
References
- Achard S, Bullmore E. Efficiency and cost of economical brain functional networks. PLoS Computational Biology. 2007;3:e17. doi: 10.1371/journal.pcbi.0030017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Achard S, Salvador R, Whitcher B, Suckling J, Bullmore E. A resilient, low-frequency, small-world human brain functional network with highly connected association cortical hubs. Journal of Neuroscience. 2006;26:63–72. doi: 10.1523/JNEUROSCI.3874-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J. The architecture of cognition. 2. Oxford: Psychology Press; 1995. [Google Scholar]
- Arnsten AF, Paspalas CD, Gamo NJ, Yang Y, Wang M. Dynamic network connectivity: A new form of neuroplasticity. Trends in Cognitive Science. 2010;14:365–375. doi: 10.1016/j.tics.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Brown JA, Deshpande V, Carlson JM, Grafton ST. Conserved and variable architecture of human white matter connectivity. Neuroimage. 2011;54:1262–1279. doi: 10.1016/j.neuroimage.2010.09.006. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET. Human brain networks in health and disease. Current Opinion in Neurology. 2009;22:340–347. doi: 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET, Verchinski BA, Mattay VS, Weinberger DR, Meyer-Lindenberg A. Hierarchical organization of human cortical networks in health and schizophrenia. Journal of Neuroscience. 2008;28:9239–9248. doi: 10.1523/JNEUROSCI.1929-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Gazzaniga MS. Understanding complexity in the human brain. Trends in Cognitive Sciences. 2011;15:200–209. doi: 10.1016/j.tics.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Greenfield DL, Meyer-Lindenberg A, Weinberger DR, Moore S, Bullmore E. Efficient physical embedding of topologically complex information processing networks in brains and computer circuits. PLoS Computational Biology. 2010;6:e1000748. doi: 10.1371/journal.pcbi.1000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Meyer-Lindenberg A, Achard S, Duke T, Bullmore E. Adaptive reconfiguration of fractal small-world human brain functional networks. Proceedings of the National Academy of Sciences, USA. 2006;103:19518–19523. doi: 10.1073/pnas.0606005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Meyer-Lindenberg A, Weinberger DR, Coppola R, Bullmore E. Cognitive fitness of cost-efficient brain functional networks. Proceedings of the National Academy of Sciences, USA. 2009;106:11747–11752. doi: 10.1073/pnas.0903641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Nelson BG, Mueller BA, Camchong J, Lim KO. Altered resting state complexity in schizophrenia. Neuroimage. 2012;59:2196–2207. doi: 10.1016/j.neuroimage.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Porter MA, Wymbs NF, Grafton ST, Carlson JM, Mucha PJ. Robust detection of dynamic community structure in networks. Chaos. 2012;23:013142. doi: 10.1063/1.4790830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Porter MA, Mucha PJ, Carlson JM, Grafton ST. Dynamic reconfiguration of human brain networks during learning. Proceedings of the National Academy of Sciences, USA. 2011;108:7641–7646. doi: 10.1073/pnas.1018985108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Wymbs NF, Rombach MP, Porter MA, Mucha PJ, Grafton ST. Core–periphery organisation of human brain dynamics. PLoS Computational Biology. 2013;9:e1003171. doi: 10.1371/journal.pcbi.1003171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassett DS, Yang M, Wymbs NF, Grafton ST. Learning-induced autonomy of sensorimotor systems. Nature Neuroscience. doi: 10.1038/nn.3993. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens TE, Johansen-Berg H, Woolrich MW, Smith SM, Wheeler-Kingshott CA, Boulby PA, et al. Non-invasive mapping of connections between human thalamus and cortex using diffusion imaging. Nature Neuroscience. 2003;6:750–757. doi: 10.1038/nn1075. [DOI] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magnetic Resonance in Medicine. 1995;34:537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Blank IA, Kanwisher N, Fedorenko E. A functional dissociation between language and multiple-demand systems revealed in patterns of bold signal fluctuations. Journal of Neurophysiology. 2014;112:1105–1118. doi: 10.1152/jn.00884.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollobás B. Random graphs. Cambridge, UK: Cambridge University Press; 2001. [Google Scholar]
- Borgatti SP, Everett MG. Models of core/periphery structures. Social Networks. 1999;21:375–395. [Google Scholar]
- Borgers CSE, Kopell N. Gamma oscillations mediate stimulus competition and attentional selection in a cortical network model. Proceedings of the National Academy of Sciences, USA. 2008;105:18023–18028. doi: 10.1073/pnas.0809511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun U, Plichta MM, Esslinger C, Sauer C, Haddad L, Grimm O, et al. Test-retest reliability of resting-state connectivity network characteristics using fMRI and graph theoretical measures. Neuroimage. 2012;59:1404–1412. doi: 10.1016/j.neuroimage.2011.08.044. [DOI] [PubMed] [Google Scholar]
- Braver TS. The variable nature of cognitive control: A dual mechanisms framework. Trends in Cognitive Sciences. 2012;16:106–113. doi: 10.1016/j.tics.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler SL, Richter CG. Interareal oscillatory synchronization in top–down neocortical processing. Current Opinion in Neurobiology. 2015;31:62–66. doi: 10.1016/j.conb.2014.08.010. [DOI] [PubMed] [Google Scholar]
- Brodmann K. Vegleichende lokalisationslehre der grosshirnde. Leipzig: Barth; 1909. [Google Scholar]
- Brown JA, Terashima KH, Burggren AC, Ercoli LM, Miller KJ, Small GW, et al. Brain network local interconnectivity loss in aging APOE-4 allele carriers. Proceedings of the National Academy of Sciences USA. 2011;108:20760–20765. doi: 10.1073/pnas.1109038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM. The evolution of distributed association networks in the human brain. Trends in Cognitive Sciences. 2013;17:648–665. doi: 10.1016/j.tics.2013.09.017. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Sepulcre J, Talukdar T, Krienen FM, Liu H, Hedden T, et al. Cortical hubs revealed by intrinsic functional connectivity: Mapping, assessment of stability, and relation to Alzheimer’s disease. Journal of Neuroscience. 2009;29:1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Barnes A, Bassett DS, Fornito A, Kitzbichler M, Meunier D, et al. Generic aspects of complexity in brain imaging data and other biological systems. Neuroimage. 2009;47:1125–1134. doi: 10.1016/j.neuroimage.2009.05.032. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. Complex brain networks: Graph theoretical analysis of structural and functional systems. Nature Review Neuroscience. 2009;10:186–198. doi: 10.1038/nrn2575. [DOI] [PubMed] [Google Scholar]
- Bullmore E, Sporns O. The economy of brain network organization. Nature Reviews Neuroscience. 2012;13:336–349. doi: 10.1038/nrn3214. [DOI] [PubMed] [Google Scholar]
- Bullmore ET, Bassett DS. Brain graphs: Graphical models of the human brain connectome. Annual Reviews of Clinical Psychology. 2011;7:113–140. doi: 10.1146/annurev-clinpsy-040510-143934. [DOI] [PubMed] [Google Scholar]
- Butts CT. Revisiting the foundations of network analysis. Science. 2009;325:414–416. doi: 10.1126/science.1171022. [DOI] [PubMed] [Google Scholar]
- Buzsaki G. Rhythms of the brain. 1. Oxford: Oxford University Press; 2011. [Google Scholar]
- Caeyenberghs K, Leemans A, Heitger MH, Leunissen I, Dhollander T, Sunaert S, et al. Graph analysis of functional brain networks for cognitive control of action in traumatic brain injury. Brain. 2012;135:1293–1307. doi: 10.1093/brain/aws048. [DOI] [PubMed] [Google Scholar]
- Caeyenberghs K, Leemans A, Leunissen I, Gooijers J, Michiels K, Sunaert S, et al. Altered structural networks and executive deficits in traumatic brain injury patients. Brain Structure and Function. 2014;219:193–209. doi: 10.1007/s00429-012-0494-2. [DOI] [PubMed] [Google Scholar]
- Calhoun VD, Pekar JJ, McGinty VB, Adali T, Watson TD, Pearlson GD. Different activation dynamics in multiple neural systems during simulated driving. Human Brain Mapping. 2002;16:158–167. doi: 10.1002/hbm.10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camchong J, MacDonald AWR, Bell C, Mueller BA, Lim KO. Altered functional and anatomical connectivity in schizophrenia. Schizophrenia Bulletin. 2011;37:640–650. doi: 10.1093/schbul/sbp131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Plichta MM, Schafer A, Haddad L, Grimm O, Schneider M, et al. Test-retest reliability of fMRI-based graph theoretical properties during working memory, emotion processing, and resting state. Neuroimage. 2014;84:888. doi: 10.1016/j.neuroimage.2013.09.013. [DOI] [PubMed] [Google Scholar]
- Chemla S, Chavane F. A biophysical cortical column model to study the multi-component origin of the vsdi signal. Neuroimage. 2010;53:420–438. doi: 10.1016/j.neuroimage.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Chen G, Ward BD, Xie C, Li W, Wu Z, Jones JL, et al. Classification of Alzheimer disease, mild cognitive impairment, and normal cognitive status with large-scale network analysis based on resting-state functional MR imaging. Radiology. 2011;259:213–221. doi: 10.1148/radiol.10100734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching S, Brown EN, Kramer MA. Distributed control in a mesoscale cortical network model: Implications for seizure suppression. Physical Review E. 2012;86:021920. doi: 10.1103/PhysRevE.86.021920. [DOI] [PubMed] [Google Scholar]
- Cisler JM, James GA, Tripathi S, Mletzko T, Heim C, Hu XP, et al. Differential functional connectivity within an emotional regulation neural network among individuals resilient and suceptible to the depressogenic effects of early life stress. Psychological Medicine. 2012;43:507. doi: 10.1017/S0033291712001390. [DOI] [PubMed] [Google Scholar]
- Cole MW, Bassett DS, Power JD, Braver TS, Petersen SE. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83:238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Laurent P, Stocco A. Rapid instructed task learning: A new window into the human brain’s unique capacity for flexible cognitive control. Cognitve, Affective, and Behavioral Neuroscience. 2013;13:1–22. doi: 10.3758/s13415-012-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Yarkoni T, Repovs G, Anticevic A, Braver TS. Global connectivity of prefrontal cortex predicts cognitive control and intelligence. Journal of Neuroscience. 2012;32:8988–8999. doi: 10.1523/JNEUROSCI.0536-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conaco C, Bassett DS, Zhou H, Arcila ML, Degnan SM, Degnan BM, et al. Functionalization of a protosynaptic gene expression network. Proceedings of the National Academy of Sciences, USA. 2012;109(Suppl 1):10612–10618. doi: 10.1073/pnas.1201890109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbetta M, Shulman GL. Control of goal-directed and stimulus-driven attention in the brain. Nature Reviews Neuroscience. 2002;3:201–215. doi: 10.1038/nrn755. [DOI] [PubMed] [Google Scholar]
- Craddock RC, Jbabdi S, Yan CG, Vogelstein JT, Castellanos FX, Di Martino A, et al. Imaging human connectomes at the macro scale. Nature Methods. 2013;10:524–539. doi: 10.1038/nmeth.2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossley NA, Mechelli A, Scott J, Carletti F, Fox PT, McGuire P, et al. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137:2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David O, Cosmelli D, Friston KJ. Evaluation of different measures of functional connectivity using a neural mass model. Neuroimage. 2004;21:659–673. doi: 10.1016/j.neuroimage.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Dawson DA, Cha K, Lewis LB, Mendola JD, Shmuel A. Evaluation and calibration of functional network modeling methods based on known anatomical connections. Neuroimage. 2013;67:331–343. doi: 10.1016/j.neuroimage.2012.11.006. [DOI] [PubMed] [Google Scholar]
- de Reus MA, van den Heuvel MP. The parcellation-based connectome: Limitations and extensions. Neuroimage. 2013;80:397–404. doi: 10.1016/j.neuroimage.2013.03.053. [DOI] [PubMed] [Google Scholar]
- Deco G, Jirsa VK, McIntosh AR. Emerging concepts for the dynamical organization of resting-state activity in the brain. Nature Reviews Neuroscience. 2011;12:43–56. doi: 10.1038/nrn2961. [DOI] [PubMed] [Google Scholar]
- Dennis EL, Jahanshad N, Toga AW, McMahon KL, de Zubicaray GI, Martin NG, et al. Test-retest reliability of graph theory measures of structural brain connectivity. Medical Image Computing and Computer-Assisted Intervention. 2012;15:305–312. doi: 10.1007/978-3-642-33454-2_38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuker L, Bullmore ET, Smith M, Christensen S, Nathan PJ, Rockstroh B, et al. Reproducibility of graph metrics of human brain functional networks. Neuroimage. 2009;47:1460–1468. doi: 10.1016/j.neuroimage.2009.05.035. [DOI] [PubMed] [Google Scholar]
- Diedrichsen J, Balsters JH, Flavell J, Cussans E, Ramnani N. A probabalistic mr atlas of the human cerebellum. Neuroimage. 2009;46:39–46. doi: 10.1016/j.neuroimage.2009.01.045. [DOI] [PubMed] [Google Scholar]
- Doron KW, Bassett DS, Gazzaniga MS. Dynamic network structure of interhemispheric coordination. Proceedings of the National Academy of Sciences, USA. 2012;109:18661–18668. doi: 10.1073/pnas.1216402109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NU, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RA, et al. Distinct brain networks for adaptive and stable task control in humans. Proceedings of the National Academy of Sciences, USA. 2007;104:11073–11078. doi: 10.1073/pnas.0704320104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosenbach NU, Nardos B, Cohen AL, Fair DA, Power JD, Church JA, et al. Prediction of individual brain maturity using fMRI. Science, USA. 2010;329:1358–1361. doi: 10.1126/science.1194144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff SB, Paus T, Caspers S, Grosbras MH, Evans A, Zilles K, et al. Assignment of functional activations to probabalistic cytoarchitectonic areas revisited. Neuroimage. 2007;36:511–521. doi: 10.1016/j.neuroimage.2007.03.060. [DOI] [PubMed] [Google Scholar]
- Ekman M, Derrfuss J, Tittgemeyer M, Fiebach CJ. Predicting errors from reconfiguration patterns in human brain networks. Proceedings of the National Academy of Sciences, USA. 2012;109:16714–16719. doi: 10.1073/pnas.1207523109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esslinger C, Walter H, Kirsch P, Erk S, Schnell K, Arnold C, et al. Neural mechanisms of a genome-wide supported psychosis variant. Science. 2009;324:605. doi: 10.1126/science.1167768. [DOI] [PubMed] [Google Scholar]
- Farbota KD, Bendlin BB, Alexander AL, Rowley HA, Dempsey RJ, Johnson SC. Longitudinal diffusion tensor imaging and neuropsychological correlates in traumatic brain injury patients. Frontiers in Human Neuroscience. 2012;6:160. doi: 10.3389/fnhum.2012.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko E. The role of domain-general cognitive control in language comprehension. Frontiers in Psychology. 2014;5:1–17. doi: 10.3389/fpsyg.2014.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorenko E, Thompson-Schill SL. Reworking the language network. Trends in Cognitive Science. 2014;18:120–126. doi: 10.1016/j.tics.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete T, Wilf M, Rubin D, Edelman S, Malach R, Mujica-Parodi LR. Combining classification with fMRI-derived complex network measures for potential neurodiagnostics. PLoS One. 2013;8:e62867. doi: 10.1371/journal.pone.0062867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Bassett D, Meunier D, Yucel M, Wood SJ, et al. Genetic influences on economical properties of human functional cortical networks. Journal of Neuroscience. 2011;31:3261–3270. doi: 10.1523/JNEUROSCI.4858-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Zalesky A, Pantelis C, Bullmore ET. Schizophrenia, neuroimaging and connectomics. Neuroimage. 2012;62:2296–2314. doi: 10.1016/j.neuroimage.2011.12.090. [DOI] [PubMed] [Google Scholar]
- Fortunato S. Community detection in graphs. Physics Reports. 2010;486:75–174. [Google Scholar]
- Friston KJ. Functional and effective connectivity in neuroimaging: A synthesis. Human Brain Mapping. 1994;2:56–78. [Google Scholar]
- Gates KM, Molenaar PC. Group search algorithm recovers effective connectivity maps for individuals in homogeneous and heterogeneous samples. Neuroimage. 2012;63:310–319. doi: 10.1016/j.neuroimage.2012.06.026. [DOI] [PubMed] [Google Scholar]
- Gerhart J, Kirschner M. The theory of facilitated variation. Proceedings of the National Academy of Sciences, USA. 2007;104(Suppl 1):8582–8589. doi: 10.1073/pnas.0701035104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giessing C, Thiel CM, Alexander-Bloch AF, Patel AX, Bullmore ET. Human brain functional network changes associated with enhanced and impaired attentional task performance. Journal of Neuroscience. 2013;33:5903–5914. doi: 10.1523/JNEUROSCI.4854-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestet CE, Nichols TE, Bullmore ET, Simmons A. Brain network analysis: Separating cost from topology using cost-integration. PLoS One. 2011;6:e21570. doi: 10.1371/journal.pone.0021570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginestet CE, Simmons A. Statistical parametric network analysis of functional connectivity dynamics during a working memory task. Neuroimage. 2011;55:e21570. doi: 10.1016/j.neuroimage.2010.11.030. [DOI] [PubMed] [Google Scholar]
- Grimm S, Beck J, Schuepbach D, Hell D, Boesiger P, Bermpohl F, et al. Imbalance between left and right dorsolateral prefrontal cortex in major depression is linked to negative emotional judgment: An fMRI study in severe major depressive disorder. Biological Psychiatry. 2008;63:369–376. doi: 10.1016/j.biopsych.2007.05.033. [DOI] [PubMed] [Google Scholar]
- Hagmann P, Cammoun L, Gigandet X, Meuli R, Honey CJ, Wedeen VJ, et al. Mapping the structural core of human cerebral cortex. PLoS Biology. 2008;6:e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson M, Tokoglu F, Sun Z, Schafer RJ, Skudlarski P, Gore JC, et al. Connectivity-behavior analysis reveals that functional connectivity between left BA39 and Broca’s area varies with reading ability. Neuroimage. 2006;31:513–519. doi: 10.1016/j.neuroimage.2005.12.040. [DOI] [PubMed] [Google Scholar]
- He H, Sui J, Yu Q, Turner JA, Ho BC, Sponheim SR, et al. Altered small-world brain networks in schizophrenia patients during working memory performance. PLoS One. 2012;7:e38195. doi: 10.1371/journal.pone.0038195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Chen Z, Gong G, Evans A. Neuronal networks in Alzheimer’s disease. Neuroscientist. 2009;15:333–350. doi: 10.1177/1073858409334423. [DOI] [PubMed] [Google Scholar]
- He Y, Chen ZJ, Evans AC. Small-world anatomical networks in the human brain revealed by cortical thickness from MRI. Cerebral Cortex. 2007;17:2407–2419. doi: 10.1093/cercor/bhl149. [DOI] [PubMed] [Google Scholar]
- Heinzle J, Wenzel MA, Haynes JD. Visuomotor functional network topology predicts upcoming tasks. Journal of Neuroscience. 2012;32:9960–9968. doi: 10.1523/JNEUROSCI.1604-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitger MH, Ronsse R, Dhollander T, Dupont P, Caeyenberghs K, Swinnen SP. Motor learning-induced changes in functional brain connectivity as revealed by means of graph-theoretical network analysis. Neuroimage. 2012;61:633. doi: 10.1016/j.neuroimage.2012.03.067. [DOI] [PubMed] [Google Scholar]
- Herculano-Houzel S. Coordinated scaling of cortical and cerebellar numbers of neurons. Frontiers in Neuroanatomy. 2010;4:1–8. doi: 10.3389/fnana.2010.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermundstad AM, Bassett DS, Brown KS, Aminoff EM, Clewett D, Freeman S, et al. Structural foundations of resting-state and task-based functional connectivity in the human brain. Proceedings of the National Academy of Sciences, USA. 2013;110:6169–6174. doi: 10.1073/pnas.1219562110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higham DJ, Rasajski M, Przulj N. Fitting a geometric graph to a protein-protein interaction network. Bioinformatics. 2008;24:1093–1099. doi: 10.1093/bioinformatics/btn079. [DOI] [PubMed] [Google Scholar]
- Holme P, Saramäki J. Temporal networks. Physics Reports. 2012;519:97–125. [Google Scholar]
- Illari PM, Williamson J. What is a mechanism? Thinking about mechanisms across the sciences. European Journal for Philosophy of Science. 2012;2:119–135. [Google Scholar]
- Jeong B, Wible CG, Hashimoto R, Kubicki M. Functional and anatomical connectivity abnormalities in left inferior frontal gyrus in schizophrenia. Human Brain Mapping. 2009;30:4138–4151. doi: 10.1002/hbm.20835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnison J, Padmala S, Choi JM, Pessoa L. Network analysis reveals increased integration during emotional and motivational processing. Journal of Neuroscience. 2012;32:8361–8372. doi: 10.1523/JNEUROSCI.0821-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnunen KM, Greenwood R, Powell JH, Leech R, Hawkins PC, Bonnelle V, et al. White matter damage and cognitive impairment after traumatic brain injury. Brain. 2011;134:440–463. doi: 10.1093/brain/awq347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner M, Gerhart J. Evolvability. Proceedings of the National Academy of Sciences, USA. 1998;95:8420–8427. doi: 10.1073/pnas.95.15.8420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzbichler MG, Henson RN, Smith ML, Nathan PJ, Bullmore ET. Cognitive effort drives workspace configuration of human brain functional networks. Journal of Neuroscience. 2011;31:8259–8270. doi: 10.1523/JNEUROSCI.0440-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klados MA, Kanatsouli K, Antoniou I, Babiloni F, Tsirka V, Bamidis PD, et al. A graph theoretical approach to study the organization of the cortical networks during different mathematical tasks. PLOS One. 2013;8:e71800. doi: 10.1371/journal.pone.0071800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenigs M, Grafman J. The functional neuroanatomy of depression: Distinct roles for ventromedial and dorsolateral prefrontal cortex. Behavioural Brain Research. 2009;201:239–243. doi: 10.1016/j.bbr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopell NJ, Gritton HJ, Whittington MA, Kramer MA. Beyond the connectome: The dynome. Neuron. 2014;83:1319–1320. doi: 10.1016/j.neuron.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kose F, Budczies J, Holschneider M, Fiehn O. Robust detection and verification of linear relationships to generate metabolic networks using estimates of technical errors. BMC Bioinformatics. 2007;8:162. doi: 10.1186/1471-2105-8-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziol LF, Budding DE. Subcortical structures and cognition. 1. New York: Springer; 2009. [Google Scholar]
- Lancichinetti A, Fortunato S. Consensus clustering in complex networks. Scientific Reports. 2012;2:336. doi: 10.1038/srep00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer N, Pedroni A, Jäncke L. The problem of thresholding in small-world network analysis. PLoS One. 2013;8:e53199. doi: 10.1371/journal.pone.0053199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley P, Laird JE, Rogers S. Cognitive architectures: Research issues and challenges. Cognitive Systems Research. 2009;10:141–160. [Google Scholar]
- Latora V, Marchiori M. Efficient behavior of small-world networks. Physical Review Letters. 2001;87:198701. doi: 10.1103/PhysRevLett.87.198701. [DOI] [PubMed] [Google Scholar]
- Latora V, Marchiori M. Economic small-world behavior in weighted networks. The European Physics Journal B. 2003;32:249–263. [Google Scholar]
- Leunissen I, Coxon JP, Geurts M, Caeyenberghs K, Michiels K, Sunaert S, et al. Disturbed cortico-subcortical interactions during motor task switching in traumatic brain injury. Human Brain Mapping. 2013;34:1254–1271. doi: 10.1002/hbm.21508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Zou Q, He Y, Yang Y. Coupling of functional connectivity and regional cerebral blood flow reveals a physiological basis for network hubs of the human brain. Proceedings of the National Academy of Sciences, USA. 2013;110:1929–1934. doi: 10.1073/pnas.1214900110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Calhoun VD, Eichele T, Du W, Adali T. Modulations of functional connectivity in the healthy and schizophrenia groups during task and rest. Neuroimage. 2012;62:1694–1704. doi: 10.1016/j.neuroimage.2012.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madden DJ, Bennett IJ, Song AW. Cerebral white matter integrity and cognitive aging: Contributions from diffusion tensor imaging. Neuropsychology Review. 2009;19:415–435. doi: 10.1007/s11065-009-9113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantzaris AV, Bassett DS, Wymbs NF, Estrada E, Porter MA, Mucha PJ, et al. Dynamic network centrality summarizes learning in the human brain. Journal of Complex Networks. 2013;1:83–92. [Google Scholar]
- Maslov S, Sneppen K. Specificity and stability in topology of protein networks. Science. 2002;296:910–913. doi: 10.1126/science.1065103. [DOI] [PubMed] [Google Scholar]
- McHaffie JG, Stanford TR, Stein BE, Coizet V, Redgrave P. Subcortical loops through the basal ganglia. Trends in Neurosciences. 2005;28:401–407. doi: 10.1016/j.tins.2005.06.006. [DOI] [PubMed] [Google Scholar]
- McKeown MJ, Makeig S, Brown GG, Jung TP, Kindermann SS, Bell AJ, et al. Analysis of fMRI data by blind separation into independent spatial components. Human Brain Mapping. 1998;6:160–188. doi: 10.1002/(SICI)1097-0193(1998)6:3<160::AID-HBM5>3.0.CO;2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier D, Fonlupt P, Saive AL, Plailly J, Ravel N, Royet JP. Modular structure of functional networks in olfactory memory. Neuroimage. 2014;95:264–275. doi: 10.1016/j.neuroimage.2014.03.041. [DOI] [PubMed] [Google Scholar]
- Meunier D, Lambiotte R, Fornito A, Ersche KD, Bullmore ET. Hierarchical modularity in human brain functional networks. Frontiers in Neuroinformatics. 2009;3:37. doi: 10.3389/neuro.11.037.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael AM, Calhoun VD, Andreasen NC, Baum SA. A method to classify schizophrenia using inter-task spatial correlations of functional brain images. Conference Proceedings—IEEE Engineering in Medicine and Biology Society. 2008:5510–5513. doi: 10.1109/IEMBS.2008.4650462. [DOI] [PubMed] [Google Scholar]
- Milenkovic T, Filippis I, Lappe M, Przulj N. Optimized null model for protein structure networks. PLoS One. 2009;4:e5967. doi: 10.1371/journal.pone.0005967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EK, Cohen JD. An integrative theory of prefrontal cortex function. Annual Review of Neuroscience. 2001;24:167–202. doi: 10.1146/annurev.neuro.24.1.167. [DOI] [PubMed] [Google Scholar]
- Mishkin M, Ungerleider LG. Contribution of striate inputs to the visuospatial functions of parieto-preoccipital cortex in monkeys. Journal of Gerontology Series A. 2014;6:57–77. doi: 10.1016/0166-4328(82)90081-x. [DOI] [PubMed] [Google Scholar]
- Miyake A, Friedman NP, Emerson MJ, Witzki AH, Howerter A. The unity and diversity of executive functions and their contributions to complex “frontal lobe” tasks: A latent variable analysis. Cognitive Psychology. 2000;41:49–100. doi: 10.1006/cogp.1999.0734. [DOI] [PubMed] [Google Scholar]
- Mokhtari F, Hossein-Zadeh GA. Decoding brain states using backward edge elimination and graph kernels in fMRI connectivity networks. Journal of Neuroscience Methods. 2013;212:259–268. doi: 10.1016/j.jneumeth.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Moretti L, Cristofori I, Weaver SM, Chau A, Portelli JN, Grafman J. Cognitive decline in older adults with a history of traumatic brain injury. Lancet Neurology. 2012;11:1103–1112. doi: 10.1016/S1474-4422(12)70226-0. [DOI] [PubMed] [Google Scholar]
- Moussa MN, Vechlekar CD, Burdette JH, Steen MR, Hugenschmidt CE, Laurienti PJ. Changes in cognitive state alter human functional brain networks. Frontiers in Human Neuroscience. 2011;5:83. doi: 10.3389/fnhum.2011.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucha PJ, Richardson T, Macon K, Porter MA, Onnela JP. Community structure in time-dependent, multiscale, and multiplex networks. Science. 2010;328:876–878. doi: 10.1126/science.1184819. [DOI] [PubMed] [Google Scholar]
- Muller AM, Meyer M. Language in the brain at rest: New insights from resting state data and graph theoretical analysis. Frontiers in Human Neuroscience. 2014;8:228. doi: 10.3389/fnhum.2014.00228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano-Saito A, Martinu K, Monchi O. Function of basal ganglia in bridging cognitive and motor modules to perform an action. Frontiers in Human Neuroscience. 2014;8:187. doi: 10.3389/fnins.2014.00187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson SM, Cohen AL, Power JD, Wig GS, Miezin FM, Wheeler ME, et al. A parcellation scheme for human left lateral parietal cortex. Neuron. 2010;67:156–170. doi: 10.1016/j.neuron.2010.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman MEJ. Networks: An introduction. Oxford: Oxford University Press; 2010. [Google Scholar]
- Newman MEJ, Girvan M. Finding and evaluating community structure in networks. Physical Review E. 2004;69:026113. doi: 10.1103/PhysRevE.69.026113. [DOI] [PubMed] [Google Scholar]
- Niogi SN, Mukherjee P, Ghajar J, Johnson C, Kolster RA, Sarkar R, et al. Extent of microstructural white matter injury in postconcussive syndrome correlates with impaired cognitive reaction time: A 3T diffusion tensor imaging study of mild traumatic brain injury. American Journal of Neuroradiology. 2008;29:967–973. doi: 10.3174/ajnr.A0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen JP, Ziv E, Bukshpun P, Pojman N, Wakahiro M, Berman JI, et al. Test-retest reliability of computational network measurements derived from the structural connectome of the human brain. Brain Connectivity. 2013;3:160–176. doi: 10.1089/brain.2012.0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios EM, Sala-Llonch R, Junque C, Roig T, Tormos JM, Bargallo N, et al. White matter integrity related to functional working memory networks in traumatic brain injury. Neurology. 2012;78:852–860. doi: 10.1212/WNL.0b013e31824c465a. [DOI] [PubMed] [Google Scholar]
- Palva JM, Monto S, Kulashekhar S, Palva S. Neural synchrony reveals working memory networks and predicts individual memory capacity. Proceedings of the National Academy of Sciences, USA. 2010;107:7580–7585. doi: 10.1073/pnas.0913113107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palva S, Monto S, Palva JM. Graph properties of synchronized cortical networks during visual working memory maintenance. Neuroimage. 2010;49:3257–3268. doi: 10.1016/j.neuroimage.2009.11.031. [DOI] [PubMed] [Google Scholar]
- Pandit AS, Expert P, Lambiotte R, Bonnelle V, Leech R, Turkheimer FE, et al. Traumatic brain injury impairs small-world topology. Neurology. 2013;80:1826–1833. doi: 10.1212/WNL.0b013e3182929f38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks EL, Madden DJ. Brain connectivity and visual attention. Brain Connectivity. 2013;3:317–338. doi: 10.1089/brain.2012.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passingham RE, Rowe JB, Sakai K. Has brain imaging discovered anything new about how the brain works? Neuroimage. 2013;66:142–150. doi: 10.1016/j.neuroimage.2012.10.079. [DOI] [PubMed] [Google Scholar]
- Plenz D, Niebur E, Schuster HG. Criticality in neural systems. Hoboken, NJ: Wiley; 2014. [Google Scholar]
- Poldrack RA. Is “efficiency” a useful concept in cognitive neuroscience? Developmental Cognitive Neuroscience. 2014;11:12–17. doi: 10.1016/j.dcn.2014.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomarol-Clotet E, Canales-Rodriguez EJ, Salvador R, Sarro S, Gomar JJ, Vila F. Medial prefrontal cortex pathology in schizophrenia as revealed by convergent findings from multimodal imaging. Molecular Psychiatry. 2010;15:823–830. doi: 10.1038/mp.2009.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter MA, Onnela JP, Mucha PJ. Communities in networks. North American Mathematical Society. 2009;56:1082–1097. 1164–1166. [Google Scholar]
- Power JD, Cohen AL, Nelson SM, Wig GS, Barnes KA, Church JA, et al. Functional network organization of the human brain. Neuron. 2011;72:665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power JD, Petersen SE. Control-related systems in the human brain. Current Opinion in Neurobiology. 2013;23:223–228. doi: 10.1016/j.conb.2012.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Wei M, Liu H, Yan R, Luo G, Yao Z, et al. Large scale brain functional networks support sentence comprehension: Evidence from both explicit and implicit language tasks. PLoS One. 2013;8:e80214. doi: 10.1371/journal.pone.0080214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Wei M, Liu H, Yan R, Luo G, Yao Z, et al. Abnormal brain anatomical topological organization of the cognitive-emotional and the frontoparietal circuitry in major depressive disorder. Magnetic Resonance in Medicine. 2014;72:1397–1407. doi: 10.1002/mrm.25036. [DOI] [PubMed] [Google Scholar]
- Raichle ME. The restless brain. Brain Connectivity. 2011;1:3–12. doi: 10.1089/brain.2011.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Kuceyeski A, Weiner M. A network diffusion model of disease progression in dementia. Neuron. 2012;73:1204–1215. doi: 10.1016/j.neuron.2011.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reijmer YD, Leemans MKLJ, Brundel A, Biessels GJ. Disruption of the cerebral white matter network is related to slowing of information processing speed in patients with type 2 diabetes. Diabetes. 2013;62:2112–2115. doi: 10.2337/db12-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richiardi J, Achard S, Bunke H, Van De Ville D. Machine learning with brain graphs: Predictive modeling approaches for functional imaging in systems neuroscience. Signal Processing Magazine, IEEE. 2013;30:58–70. [Google Scholar]
- Richiardi J, Eryilmaz H, Schwartz S, Vuilleumier P, Van De Ville D. Decoding brain states from fMRI connectivity graphs. Neuroimage. 2011;56:616–626. doi: 10.1016/j.neuroimage.2010.05.081. [DOI] [PubMed] [Google Scholar]
- Richiardi J, Gschwind M, Simioni S, Annoni JM, Greco B, Hagmann P, et al. Classifying minimally disabled multiple sclerosis patients from resting state functional connectivity. Neuroimage. 2012;62:2021–2033. doi: 10.1016/j.neuroimage.2012.05.078. [DOI] [PubMed] [Google Scholar]
- Ritter P, Schirner M, McIntosh AR, Jirsa VK. The virtual brain integrates computational modeling and multimodal neuroimaging. Brain Connectivity. 2013;3:121–145. doi: 10.1089/brain.2012.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RE, Anderson EJ, Husain M. White matter microstructure and cognitive function. Neuroscientist. 2013;19:8–15. doi: 10.1177/1073858411421218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rombach MP, Porter MA, Fowler JH, Mucha PJ. Core–periphery structure in networks. 2012 arXiv, 1202.2684. [Google Scholar]
- Ronan L, Voets NL, Hough M, Mackay C, Roberts N, Suckling J, et al. Consistency and interpretation of changes in millimeter-scale cortical intrinsic curvature across three independent datasets in schizophrenia. Neuroimage. 2012;63:611–621. doi: 10.1016/j.neuroimage.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybarsch M, Bornholdt S. Binary threshold networks as a natural null model for biological networks. Physical Review E. 2012;86:026114. doi: 10.1103/PhysRevE.86.026114. [DOI] [PubMed] [Google Scholar]
- Sanabria-Diaz G, Melie-García L, Iturria-Medina Y, Alemán-Gómez Y, Hernández-González G, Valdés-Urrutia L, et al. Surface area and cortical thickness descriptors reveal different attributes of the structural human brain networks. Neuroimage. 2010;50:1497–1510. doi: 10.1016/j.neuroimage.2010.01.028. [DOI] [PubMed] [Google Scholar]
- Schapiro AC, Rogers TT, Cordova NI, Turk-Browne NB, Botvinick MM. Neural representations of events arise from temporal community structure. Nature Neuroscience. 2013;16:486–492. doi: 10.1038/nn.3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiff S. Neural control engineering. Cambridge, MA: MIT Press; 2012. [Google Scholar]
- Schwarz AJ, McGonigle J. Negative edges and soft thresholding in complex network analysis of resting state functional connectivity data. Neuroimage. 2011;55:1132–1146. doi: 10.1016/j.neuroimage.2010.12.047. [DOI] [PubMed] [Google Scholar]
- Seeley WW, Menon V, Schatzberg A, Keller J, Glover GH. Dissociable intrinsic connectivity networks for salience processing and executive control. Journal of Neuroscience. 2007;27:2349–2356. doi: 10.1523/JNEUROSCI.5587-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J. Stepwise connectivity of the modal cortex reveals the multimodal organization of the human brain. Journal of Neuroscience. 2012;32:10649–10661. doi: 10.1523/JNEUROSCI.0759-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre J. Integration of visual and motor functional streams in the human brain. Neuroscience Letters. 2014;567:68–73. doi: 10.1016/j.neulet.2014.03.050. [DOI] [PubMed] [Google Scholar]
- Shamay-Tsoory SG, Aharon-Peretz J, Perry D. Two systems for empathy: A double dissociation between emotional and cognitive empathy in inferior frontal gyrus versus ventromedial prefrontal lesions. Brain. 2008;201:617–627. doi: 10.1093/brain/awn279. [DOI] [PubMed] [Google Scholar]
- Shen H, Wang L, Liu Y, Hu D. Discriminative analysis of resting-state functional connectivity patterns of schizophrenia using low dimensional embedding of fMRI. Neuroimage. 2010;49:3110–3121. doi: 10.1016/j.neuroimage.2009.11.011. [DOI] [PubMed] [Google Scholar]
- Shenoy KV, Kaufman MT, Sahani M, Churchland MM. A dynamical systems view of motor preparation: Implications for neural prosthetic system design. Progress in Brain Research. 2011;192:33–58. doi: 10.1016/B978-0-444-53355-5.00003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirer WR, Ryali S, Rykhlevskaia E, Menon V, Greicius MD. Decoding subject-driven cognitive states with whole-brain connectivity patterns. Cerebral Cortex. 2012;22:158–165. doi: 10.1093/cercor/bhr099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenhuhner F, Weiss SA, Coppola R, Weinberger DR, Bassett DS. Intra- and inter-frequency brain network structure in health and schizophrenia. PLoS One. 2013;8:e72351. doi: 10.1371/journal.pone.0072351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver JM, McAllister TW, Arciniegas DB. Depression and cognitive complaints following mild traumatic brain injury. American Journal of Psychiatry. 2009;166:653–661. doi: 10.1176/appi.ajp.2009.08111676. [DOI] [PubMed] [Google Scholar]
- Simpson SL, Hayasaka S, Laurienti PJ. Exponential random graph modeling for complex brain networks. PLoS One. 2011;6:e20039. doi: 10.1371/journal.pone.0020039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SL, Moussa MN, Laurienti PJ. An exponential random graph modeling approach to creating group-based representative whole-brain connectivity networks. Neuroimage. 2012;60:1117–1126. doi: 10.1016/j.neuroimage.2012.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, et al. Correspondence of the brain’s functional architecture during activation and rest. Proceedings of the National Academy of Sciences, USA. 2009;106:13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SM, Miller KL, Salimi-Khorshidi G, Webster M, Beckmann CF, Nichols TE, et al. Network modelling methods for fMRI. Neuroimage. 2011;54:875–891. doi: 10.1016/j.neuroimage.2010.08.063. [DOI] [PubMed] [Google Scholar]
- Song S, Liu Y, Zhou Y, Wang K, Yu C, Jiang T. Default network and intelligence difference. Conference Proceedings—IEEE Engineering in Medicine and Biology Society. 2009;2009:2212–2215. doi: 10.1109/IEMBS.2009.5334874. [DOI] [PubMed] [Google Scholar]
- Sporns O. Small-world connectivity, motif composition, and complexity of fractal neuronal connections. Biosystems. 2006;85:55–64. doi: 10.1016/j.biosystems.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Sporns O. Networks of the brain. Cambridge, MA: MIT Press; 2010. [Google Scholar]
- Sporns O. The human connectome: A complex network. Annals of the New York Academy of Sciences. 2011;1224:109–125. doi: 10.1111/j.1749-6632.2010.05888.x. [DOI] [PubMed] [Google Scholar]
- Sporns O. From simple graphs to the connectome: Networks in neuroimaging. Neuroimage. 2012;62:881–886. doi: 10.1016/j.neuroimage.2011.08.085. [DOI] [PubMed] [Google Scholar]
- Sporns O. Contributions and challenges for network models in cognitive neuroscience. Nature Neuroscience. 2014;17:652–660. doi: 10.1038/nn.3690. [DOI] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kötter R. The human connectome: A structural description of the human brain. PLoS Computational Biology. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffens DC, Taylor WD, Denny KL, Bergman SR, Wang L. Structural integrity of the uncinate fasciculus and resting state functional connectivity of the ventral prefrontal cortex in late life depression. PLoS One. 2011;6:e22697. doi: 10.1371/journal.pone.0022697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens AA, Tappon SC, Garg A, Fair DA. Functional brain network modularity captures inter- and intra-individual variation in working memory capacity. PLoS One. 2012;7:e30468. doi: 10.1371/journal.pone.0030468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoodley CJ, Schmahmann JD. Functional topography in the human cerebellum: A meta-analysis of neuroimaging studies. Neuroimage. 2009;44:489–501. doi: 10.1016/j.neuroimage.2008.08.039. [DOI] [PubMed] [Google Scholar]
- Sugranyes G, Kyriakopoulos M, Dima D, O’Muircheartaigh J, Corrigall R, Pendelbury G, et al. Multimodal analyses identify linked functional and white matter abnormalities within the working memory network in schizophrenia. Schizophrenia Research. 2012;138:136–142. doi: 10.1016/j.schres.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD. Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Computational Biology. 2008;4:e1000100. doi: 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczepanski SM, Knight RT. Insights into human behavior from lesions to the prefrontal cortex. Neuron. 2014;83:1–17. doi: 10.1016/j.neuron.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleb N. Black swans and the domains of statistics. The American Statistician. 2007;61:198–200. [Google Scholar]
- Telesford QK, Morgan AR, Hayasaka S, Simpson SL, Barret W, Kraft RA, et al. Reproducibility of graph metrics in fMRI networks. Frontiers in Neuroinformatics. 2010;4:117. doi: 10.3389/fninf.2010.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson-Schill SL, Swick D, Farah MJ, D’Esposito M, Kan IP, Knight RT. Verb generation in patients with focal frontal lesions: A neuropsychological test of neuroimaging findings. Proceedings of the National Academy of Sciences, USA. 1998;95:15855–15860. doi: 10.1073/pnas.95.26.15855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne T, Stumpf MP. Generating confidence intervals on biological networks. BMC Bioinformatics. 2007;8:467. doi: 10.1186/1471-2105-8-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk-Browne NB. Functional interactions as big data in the human brain. Science. 2013;342:580–584. doi: 10.1126/science.1238409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnavi SN, Vlassenko AG, Rundle MM, Snyder AZ, Mintun MA, Raichle ME. Regional aerobic glycolysis in the human brain. Proceedings of the National Academy of Sciences, USA. 2010;107:17757–17762. doi: 10.1073/pnas.1010459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente TW, Coronges K, Lakon C, Costenbader E. How correlated are network centrality measures? Connect (Tor) 2008;28:16–26. [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Mandl RC, Stam CJ, Kahn RS, Hulshoff Pol HE. Aberrant frontal and temporal complex network structure in schizophrenia: A graph theoretical analysis. Journal of Neuroscience. 2010;30:15915–15926. doi: 10.1523/JNEUROSCI.2874-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. Journal of Neuroscience. 2011;31:15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends in Cognitive Sciences. 2013;17:683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- van Wijk BC, Stam CJ, Daffertshofer A. Comparing brain networks of different size and connectivity density using graph theory. PLoS One. 2010;5:e13701. doi: 10.1371/journal.pone.0013701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang L, Zang Y, Yang H, Tang H, Gong Q, et al. Parcellation-dependent small-world brain functional networks: A resting-state fMRI study. Human Brain Mapping. 2009;30:1511–1523. doi: 10.1002/hbm.20623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JH, Zuo XN, Gohel S, Milham MP, Biswal BB, He Y. Graph theoretical analysis of functional brain networks: Test-retest evaluation on short- and long-term resting-state functional MRI data. PLoS One. 2011;6:e21976. doi: 10.1371/journal.pone.0021976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Li Y, Metzak P, He Y, Woodward TS. Age-related changes in topological patterns of large-scale brain functional networks during memory encoding and recognition. Neuroimage. 2010;50:862–872. doi: 10.1016/j.neuroimage.2010.01.044. [DOI] [PubMed] [Google Scholar]
- Warren DE, Power JD, Bruss J, Denburg NL, Waldron EJ, Sun H, et al. Network measures predict neuropsychological outcome after brain injury. Proceedings of the National Academy of Sciences, USA. 2014;111:14247–14252. doi: 10.1073/pnas.1322173111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Hirose S, Wada H, Imai Y, Machida T, Shirouzu I, et al. A pairwise maximum entropy model accurately describes resting-state human brain networks. Nature Communications. 2013;4:1370. doi: 10.1038/ncomms2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Woodward J. Scientific explanation. Zalta EN, editor. The Stanford encyclopedia of philosophy. 2014 Available at plato.stanford.edu/archives/win2014/entries/scientific-explanation/
- Wu TC, Wilde EA, Bigler ED, Yallampalli R, McCauley SR, Troyanskaya M, et al. Evaluating the relationship between memory functioning and cingulum bundles in acute mild traumatic brain injury using diffusion tensor imaging. Journal of Neurotrauma. 2010;27:303–307. doi: 10.1089/neu.2009.1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wymbs NF, Bassett DS, Mucha PJ, Porter MA, Grafton ST. Differential recruitment of the sensorimotor putamen and frontoparietal cortex during motor chunking in humans. Neuron. 2012;74:936–946. doi: 10.1016/j.neuron.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Zhang H, Hui M, Long Z, Jin Z, Liu Y, et al. Dynamic reconfiguration of human brain betwork during learning. Neuroscience. 2014;261:7641–7646. [Google Scholar]
- Yeo BT, Krienen FM, Sepulcre J, Sabuncu MR, Lashkari D, Hollinshead M, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. Journal of Neurophysiology. 2011;106:1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Sui J, Liu J, Plis SM, Kiehl KA, Pearlson G, et al. Disrupted correlation between low frequency power and connectivity strength of resting state brain networks in schizophrenia. Schizophrenia Research. 2013;143:165–171. doi: 10.1016/j.schres.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore E. On the use of correlation as a measure of network connectivity. Neuroimage. 2012;60:2096–2106. doi: 10.1016/j.neuroimage.2012.02.001. [DOI] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Harding IH, Cocchi L, Yucel M, Pantelis C, et al. Whole-brain anatomical networks: Does the choice of nodes matter? Neuroimage. 2010;50:970–983. doi: 10.1016/j.neuroimage.2009.12.027. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Fan L, Zhang Y, Wang J, Zhu M, Zhang Y, et al. Connectivity-based parcellation of the human posteromedial cortex. Cerebral Cortex. 2014;24:719–727. doi: 10.1093/cercor/bhs353. [DOI] [PubMed] [Google Scholar]
- Zhou J, Gennatas ED, Kramer JH, Miller BL, Seeley WW. Predicting regional neurodegeneration from the healthy brain functional connectome. Neuron. 2012;73:1216–1227. doi: 10.1016/j.neuron.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo XN, Ehmke R, Mennes M, Imperati D, Castellanos FX, Sporns O, et al. Network centrality in the human functional connectome. Cerebral Cortex. 2012;22:1862–1875. doi: 10.1093/cercor/bhr269. [DOI] [PubMed] [Google Scholar]